Summary
We show that amino acid co-variation in proteins, extracted from the evolutionary sequence record, can be used to fold transmembrane proteins. We use this technique to predict previously unknown, 3D structures for 11 transmembrane proteins (with up to 14 helices) from their sequences alone. The prediction method (EVfold_membrane), applies a maximum entropy approach to infer evolutionary co-variation in pairs of sequence positions within a protein family and then generates all-atom models with the derived pairwise distance constraints. We benchmark the approach with blinded, de novo computation of known transmembrane protein structures from 23 families, demonstrating unprecedented accuracy of the method for large transmembrane proteins. We show how the method can predict oligomerization, functional sites, and conformational changes in transmembrane proteins. With the rapid rise in large-scale sequencing, more accurate and more comprehensive information on evolutionary constraints can be decoded from genetic variation, greatly expanding the repertoire of transmembrane proteins amenable to modelling by this method.
Introduction
Membrane proteins allow cells to interact with the extracellular environment and to communicate with other cells. More than 25% of all human proteins have integral membrane domains; many of these are medically important, with nearly half of all drug targets containing a membrane domain (Bakheet and Doig, 2009; Overington et al., 2006). Knowing the three-dimensional (3D) structure of a membrane protein facilitates the characterizations its molecular mechanism and accelerates the development of pharmacological agents targeting its (Katritch et al., 2012). Despite great progress in determining structures by experimental methods (Chen et al., 2010; Cherezov et al., 2007; Choe et al., 2011; Long et al., 2007; Miller and Long, 2012; Rasmussen et al., 2011; Rasmussen et al., 2007), the 3D structures of most transmembrane proteins remain unknown and comparative modeling maximally covers 10% of all human transmembrane proteins. Efficient and accurate computational approaches that predict 3D-structures of membrane proteins would be a valuable tool to complement existing experimental approaches.
Well-established methods of structure prediction, such as energy minimization and database fragment searches, have previously addressed the problem of prediction of transmembrane protein structures. However, these calculations were limited both in protein size (≤ 7 transmembrane helices) as well as accuracy, despite added information on helix-helix contact predictions from experimentally non-homologous structures and a few known experimentally determined contacts (Barth et al., 2009; Yarov-Yarovoy et al., 2006).
It is possible that constraints on the function and structure of proteins are reflected in conserved interactions between pairs, or groups, of amino acids. If so, then evolutionary correlations may be observed between specific sequence positions. Previous work has attempted to use correlations between residues, amongst other methods, to predict structural proximity and functional features (Fuchs et al., 2007) (Cronet et al., 1993; Fatakia et al., 2009; Horn et al., 1998; Nemoto et al., 2004). The most accurate of these strategies use global statistical methods, such as maximum entropy (Marks et al., 2011; Morcos et al., 2011), Bayesian networks (Burger and van Nimwegen, 2010) or covariance estimation (Jones et al., 2012; Meinshausen and Buhlmann, 2006). However, only recently it was reported that a maximum-entropy analysis of residue correlations in sequence families could provide sufficient information about proximity of residues in 3D to compute correct folds of protein structures in 15 example cases, using EVfold (Marks et al., 2011).
Here, we report the development of an algorithm EVfold-membrane that enables de novo prediction of 3D structures of unknown alpha-helical transmembrane proteins from evolutionary constraints, using neither fragments, threading, nor homologous 3D structures. We predict the structures of 11 transmembrane proteins of unknown structure, including six pharmacological targets (Figure 1, Table 1). To verify that our predicted structures are plausible, we systematically test our ability to predict, in blinded fashion, the structures of a diverse set of 25 transmembrane proteins with known 3D structures (Table 1) and find an unprecendented level of agreement with the cognate crystal structures (TM-scores > 0.5 for 22/25 of the benchmarked proteins). We find that functionally important regions of each protein tend to be more accurately predicted than the protein as a whole and that residues subject to multiple pair constraints tend to be in substrate binding pockets, oligomerization interfaces, and/or involved in conformational changes.
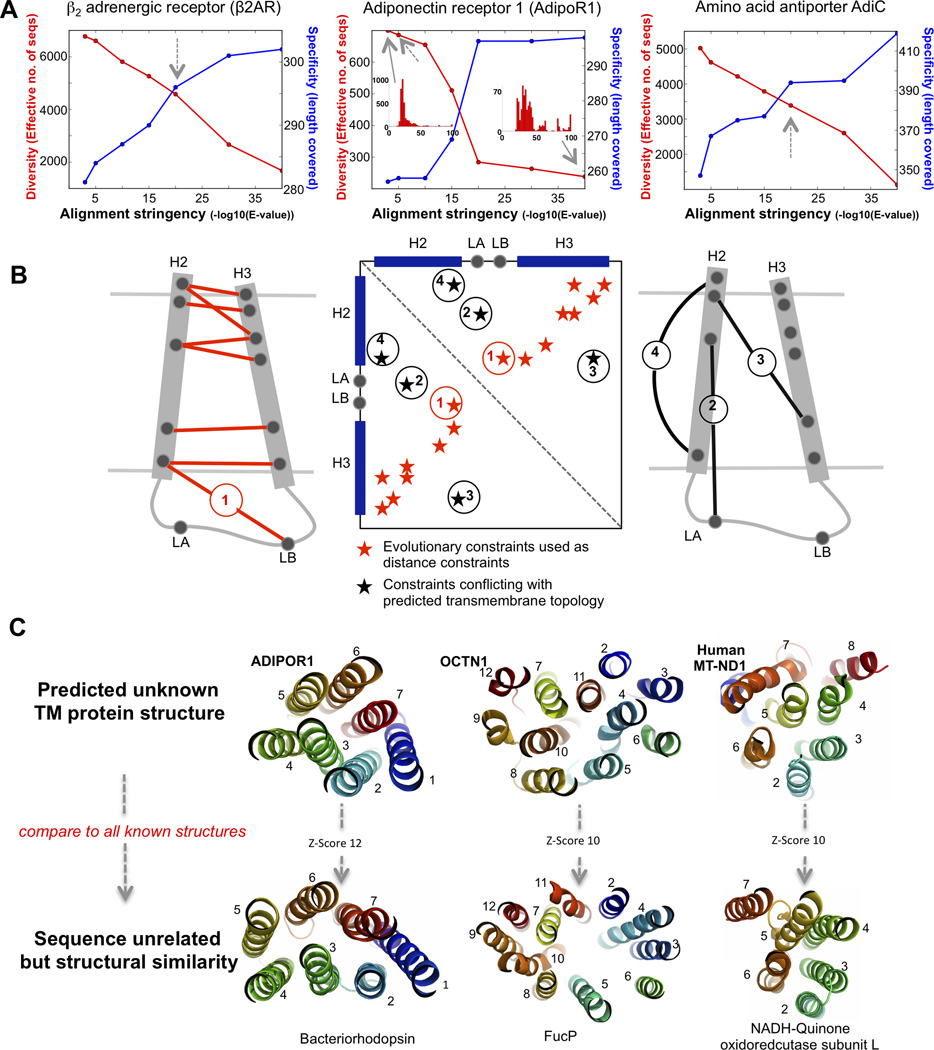
Figure 1. De novo predicted 3D models of membrane proteins with no known structure (related to Figure S1).
Cartoon shows evolutionary couplings as calculated by EVfold – membrane placed as distance constraints on extended polypeptide before folding. Top ranked models of a representative set of 6 transmembrane proteins from diverse families, which have no members with known 3D structures. Models are cartoon representation with rainbow coloring blue N terminal to red C terminal, seen from the side (left) and non-cytoplasmic side (right). Naming conventions, 3D coordinates and input files in Tables 1, S1 Data S1, Data S2–5.
Table 1.
Predicted proteins of known and unknown experimental structure
|
Uniprot name known structure |
length | TMH1 | E-val2 | model length |
#seq3 | top #4 | TM5 | Ca-rmsd6 | best #4 | TM5 | Ca-rmsd6 | PDB7 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADIC_SALTY | 445 | 12 | E-20 | 394 | 24284 | 240_15 | 0.67 | 4.2 (300) | 240_15 | 0.67 | 4.2 (300) | 3ncyA |
| ADRB2_HUMAN | 413 | 7 | E-20 | 296 | 35593 | 160_5 | 0.67 | 3.3 (201) | 160_5 | 0.67 | 3.3 (201) | 2rh1A |
| ADT1_BOVIN | 298 | 6 | E-40 | 285 | 9828 | 200_20 | 0.48 | 3.8 (136) | 270_17 | 0.51 | 4.0 (152) | 1okcA |
| AMTB_ECOLI | 428 | 10 | E-5 | 396 | 4407 | 270_17 | 0.67 | 3.9 (262) | 280_5 | 0.67 | 3.6 (260) | 1xqfA |
| AQP4_HUMAN | 323 | 6 | E-10 | 215 | 6469 | 80_19 | 0.50 | 2.9 (100) | 100_14 | 0.51 | 3.4 (110) | 3gd8A |
| BTUC_ECOLI | 326 | 10 | E-10 | 299 | 12926 | 250_19 | 0.67 | 3.2 (209) | 250_19 | 0.67 | 3.2 (209) | 1l7vA |
| C3NQD8_VIBCJ | 461 | 12 | E-20 | 431 | 13864 | 250_11 | 0.62 | 4.6 (306) | 290_8 | 0.63 | 4.3 (305) | 3mktA |
| C6E9S6_ECOBD | 485 | 14 | E-10 | 412 | 63730 | 180_9 | 0.63 | 4.2 (299) | 180_9 | 0.63 | 4.2(299) | 3rkoN |
| COX1_BOVIN | 514 | 12 | E-40 | 486 | 73822 | 150_6 | 0.66 | 4.5 (360) | 150_11 | 0.66 | 4.4 (354) | 1occA |
| COX3_BOVIN | 261 | 7 | E-3 | 182 | 10705 | 50_9 | 0.69 | 2.8 (151) | 50_9 | 0.69 | 2.8 (151) | 1occC |
| CYB_BOVIN | 379 | 8 | E-3 | 335 | 43891 | 120_4 | 0.58 | 4.1 (203) | 100_9 | 0.64 | 3.7 (231) | 1pp9B |
| FIEF_ECOLI | 300 | 6 | E-5 | 197 | 9722 | 200_10 | 0.59 | 2.8 (119) | 40_7 | 0.63 | 2.8 (131) | 3h90A |
| GLPG_ECOLI | 276 | 6 | E-5 | 169 | 5263 | 120_11 | 0.64 | 2.6 (126) | 120_11 | 0.64 | 2.6 (126) | 3b45A |
| GLPT_ECOLI | 452 | 12 | E-30 | 402 | 24912 | 330_12 | 0.67 | 3.8 (283) | 330_13 | 0.67 | 4.0 (297) | 1pw4A |
| METI_ECOLI | 217 | 6 | E-15 | 206 | 30400 | 120_17 | 0.46 | 3.5 (93) | 120_6 | 0.48 | 3.4 (94) | 3dhwA |
| MIP_BOVIN | 263 | 6 | E-10 | 212 | 6468 | 150_12 | 0.55 | 3.1 (116) | 130_20 | 0.58 | 2.9 (124) | 1ymgA |
| MSBA_SALTY | 330 | 6 | E-3 | 310 | 29034 | 100_12 | 0.57 | 3.3 (180) | 110_12 | 0.61 | 3.5 (208) | 3b60A |
| O67854_AQUAE | 513 | 12 | E-3 | 463 | 4500 | 280_4 | 0.55 | 5.1 (274) | 170_20 | 0.58 | 4.8 (286) | 2a65A |
| OPSD_BOVIN | 348 | 7 | E-20 | 274 | 35901 | 110_16 | 0.70 | 3.3 (214) | 110_16 | 0.70 | 3.3 (214) | 1hzxA |
| Q87TN7_VIBPA | 485 | 8 | E-10 | 407 | 4097 | 270_12 | 0.59 | 4.0 (242) | 260_19 | 0.60 | 4.2 (258) | 3pjzA |
| Q8EKT7_SHEON | 516 | 12 | E-10 | 447 | 12063 | 100_14 | 0.40 | 4.6 (160) | 240_19 | 0.43 | 4.8(183) | 2xutA |
| Q9K0A9_NEIMB | 315 | 10 | E-10 | 297 | 4244 | 270_9 | 0.44 | 3.6 (131) | 120_9 | 0.49 | 3.9 (138) | 3zuxA |
| SGLT_VIBPA | 543 | 14 | E-5 | 487 | 9563 | 310_11 | 0.49 | 4.6 (214) | 340_10 | 0.53 | 4.8 (264) | 2xq2A |
| TEHA_HAEIN | 328 | 10 | E-3 | 304 | 1861 | 70_15 | 0.51 | 4.1 (154) | 210_17 | 0.56 | 4.0 (175) | 3m71A |
| URAA_ECOLI | 429 | 14 | E-3 | 393 | 14992 | 250_12 | 0.50 | 4.8 (194) | 250_5 | 0.50 | 4.5 (189) | 3qe7A |
| unknown structure | structural similarity to | Z8 | Ca-rmsd6 | PDB7 | ||||||||
| ADR1_HUMAN | 375 | 7 | E-5 | 223 | 3410 | 150_14 | bacteriorhodopsin | 12 | 4.5 (204) | 3haoA | ||
| NU1M_HUMAN | 318 | 8 | E-10 | 282 | 17558 | 210_18 | Mit. complex1 subunit L | 10 | 5.0 (170) | 3rkoL | ||
| S22A4_HUMAN | 551 | 12 | E-30 | 373 | 21704 | 220_11 | L-fucose permease FucP | 10 | 6.0 (267) | 3o7qA | ||
| ABCG2_HUMAN | 655 | 7 | E-10 | 274 | 5404 | 210_3 | - | - | - | - | ||
| ELOV4_HUMAN | 314 | 7 | E-3 | 233 | 1436 | 190_6 | - | - | - | - | ||
| SL9A1_HUMAN | 815 | 13 | E-10 | 367 | 6020 | 210_17 | Acriflavine res. prot. AcrB | 4 | 4.7 (165) | 2gifA | ||
| MSMO1_HUMAN | 293 | 5 | E-20 | 220 | 897 | 70_13 | - | - | - | - | ||
| S13A1_HUMAN | 595 | 15 | E-20 | 543 | 1836 | none9 | - | - | - | - | ||
| EAMA_ECOLI | 299 | 10 | E-5 | 276 | 31753 | 250_10 | - | - | - | - | ||
| LIVH_ECOLI | 308 | 8 | E-3 | 282 | 23968 | 230_16 | Permease protein BtuC | 6 | 4.1 (140) | 1l7vA | ||
| GABR1_HUMAN | 961 | 7 | E-5 | 298 | 2871 | 190_19 | β2 adrenergic receptor | 6 | 6.0 (191) | 3p0gA | ||
number of transmembrane helices
E-value for HHblits sequence search
number of sequences in multiple sequence alignment
number of evolutionary constraints used and model number of blind top ranked and best generated model, respectively
TM score
Ca-root mean square deviation in Å
accession code and chain of PDB structure
DALI Z-score
no model looks plausible (large protein, few sequences)
Results
Global statistical approach for protein structures from sequences
Our hypothesis is that evolution conserves interactions between residues that are important to maintaining structure and function by constraining the sets of mutations accepted at interacting sites. To find these constraints couplings for each membrane protein, we build a multiple sequence alignment (Remmert et al., 2011) with sufficiently diverse sequences to detect evolutionary co-variation and minimize statistical noise. To maximize the power of detection, we developed a method to optimize the trade-off between the number of sequences aligned (i.e., depth) and alignment specificity, a proxy for functional similarity to the query sequence, which is quantified by the sequence range (i.e., breadth) covered by the alignment (Figures 2A and S2, Experimental Procedures). For example, for bovine Adt1, which catalyzes the exchange of cytoplasmic ADP with mitochondrial ATP, we use a stringency value (E) of 10−40, ensuring that 70% of its residues in the sequence are covered by the alignment. In general, for a protein of length L, we require at least 3L sequences and to cover at least 0.7*L of the residues in the sequence of interest.
Figure 2. From sequence alignment to folded structures. (Related to Figure S2).
A. Building the alignment for the EC calculation for the specific query protein requires a trade-off between specificity and diversity. To investigate this blindly, we scan a range of alignment depths using different expectation values, calculate the effective number of sequences returned (diversity) and the number of residues in our query protein sequence which do not have more than 30% gaps in the alignment column of the alignment (coverage); Dashed arrows point to chosen stringency for folding. Contrast in the distribution of sequence space at different alignment depths in histograms of the range of number of sequences with the 0–100 % identity to query protein sequence (Insets, middle panel) (related to Figure S2).
B. Schematic showing constraint conflict resolution between predicted co-evolution and predicted secondary structure/membrane topology. In all cases we follow the predicted membrane topology and discard co-evolving residue pairs that conflict with this prediction. The predicted toy contact map (middle panel), shows evolutionary constraints that conflict with the predicted membrane topology that are removed (black stars). evolutionary constraints that do not conflict with the predicted membrane topology are not removed, irrespective of any knowledge about their distance in 3D space (constraint 1)
C. The top ranked model from the set of each de novo predicted structures was compared to the entire PDB using the structural alignment program DALI (Holm and Sander, 1995). 3 of the 6 predicted 3D TM protein structures with significant structural similarities to known transmembrane protein folds are shown.
To discover residue interactions that are conserved by evolution we developed an algorithm that extracts patterns of amino acid co-evolution from these sequence alignments (Lapedes et al., 1997; Marks et al., 2011; Morcos et al., 2011). The algorithm, using entropy maximization. reduces the set of all correlations between pairs of positions in the sequence to an essential set, which best explains all the other correlations and are therefore likely to be causative (i.e., likely to reflect residue interactions constrained in evolution). Our statistical approach is thus in a class of algorithms addressing the classic problem of deriving 'causation from correlation'. Our ‘global’ statistical approach is different from ‘local’ approaches, such as mutual information (MI), and variants thereof (Fodor and Aldrich, 2004; Livesay et al., 2012). The MI of pairs of columns in a sequence alignment is ‘local’ in that it quantifies co-variation for each pair independently of all other pairs, potentially leading to inconsistencies. The simplest inconsistencies in local models are transitive correlations, e.g., correlations between a non-contact pair A–C in a triplet A–B–C that arise from transitive influence in contact pairs A–B and B–C. Thus, pairs with high MI scores are not necessarily constrained by a direct interaction effect, even if they are correlated.
In contrast, our entropy maximization approach builds a probability model for the entire sequence, such that the scores for each pair of residues are consistent with other pairs, thereby preventing high scoring from transitive relationships in the data. Starting with a simple covariance matrix between all pairs of columns in the alignment, entropy maximization gives rise to a formalism similar to the well-known inverse Ising model of ferromagnetism (in which there are two states) except that for protein sequences each site (i.e., sequence position) can be assigned to one of 21 discrete states (20 amino acids or a gap), as in the Potts model in physics. The numerical parameters in the entropy maximization method (analogous to the spin-spin interactions in the Ising model Hamiltonian) can be computed efficiently by inverting a covariance matrix. This algorithmic entropy maximization solution is similar to partial correlation methods in Gaussian Graphical Models for continuous distributions (Dempster, 1972). In entropy maximization, after the covariance matrix inversion, the residue pair scores, or evolutionary coupling scores, are consistent with the correlation data between pairs of positions and single column data, including conservation, while making a minimum set of other assumptions. While constrained interactions can arise from diverse evolutionary requirements, we find that many reflect interactions between residues close in space and are thus highly productive as distance constraints for protein folding (Marks et al., 2011).
The structure of transmembrane proteins is additionally constrained by the presence of the membrane. Hence we can blindly remove predicted coevolved pairs for which 3D proximity is unlikely (Figures 2B, and S3, Experimental Procedures). The resulting set of evolutionary constraints and the predicted secondary structure are interpreted as distance constraints on extended polypeptide chains (Data S1). Distance geometry and out-the-box simulated annealing using the CNS software (Brunger et al., 1998) are used to fold the chain ab initio to produce around 500 3D all-atom coordinate models for each protein. To assess the set of predicted structures for each protein, we apply an automated membrane-specific ranking of the computed models, which combines the quality of secondary structure formation, lipid accessibility of the residues, and a measure of violation of the evolutionary constraints and cluster the structures, excluding predictions not represented in the larger clusters (Experimental Procedures).
Prediction of unknown 3D structures of alpha-helical transmembrane proteins
A survey of targets in the DrugBank database (Knox et al., 2011) for transmembrane proteins yielded 20 non-redundant families with more than 1000 sequences, ≥5 predicted transmembrane helices, and without a known 3D structure for any family member (Tables 1 and S1, Experimental Procedures). We selected 11 of these targets for detailed analysis, covering diverse sizes and functional types, with several of these having more than one drug target. Coordinates for the remaining 9 families are available at evfold.org/transmembrane. These proteins are implicated in many diseases including diabetes, obesity, Crohns disease, breast cancer, Leber hereditary optic neuropathy, Alzheimer’s disease, and Parkinson’s disease (Holland et al., 2011; Pei et al., 2011; Peltekova et al., 2004; Yamauchi et al., 2003) (Doyle et al., 1998) (Natarajan et al., 2012); (Howell et al., 1991; Jaksch et al., 1996) (Aldahmesh et al., 2011; Zhang et al., 2001). We predicted 400–600 all-atom 3D models for each protein (Data S2 and Experimental Procedures). The predicted structures of five of the proteins had similar folds to other known 3D membrane protein structures (Figure 2C) despite negligible sequence similarity, a recurring theme seen in structural genomics and earlier work (Holm and Sander, 1993; Murzin, 1993). Predicted structures of three membrane proteins show some structural similarities to those of other sequence-distant members of the same PFAM clan. A search against all known 3D structures with our top-ranked model of the human Octn1, a 12-helical transporter sugar transporter, yields several significant hits to structures in the Major Facilitator Superfamily, including FucP (PDB:3o7q (Dang et al., 2010), and GlpT (PDB:1pw4 (Law et al., 2008), (Figures 1 and 2C, Tables 1 and S1). FucP and GlpT sequences were not in our alignment and have only 10% and 7% sequence identity to Octn1 respectively, below the level allowing inference of structural homology. Similarly, the 8 transmembrane helical E. Coli LIVH, a high affinity leucine transporter, is structurally similar to the bacterial B12 uptake protein BtuC (PDB:1l7v (Locher et al., 2002), despite only 8% sequence identity between the proteins (Tables 1 and S1). Thirdly, the predicted structures of the GABA receptor 1, a protein involved in synaptic inhibition and a pharmacological target, are structurally similar to other GPCRs despite a negligible sequence identity (10%), (Figures 1 and S1, Table 1). Although this result is not so surprising, the sequence diversity in GPCRs is sufficiently high that de novo computation of the 3D structure from evolutionary couplings may be of interest, in addition to model building by remote homology (Katritch et al., 2012). (Figure 1, Figure S1, Table 1). In the predicted models of the GABA receptor, a lack of well-ordered structure formed by the extracellular loops, and a lack of β-sheet formation by the predicted β-strands, indicate potential model errors. Nevertheless, high scoring predicted residue pair interactions in the extracellular region, specifically between loops 2/3 and 3/4, are located close to the putative extracellular ligand binding domain. Given the moderate number of sequences in this GABA receptor family, we expect the current accuracy to be limited, but the models may serve as a useful starting point for further iteration using hybrid approaches and different alignment depths.
The five top ranked predicted adiponectin receptor 3D structures are surprisingly similar (~ 4.5 Å Cα-rmsd over 204 residues) to the bacteriorhodopsin crystal structure (PDB:3hao), with highly significant Dali (Holm and Sander, 1995) Z-scores between 7 and 13, despite negligible sequence identity (8%) (Figures 1 and 2C, Data S2). Although adiponectin receptor is a 7-transmembrane protein, it was not previously thought to have structural or functional similarity to G-protein coupled receptors and is inverted with respect to the membrane (Yamauchi et al., 2003). Assuming our predictions are accurate, it remains an open question whether the similarity of AdipoR1 to the GPCR fold is an example of divergent evolution or the result of convergent evolution to an exceptionally robust 7-helical fold.
We also find significant structural similarity of predicted structures of the human MT-ND1 subunit to the recently-solved structure of one of the major membrane subunits of Respiratory Complex I (E. coli, 3rko-C, (NuoL subunit) (Efremov and Sazanov, 2011); again, the sequences of MT-ND1 and the NuoL subunit are unrelated with <8% identity. However, we do not find high topological similarity to the coarse grained model of the bacterial NuoL subunit (homologous to MT_ND1), which was solved at low resolution without residue assignment (Efremov et al., 2010), and the NuoL subunit is almost double the size of our modeled protein. Nevertheless, our MT-ND1 structures overlay optimally on precisely the regions of bacterial subunits that are structurally duplicated within each protein (in NuoL, TM helices 3–7 and 8–15), further supporting the idea that this is a repeating structural evolutionary module (Efremov and Sazanov, 2011). Since these mitochondrial subunits are functionally related and spatially coincident throughout evolution, the structural relationship between them may plausibly result from divergent evolution of the sequence. Taken together, these examples of structural relationships between the predicted models and the structures of functionally related, but sequence-distant proteins, provides support for the accuracy of the de novo prediction.
Benchmark: blinded prediction of known 3D structure transmembrane proteins
To evaluate the performance of the prediction protocol, we computed the 3D-structures of α-helical membrane proteins of known structure from the proteins’ sequences alone, i.e. ignoring all aspects of known 3D structures, including sequence-similar fragments. We selected all α - helical membrane proteins from all Pfam families that have more than 1000 sequences, sufficient sequence coverage and more than 4 helices. This resulted in a set of 25 membrane proteins with up to 487 residues (up to 14 transmembrane helices) in 23 structurally diverse families. This set includes the human β2 adrenergic receptor (GPCR family), the S. typhimurium arginine/agmatine antiporter ADIC (amino acid/polyamine transporter superfamily), and the E. coli Glycerol-3-phosphate transporter (GlpT) (Major Facilitator Superfamily) (Table 1, Data S3–5).
The EVfold-membrane protocol provides a ranked set of predicted structures for each protein, which we then compare to a cognate crystal structure. The combined score used for ranking the generated models reliably identifies structures of high accuracy and in some cases even the best model in the top 10 (Tables 1 and S1 and Figure S4).. Overall, 21 of our test set of 23 diverse α-helical transmembrane proteins are reliably predicted with template modeling (TM) scores of 0.5–0.7 and Cα-rmsd 2.6–4.8Å over > 70% of the length (Figure 3A and 3B, Tables 1 and S1). Template modeling score (range 0.0–1.0), is considered reasonable when >0.5 and is comparable across proteins of varying lengths (Zhang and Skolnick, 2004). This blindly predicted set allows assessment of the relationship between the number of evolutionary constraints that are not spatially close in the cognate crystal structure (false positives) and the accuracy of our 3D structure prediction. The highest ranked evolutionary constraints (1–20) contain ~ 2% false positives, while the proportion of true positives decreases monotonically as a function of the number of constraints (Figure S3). However, the accuracy of folding, as measured by TM score, is remarkably robust to variation in the proportion of true positives and is stable over many different folding experiments, in which the numbers of constraints is steadily increased (Figure 3C). Details of the distribution of predicted contacts along the protein chain and the precise nature of false positives, such as mutual effective cancellation, may contribute to this robustness.
Figure 3. Accuracy of blinded 3D structure prediction for candidates with known structure. (Related to Figure S3, Figure S4).
A. Structural superpositions of predicted structures (blue) onto experimental structures (grey). First panel for each protein: side view from within the membrane; second panel: top-down view from non-cytoplasmic side. All figures rendered with PyMOL.
B. Accuracy of 3D structure prediction for candidates with known structure: Template modeling score (TM score) (Zhang and Skolnick, 2004) of the best model for each protein plotted against the number of sequences in the multiple sequence alignment, normalized by modeled protein length C. 3D prediction accuracy is surprisingly stable as the true positive rate of evolutionary constraints decreases, going down the list of ranked EC's. The TM score of the best prediction (blue) and the true positive rate (red) are plotted for increasing numbers of evolutionary constraints (divided by the number of residues in the protein to allow comparison between proteins). Distance cutoffs to define true contacts of true positive rate are 5Å (red dots), 7Å (red dashes) and 8Å (red) (Figure S3, Figure S4 and Data S2–5)
Currently, state-of-the-art approaches for de novo folding are based primarily on searching for sequence-similar fragments in 3D structure databases followed by fragment assembly using specially designed empirical force fields. The key limitation is the enormous size of the conformational search space. Our approach overcomes this limitation using the information in the evolutionary constraints and its direct translation to 3D coordinates via distance geometry translates, leading to a considerable performance advantage relative to earlier methods. The advantage is apparent in terms of (1) protein size range, (2) prediction accuracy, (3) efficiency of conformational search and lack of dependence on fragments and helix-helix contacts from previously solved 3D structures. (1) More than 50% of membrane proteins have 8–14 transmembrane helices. Here, we report models of proteins with up to 14 helices and anticipate that our method will allow the prediction of even larger membrane proteins, as we see no deterioration of accuracy with size (up to almost 500 residues) and obtain accurate 3D fold with as little as one constraint per residue over the entire size range. In contrast, previous prediction tools have been used to generate models for proteins with only 4–7 helices reported (Barth et al. 2009). (2) To compare accuracy, we predicted structures for 5 of the same proteins predicted by Barth et al. (2009) (Table S2). Our method reached the threshold coordinate accuracy of 4Å over comparable or significant larger regions (e.g., 89% rather than 40% of residues for bovine rhodopsin), and (3) it explored conformational search space more efficiently (e.g., ~500 candidate models compared to 200,000 generated in (Barth et al., 2009)). As a result of this efficiency gain, in current practice, EVfold all-atom models can be generated on a laptop in a few minutes per structure, without the need for supercomputers. A possible conceptual and practical advantage with the EVfold-membrane method is information about the roles of residues and residue interactions in protein function, as a result of extracting coupling information at the protein level filtered through functional selection over a myriad of evolutionary experiments.
While the results from our validation set of proteins are encouraging, they raise the question of whether we can predict the success of our approach for any given protein of interest, based on sequence information alone. In general, the accuracy of the predicted model increases with the number of sequences in the alignment normalized for the length of the protein (Figure 3B). For instance, the predicted structures of two proteins, a proton/peptide symporter and a bile acid symporter, have the lowest TM scores (0.4–0.5) compared to their cognate crystal structures, and have amongst the lowest number of sequences per residue in their input alignments (26 and 3, Table S1). Conversely, the predicted structure of bovine rhodopsin has 131 sequences per residue, and an excellent TM score of 0.7. Thus the number of sequences, the diversity of sequences, and the coverage of the length of the protein will no doubt be important metrics in estimating the likely accuracy of predictions and will be used to develop metrics for more accurate and more subfamily-specific structure calculations.
Evolutionary constraints include homo-oligomer contacts
Not all residue interactions that are strongly constrained by evolution are close in the 3D structure of the monomeric protein. Residue pairs close in transmembrane protein homoligomers may thus appear in conflict with other monomer constraints and/or the predicted 3D fold. For example, in the computed structure of the ABC transporter, S. typhimurium MsbA, evolutionary couplings between transmembrane helix 2 and transmembrane helices 5 and 6, are false positives with respect to monomer structure, but true positives with respect to the crystal structure dimer interface, (PDB:3b60 (Ward et al., 2007), (Figure 4A). Similarly, E. coli MetI has a cluster of evolutionary couplings with residues that are not in contact in the monomer but form contacts in the dimer (PDB:3dhw (Kadaba et al., 2008)). If successfully identified, the removal of the conflicting oligomer evolutionary couplings from the folding calculation improves the accuracy of prediction for the monomer (blinded test done in MsbA and MetI, data not shown).
Figure 4. Evolutionary constraints on residue pairs in oligomerization interfaces. (Related to Figure S5).
Contact maps of top ranked predicted EC's (red stars in A and B) overlaid on crystal structure contacts (grey, known only in A). Residue pairs coevolving due to inter-monomer contacts in the homo-oligomer (black circles) in an overlay of top ranked predicted evolutionary constraints (red) experimental structure contacts (grey), where known, on contact maps for each protein. In the monomer (blue or green ribbon with blue or green residue balls), the corresponding residue pairs would be false positive contacts (blue with blue or green with green do not make contact in the monomer), but would be true positives in the homo-oligomer structure (contacting blue-green pairs). A. Four examples of inference of oligomer contacts from EC's of known 3D structures (also Figure S5). B. Predicted dimer contacts of AdipoR1, shown on predicted monomer structures. EC pairs (black circles) at a large distance in monomer structure (~ 23A, green with green, blue with blue) are close (green-blue contact pair) in predicted dimers. Predicted dimer cartoon (right) is a rough estimate, produced by manual-visual docking of monomers, satisfying the majority of predicted dimer interface EC pairs (middle panel).
We also predict oligomer contacts for proteins of unknown 3D structures, such as AdipoR1. To identify potential dimerization contacts, we noted that some evolutionary constraints are inconsistent with the monomer predicted structure, and may therefore be involved in the putative dimerization interface. Interpreting these evolutionary constraints as distance constraints between residues in two separate monomer structures, shows that the AdipoR1 dimer interface involves contacts between the loop from helices 4 to 5 and both helices 1 and 7 (Figure 4B). Consistent with our prediction of the dimerization region are experimental observations that mutations in the GXXXG motif on transmembrane helix 5 of AdipoR1 disrupt dimerization (Kosel et al., 2010). Q335 on the transmembrane helix 7 is unusually strongly constrained, in spite of a low 19% conservation level as a single residue, as a partner in over 11 evolutionary couplings, some of which may be across this putative interface (Figure 4B). These examples suggest that homo-oligomer contact detection using evolutionary coupling pairs, may yield valuable testable information. It remains an algorithmic challenge to identify such evolutionary couplings between the components of oligomers in a more automated fashion.
Evolutionary constraints reflect conformational change
Many proteins can adopt different distinct conformations as part of their function (Tokuriki and Tawfik, 2009). Can we correctly predict more than one 3D conformation of a protein by extracting and analyzing evolutionary couplings from one set of protein sequences? We investigated this challenge by an analysis of known structures and genuine prediction., GlpT and Octn1 belong to the functionally diverse sub-families of the large Major Facilitator Superfamily, secondary membrane transporters which move substrate across the membrane by the alternating between two alternative conformations of the channel, one open to the cytoplasm, and the other open to the periplasm or extracellular space (Boudker and Verdon, 2010; Huang et al., 2003).
Comparing the predicted model of Glpt to the crystal structure 1pw4 (cytoplasm-open conformation), we noticed that the predicted cytoplasmic side of the transporter channel is not as open as in the crystal structure (Figure 3A). The Glpt evolutionary couplings differ from contacts made in the GlpT crystal structure in an apparently false positive set which would, however, be in contact in the suspected alternative cytoplasm-closed conformation (Figure 5A). Similarly, a set of contacts can be identified that are consistent only with the cytoplasm-open conformation (selection rules in Supplement). To test whether the two alternative sets of evolutionary couplings for GlpT protein would be sufficient to predict the two different conformations, we refolded GlpT with both sets separately (Figure 5A, Table S3). As expected, when we exclude evolutionary coupling pairs between the domains on the periplasmic side, we obtain models in a closed-to-cytoplasm conformation, similar in overall structure to the known closed conformation structure of the L-fucose-proton symporter FucP (PDB:3o7q) and to a homology model of LacY (Radestock and Forrest, 2011), but distinct from the known open GlpT structure of GlpT (PDB:1pw4). The arrangements of transmembrane helices 5 and 8 and transmembrane helices 2 and 11 in the two folded models differ as expected for ‘rocking’ changes between alternative transporter conformations (Lemieux et al., 2004). Therefore, plausibly, the evolutionary constraints in the sequence family of GlpT, when decomposed into two overlapping sets, reflect two alternative conformations of the channel.
Figure 5. Co-evolved pairs consistent with open and closed conformations of proteins in the major facilitator family. (Related to Table S3).
A. Center panel: contact map for E. coli GlpT, residues less than 5Å apart in the crystal structure (grey circles, PDB:1pw4) overlaid with the top 350 EC's (red stars). The similarity of the upper left and lower right quadrants reflect the similarity of the structure and sequences of the two domains. Upper right and lower left quadrants show the predicted inter-domain contacts (all stars). Stripes in lower left and upper right quadrants cover inter-domain contacts involving the periplasmic end of the helices/loops (green strips, lower left) and the cytoplasmic ends of the helices/loops (blue strips, upper right). Predicted EC's located where stripes of the same colour cross each are likely inter-domain contacts, green and blue stars (Table S3). Right and left bottom panels: Refolded GlpT from extended polypeptide excluding blue constraints for cytoplasmic side open (right), and excluding green constraints for cytoplasmic side closed (left). The schematics (right and left top) indicate contacts used (arrows) and not used (scissors) in re-folding to get the two alternative conformations. Open conformation (right) is similar to crystal structure (Table 1) and is reproduced via re-folding, closed conformation structure (left) is previously unknown and predicted here via re-folding.
B. Details from the models in A: the two pairs of helices (H5/8 and H2/11) in the predicted models of GlpT are thought to change conformation dependent on state of substrate binding (closed at cytoplasm, green ribbons, left; open at cytoplasm, blue ribbons, right). Differences in interhelical angles are driven by the alternative use of top (green) or bottom (blue) contact pairs derived from EC's in re-folding (Table S3). C. Predicted EC pairs of human OCTN1 (red stars on contact map) determine the overall fold. Stripes in lower left and upper right quadrants cover the predicted periplasmic end of the helices/loops (green) and the cytoplasmic ends of the helices/loops (blue). Predicted evolutionary constraints (not differentiated by star color) located where stripes of the same colour cross each other are predicted inter-domain contacts. 3D structures of alternative conformations of OCTN1 not shown here. Predicted OCTN1 structure details see Figure 1, Table 1 and Data S2.
As human Octn1 (unknown structure) is also from the Major Facilitator Superfamily, we wondered if evolutionary couplings in Octn1 also contained information about alternative conformations. We compared our top-ranked model of Octn1 to all structures in the PDB and found significant hits to known structures in the Major Facilitator Superfamily, including those of GlpT and FucP. The predicted Octn1 models, as above for GlpT, looked like an intermediate conformation between outward-open and inward-open, consistent with the expectation that both states are constrained by evolution (Figure 1 and Data S2). Examination of the distribution of EC pairs suggests that they contain information for two conformations of the transporter (Figure 5C).
Given that our evolutionary constraints contain information about the different states of members of the Major Facilitator Superfamily, we anticipate that evolutionary constraints might help to unravel the precise conformational changes upon substrate binding and transport.
Evolutionary constraints mark functional residues
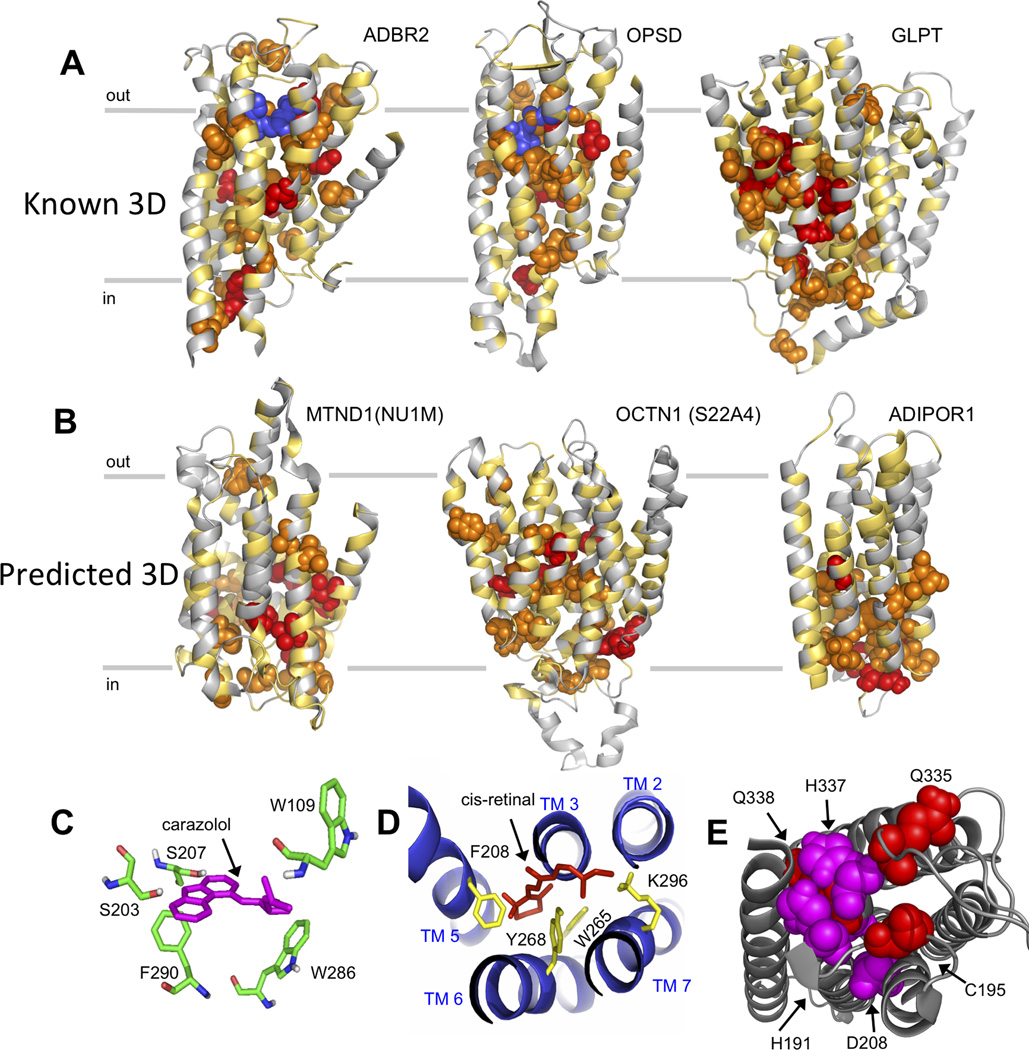
Conservation of amino acids in proteins in single columns is routinely used to infer functional importance of the site and assess the consequences of genetic variation. As our evolutionary analysis reflects both residue-residue correlations and single residue terms, we wondered if the strength of evolutionary couplings on a residue is an indication of its general functional importance for the protein. To assess this, we calculate the total evolutionary coupling score for a given residue by summing the evolutionary coupling values over all high-ranking pairs involving that residue (Experimental Procedures, Table S4, Data S6). We find that in Adrb2, Opsd and GlpT, residues with high total coupling scores line the substrate binding sites and affect signaling or transport; for instance, W109, D113, Y141 in Adrb2; K296, W265 and H211 in Opsd; and Y393, H165 and K90 in GlpT (Huang et al., 2003) (Law et al., 2008) (Valiquette et al., 1995) (Figure 6A). Higher prediction accuracy of atomic coordinates near the active sites for Adbr2 and Opsd than for the average of the protein reflects the multiple constraints, i.e., high total coupling score, on these sites (Figure 6C).
Figure 6. Known functional sites contain residues strongly involved in evolutionary constraints. (Related to Table S4).
A, B. The total evolutionary coupling score on individual residues reflects likely functional involvement (top 5% (red spheres), top 6–15% (orange spheres), all others (yellow ribbon)); scores as in Table S4. A. The ligands carazolol in Adrb2 and retinal in OPSD (blue spheres) were positioned in the predicted structure by globally superimposing the most accurate predicted model and the experimental structure plus ligand (experimental structures not shown, no docking was performed). B. Residues with high evolutionary coupling scores mapped on the predicted structures of unknown-structure transmembrane proteins.
C,D. Above average accuracy of blinded prediction of atomic positions of the binding site of Adrb2 (1.6 Å Cα-rmsd over 9 residues, C) and bovine rhodopsin (1.8Å Cα-rmsd over 10 residues, D). E. Likely functional residues (high evolutionary coupling scores) in AdipoR1 on the predicted cytoplasmic side (known functional residues in magenta, predicted functional residues in red).
In the unknown structure AdipoR1, residues with a high total coupling score include putative enzymatic residues S187, H191, D208, H337, H341 (Holland et al., 2011) (Pei et al., 2011) together with the top 3 high scoring residues, C195, A235, and Q335, which cluster together (within ~4 Å) in the predicted 3D structure, indicating that they are important in the activity of AdipoR1 (Figure 6B). Similarly, clusters of residues with high scoring in Octn1 make potential salt bridges at the cytoplasmic side of the domains (169R-220E, 397R-450E), cluster in the central transport pore (N210, Y211, C236, E381, and R469) and are potentially involved in conformational changes. Residues with high total coupling scores in our predicted models of human MT-ND1, are clustered in a periplasmic-oriented pocket and along the mitochondrial interface with the hydrophilic domain and the putative quinone binding site (Figure 6B) (Efremov and Sazanov, 2011). Mutations in MT-ND1 at residues Y30 and M31 are associated with Alzheimer’s disease and Leber’s hereditary optic neuropathy (LHON) (Johns et al., 1992), and these two residues have particularly high total coupling scores, suggesting that they are functionally constrained by interactions with several other residues.
We hypothesize that many evolutionary coupling pairs, whether or not close in the 3D structure, may be functionally important. The examples presented here, however, are not the result of an exhaustive analysis. Therefore, reliable functional interpretation of evolutionary constraints, whether indicative of intra-monomer contacts or not, remains a challenge. Our results here provide some confidence in the validity of the conceptual link between the strength of evolutionary constraints on a residue and its functional importance, whether through location in binding sites or involvement in conformational changes.
Discussion
The process of evolution and the massive sequencing of diverse species have provided the opportunity to compute an important aspect of molecular phenotype, protein 3D structure, and the EVfold method appears to achieve a useful level of accuracy. However, a serious gap remains between predicted and experimental structures. While an overall Cα-rmsd of 4–5 Å across hundreds of residues does imply the correct identification of the overall fold, it also implies that particular atomic positions, the interdigitation of packed side chains and loop conformations can be incorrect in detail, although they appear more accurate near heavily constrained binding sites. To improve the quality of the predicted contacts and resulting atomic structures, four areas of focus hold particular promise: (1) improved information handling in sequence space, such as improvements in weighting schemes for sequences, evaluation of alignment diversity, inclusion of higher order terms, and consistency filters to reduce the number of false positive pairs; (2) automated procedures to distinguish between internal and homo-oligomer pair contacts and to identify contacts reflecting alternative conformations; (3) the use of fragments imported from known structures; and, (4) the use of advanced energy refinement methods, including molecular dynamics and Monte-Carlo simulations (Dror et al., 2011; MacCallum et al., 2011)
Even at the current level of accuracy, a number of applications may have immediate benefit. One is the development of hybrid methods of structure determinations. In NMR spectroscopy, inclusion of evolutionary constraints from sequences may permit structure determination with a smaller number of chemical shifts and NOEs, saving machine time, or permitting the solution of larger protein structures than previously reachable. In protein crystallography, the solution of a 3D structure from a native data set alone may become possible, without the need for heavy atom derivatives or MAD phasing, via molecular replacement searches starting with predicted 3D structures. If successful in future work, such methods would significantly increase the productivity of structural biology and the rate of solving new structures.
Beyond structure determination, the predicted models may be useful for pharmacological selection of compounds via docking calculations. The observation of exceptionally strong evolutionary constraints near active sites, as reported here for a few proteins, is a favorable starting point, as the accuracy of protein coordinates in active sites and binding sites is an important requirement for computational drug screening. In molecular biology in general, the placement of constrained pairs in the context of known or predicted 3D structures may also provide useful information to guide functional mutational experiments. Similarly, evolutionarily coupled pairs may be excellent design elements for engineering new proteins in synthetic biology (Russ et al., 2005) and may have a strategic role in the protein folding process (Fersht, 2008).
Inferred evolutionary constraints may also help guide the computational assembly of protein monomers into complexes, with or without low-resolution information from electron diffraction or similar methods. The computational extension to predict the structure of protein complexes is a straightforward generalization using pairwise sequence alignments, with a homologous pair of sequences in place of a single sequence and derivation of evolutionary couplings not within a protein but between two potentially interacting proteins (Pazos and Valencia, 2002; Skerker et al., 2008; Weigt et al., 2009). We see no practical limit in the size of complexes accessible to such computation, provided sufficiently diverse sequence information is available, as the configuration of even large complexes with tens of constituents effectively can be deduced from calculation of all pairwise protein interactions in the complex. The nuclear pore complex, as solved by computational assembly from protein-protein pair information determined experimentally, would be an excellent test case (Fernandez-Martinez et al., 2012).
Looking forward, how much information about 3D folds of transmembrane proteins can be gained if this kind of method is broadly and successfully applied? A current snapshot of protein families, as organized in the PFAM 26.0 database, has about 2 million transmembrane proteins in 1259 protein families, of which 107 families have one or more 3D structures. An additional 150 families appear to have sufficient sequences to be modeled using evolutionary couplings, including those with β-sheet folds (not tested here). Given the current efficiency and rapid development of DNA sequencing technology, perhaps another 500 families would accrue similar levels of sequence information to have their folds determined in about two years, with subsequent rapid growth likely.
On a practical level, the simplicity of the theoretical approach and efficiency of the computational implementation, with computation in about an hour for proteins up to 500 residues, will allow availability of the EVfold procedure to a broad community of researchers, not limited to structural biologists, in either pre-computed or server mode. For the proteins described here, detailed data, such as 3D coordinates and evolutionary constraints, as well as software code for their calculation, are available at evfold.org/transmembrane. Computational protein folding using evolutionary constraints may thus drive new experimental approaches that will harness the massive explosion in genomic sequencing by reading the evolutionary footprints of protein structure and function.
Experimental Procedures
Full methods are described in Supplemental Experimental Procedures. All EC scores and residue name mappings are in Data S1, all 3D model coordinates, input files, analysis in Data S2–S5 and on the web at www.evfold.org/transmembrane..
Selection of membrane proteins
To test our ability to predict the 3D structure of alpha-helical multipass membrane proteins (>=5 helices), we compiled a set of 25 proteins from 23 different Pfam families from the database of membrane proteins of known 3D structure (http://blanco.biomol.uci.edu/mpstruc/listAll/list). We optimized the set for non-redundancy and depth of sequence alignment. The set of interesting membrane protein families with no known representative structure was chosen by selecting transmembrane proteins that are drug targets, using the DrugBank (Knox et al., 2011), Pfam (Punta et al., 2012) and CAMPS (Neumann et al., 2011) databases. For this initial study we selected proteins with at least 2*L sequences in the family alignment (L = protein length) and with more than 70% coverage (breadth).
Multiple sequence alignments
Multiple sequence alignments for each candidate proteins were obtained using HHblits (Remmert et al., 2011) sequence searches against the UniProt database at a range of different E-values. The alignment used for constraint inference was selected by choosing the E-value giving the best trade-off between a maximum number of sequences in the alignment, and sufficient coverage of the entire transmembrane domain by most sequences in the alignment (all alignments available at evfold.org/transmembrane) (Figure 2A, Figure S2).
Inference of evolutionary constraints from sequence variation
To predict the 3D structure of membrane proteins, we devised a membrane protein specific version of the original EVfold method (Marks et al., 2011) and name it EVfold-membrane. First, a set of evolutionary couplings between residue pairs is inferred by computing the parameters in a global maximum entropy probability model of the multiple sequence alignment (Data S1). This set is ranked according to coupling strength and filtered for inconsistency with predicted membrane topology and predicted secondary structure.
Ab initio folding from membrane protein sequence
All predictions started from fully extended polypeptide using increasing numbers of evolutionary constraints, from 40 to L constraints (L = length of modeled sequence) in steps of 10, with 20 models generated for each EC bin. Additionally, we added distance and dihedral angle constraints consistent with predicted secondary structure. The folding protocol uses default modules from the CNS software suite (Brunger, 2007) which consists of distance geometry, simulated annealing and energy minimization stages. Each model takes about 1–2 minutes of computing time on a single CPU for a protein of average size.
Clustering and ranking of predicted models
Although only a small number of models is generated, we devised a ranking scheme based on simple intuitive requirements for membrane proteins, such as satisfaction of unused constraints (adapted from (Miller and Eisenberg, 2008)), predicted secondary structure and predicted lipid exposure agreement in the folded models. Structures are additionally clustered using MaxCluster (Siew et al., 2000) single-linkage clustering to eliminate high-ranked outliers belonging to small clusters.
Assessment of evolutionary constraints and prediction quality
Predicted evolutionary constraints were compared to observed contacts from crystal structures using contact maps and false positive rate plots (Data S2–5 and EVfold.org/transmembrane). Predicted models of known structures were compared to a representative crystal structure (Table S1) using the Cα-rmsd, TM and GDT scores calculated with MaxCluster (Zemla, 2003; Zhang and Skolnick, 2005). Predicted 3D structures for transmembrane proteins of unknown structure, for which no family member structure has been solved yet, are compared for structural homology against a representative set of proteins in the Protein Data Bank (PDB) using structural alignments with DALI (Holm and Sander, 1993) and FATCAT (Ye and Godzik, 2004).
Residues with high total evolutionary coupling scores
To quantify the strength of evolutionary constraints on a residue, we calculated the total strength of evolutionary constraints per residue. For each residue, we sum the pair scores obtained from the maximum entropy model over all high-ranking pairs it is involved in, down to a predefined cutoff (Data S1). The score for each residue is normalized by the average score for all residues in the full sequence (Table S4).
Supplementary Material
Highlights.
3D structure prediction of 11 membrane proteins with no known structure
Algorithm finds evolutionary couplings from genetic variation and massive sequencing
Accuracy tested by blind prediction of known 3D protein structures from 23 families
Computation is fast; provides clues for functional sites, conformations, and oligomers
Acknowledgements
We thank Guy Montelione, Rebecca Ward, Nicholas Stroustrup, Steven Long, Nikola Pavletich, Anil Korkut, Edda Kloppman, Marc Offman, Yitzak Pilpel, Andrea Pagnani, Richard Stein, James Thompson and Michael Lappe for scientific discussions; Johannes Soeding and Michael Remmert for help with HHblits; Marco Punta and Jaina Mistry for help with PFAM numbers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldahmesh MA, Mohamed JY, Alkuraya HS, Verma IC, Puri RD, Alaiya AA, Rizzo WB, Alkuraya FS. Recessive mutations in ELOVL4 cause ichthyosis, intellectual disability, and spastic quadriplegia. Am J Hum Genet. 2011;89:745–750. doi: 10.1016/j.ajhg.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakheet TM, Doig AJ. Properties and identification of human protein drug targets. Bioinformatics. 2009;25:451–457. doi: 10.1093/bioinformatics/btp002. [DOI] [PubMed] [Google Scholar]
- Barth P, Wallner B, Baker D. Prediction of membrane protein structures with complex topologies using limited constraints. Proc Natl Acad Sci U S A. 2009;106:1409–1414. doi: 10.1073/pnas.0808323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudker O, Verdon G. Structural perspectives on secondary active transporters. Trends Pharmacol Sci. 2010;31:418–426. doi: 10.1016/j.tips.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burger L, van Nimwegen E. Disentangling direct from indirect co-evolution of residues in protein alignments. PLoS Comput Biol. 2010;6:e1000633. doi: 10.1371/journal.pcbi.1000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Hu L, Punta M, Bruni R, Hillerich B, Kloss B, Rost B, Love J, Siegelbaum SA, Hendrickson WA. Homologue structure of the SLAC1 anion channel for closing stomata in leaves. Nature. 2010;467:1074–1080. doi: 10.1038/nature09487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin, II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- Cronet P, Sander C, Vriend G. Modeling of transmembrane seven helix bundles. Protein Eng. 1993;6:59–64. doi: 10.1093/protein/6.1.59. [DOI] [PubMed] [Google Scholar]
- Dang S, Sun L, Huang Y, Lu F, Liu Y, Gong H, Wang J, Yan N. Structure of a fucose transporter in an outward-open conformation. Nature. 2010;467:734–738. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- Dempster AP. Covariance Selection. Biometrics. 1972;28:157–175. [Google Scholar]
- Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, Ross DD. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci U S A. 1998;95:15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror RO, Pan AC, Arlow DH, Borhani DW, Maragakis P, Shan Y, Xu H, Shaw DE. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc Natl Acad Sci U S A. 2011;108:13118–13123. doi: 10.1073/pnas.1104614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex, I. Nature. 2010;465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- Efremov RG, Sazanov LA. Structure of the membrane domain of respiratory complex, I. Nature. 2011;476:414–420. doi: 10.1038/nature10330. [DOI] [PubMed] [Google Scholar]
- Fatakia SN, Costanzi S, Chow CC. Computing highly correlated positions using mutual information and graph theory for G protein-coupled receptors. PLoS One. 2009;4:e4681. doi: 10.1371/journal.pone.0004681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Martinez J, Phillips J, Sekedat MD, Diaz-Avalos R, Velazquez-Muriel J, Franke JD, Williams R, Stokes DL, Chait BT, Sali A, et al. Structure-function mapping of a heptameric module in the nuclear pore complex. J Cell Biol. 2012;196:419–434. doi: 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor AA, Aldrich RW. Influence of conservation on calculations of amino acid covariance in multiple sequence alignments. Proteins. 2004;56:211–221. doi: 10.1002/prot.20098. [DOI] [PubMed] [Google Scholar]
- Fuchs A, Martin-Galiano AJ, Kalman M, Fleishman S, Ben-Tal N, Frishman D. Co-evolving residues in membrane proteins. Bioinformatics. 2007;23:3312–3319. doi: 10.1093/bioinformatics/btm515. [DOI] [PubMed] [Google Scholar]
- Holland WL, Miller RA, Wang ZV, Sun K, Barth BM, Bui HH, Davis KE, Bikman BT, Halberg N, Rutkowski JM, et al. Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nature medicine. 2011;17:55–63. doi: 10.1038/nm.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Horn F, Bywater R, Krause G, Kuipers W, Oliveira L, Paiva AC, Sander C, Vriend G. The interaction of class B G protein-coupled receptors with their hormones. Receptors Channels. 1998;5:305–314. [PubMed] [Google Scholar]
- Howell N, Bindoff LA, McCullough DA, Kubacka I, Poulton J, Mackey D, Taylor L, Turnbull DM. Leber hereditary optic neuropathy: identification of the same mitochondrial ND1 mutation in six pedigrees. Am J Hum Genet. 1991;49:939–950. [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- Jaksch M, Hofmann S, Kaufhold P, Obermaier-Kusser B, Zierz S, Gerbitz KD. A novel combination of mitochondrial tRNA and ND1 gene mutations in a syndrome with MELAS, cardiomyopathy, and diabetes mellitus. Hum Mutat. 1996;7:358–360. doi: 10.1002/(SICI)1098-1004(1996)7:4<358::AID-HUMU11>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Johns DR, Neufeld MJ, Park RD. An ND-6 mitochondrial DNA mutation associated with Leber hereditary optic neuropathy. Biochem Biophys Res Commun. 1992;187:1551–1557. doi: 10.1016/0006-291x(92)90479-5. [DOI] [PubMed] [Google Scholar]
- Jones DT, Buchan DW, Cozzetto D, Pontil M. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012;28:184–190. doi: 10.1093/bioinformatics/btr638. [DOI] [PubMed] [Google Scholar]
- Kadaba NS, Kaiser JT, Johnson E, Lee A, Rees DC. The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation. Science. 2008;321:250–253. doi: 10.1126/science.1157987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, et al. DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosel D, Heiker JT, Juhl C, Wottawah CM, Bluher M, Morl K, Beck-Sickinger AG. Dimerization of adiponectin receptor 1 is inhibited by adiponectin. J Cell Sci. 2010;123:1320–1328. doi: 10.1242/jcs.057919. [DOI] [PubMed] [Google Scholar]
- Lapedes AS, Giraud BG, Liu LC, Stormo GD. Correlated Mutations in Protein Sequences: Phylogenetic and Structural Effects (Santa Fe Institute) 1997 [Google Scholar]
- Law CJ, Almqvist J, Bernstein A, Goetz RM, Huang Y, Soudant C, Laaksonen A, Hovmoller S, Wang DN. Salt-bridge dynamics control substrate-induced conformational change in the membrane transporter GlpT. J Mol Biol. 2008;378:828–839. doi: 10.1016/j.jmb.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux MJ, Huang Y, Wang DN. The structural basis of substrate translocation by the Escherichia coli glycerol-3-phosphate transporter: a member of the major facilitator superfamily. Curr Opin Struct Biol. 2004;14:405–412. doi: 10.1016/j.sbi.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Livesay DR, Kreth KE, Fodor AA. A critical evaluation of correlated mutation algorithms and coevolution within allosteric mechanisms. Methods Mol Biol. 2012;796:385–398. doi: 10.1007/978-1-61779-334-9_21. [DOI] [PubMed] [Google Scholar]
- Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- MacCallum JL, Perez A, Schnieders MJ, Hua L, Jacobson MP, Dill KA. Assessment of protein structure refinement in CASP9. Proteins. 2011;79(Suppl 10):74–90. doi: 10.1002/prot.23131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, Sander C. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011;6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinshausen N, Buhlmann P. High-dimensional graphs and variable selection with the Lasso. Ann Stat. 2006;34:1436–1462. [Google Scholar]
- Miller AN, Long SB. Crystal structure of the human two-pore domain potassium channel K2P1. Science. 2012;335:432–436. doi: 10.1126/science.1213274. [DOI] [PubMed] [Google Scholar]
- Miller CS, Eisenberg D. Using inferred residue contacts to distinguish between correct and incorrect protein models. Bioinformatics. 2008;24:1575–1582. doi: 10.1093/bioinformatics/btn248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, Zecchina R, Onuchic JN, Hwa T, Weigt M. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci U S A. 2011;108:E1293–E1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. Embo J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Xie Y, Baer MR, Ross DD. Role of breast cancer resistance protein (BCRP/ABCG2) in cancer drug resistance. Biochem Pharmacol. 2012 doi: 10.1016/j.bcp.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto W, Imai T, Takahashi T, Kikuchi T, Fujita N. Detection of pairwise residue proximity by covariation analysis for 3D-structure prediction of G-protein-coupled receptors. Protein J. 2004;23:427–435. doi: 10.1023/b:jopc.0000039556.95629.cf. [DOI] [PubMed] [Google Scholar]
- Neumann S, Hartmann H, Martin-Galiano AJ, Fuchs A, Frishman D. Camps 2.0: Exploring the sequence and structure space of prokaryotic, eukaryotic, and viral membrane proteins. Proteins. 2011 doi: 10.1002/prot.23242. [DOI] [PubMed] [Google Scholar]
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- Pazos F, Valencia A. In silico two-hybrid system for the selection of physically interacting protein pairs. Proteins. 2002;47:219–227. doi: 10.1002/prot.10074. [DOI] [PubMed] [Google Scholar]
- Pei J, Millay DP, Olson EN, Grishin NV. CREST--a large and diverse superfamily of putative transmembrane hydrolases. Biol Direct. 2011;6:37. doi: 10.1186/1745-6150-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radestock S, Forrest LR. The alternating-access mechanism of MFS transporters arises from inverted-topology repeats. J Mol Biol. 2011;407:698–715. doi: 10.1016/j.jmb.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Remmert M, Biegert A, Hauser A, Soding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- Russ WP, Lowery DM, Mishra P, Yaffe MB, Ranganathan R. Natural-like function in artificial WW domains. Nature. 2005;437:579–583. doi: 10.1038/nature03990. [DOI] [PubMed] [Google Scholar]
- Siew N, Elofsson A, Rychlewski L, Fischer D. MaxSub: an automated measure for the assessment of protein structure prediction quality. Bioinformatics. 2000;16:776–785. doi: 10.1093/bioinformatics/16.9.776. [DOI] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- Valiquette M, Parent S, Loisel TP, Bouvier M. Mutation of tyrosine-141 inhibits insulin-promoted tyrosine phosphorylation and increased responsiveness of the human beta 2-adrenergic receptor. Embo J. 1995;14:5542–5549. doi: 10.1002/j.1460-2075.1995.tb00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci U S A. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigt M, White RA, Szurmant H, Hoch JA, Hwa T. Identification of direct residue contacts in protein-protein interaction by message passing. Proc Natl Acad Sci U S A. 2009;106:67–72. doi: 10.1073/pnas.0805923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, Kita S, Sugiyama T, Miyagishi M, Hara K, Tsunoda M, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423:762–769. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006;62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Godzik A. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004;32:W582–W585. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Kniazeva M, Han M, Li W, Yu Z, Yang Z, Li Y, Metzker ML, Allikmets R, Zack DJ, et al. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet. 2001;27:89–93. doi: 10.1038/83817. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.