Abstract
The extraction of information from the scientific literature is a complex task—for researchers doing manual curation and for automatic text processing solutions. The identification of protein–protein interactions (PPIs) requires the extraction of protein named entities and their relations. Semi-automatic interactive support is one approach to combine both solutions for efficient working processes to generate reliable database content. In principle, the extraction of PPIs can be achieved with different methods that can be combined to deliver high precision and/or high recall results in different combinations at the same time. Interactive use can be achieved, if the analytical methods are fast enough to process the retrieved documents. PCorral provides interactive mining of PPIs from the scientific literature allowing curators to skim MEDLINE for PPIs at low overheads. The keyword query to PCorral steers the selection of documents, and the subsequent text analysis generates high recall and high precision results for the curator. The underlying components of PCorral process the documents on-the-fly and are available, as well, as web service from the Whatizit infrastructure. The human interface summarizes the identified PPI results, and the involved entities are linked to relevant resources and databases. Altogether, PCorral serves curator at both the beginning and the end of the curation workflow for information retrieval and information extraction.
Database URL: http://www.ebi.ac.uk/Rebholz-srv/pcorral.
Introduction
Protein–protein interactions (PPIs) are essential for biomedical research, as PPIs initiate functions and processes in biological systems (1, 2). Furthermore, single PPIs can be composed to describe complete protein interaction networks and complex regulatory events forming the core to the genetic regulation (3–5).
Several databases contain information about PPIs in different ways. Examples of such databases are IntAct (6), STRING (7), Mint (8), BioGRID (9) and MIPS (10). The development of these resources requires thoroughly analysing the scientific literature and identifying all relevant information (11, 12). This ongoing work is outperformed by the continuous increase of newly published biomedical literature leading into the growth of resources such as MEDLINE®, and both processes are central to the development of support tools for database curation work.
Biocuration workflows are composed of the following main processing tasks (13): (i) collecting related documents, (ii) identifying and indexing entities of interest and (iii) collecting information for curating specific relations. In more detail, the curation work is usually initiated by accumulating information (called ‘information retrieval’ or IR). In this part, no limitation is put on the gathering process to achieve a comprehensive search and to avoid unnecessary biases linked to any restrictions to the size of the data sample. Subsequently, the document collection has to be narrowed down to focus the results to specific information for example to the identification of relations between entities (called ‘information extraction’ or IE).
Solutions and tools have been suggested and published by the research community for the identification of PPIs from the scientific literature (14, 15). Such solutions comprise machine-learning approaches and rule-base systems for the identification of gene mentions, but also full parsing solutions for the scientific documents to identify interactions between the entity mentions, where these solutions have been optimized for high-precision results in the relation extraction (16, 17, 19, 20). IE based on syntactic parsing requires efficient processing means owing to the high computational overhead. Such solutions exploit well-defined grammatical relations between co-located entities and, as a result of its high specificity, frequently miss a significant portion of molecular interactions in text, especially for complex interactions, e.g. binding, regulation (19). Machine learning is getting popular for interpreting extracted grammatical relations. As anticipated, the systems based on machine learning usually perform better on the set of articles with similar distribution of terms. Therefore, the evaluation result against gold standard corpora could be over optimistic.
Only a few solutions are currently available that identify PPIs from the scientific literature on delivery of a specific gene name to initiate the retrieval: two solutions are, for instance, iHOP (20) and PPI finder (21). These solutions allow exploring the identified PPIs, but the user is limited to navigating through many of the already known PPIs that have been identified at a high frequency rate. This is due to the fact that these systems analyse the complete MEDLINE repository; therefore, the selection is not focused on a specific subset of the literature repository for the curation task. Other tools do allow identifying pairs of entities based on a specific MEDLINE query, and thus these tools enable targeting a specific topic, e.g. FACTA (22), but in this case, the relation extraction is not targeting PPIs; therefore, the curator ends up skimming a large number of entity pairs for PPI mentions.
As a conclusion, the available approaches only partially cover the needs that are required for a complete biomedical curation workflow setup, as they either satisfy the needs of the first step only, i.e. collecting related publications, or the third step, i.e. identifying the parts of a specific interaction. We have developed PCorral (Protein Corral, http://www.ebi.ac.uk/Rebholz-srv/pcorral) that combines IR and IE in a single application. It produces results from different extraction methods in a single approach enabling curators to focus on high recall only, or high precision only in the same processing step. The interactive interface of PCorral supports curation work and interactive exploration of the full set of MEDLINE, and curators may integrate the text processing services from Whatizit into their own curation infrastructure.
Methods
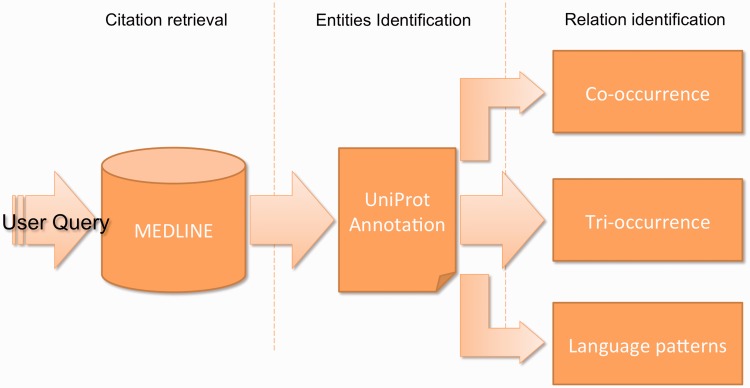
Figure 1 gives a schematic overview on the infrastructure and workflow of PCorral, which demonstrates its suitability for the biological curation routine work. The front end of PCorral gathers and organizes the results in a tabular view (c.f. ‘Results’ section). Using a keyword query interface, the user submits his query and retrieves all relevant documents from MEDLINE, and then all documents and statements are processed on-the-fly in a short period, and the extracted findings are delivered to the user.
Figure 1.
PCorral back end workflow. The processing is split into three main parts: collection of relevant citations querying an index on MEDLINE, identification of gene mentions and normalization to UniProt identifiers and extraction of relations among the identified genes.
The first step in PCorral’s workflow consists of collecting publications specified by the user’s query; e.g. carotenoid pathway or breast cancer. The articles are retrieved through the MEDLINE index; citations are ranked according to their similarity to the query as determined by Lucene’s (http://lucene.apache.org/core/) scoring algorithm. This algorithm identifies which MEDLINE fields, if any, are specified in the query and the syntax of the query, which allows delimiting the terms in the query. Each term is scored according to its relevance to the documents in MEDLINE. The MEDLINE index is the same one used by EBIMed (23) and Whatizit (24), and all three systems share the same query syntax (http://www.ebi.ac.uk/Rebholz-srv/ebimed/help.jsp#querysyntax). The text from the recovered citations is processed to identify sentence boundaries and protein/gene mentions (Whatizit-UniProt), which are then mapped to UniProt identifiers. Basic disambiguation uses the term frequencies from the British National Corpus to distinguish between terms (and entities) that are part of general English (e.g. insulin) in contrast to the specific terminology from UniProtKB (25).
PPIs are annotated using three related methods: co-occurrence (CO), tri-occurrence (CO3) and language patterns (SynP). All three methods solve a specific extraction task (see later in the text) and—according to the specification of the tasks—the results from the three methods form proper subsets of each other: the results from SynP are a subset of the results from CO3, and the same for CO3 in comparison to CO. The first method (CO) is based on COs and is the same one used in EBIMed. These interactions are based on abstract and sentence level COs. The method delivers the highest recall and is appropriate for exploratory purposes.
The CO3 is more restrictive than the CO method. In addition to two proteins co-occurring in the set, an interaction verb has to be identified from the context of the identified interaction partners. Any triplet of two proteins/genes (PGN) and a verb mention combined in one of the following forms is accepted: (i) ‘PGN VP PGN’, (ii) ‘nomVP PGN PGN’ and (iii) ‘PGN PGN nomVP’, where VP is the verb phrase that represents all the conjugational verb forms and nomVP is the nominalization of a verb form. Only the pre-selected verbs are considered and, in the case of coordination of two such verbs, both are considered.
The module that identifies and highlights PPIs searches for phrases that contain a verb or a nominal form describing an interaction like binding or dimerization; the list of verbs is displayed in Table 1. The upper set in Table 1 comprises all verbal forms that denote chemical alterations of a protein. The second set of verbs consists of forms that report on interaction and regulation events. ‘Associate’ does not denote any specific binding or transformation event (26).
Table 1.
List of verbs used in PCorral split into groups defining the interaction type
| Verbs denoting protein chemical modification | acetylate, acylate, amidate, brominate, biotinylate, carboxylate, cysteinylate, farnesylate, formylate, ‘hydrox[iy]late’, methylate, demethylate, ‘myristo?ylate’, ‘palmito?ylate’, phosphorylate, dephosphorylate, pyruvate, nitrosylate, sumoylate, ‘ubiquitin(yl)?ate’ |
| Verbs denoting interaction and regulation events | associate, dissociate, assemble, attach, bind, complex, contact, couple, ‘(multi|di)meri[zs]e’, link, interact, precipitate, regulate, inhibit, activate, ‘down[-]regulate’, express, suppress, ‘up[-]regulate’, block, contain, inactivate, induce, modify, overexpress, promote, stimulate, substitute, catalyze, cleave, conjugate, disassemble, discharge, mediate, modulate, repress, transactivate |
The verb forms are given in a regular expression form also including morphological variants of verb forms.
If two different verbs have been identified in the context of a gene pair, then both occurrences have been counted. This is also the case for gene pairs that have been identified with syntactical patterns (see later in the text), but this case only occurs at a low frequency.
The approach using syntactical SynPs is more specific, i.e. adds further restrictions to the relation extraction approach in comparison to the solutions called CO and CO3. It extracts PPIs at the highest precision levels but does miss a number of interactions (lower recall). This approach makes use of the following components:
First, one module identifies single adjectives (‘adj’), combinations of adjectives and adverbs and the coordination of adverbs. The second module selects the conjugational forms of ‘to be’, also in combination with leading, interleaving and trailing adverbs (‘beForm’; see Figure 2). The next module, seeks phrases like ‘were initially observed’ to be combined with ‘to’ and the infinitive of an interaction verb (‘shownForm’). In the same sense, modal verbs with optional trailing adverbs, where modal verbs are any of the following: can, could, cannot, do, may, might, must, need, ought, shall, should and would.
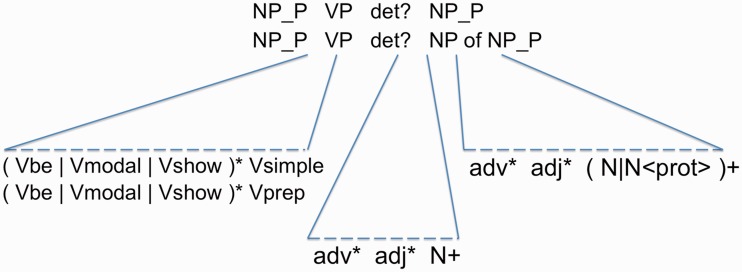
Figure 2.
(Syntactical patterns) The diagram explains the composition of the SynPs. The verb phrase (VP) is composed of several subcomponents that enable the identification of modal verbs (Vmodal), forms of to be (Vbe) and common forms of hedging (Vshown). NP_P is an NP containing a protein mention.
Then, the identification of verb phrases is composed of five modules: Vsimple covers the verb itself with only optional leading or trailing adverbs. Vprep extends Vsimple by a trailing preposition to catch expressions such as ‘bound to’ or ‘interact with’. Vbe extends the previous modules by allowing any of the matches produced by the ‘beForm’ stage in front of them and thus targets phrases such as ‘is regulated’ or ‘are positively regulated by’, Vshown allows a match for SynPs that denote expressions like ‘has been shown’ followed by ‘to’ and a match of beForms in front of Vsimple and Vprep. This will tag phrases like ‘have been shown to be phosphorylated’. Finally, Vmodal works like Vshown but uses a modal verb from the ‘shownForm’ stage. It will catch phrases like ‘may be linked to’.
Last, the module for noun phrases (NP) identification selects single and multiple nouns in combination with leading adjective modifiers, including coordination of adjective modifier elements leading the sequence of nouns. PGNs are identified as nouns. NPs do not include determiners (e.g. ‘novel orphan receptor TAK1’). Finally, the module for the PPI syntactical patterns identifies combinations of the previously identified components, such as NP_P VP det? NP_P and NP_P VP det? NP of NP_P, where NP_P is an NP that contains an identified PGN.
These construction rules for syntactical patterns lead to the selection of structures that are similar to CO3 representations, that form a subset of the CO3 representations and that produce results with highest precision. Similar structures have been proposed by (25). The syntactical patterns preserve the word order that has been used in the CO3 extraction method, but as additional feature better specifies the verb phrases that are accepted for the extraction of PPIs, and thus generates higher precision results.
Further effort has been spent on the resolution of hedging forms used by authors, i.e. the common use of expressions such as ‘PGN has been shown to’ (‘shownForm’ syntactical phrase patterns), to increase the recall of the extraction method. In the same vein, the use of syntactical patterns denoting nominalizations improved the recall for the identification of PPIs and follows the representation VP_NP ‘(of | with | between | through | from)’ det? NP_P ‘(and | with | within | via | through | by)’ det? NP_P, where VP_NP is the nominalization of the verb form.
The PPI modules have been assessed using publicly available corpora. Comparative results with a focus to the performance of the different verbs used are available from (26). The IE pipeline can also be applied as a Whatizit (http://www.ebi.ac.uk/webservices/whatizit/info.jsf) Web service (whatizitProteinInteraction, WhatizitProteinInteractionPMID) for the processing of scientific literature for the identification of PPIs from the text. The system delivers the MEDLINE citations with appended information about the method that identified the PPI, a reference to the matched text and the Uniprot identifiers of the related proteins.
Results
The simple search of PCorral (c.f. Figure 3) interprets a user query to retrieve the documents from MEDLINE that have to be processed. By default, PCorral retrieves the top 500 most relevant citations. Advanced search offers more complex queries to limit or increase the coverage of MEDLINE abstracts for the analysis. In addition, the advanced search allows selecting a specific organism from a predefined list, and this choice restricts the annotation of proteins to those UniProtKB identifiers that belong to the selected organism leading to organism-specific results. The same approach is used by EBIMed.
Figure 3.
PCorral query interface.
The query interface complies with the document retrieval features that are standards in publicly available search engines, such as PubMed®, and follows the specifications of Apache Lucene: e.g. ‘AND’ and ‘OR’ queries, keyword mentions and combinations of text features, query language for term and token variability.
Once the citations have been retrieved and fully processed, which may take from only a few seconds up to several minutes (visualized in a progress bar), the interface provides the content as a table containing the extracted PPIs (c.f. Figure 4). The list of identified PPI pairs are ranked according to the frequency of the PGN mentions across the whole selected document set, and the most frequent proteins are listed in the top ranks. The related parts of the table show the proteins that the primary protein is interacting with considering the different PPIs extraction methods. The display offers further information such as the frequency counts of abstracts and sentences that make reference to the identified PPIs sorted according to the three methods into different columns. Further information is available for each interaction, as the verb has been identified and displayed that is relevant for the interactions. All results are interlinked with the underlying biomedical reference databases and also with the MEDLINE documents from which the evidence has been extracted (c.f. Figure 5).
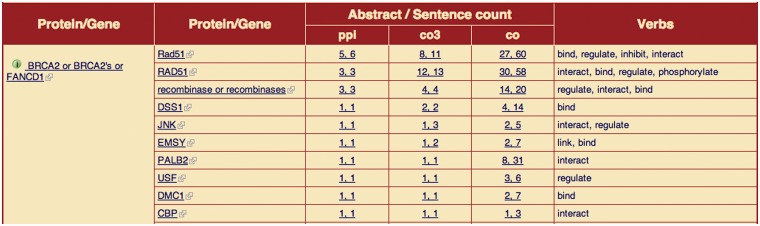
Figure 4.
PPI summary table. The screenshot displays in the top ranks those proteins that interact frequently with BRCA2 (using the query ‘Breast cancer’): amongst all proteins, RAD51 is most frequently linked to BRCA2 across the selection of documents. The frequency of findings per abstract and per sentence listed for each method is present as well [language pattern (ppi), tri-occurrence (co3) and co-occurrence (co)], including the interaction verbs.
Figure 5.
Example annotation sentences with PPIs. Highlighting of the evidences that allow better identification and curation of the PPIs. Each highlighted protein/gene is linked back to UniProt. Interaction verbs are denoted in square brackets.
In a more comprehensive evaluation, we have analysed which results can be produced from the biomedical literature, namely from MEDLINE abstracts, in comparison with results from full text articles, which are referenced in curated databases. IntAct provides a collection of text from full text articles and the extracted results. These were made available in BioCreative II and can be used for direct comparisons.
In a second evaluation, we have compared the performance of the SynPs considering the different types of verb forms on full text data in comparison with the BioCreative II PPI data set. This evaluation measures the performance of the openly accessible extraction methods against the publicly available benchmark data set.
Table 2 shows the results of running the extraction algorithms on the IntAct text mining corpus (IntAct sentences for text-mining, ftp://ftp.ebi.ac.uk/pub/databases/intact/current/various/data-mining). The corpus contains 9719 manually curated molecular interactions from 1551 publications. We ran PCorral’s extractors on the abstracts of the same set of the publications and then compared the extractions of each publication with the same publication’s interactions in the corpus. When all entities of an extracted interaction match those of an interaction from the same document in the corpus, a true positive is counted. With CO method, 17.54% interactions from the corpus are correctly identified, and 28.73% of overall predictions are correct. The precision increased when the interaction identification was based on CO3, however, with a significant drop on the recall. The extraction based on the SynPs achieved the highest precision, but largely sacrificing the recall.
Table 2.
Evaluation of COs, CO3, SynP for PPIs on MEDLINE abstracts
| Method | Predictions | Correct predictions | Precision (%) | Recall (%) | F-measure (%) |
|---|---|---|---|---|---|
| CO | 5934 | 1705 | 28.73 | 17.54 | 21.78 |
| CO3 | 1461 | 454 | 31.07 | 4.67 | 8.12 |
| SynP | 370 | 142 | 38.38 | 1.46 | 2.81 |
Table 3 shows the results of running the extraction algorithms on the BioCreative II (27) PPI full text sentences. We find that the recall on full text is higher compared with MEDLINE citations. On the other hand, the precision of MEDLINE information is much higher. We find that MEDLINE COs already deliver a large number of relations, which are reliable in terms of reproducibility of the results in the IntAct database.
Table 3.
Evaluation of CO, CO3, SynP for PPIs on the BioCreative II sentences
| Method | Predictions | Correct predictions | Precision (%) | Recall (%) | F-measure (%) |
|---|---|---|---|---|---|
| CO | 52 136 | 785 | 1.5 | 33.2 | 2.9 |
| CO3 | 15 823 | 609 | 3.8 | 28.8 | 6.8 |
| SynP | 2078 | 358 | 17.2 | 17.0 | 17.1 |
CO3 and SynP rely on verbs that we have collected from the research work using different experiments and then published as reference work (26). We now compare the performance of the different verbs against the content from the corpus to better understand their contributions to the correct predictions (c.f. Table 4). Only verbs from Table 1 that have contributed to PPI identification in the BioCreative II corpus have been listed in Table 4.
Table 4.
List of verbs that contributed to a correct prediction of related proteins
| Regulate | 179 | 12 | 6.7 | 0.6 | 1.0 |
| Contain | 286 | 12 | 4.2 | 0.6 | 1.0 |
| Inhibit | 130 | 9 | 6.9 | 0.4 | 0.8 |
| Mediate | 136 | 7 | 5.1 | 0.3 | 0.6 |
| Activate | 165 | 7 | 4.2 | 0.3 | 0.6 |
| Modulate | 31 | 5 | 16.1 | 0.2 | 0.5 |
| Precipitate | 31 | 4 | 12.9 | 0.2 | 0.4 |
| Express | 218 | 4 | 1.8 | 0.2 | 0.3 |
| Promote | 42 | 3 | 7.1 | 0.1 | 0.3 |
| Induce | 110 | 3 | 2.7 | 0.1 | 0.3 |
| Modify | 6 | 2 | 33.3 | 0.1 | 0.2 |
| Dephosphorylate | 8 | 2 | 25.0 | 0.1 | 0.2 |
| Complex | 15 | 2 | 13.3 | 0.1 | 0.2 |
| Stimulate | 41 | 2 | 4.9 | 0.1 | 0.2 |
| Downregulate | 6 | 2 | 33.3 | 0.1 | 0.2 |
| Methylate | 6 | 1 | 16.7 | 0.0 | 0.1 |
| Substitute | 7 | 1 | 14.3 | 0.0 | 0.1 |
| Assemble | 11 | 1 | 9.1 | 0.0 | 0.1 |
| Block | 30 | 1 | 3.3 | 0.0 | 0.1 |
| Suppress | 40 | 1 | 2.5 | 0.0 | 0.1 |
They are sorted according to their F-measure. The list can be used to tune an IE system for performance (e.g. for precision, recall, speed).
Amongst these verbs are the following: upregulate, dissociate, couple, link, overexpress, repress, inactivate, cleave and acetylate. When comparing the list of verbs from Table 4 to the proposed verbs from other authors (see Table 1), we identify that the verbs ‘downregulate’, ‘upregulate’, ‘inactivate’ and ‘stimulate’ do not play an important role, whereas ‘associate’ and ‘contain’ play an important role for the predictions.
The entries in Table 4 can be used to optimize the performance of an IE solution, i.e. selection of verbs with a high F-measure to improve the precision/recall ratio of the IE solution and integration of the best performing verbs to improve the overall coverage of the solution. Certainly, more knowledge about the subframe categorizations of the listed verbs will help to further optimize any IE solution and will give contributions to the event identification overall.
Discussion
We present a solution for the identification of PPIs from the scientific literature, which is unique in the sense that it combines IR and IE for PPIs and delivers high recall versus high precision results from the same distribution of documents. We argue that this approach supports curators in their work, as they can oversee results for PPIs at different levels of quality.
The curation of PPIs requires evidences from the scientific literature or other resources, PCorral produces such references. In addition, PCorral automatically interlinks the results with primary data resources from the biomedical research community, which enables further exploration and thus eases the curation process. The entity recognition in combination with the proposed extraction methods fulfils most of the relevant tasks for interactive curation. PCorral does not rely on syntactic parsing, therefore allowing fast processing on any input text that has not been processed before, e.g. grammatical relation analysis based on syntactic parsing on MEDLINE. Unlike machine-learning approach, its performance is independent from any gold standards.
We find as well that processing MEDLINE is different to processing full text articles. In the BioCreative II results in Table 3, we find that COs produce a combinatorial explosion of PPIs that we do not find when processing MEDLINE abstracts. This combinatorial explosion is mitigated by the SynPs. This result is important when we process documents that do not come from MEDLINE using the web service interface.
We have presented ways to extract relations from the scientific literature that can be combined into a single retrieval and extraction engine. From the methods used by PCorral, CO is the most general approach followed by CO3 and then the SynPs. We will further explore the integration of full parsing into the retrieval engine without compromising the retrieval throughput (i.e. recall through COs).
PPIs differ from other types of interactions, e.g. chemical–protein interaction, as products of interactions are often macromolecular complexes. This motivates that the retrieval is steering the choice of citations and the extraction of PPIs. In addition, this proposes challenges on anaphoric and metonymic co-reference, which we are willing to study to improve the performance of PCorral.
The current implementation of the PCorral interface works on MEDLINE, as there is limited access to full text articles. As we have seen in the evaluation, this is a drawback for the recall that can be currently achieved. We are planning to add full text capabilities to it as soon as more full text articles become available.
As highlighted in the ‘Methods’ section, PCorral pipeline is accessible from Whatizit, so ad-hoc documents can be processed on-the-fly using its web service capabilities. In addition, Whatizit is integrated into the Taverna bioinformatics workflow management system (http://www.taverna.org.uk/introduction/taverna-in-use/bioinformatics), which allows integrating PCorral with other workflow components for automatic processing.
Funding
The work has been funded by the Network of Excellence ‘Semantic Interoperability and Data Mining in Biomedicine' (NoE 507505), the EC's FP6 Strep project ‘BOOTStrep' (FP6 - 028099) and the EC's FP7 ICT Strep project ‘Mantra' (FP7-ICT-2011-4.1 - 296410). Chen Li is funded by the Cambridge Overseas Trust and the European Molecular Biology Laboratory (EMBL-EBI). Funding for open access charge: EC's FP7 ICT Strep project ‘Mantra' (FP7-ICT-2011-4.1 - 296410).
Conflict of interest. None declared.
References
- 1.Jaeger S, Gaudan S, Leser U, et al. Integrating protein-protein interactions and text mining for protein function prediction. BMC Bioinformatics. 2008;9:S8. doi: 10.1186/1471-2105-9-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kafkas S, Varouglu E, Rebholz-Schuhmann D, et al. Functional variation of alternative splice forms in their protein interaction networks: a literature mining approach. BMC Bioinformatics. 2010;11:S5. [Google Scholar]
- 3.Saric J, Jensen LJ, Ouzounova R, et al. Extraction of regulatory gene/protein networks from Medline. Bioinformatics. 2006;22:645–650. doi: 10.1093/bioinformatics/bti597. [DOI] [PubMed] [Google Scholar]
- 4.Kim JJ, Rebholz-Schuhmann D. Improving the extraction of complex regulatory events from scientific text by using ontology-based inference. J. Biomed. Semantics. 2011;2:S3. doi: 10.1186/2041-1480-2-S5-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Haagen HH, ‘t Hoen PA, Botelho Bovo A, et al. Novel protein-protein interactions inferred from literature context. PLoS One. 2009;4:e7894. doi: 10.1371/journal.pone.0007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerrien S, Aranda B, Breuza L, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkq973. D561–D568. doi:10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Licata L, Briganti L, Peluso D, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, et al. The BioGRID interaction database: 2011 update. Nucleic Acids Res. 2011;39(Suppl. 1):D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagel P, Kovac S, Oesterheld M, et al. The MIPS mammalian protein–protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 11.Ananiadou S, Pyysalo S, Tsujii J, et al. Event extraction for systems biology by text mining the literature. Trends Biotechnol. 2010;28:381–390. doi: 10.1016/j.tibtech.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Rebholz-Schuhmann D, Oellrich A, Hoehndorf R. Text-mining solutions for biomedical research: enabling integrative biology. Nat. Rev. Genet. 2012;13:829–839. doi: 10.1038/nrg3337. [DOI] [PubMed] [Google Scholar]
- 13.Hirschman L, Burns GA, Krallinger M, et al. Text mining for the biocuration workflow. Database. 2012;2012 doi: 10.1093/database/bas020. article ID bas020; doi:10.1093/database/bas020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakenberg J, Leaman R, Vo NH, et al. Efficient extraction of protein-protein interactions from full-text articles. IEEE/ACM Trans Comput Biol Bioinform. 2010;7:481–494. doi: 10.1109/TCBB.2010.51. [DOI] [PubMed] [Google Scholar]
- 15.Daraselia N, Yuryev A, Egorov S, et al. Extracting human protein interactions from MEDLINE using a full-sentence parser. Bioinformatics. 2004;20:604–611. doi: 10.1093/bioinformatics/btg452. [DOI] [PubMed] [Google Scholar]
- 16.Rinaldi F, Schneider G, Kaljurand K, et al. Mining of relations between proteins over biomedical scientific literature using a deep-linguistic approach. Artif. Intell. Med. 2007;39:127–136. doi: 10.1016/j.artmed.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Miwa M, Saetre R, Miyao Y, et al. Protein-protein interaction extraction by leveraging multiple kernels and parsers. Int. J Med. Inform. 2009;78:e39–e46. doi: 10.1016/j.ijmedinf.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Hao Y, Zhu X, Huang M, et al. Discovering patterns to extract protein-protein interactions from the literature: part II. Bioinformatics. 2005;21:3294–3300. doi: 10.1093/bioinformatics/bti493. [DOI] [PubMed] [Google Scholar]
- 19.Kim JD, Wang Y, Takagi T, et al. Overview of Genia Event task in BioNLP shared task 2011. ACL HLT. 2011;2011 doi: 10.1186/1471-2105-13-S11-S1. 7–15. http://aclweb.org/anthology/W/W11/W11-18.pdf#page=19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann R, Valencia A. Implementing the iHOP concept for navigation of biomedical literature. Bioinformatics. 2005;21(Suppl 2) doi: 10.1093/bioinformatics/bti1142. ii252–i258. http://www.ncbi.nlm.nih.gov/pubmed/16204114. [DOI] [PubMed] [Google Scholar]
- 21.He M, Wang Y, Li W. 2009 PPI Finder: a mining tool for human protein-protein interactions (K. Selvarajoo, Ed.). PLoS One, 4, 6. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2641004&tool=pmcentrez&rendertype=abstract. [Google Scholar]
- 22.Tsuruoka Y, Tsujii J, Ananiadou S. FACTA: a text search engine for finding associated biomedical concepts. Bioinformatics. 2008;24:2559–2560. doi: 10.1093/bioinformatics/btn469. http://bioinformatics.oxfordjournals.org/cgi/content/abstract/24/21/2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rebholz-Schuhmann D, Kirsch H, Arregui M, et al. EBIMed–text crunching to gather facts for proteins from Medline. Bioinformatics. 2007;23:e237–e244. doi: 10.1093/bioinformatics/btl302. http://www.ncbi.nlm.nih.gov/pubmed/17237098. [DOI] [PubMed] [Google Scholar]
- 24.Rebholz-Schuhmann D, Arregui M, Gaudan S, et al. Text processing through Web services: calling Whatizit. Bioinformatics. 2008;24:296–298. doi: 10.1093/bioinformatics/btm557. http://www.ncbi.nlm.nih.gov/pubmed/18006544. [DOI] [PubMed] [Google Scholar]
- 25.Huang M, Zhu X, Hao Y, et al. Discovering patterns to extract protein-protein interactions from full texts. Bioinformatics. 2004;20:3604–3612. doi: 10.1093/bioinformatics/bth451. http://www.ncbi.nlm.nih.gov/pubmed/15284092. [DOI] [PubMed] [Google Scholar]
- 26.Rebholz-Schuhmann D, Jimeno-Yepes A, Arregui M, et al. Measuring prediction capacity of individual verbs for the identification of protein interactions. J. Biomed. Inform. 2010;43:200–207. doi: 10.1016/j.jbi.2009.09.007. http://linkinghub.elsevier.com/retrieve/pii/S153204640900135X. [DOI] [PubMed] [Google Scholar]
- 27.Krallinger M, Leitner F, Rodriguez-Penagos C, et al. Overview of the protein-protein interaction annotation extraction task of BioCreative II. Genome Biol. 2008;9(Suppl. 2):S4. doi: 10.1186/gb-2008-9-s2-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C, Liakata M, Rebholz-Schuhmann D. Biological network extraction from scientific literature. Briefings in Bioinformatics. 2013 doi: 10.1093/bib/bbt006. [DOI] [PubMed] [Google Scholar]