Abstract
Bovine milk oligosaccharides (BMOs) are recognized by the dairy and food industries, as well as by infant formula manufacturers, as novel, high-potential bioactive food ingredients. Recent studies revealed that bovine milk contains complex oligosaccharides structurally related to those previously thought to be present in only human milk. These BMOs are microbiotic modulators involved in important biological activities, including preventing pathogen binding to the intestinal epithelium and serving as nutrients for a selected class of beneficial bacteria. Only a small number of BMO structures are fully elucidated. To better understand the potential of BMOs as a class of biotherapeutics, their detailed structure analysis is needed. This study initiated the development of a structure library of BMOs and a comprehensive evaluation of structure-related specificity. The bovine milk glycome was profiled by high-performance mass spectrometry and advanced separation techniques to obtain a comprehensive catalog of BMOs, including several novel, lower abundant neutral and fucosylated oligosaccharides that are often overlooked during analysis. Structures were identified using isomer-specific tandem mass spectroscopy and targeted exoglycosidase digestions to produce a BMO library detailing retention time, accurate mass and structure to allow their rapid identification in future studies.
Keywords: bovine colostrum, high-performance liquid chromatography, oligosaccharides, tandem mass spectrometry
Introduction
Free oligosaccharides are a dynamic and structurally diverse class of carbohydrates representing the third most abundant component in mammalian milk after lactose and lipids (Kunz et al. 2000; Boehm and Stahl 2007; Fong et al. 2011). Human milk oligosaccharides (HMOs) participate in several protective and physiological roles, including immunoregulation and inhibition of pathogen adhesion in the gastrointestinal tract of infants (Klein et al. 2000; Martin-Sosa et al. 2002; Hakkarainen et al. 2005; Coppa et al. 2006). Human milk is a well-established source of prebiotic oligosaccharides, an indigestible form of carbohydrates that plays a critical role in establishing the intestinal flora of infants by stimulating growth of beneficial bacteria (Coppa et al. 2004; LoCascio et al. 2007). Comprehensive studies characterizing these carbohydrates support the idea that their structural diversity is the basis for a multitude of biological functions. There is increasing interest in finding a source of complex oligosaccharides for industrial-scale extraction.
Previous studies on bovine colostrum focused principally on the highly abundant acidic oligosaccharides. Recently, the lower abundant oligosaccharides from bovine milk were shown to have complex structures closely related to those from human milk (Tao et al. 2008, 2009; Barile et al. 2010, 2011). Milk oligosaccharides are a complex class of glycans defined as carbohydrates that contain 3–10 monosaccharides covalently linked through glycosidic bonds (Tao et al. 2008; Barile et al. 2010). The monosaccharides that make up bovine milk oligosaccharides (BMOs) include glucose (Glc), galactose (Gal), N-acetylglucosamine (GlcNAc), fucose (Fuc), N-acetylneuraminic acid (NeuAc), and N-glycolylneuraminic acid (NeuGc) (Chai et al. 2005; Mehra and Kelly 2006; Boehm and Stahl 2007; Wu, Tao, et al. 2010). The majority of BMOs contain a lactose core consisting of a Glc β1-4 linked to Gal (Urashima et al. 2001; Mehra and Kelly 2006). BMOs can also possess a lactosamine core consisting of GlcNAc β1-4 linked to Gal (Saito et al. 1981; Veh et al. 1981; Gopal and Gill 2000; Urashima et al. 2001).
Recent studies have demonstrated beneficial functions of milk oligosaccharides in vitro, which suggest that BMOs have potential as a new source of microbiotic modulators with the potential to mimic the more complex oligosaccharides of human milk (Hakkarainen et al. 2005; Zivkovic and Barile 2011). It is becoming increasingly apparent that the highly specific oligosaccharide structures have the direct control of their biological function. In an effort to better understand the potential of BMOs as a new source of biotherapeutics, detailed structure analysis is needed. The goals of this study were 2-fold: first, we aimed to develop a comprehensive BMO structure library for rapid structural identification, and second, we aimed to perform a comparative analysis of the oligosaccharides present in bovine and human milk to estimate the overlap between the two sample sets and evaluate their structure specificity.
Bovine colostrum is currently being used in a variety of health-promoting supplements worldwide, so a comparison between the oligosaccharides found in bovine colostrum and human milk is appropriate. Oligosaccharides in bovine milk are 20 times less concentrated than human milk; however, in bovine colostrum, the concentration of sialylated oligosaccharides is exceptionally high (Veh et al. 1981; Tao et al. 2009). HMOs are produced at nearly constant amounts during the lactation process (Ninonuevo et al. 2008), whereas BMOs decrease considerably even during the first few days of lactation (Tao et al. 2009). Furthermore, HMOs are highly fucosylated, with as much as 70% fucosylation (Wu, Tao, et al. 2010), whereas BMOs do not contain fucosylation at any appreciable levels (Gopal and Gill 2000; Tao et al. 2009; Wu, Tao, et al. 2010). Conversely, BMOs are as much as 50% sialylated (Gopal and Gill 2000; Tao et al. 2008), and human milk oligosaccharides are ∼20% sialylated (Ninonuevo et al. 2006; Wu, Grimm, et al. 2010). Human milk does not contain the NeuGc monosaccharide residue that is found in bovine colostrum and other mammalian milks (Tao et al. 2008; Wu, Grimm, et al. 2010).
The ability to detect and analyze milk oligosaccharides, especially the lower abundant components, was largely hindered until recent advancements were made in analytical techniques, including nuclear magnetic resonance spectroscopy (Guerardel et al. 1999; Chai et al. 2005), chromatography such as high pH anion-exchange chromatography (Kunz and Rudloff 1996; Stiasny et al. 1996; Leo et al. 2009; Mariño et al. 2011), capillary electrophoresis (Shen et al. 2000; Albrecht et al. 2010; Huhn et al. 2010) and mass spectrometry (MS; Stahl et al. 1994; Finke et al. 1999; Pfenninger et al. 2002; Rohmer et al. 2011; Yang et al. 2011; Blank et al. 2012). Our laboratory has reported extensively on the characterization of HMOs and BMOs by microfluidic chip-based nanoflow liquid chromatography (nano-LC)/MS (Ninonuevo et al. 2005, 2006; Tao et al. 2008; Wu, Grimm, et al. 2010; Wu, Tao, et al. 2010; Wickramasinghe et al. 2011). This current work is a continuation of previous studies from our laboratory where we match previously published and newly identified structures in bovine milk to their unique retention time to build a structure library based on isomer-specific tandem MS and exoglycosidase digestions. The nano-LC chip employs a porous graphitized carbon stationary phase for reproducible separation of isomeric oligosaccharide structures (Chu et al. 2009; Hua et al. 2011). Post-separation, a quadrupole time-of-flight (Q-TOF) mass spectrometer provides high mass accuracy detection as well as isomer-specific tandem MS. This strategy is both highly reproducible and sensitive, enabling the identification of a variety of oligosaccharide structures from complex biological mixtures.
Although BMOs are less abundant than HMOs, current manufacturing capabilities make it possible to easily enrich for these oligosaccharides from bovine colostrum in significant quantities. Additionally, major cheese manufacturing companies process millions of gallons of dairy byproducts and cheese whey each day which can be used as a potential oligosaccharide source. Cheese whey has long been considered a waste byproduct; however, the increasing ability to reclaim BMOs from whey by membrane filtration makes even the least abundant components of potential commercial significance.
For the present study, a strategy for characterizing reduced BMO structures using nano-LC/MS, isomer-specific tandem MS and strategic exoglycosidase digestions for the linkage assignment was employed to develop a comprehensive oligosaccharide structure library. The sensitive and robust method identified novel neutral and fucosylated BMOs as well as structures with a lactosamine core. Over 50 oligosaccharides, many previously unreported, were identified in a pooled bovine colostrum sample. These oligosaccharides were compiled into a structure library detailing accurate mass, monosaccharide composition, structure, abundance and retention time. Additionally, a qualitative comparison of the oligosaccharides present in pooled bovine colostrum and human milk samples examined the implication of bovine colostrum as a source of HMO analogs.
Results
BMO structural determination via exoglycosidase digestion
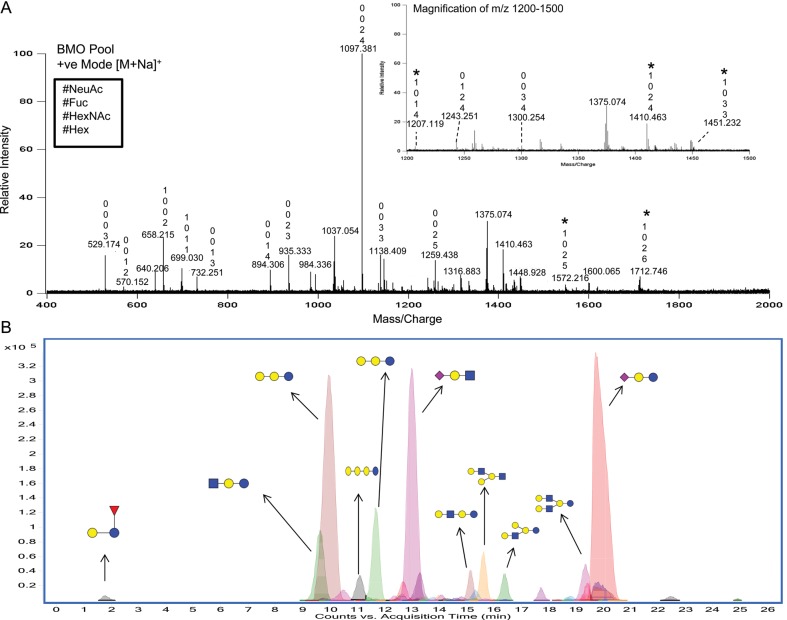
A representative oligosaccharide profile from pooled bovine colostrum analyzed via matrix-assisted laser desorption ionization (MALDI) Fourier transform ion cyclotron resonance (FT-ICR) MS is shown in Figure 1A. A total of 17 reduced oligosaccharide compositions from the pooled bovine colostrum sample were identified after detection in the positive-ion mode. Because there is no separation, this number does not include the isomers associated with each oligosaccharide composition. The compositions of the oligosaccharides are denoted in Figure 1 above the peaks according to the number of residues of Hex, HexNAc, Fuc and NeuAc (from bottom to top). Interestingly, MALDI MS in the positive mode tends to favor neutral oligosaccharides, which makes the abundance of the anionic oligosaccharides appear low (Chu and Lebrilla 2010). However, these structures actually comprise nearly 70% of the total oligosaccharide abundances (Gopal and Gill 2000; Tao et al. 2008). In the negative-ion mode, the MALDI profile shown in Supplementary data, Figure S1, the base peak ion at m/z 656.2 [(M+Na)-2H]−, corresponded to the well-known oligosaccharide sialyllactose (Schneir and Rafelson 1966). It is important to note that siaylated compounds readily undergo free exchange of the carboxylic acid proton with a sodium cation resulting in an additional 22 Da in their mass assignment. The combined positive- and negative-ion mode MALDI MS analysis provided a rapid and semi-quantitative measure of the glycan pool. Based on these MALDI MS results, a total of 22 distinct compositions were detected.
Fig. 1.
(A) The MALDI FT-ICR MS profile of a reduced BMO pool in the positive-ion mode. The numbers shown above the peaks represent, from bottom to top, the number of Hex, HexNAc, Fuc and NeuAc residues. Most ions are [M+Na]+ and the [M-H+2Na]+ ions are denoted with a star. (B) The -HPLC-Chip/TOF MS profile of the reduced BMO pool. Major peaks are labeled with putative structures, where blue circles denote Glc, blue squares denote GlcNAc, yellow circles denote Gal, purple diamonds denote NeuAc and red triangles denote Fuc. (These symbols are used throughout all figures.)
Chromatographic separation coupled with MS yields a comprehensive profile that provides isomer separation and detection of both neutral and anionic components. The extracted compound chromatogram (Figure 1B) from the LC/MS demonstrates the diversity of oligosaccharides in the pooled colostrum sample and shows numerous lower abundant neutral species in the presence of higher abundant sialylated species. Interestingly, the majority of anionic oligosaccharides in bovine milk are sialyllactose and sialyllactosamine, which are simple trisaccharides with well-characterized structures (Schneir and Rafelson 1966; Veh et al. 1981). Extensive studies from this laboratory (Tao et al. 2008, 2009) and those from other laboratories (Newburg and Neubauer 1995; Gopal and Gill 2000; Mariño et al. 2011) further support that indeed the most abundant acidic oligosaccharide in bovine milk is sialyllactose. There are larger and more complex acidic BMOs (Tao et al. 2008; Barile et al. 2010; Mariño et al. 2011); however, these oligosaccharides are of relatively low abundances and typically require enrichment for characterization. This present study focused on profiling and characterizing the less studied neutral oligosaccharides in bovine colostrum.
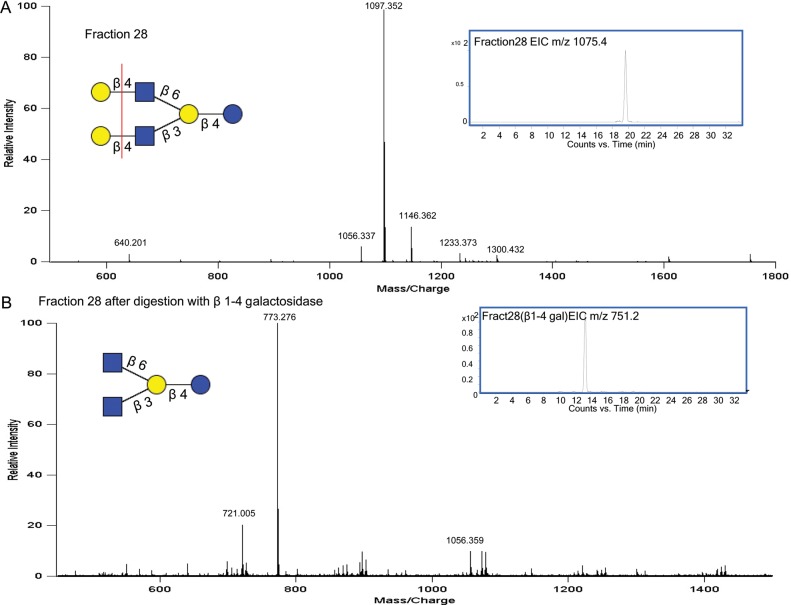
The reduced BMO-pooled sample was fractionated using an off-line high-performance liquid chromatography (HPLC) to isolate structures of interest for structural analysis using targeted exoglycosidase digestions (Wu, Grimm, et al. 2010; Wu, Tao, et al. 2010; Aldredge et al. 2012). MALDI MS analysis, which produced mainly sodiated ions, was used to identify the number of compositions present in each fraction. Chip/Q-TOF MS analysis, which largely produced protonated ions, was used to identify the number of isomers associated with each composition. Fraction 28, as analyzed by MALDI and shown in Figure 2A, yielded one dominant peak, m/z 1097.4 [M+Na]+, corresponding to four Hex and two HexNAc and is known to have two main isomers lacto-N-hexaose and LNnH (lacto-N-neohexaose). After HPLC fractionation, an aliquot from each fraction of interest was analyzed via Chip/Q-TOF MS to determine the number of isomers present. The extracted ion chromatogram (EIC) of m/z 1075.4 [M+H]+ (Figure 2A) confirmed the isolation of a single isomer for that composition. To verify that this isomer corresponded to the well-known LNnH structure (Tadasu et al. 1991), the sample was treated with a β1-4 galactosidase. After the digestion, a peak in MALDI MS appeared at m/z 773.3 [M+Na]+ (Figure 2B), which corresponded to the loss of two terminal hexose monosaccharides from m/z 1097.4 [M+Na]+. This mass shift confirmed the presence of two terminal β1-4 linked Gal monosaccharides as depicted in the structure shown in Figure 2A. Similar treatment of enzymatic digestion coupled with MS was used to determine additional constituents isolated from the pooled BMO sample.
Fig. 2.
(A) MALDI FT-ICR MS profile of fraction 28 (from the off-line BMO pool separation) showing the isolation of m/z 1097.4 with EIC inset. (B) MS profile of fraction 28 after digestion with β1-4 galactosidase showing loss of two hexose mass shift with the EIC inset.
Terminal Gal linkage determination
An additional structural elucidation strategy involved the use of an enzyme-MS method on pooled samples without prior off-line purification. Using this technique, enzymes were added to aliquots of the BMO pool and the resulting EICs were monitored for changes. This method allowed a single enzyme to probe multiple compounds simultaneously, and multiple EICs enabled monitoring of multiple products in a high-throughput and cost-effective manner.
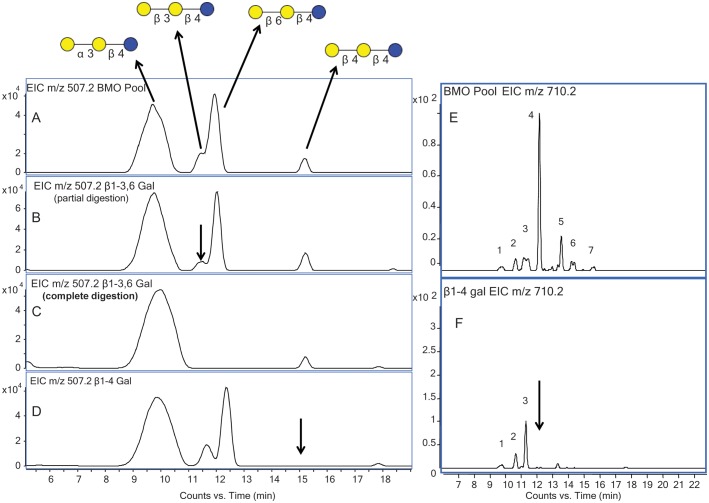
As an example, the ion, m/z 507.2 [M+H]+, corresponds to the composition of three Hex. This small trisaccharide structure consists of galactosyllactose and is often overlooked in the analysis of milk oligosaccharides because trisaccharides elute rapidly during HPLC separation. These compounds are significant as they represent homologs of galacto-oligosaccharides (GOS), which are currently produced in bulk amounts for use as food supplements in dietary products. These trisaccharides support the growth of the beneficial bifidobacteria and lactobacilli (Salvini et al. 2011). Although small in size, they have diverse structures with as many as six isomers from only three monosaccharides (Urashima et al. 1991).
Four isomers were observed with LC/MS in bovine colostrum for m/z 507.2 [M+H]+ (Figure 3A). The pooled sample was digested with linkage-specific galactosidases to determine the linkage of the terminal Gal attached to the lactose core. A commercially available β1-3 galactosidase was used, which catalyzes the β1-6 Gal linkage at a much lower rate than the β1-3 Gal linkage. Kinetic data from the enzyme supplier showed a >100-fold preference for β1-3 over β1-6 linkages (Monks, Unpublished results). Using this information, the digestion was monitored over two time points. Figure 3B shows the EIC of the digestion at the half-way point, and Figure 3C shows the full digestion. At the digestion half-time point, the second eluting isomer (highlighted with an arrow in Figure 3B) was preferentially digested, suggesting that this isomer had a terminal β1-3 Gal. After complete digestion, a third eluting isomer revealed that had a terminal β1-6 linked Gal. Figure 3D is an EIC of pooled bovine colostrum fully digested with a highly specific β1-4 galactosidase, where the last eluting isomer confirmed the presence of a terminal β1-4 Gal. The sample was also treated with a general α-galactosidase (data not shown) to confirm that the structure of the first eluting isomer, was resistant to digestion by the beta-galactosidase enzymes. The digestion confirmed the presence of an alpha terminal Gal linkage which could correspond to a previously reported structure consisting of the lactose core with a terminal α1-3 Gal (Urashima et al. 1991). However, in the absence of a specific enzyme, this linkage was not fully resolved. These experiments enabled us to determine the structure of the four triose oligosaccharides in the pooled colostrum sample. Interestingly, the more abundant isomer, here potentially identified as Galα1–3Galβ1–4Glc, inhibits the binding of pathogenic organisms (e.g., Clostridium difficile) to the intestinal mucosa of newborn calves (Urashima et al. 1991) and may exert the same protective activity in humans.
Fig. 3.
(A) EIC of m/z 507.2 from the BMO pool showing four isomers with their respective structures labeled. (B) EIC of m/z 507.2 at the half-way point of digestion with β1-3,6 galactosidase. (C) EIC of m/z 507.2 after full-time digestion with β1-3,6 galactosidase. (D) EIC of m/z 507.2 after digestion with β1-4 galactosidase. (E) EIC of m/z 710.2 from the BMO pool showing seven isomers. (F) EIC of m/z 710.2 after digestion with β1-4 galactosidase.
Two of the major HMOs are LNT (lacto-N-tetraose) and LNnT (lacto-N-neotetraose) (Ninonuevo et al. 2006; Wu, Tao, et al. 2010), at m/z 710.2 [M+H]+, which correspond to the composition three Hex and one HexNAc. The only difference between these two structures is the linkage of the terminal Gal. Where LNT contains a terminal β1-3 linkage, LNnT contains a terminal β1-4 linkage. The presence of LNnT in bovine milk was previously confirmed with the use of standards and tandem MS (Tao et al. 2008). The EIC of m/z 710.2 [M+H]+ from the BMO-pooled sample (Figure 3E) displays one major isomer with six lower abundant species, demonstrating the diversity of structures associated with the composition three Hex and one HexNAc. These seven isomers have been labeled with numbers 1–7 for clarity. After the digestion of the pooled BMO sample with β1-4 galactosidase (Figure 3F), four of the peaks were digested, including those of the two most abundant isomers, which confirmed the presence of a terminal Gal with a β1-4 linkage. The most abundant isomer was confirmed as LNnT by comparing the fragmentation profile and retention time with those of a standard. Although the digested oligosaccharide isomers shared a terminal Gal with the same linkage, the remainder of their core structure differed. The peaks unchanged after the enzyme digestion corresponded to the remaining isomers that did not possess a terminal β1-4 Gal. The retention time of the digestion product, or core structure, can be compared with the previously characterized structures to rapidly identify the remainder of the structure.
Lactose vs lactosamine core determination via tandem MS
The majority of BMOs contain a lactose core comprised of a reducing end Glc attached to a Gal with a β1-4 linkage (Urashima et al. 2001). In fewer instances, BMO structures have a lactosamine core comprised of a GlcNAc attached to a Gal with a β1-4 linkage. Only five BMOs with a lactosamine core are reported so far (Saito et al. 1981, 1987; Veh et al. 1981). This study identified two previously unreported structures that contained the lactosamine core.
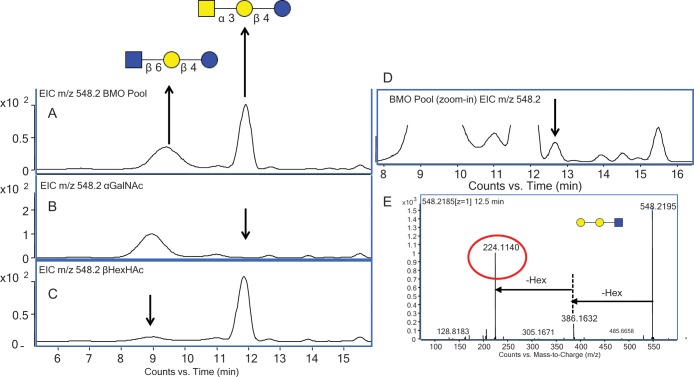
Given that the EIC of m/z 548.2 [M+H]+ corresponds to the composition two Hex and one HexNAc, two main isomers were observed, one at 9.6 min and the second at 11.2 min (Figure 4A). This composition has two previously reported structures corresponding to a lactose core and a terminal HexNAc (Urashima et al. 2001). Figure 4B shows the EIC m/z 548.2 [M+H]+ of the isomer eluted at 11.2 min after digestion with a α N-acetylgalactosaminidase, and Figure 4C shows the EIC m/z 548.2 [M+H]+ after the digestion of the isomer eluted at 9.6 min with a β N-acetylhexosaminidase. The enzyme digestions of these two structures corresponded to previously reported structures for the two Hex and one HexNAc composition (Saito et al. 1987; Tadasu et al. 1991).
Fig. 4.
(A) EIC of m/z 548.2 from the BMO pool showing two main isomers. (B) EIC of m/z 548.2 after digestion with α N-acetylgalactosaminidase. (C) EIC of m/z 548.2 after digestion with β N-acetylhexosaminidase. (D) Magnification on base line of m/z 548.2 from the BMO pool showing additional isomers. (E) Isomer-specific fragmentation of EIC m/z 548.2 at 12.5 min, confirming reducing end HexNAc.
After amplifying the baseline of the EIC m/z 548.2 [M+H]+, five additional lower abundant isomers were observed (Figure 4D). Isomer-specific tandem MS was used to obtain structural information and determine if a lactosamine core was present. When a lactosamine core is present, an m/z 224.1 peak, which corresponds to the reducing end GlcNAc, is diagnostic. Upon examining the tandem mass spectrum at the time point highlighted with an arrow in Figure 4D, the diagnostic fragment for a reducing end HexNAc was observed, confirming that this compound had a lactosamine core. This is the first report, to our knowledge, of an isomer for this composition with the lactosamine core.
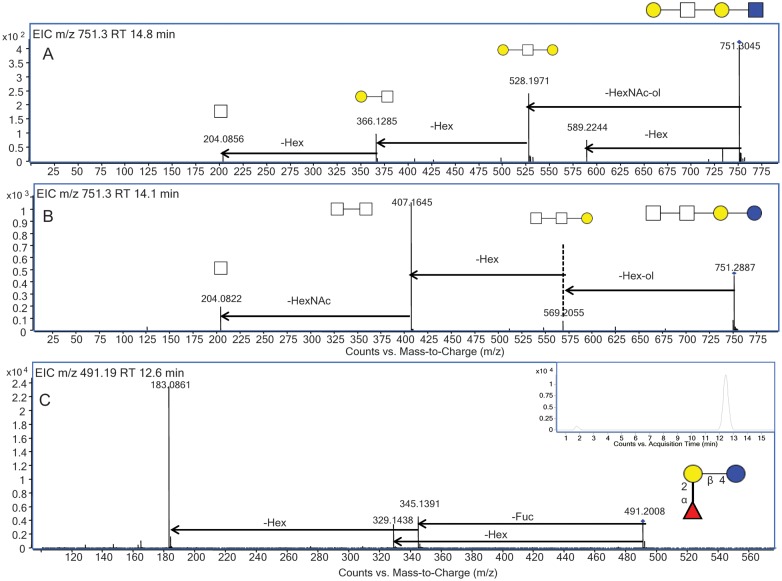
Following analysis of a BMO pool, the distinctive isomer-specific tandem MS profiles for the two isomers for m/z 751.3 at 14.1 and 14.8 min inset in Figure 5A and B, with the composition two Hex and two HexNAc, were further examined. As shown in the tandem MS profile for the isomer eluted at 14.1 min, the reducing end Hex was lost, followed by the loss of another Hex. The peak at m/z 407.16 corresponds to two N-acetylhexosamine monosaccharides, suggesting that these two monosaccharides were attached as shown in the putative structure inset in Figure 5A. The isomer eluted at 14.8 min lost a Hex from the precursor ion as well as a reducing end N-acetylhexosamine, suggesting that structure contained a lactosamine core and a terminal Hex.
Fig. 5.
(A) Isomer-specific fragmentation profile of EIC m/z 751.3 from the BMO pool at 14.8 min, with the putative structure inset. (B) Isomer-specific fragmentation profile of EIC m/z 751.3 from the BMO pool at 14.1 min, with the putative structure inset. (C) Isomer-specific fragmentation profile of EIC m/z 491.19 from the BMO pool at 12.6 min, with the structure and EIC inset.
Identification and structural characterization of fucosylated BMOs
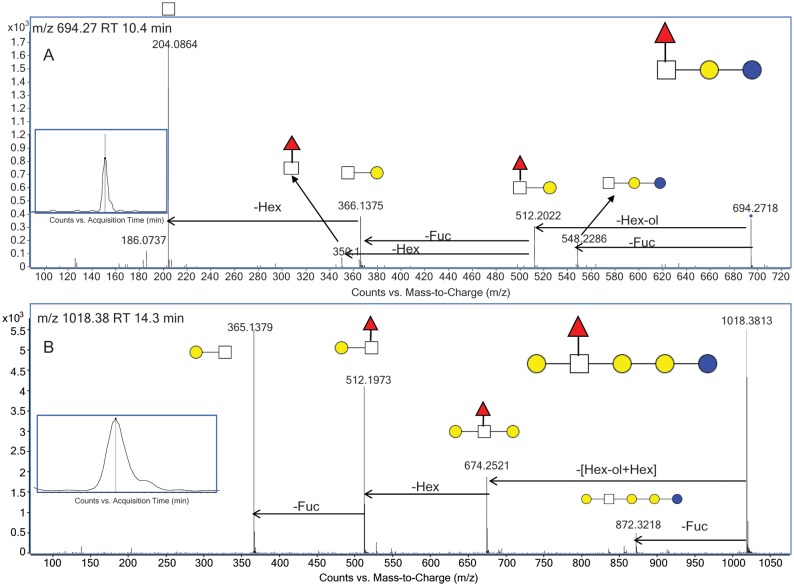
There are few reports of the presence of Fuc in BMOs. Until recently, the only confirmed fucosylated BMO structure is 3-fucosyllactosamine (Urashima et al. 2001). In the present study, a targeted approach was used to identify novel fucosylated BMOs through tandem MS. Figure 5C shows the CID tandem mass spectrum for m/z 491.19 [M+H]+ at 12.6 min, with the EIC for this mass inset. There were two isomers for the oligosaccharide detected at this m/z with the first isomer corresponding to the 3-fucosyllatose isomer, and the later eluting and more abundant isomer corresponding to 2-fucosyllactose. The retention time and tandem MS profile of these structures is in agreement with that from a comprehensive human milk study from this laboratory where these structures were previously elucidated and confirmed using standards (Wu, Tao, et al. 2010). When examining the tandem MS in Figure 5C, the peak observed at m/z 329 is a result of Fuc rearrangement (Broberg 2007; Ernst et al. 1997; Ma et al. 2000; Wuhrer et al. 2006). 2-Fucosyllactose is also an abundant oligosaccharide observed in human milk and it is known for having antipathogenic activities (Chaturvedi et al. 2001). The fucosylated oligosaccharide, previously reported in bovine and goat colostrum (Mariño et al. 2011), was observed as m/z 694.27 [M+H]+, with the composition two Hex, one HexNAc and one Fuc. Only one isomer was identified for this composition. The CID tandem mass spectrum for this oligosaccharide is shown in Figure 6A. The fragmentation profile of this oligosaccharide showed the loss of 182 Da, which corresponded to a reducing end Hex and confirmed the presence of a lactose core. The peak at m/z 512.2 corresponded to a trisaccharide consisting of a lactosamine unit (Hex and HexNAc) with a Fuc, which indicated that the Fuc was positioned on this terminal disaccharide. The tandem MS profile was to determine if the Fuc was located on the Hex or HexNAc. An m/z 350.1, which corresponded to the disaccharide composition HexNAc and Fuc, confirmed the presence of Fuc on the terminal HexNAc.
Fig. 6.
(A) Isomer-specific fragmentation of EIC m/z 694.3 at 10.4 min, with the putative structure inset. (B) Isomer-specific fragmentation of EIC m/z 1018.4 at 14.3 min, with the putative structure inset.
An additional fucosylated structure, not previously reported in bovine milk, was identified as m/z 1018.37 [M+H]+, corresponding to the composition three Hex, one HexNAc and one Fuc. The connectivity of the oligosaccharide was determined from observing the loss of the core, which was comprised of a reducing end Hex plus an additional Hex. The loss of another Hex indicated that these three monosaccharides were linear in arrangement, which meant that the terminal disaccharide was composed of a Hex, HexNAc and Fuc. There were no informative peaks in the tandem mass spectrum to determine the connectivity of these terminal components. From the above analysis based on the tandem MS, the putative structure was proposed; however, further analysis with higher energy tandem MS would allow full structural annotation.
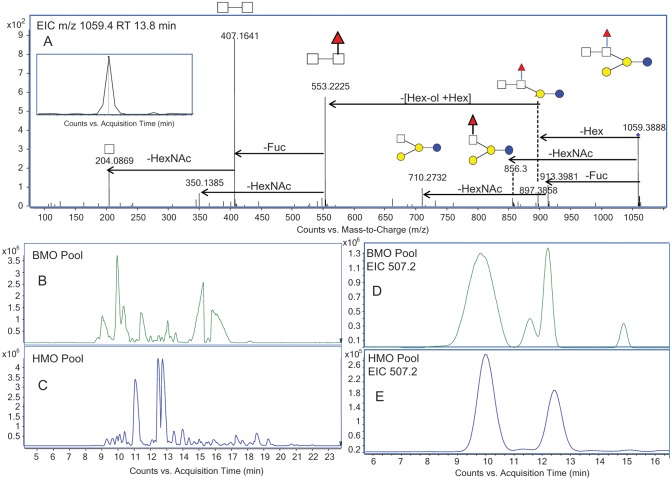
Another new fucosylated BMO was observed at m/z 1059.4 at 13.8 min, with the composition three Hex, two HexNAc and one Fuc. This oligosaccharide had two fragmentation pathways, one with a terminal HexNAc and the other with a terminal Hex. This observation suggested the branching type structure as shown in Figure 7A. The loss from m/z 879.4 to 553.2 corresponded to a reducing end Hex plus Hex, which indicated a lactose core. The peak at m/z 407.1 suggested that the two HexNAc monosaccharides were connected, whereas the peak at m/z 350.1 suggested that the Fuc was attached to a HexNAc. The peak at 856.3 corresponded to the loss of a HexNAc from the precursor and suggested that the terminal HexNAc did not have the Fuc attached to it. Based on this informative fragmentation profile, the structure was proposed.
Fig. 7.
(A) Isomer-specific fragmentation of EIC m/z 1059.4 at 13.8 min, with the putative structure inset. (B) BPC of the BMO pool. (C) BPC of the HMO pool. (D) EIC of m/z 507.2 from the BMO pool showing four isomers. (E) EIC of m/z 507.2 from the HMO pool showing two isomers.
Qualitative comparison of BMO and HMO
Bovine milk was found to contain 20 times lower oligosaccharide content than human milk. The majority of BMOs have simpler oligosaccharides that do not possess the structural complexity and diversity of human milk (Zivkovic and Barile 2011). However, lower abundant BMOs with structures similar to the more complex HMOs were reported (Zivkovic and Barile 2011). Although bovine milk differs in oligosaccharide content and abundances from those in human milk, there are many similarities. With the development of new analytical techniques, less abundant components are of potential commercial significance, making a comparison of bovine and human milk interesting and useful for future studies. Yet, to date, a comprehensive comparison of bovine milk to human milk has not been cataloged.
The base peak chromatograms from bovine milk (Figure 7B) and human milk (Figure 7C) analyses show different profiles with seemingly minimal overlap. However, upon the closer examination of the EICs for each component, the isomers in common are visible. For example, there were four isomers of a GOS-like trisaccharide in the BMO colostrum EIC of m/z 507.2 (Figure 7D). Interestingly, two of those isomers were also in the HMO pool (Figure 7E). This is the first report of GOS-like trisaccharides in human milk.
A qualitative comparison of the oligosaccharides present in both BMO and HMO pooled samples on an isomer-specific level are summarized in Table I. One of the most abundant oligosaccharides identified in the BMO pooled sample that was also found in pooled HMO was the well-characterized oligosaccharide, LNnT. The well-characterized oligosaccharide LNnH was found in both the pooled BMO colostrum samples and the HMO pool. The BMO colostrum pooled sample and the HMO pool both contained 3′-siayllactose and 2′-siayllactose isomers, as well as the corresponding siayllactosamine isomers. The study also revealed several fucosylated oligosaccharides with greater structural complexity than fucosyllactose in both the HMO pool and the bovine colostrum pool.
Table I.
List of oligosaccharides common between BMOs and HMOs
| Name | Mass | Structure | Abundance (BMO; %) | Abundance (HMO; %) |
|---|---|---|---|---|
| 2'FL | 490.190 | 0.3 | 21.73 | |
| 3'FL | 490.190 | 0.16 | 0.02 | |
| Triose A | 506.185 | 9.34 | 0.95 | |
| Triose C | 506.185 | 16.09 | 0.53 | |
| 3'SL | 635.227 | 24.81 | 1.06 | |
| 6'SL | 635.227 | 0.83 | 0.76 | |
| 3'SLN | 676.254 | 0.31 | — | |
| 6'SLN | 676.254 | 9.79 | — | |
| H2N1F1 | 693.269 | 0.13 | 0.03 | |
| LNnT | 709.264 | 3.78 | 14.40 | |
| H4N1F1 | 1017.375 | 0.04 | 0.18 | |
| H3N2F1 | 1059.4092 | 0.11 | 0.12 | |
| LNnH | 1074.396 | 1.34 | 0.41 |
Discussion
We have performed a comprehensive structure analysis on colostrum BMOs resulting in a detailed structure library. This study identified over 50 BMOs, with eight previously unreported features, each with its own unique retention time. The structure library detailing the most abundant BMOs identified is shown in Supplementary data, Table SI. Each entry includes retention time, accurate mass, oligosaccharide composition, intensity and full structure with structural linkages when possible. If the full structure was not determined in the study, a partial structure is provided including the connectivity of the monosaccharide units based on the fragmentation profile of the oligosaccharides. The library of structures was constructed on the basis of reproducible retention time and accurate masses obtained from the chip-based nano-LC coupled to MS, targeted tandem analysis and exoglycosidase digestions (Wu, Grimm, et al. 2010; Wu, Tao, et al. 2010; Aldredge et al. 2012). This database of structures will enable the rapid identification of BMOs in future studies.
Typically, BMOs are thought to contain a lactose core; however, several new oligosaccharides containing a lactosamine core were identified. Although these newly identified components were not the major oligosaccharides in bovine milk colostrum, this alternate core may be more common in BMOs than reported previously. This study confirmed the presence of the α1-2 and α1-3 fucosyllactose isomers and also identified two novel fucosylated oligosaccharides unique to bovine milk. The total amount of fucosylation was <1% of the total oligosaccharide pool, which is consistent with previous reports (Gopal and Gill 2000; Tao et al. 2008, 2009).
We identified 13 oligosaccharides common between bovine colostrum and human milk. This indicates significant overlap between these two sample sets, which has great implications for the future uses of BMOs. Further studies are in progress to identify additional structures that are common between bovine colostrum and HMOs and to quantify the amount of overlap between the two. These results validate the proposed use of bovine milk as a potential source of oligosaccharides similar in bioactivity to those in human milk. Future studies will be aimed at isolating and using these new oligosaccharides in functional studies to determine their bioactivity.
Materials and methods
Materials and reagents
Milk samples
Bovine colostrum milk samples were collected from Jersey and Holstein cows (n = 3, from each species) within 12 h of calving. The samples were pooled (BMO pool) and frozen at −80°C until further processing. Human milk samples were obtained from milk banks in San Jose, CA, and Austin, TX, and oligosaccharides were extracted as reported previously (Ninonuevo et al. 2006; Wu, Tao, et al. 2010). Non-porous graphitized carbon cartridges were obtained from Alltech Associated (Deerfield, Il). Sodium borohydride (98%) and 2,5-dihydroxybenzoic acid were purchased from Sigma-Aldrich (St Louis, MO). Recombinant β1-3,6 galactosidase, α1-3,6 galactosidase, β N-acetylhexosaminidase, α N-acetylgalactosaminidase, β N-acetylhexosaminidase, β N-acetylglucosaminidase and α2-3 neuraminidase were purchased from New England Biolabs (Ispwich, MA). β1-4 galactosidase was purchased from Prozyme (Hayward, CA). All other reagents were of the analytical or HPLC grade.
Enrichment of oligosaccharides from whole colostrum
A 500-µL aliquot of whole colostrum was added to 100 µL of water and centrifuged for 30 min at 15,000 × g and 4°C. The majority of fat (top layer) was removed, leaving a protein- and an oligosaccharide-rich fraction (bottom layer). A Folch solution of 67% chloroform and 33% methanol (v/v) was added to the defatted colostrum at a 4:1 ratio. The mixture was centrifuged for 30 min at 4000 × g and 4°C. The coagulated protein (middle layer) and remaining lipids (bottom layer) were removed, leaving an oligosaccharide-rich fraction (top layer). Ethanol was added to the enriched oligosaccharides at a 2:1 ratio. The mixture was frozen at −80°C for 1 h to precipitate remaining protein, and then centrifuged for 30 min at 4000 × g and 4°C. The oligosaccharide-rich fraction (top layer) was dried in vacuo. The dried oligosaccharides were resolubilized in 2 mL of 1.0 M sodium borohydride and kept at 65°C for 2 h to reduce the oligosaccharides from aldehydes to alditols (Aldredge et al. 2012).
Purification of milk oligosaccharides by graphitized carbon solid-phase extraction
Oligosaccharide alditols were purified by graphitized carbon solid-phase extraction. The cartridges were washed with 80% acetonitrile/0.10% trifluoroacetic acid (v/v) in water, followed by conditioning with pure water. Aqueous solutions of reduced oligosaccharides were loaded onto the cartridge and washed with pure water at a flow rate of ∼1 mL/min to remove salts and buffer. Oligosaccharides were eluted with a solution of 40% acetonitrile and 0.05% trifluoroacetic acid (v/v) in water. Samples were dried in vacuo and reconstituted in water prior to MS analysis.
Separation of BMOs with off-line HPLC
The reduced BMOs were fractionated off-line with an Agilent Hewlett-Packard Series 1100 HPLC system using a Hypercarb porous graphitized carbon (Thermoquest, Hypersil Division, Runcorn, UK) column (100 mm × 2.1 mm and 5-μm particle size). The oligosaccharides were eluted with a solvent system consisting of nanopure water (A) and acetonitrile (B) with a flow rate of 0.25 mL/min and a gradient of 0.0–25.0 min, 0–15% B; 25.0–50.0 min, 15–40% B; 50.0–70.0 min, 40–100% B; 70.0–80.0 min, 100–0% B. A total of 80 fractions were collected, dried and reconstituted with 15 μL of nanopure water prior to MS analysis.
Structural characterization using exoglycosidase digestion
Detailed procedures for exoglycosidase digestions were reported previously (Xie et al. 2001; Aldredge et al. 2012). Briefly, the digestions were carried out in a 37°C water bath with 3 μL of 0.1 M ammonium acetate buffer, 1 μL of sample and 0.5 μL of enzyme. Digestion conditions (digestion time, buffer pH etc.) were optimized for each exoglycosidase according to enzyme activity and concentration of oligosaccharides in the specific fraction.
MALDI FT-ICR MS and HPLC-Chip/Q-TOF MS analysis
Reduced and purified oligosaccharides were first analyzed using an IonSpec Hi Res MALDI FT-ICR MS (IonSpec, Lake Forest, CA) equipped with a 355-nm pulsed Nd:YAG laser, a hexapole ion guide, an ultrahigh vacuum system maintained by two turbo pumps, a cryopump and a 7.0 T-shielded superconducting magnet. The samples were spotted on a stainless steel MALDI plate with an equal volume of 2,3-dihydroxybenzoic acid matrix made up of 0.05 mg/mL of 2,3-dihydroxybenzoic acid in 50% acetonitrile/water (v/v). Oligosaccharides were analyzed in positive- and negative-ion modes and identified within a 5 ppm accurate mass criterion.
The oligosaccharides were also analyzed using an Agilent HPLC-Chip/Q-TOF (Chip/Q-TOF, Agilent Technologies, Santa Clara, CA) MS system equipped with a microwell-plate autosampler (maintained at 6°C), capillary sample loading pump, nanopump, HPLC-Chip/MS interface and the Agilent 6210 TOF MS detector. The chip consisted of a 9 × 0.075 mm i.d. enrichment column and a 43 × 0.075 mm i.d. analytical column, both packed with 5 μm porous graphitized carbon as the stationary phase. For sample loading, the capillary pump delivered 0.1% formic acid in 3.0% acetonitrile/water (v/v) isocratically at 4.0 μL/min. Injection volume was 2.0 μL for each sample. The nanopump gradient was delivered at 0.4 μL/min using (A) 0.1% formic acid in 3.0% acetonitrile/water (v/v) and (B) 0.1% formic acid in 90.0% acetonitrile/water (v/v). Samples were eluted with 0% B, 0.00–2.50 min; 0–16% B, 2.50–20.00 min; 16–44% B, 20.00–30.00 min; 44–100% B, 30.00–35.00 min; and 100% B, 35.00–45.00 min. The elution gradient was followed by a column re-equilibration at 0% B for 20 min. The drying gas temperature was 325°C and the flow rate was 4 L/min (from a mixture of 2 L of filtered nitrogen gas and 2 L of filtered dry compressed air). MS spectra were acquired in the positive-ion mode over a mass range of m/z 400–2000 with an acquisition time of 1.5 s per spectrum. Mass correction was enabled using reference masses of m/z 622.029, 922.010, 1221.991 and 1521.971 (ESI-TOF Calibrant Mix G1969-85000, Agilent Technologies).
Identification and relative quantification of BMOs
To obtain oligosaccharide profiles for each pooled milk sample, computerized algorithms extracted a comprehensive list of compound peaks in a sample and then identified the oligosaccharide compositions by accurate mass within a 20-ppm accurate mass criterion. Raw LC/MS data were filtered with a signal-to-noise ratio of 5.0 and analyzed using the Molecular Feature Extractor algorithm in the MassHunter Qualitative Analysis software (Version B.03.01, Agilent Technologies). Taking into account the expected charge carriers, potential neutral mass losses and a predicted isotopic distribution, the total ion chromatogram was divided into individual extracted compound chromatograms. Each extracted compound chromatogram represented the summed chromatograms of all ion species associated with a single compound (e.g., the singly protonated, doubly protonated, singly dehydrated etc. ions). Thus, each individual extracted compound chromatogram peak represented the total ion count associated with a distinct compound.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by the University of California Discovery Fund, the California Dairy Research Foundation (10GEB-02NH) and the National Institutes of Health (Bethesda, MD) through grants from the Eunice K. Shriver National Institute of Child Health and Human Development Grant (HD059127) and the National Center for Research Resources, a component of the National Institutes of Health (UL1 RR024146). The project described was partially supported by the National Center for Complementary & Alternative Medicine (1R01AT007079-01). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Complementary & Alternative Medicine or the National Institutes of Health.
Conflict of interest
None declared.
Abbreviations
BMO, bovine milk oligosaccharide; EIC, extracted ion chromatogram; FT-ICR, Fourier transform ion cyclotron resonance; Fuc, fucose; Gal, galactose; Glc, glucose; GlcNAc, N-acetylglucosamine; GOS, galacto-oligosaccharides; HMO, human milk oligosaccharide; HPLC, high-performance liquid chromatography; LNnH, lacto-N-neohexaose; LNnT, lacto-N-neotetraose; LNT, lacto-N-tetraose; MALDI, matrix-assisted laser desorption ionization; MS, mass spectrometry; nano-LC, nanoflow liquid chromatography; NeuAc, N-acetylneuraminic acid; NeuGc, N-glycolylneuraminic acid; Q-TOF, quadrupole time of flight.
Supplementary Material
Acknowledgements
The authors thank CJ Dillard for critical reading of the manuscript and Agilent Technology for assistance with the instrumentation.
References
- Albrecht S, Schols HA, van den Heuvel EGHM, Voragen AGJ, Gruppen H. CE-LIF-MSn profiling of oligosaccharides in human milk and feces of breast-fed babies. Electrophoresis. 2010;31:1264–1273. doi: 10.1002/elps.200900646. [DOI] [PubMed] [Google Scholar]
- Aldredge D, An HJ, Tang N, Waddell K, Lebrilla CB. Annotation of a serum N-glycan library for rapid identification of structures. J Proteome Res. 2012;11:1958–1968. doi: 10.1021/pr2011439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile D, Marotta M, Chu C, Mehra R, Grimm R, Lebrilla CB, German JB. Neutral and acidic oligosaccharides in Holstein-Friesian colostrum during the first 3 days of lactation measured by high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. J Dairy Sci. 2010;93:3940–3949. doi: 10.3168/jds.2010-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile D, Meyrand M, Lebrilla CB, German JB. Examining bioactive components of milk sources of complex oligosaccharides (part 2) Agro Food Ind Hi Tech. 2011;22:37–39. [Google Scholar]
- Blank D, Dotz V, Geyer R, Kunz C. Human milk oligosaccharides and Lewis blood group: Individual high-throughput sample profiling to enhance conclusions from functional studies. Adv Nutr. 2012;3:440S–449S. doi: 10.3945/an.111.001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm G, Stahl B. Oligosaccharides from milk. J Nutr. 2007;137:847S–849S. doi: 10.1093/jn/137.3.847S. [DOI] [PubMed] [Google Scholar]
- Broberg A. High-performance liquid chromatography/electrospray ionization ion-trap mass spectrometry for analysis of oligosaccharides derivatized by reductive amination and N,N-dimethylation. Carbohydr Res. 2007;342:1462–1469. doi: 10.1016/j.carres.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Chai W, Piskarev VE, Zhang Y, Lawson AM, Kogelberg H. Structural determination of novel lacto-N-decaose and its monofucosylated analogue from human milk by electrospray tandem mass spectrometry and 1H NMR spectroscopy. Arch Biochem Biophys. 2005;434:116–127. doi: 10.1016/j.abb.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Chaturvedi P, Warren C, Altaye M, Morrow A, Ruiz-Palacios G, Pickering L, Newburg D. Fucosylated human milk oligosaccharides vary between individuals and over the course of lactation. Glycobiology. 2001;11:365–372. doi: 10.1093/glycob/11.5.365. [DOI] [PubMed] [Google Scholar]
- Chu CS, Lebrilla CB. Introduction to modern techniques in mass spectrometry. In: Jue T, editor. Biomedical Applications of Biophysics. New York: Humana Press; 2010. [Google Scholar]
- Chu CS, Niñonuevo MR, Clowers BH, Perkins PD, An HJ, Yin H, Killeen K, Miyamoto S, Grimm R, Lebrilla CB. Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics. 2009;9:1939–1951. [Google Scholar]
- Coppa GV, Bruni S, Morelli L, Soldi S, Gabrielli O. The first prebiotics in humans: Human milk oligosaccharides. J Clin Gastroenterol. 2004;38:S80–S83. doi: 10.1097/01.mcg.0000128926.14285.25. [DOI] [PubMed] [Google Scholar]
- Coppa GV, Zampini L, Galeazzi T, Facinelli B, Ferrante L, Capretti R, Orazio G. Human milk oligosaccharides inhibit the adhesion to Caco-2 cells of diarrheal pathogens: Escherichia coli, Vibrio cholerae, and Salmonella fyris. Pediatr Res. 2006;59:377–382. doi: 10.1203/01.pdr.0000200805.45593.17. [DOI] [PubMed] [Google Scholar]
- Ernst B, Müller DR, Richter WJ. False sugar sequence ions in electrospray tandem mass spectrometry of underivatized sialyl-Lewis-type oligosaccharides. Int J Mass Spectrom Ion Processes. 1997;160:283–290. [Google Scholar]
- Finke B, Stahl B, Pfenninger A, Karas M, Daniel H, Sawatzki G. Analysis of high-molecular-weight oligosaccharides from human milk by liquid chromatography and MALDI-MS. Anal Chem. 1999;71:3755–3762. doi: 10.1021/ac990094z. [DOI] [PubMed] [Google Scholar]
- Fong B, Ma K, McJarrow P. Quantification of bovine milk oligosaccharides using liquid chromatography, selected reaction monitoring, and mass spectrometry. J Agric Food Chem. 2011;59:9788–9795. doi: 10.1021/jf202035m. [DOI] [PubMed] [Google Scholar]
- Gopal PK, Gill HS. Oligosaccharides and glycoconjugates in bovine milk and colostrum. Br J Nutr. 2000;84(Suppl 1):S69–S74. doi: 10.1017/s0007114500002270. [DOI] [PubMed] [Google Scholar]
- Guerardel Y, Morelle W, Plancke Y, Lemoine J, Strecker G. Structural analysis of three sulfated oligosaccharides isolated from human milk. Carbohydr Res. 1999;320:230–238. doi: 10.1016/s0008-6215(99)00153-6. [DOI] [PubMed] [Google Scholar]
- Hakkarainen J, Toivanen M, Leinonen A, Frangsmyr L, Stromberg N, Lapinjoki S, Nassif X, Tikkanen-Kaukanen C. Human and bovine milk oligosaccharides inhibit neisseria meningitidis pili attachment in vitro. J Nutr. 2005;135:2445–2448. doi: 10.1093/jn/135.10.2445. [DOI] [PubMed] [Google Scholar]
- Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, Lebrilla CB. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst. 2011;136:3663–3671. doi: 10.1039/c1an15093f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhn C, Ramautar R, Wuhrer M, Somsen G. Relevance and use of capillary coatings in capillary electrophoresis–mass spectrometry. Anal Bioanal Chem. 2010;396:297–314. doi: 10.1007/s00216-009-3193-y. [DOI] [PubMed] [Google Scholar]
- Klein N, Schwertmann A, Peters M, Kunz C, Strobel S. Immunomodulatory effects of breast milk oligosaccharides. Adv Exp Med Biol. 2000;478:251–259. doi: 10.1007/0-306-46830-1_23. [DOI] [PubMed] [Google Scholar]
- Kunz C, Rudloff S. Structural and functional aspects of oligosaccharides in human milk. Z Ernahrungswiss. 1996;35:22–31. doi: 10.1007/BF01612024. [DOI] [PubMed] [Google Scholar]
- Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: Structural, functional, and metabolic aspects. Annu Rev Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- Leo F, Asakuma S, Nakamura T, Fukuda K, Senda A, Urashima T. Improved determination of milk oligosaccharides using a single derivatization with anthranilic acid and separation by reversed-phase high-performance liquid chromatography. J Chromatogr A. 2009;1216:1520–1523. doi: 10.1016/j.chroma.2009.01.015. [DOI] [PubMed] [Google Scholar]
- LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA, German JB. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- Ma Y-L, Vedernikova I, Van den Heuvel H, Claeys M. Internal glucose residue loss in protonated O-diglycosyl flavonoids upon low-energy collision-induced dissociation. J Am Soc Mass Spec. 2000;11:136–144. doi: 10.1016/S1044-0305(99)00133-6. [DOI] [PubMed] [Google Scholar]
- Mariño K, Lane JA, Abrahams JL, Struwe WB, Harvey DJ, Marotta M, Hickey RM, Rudd PM. Method for milk oligosaccharide profiling by 2-aminobenzamide labeling and hydrophilic interaction chromatography. Glycobiology. 2011;21:1317–1330. doi: 10.1093/glycob/cwr067. [DOI] [PubMed] [Google Scholar]
- Martin-Sosa S, Martinn M, Hueso P. The sialylated fraction of milk oligosaccharides is partially responsible for binding to enterotoxigenic and uropathogenic Escherichia coli human strains. J Nutr. 2002;132:3067–3072. doi: 10.1093/jn/131.10.3067. [DOI] [PubMed] [Google Scholar]
- Mehra R, Kelly P. Milk oligosaccharides: Structural and technological aspects. Int Dairy J. 2006;16:1334–1340. [Google Scholar]
- Monks B. New England Biolabs, Inc. Unpublished results.
- Newburg DS, Neubauer S. Handbook of Milk Composition. Orlando: Academic Press; 1995. Carbohydrates in milks, analysis, quantities, and significance. In: Jensen RG, editor; pp. 273–349. [Google Scholar]
- Ninonuevo M, An H, Yin H, Killeen K, Grimm R, Ward R, German B, Lebrilla C. Nanoliquid chromatography-mass spectrometry of oligosaccharides employing graphitized carbon chromatography on microchip with a high-accuracy mass analyzer. Electrophoresis. 2005;26:3641–3649. doi: 10.1002/elps.200500246. [DOI] [PubMed] [Google Scholar]
- Ninonuevo MR, Park Y, Yin H, Zhang J, Ward RE, Clowers BH, German JB, Freeman SL, Killeen K, Grimm R. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- Ninonuevo MR, Perkins PD, Francis J, Lamotte LM, LoCascio RG, Freeman SL, Mills DA, German JB, Grimm R, Lebrilla CB. Daily variations in oligosaccharides of human milk determined by microfluidic chips and mass spectrometry. J Agric Food Chem. 2008;56:618–626. doi: 10.1021/jf071972u. [DOI] [PubMed] [Google Scholar]
- Pfenninger A, Karas M, Finke B, Stahl B. Structural analysis of underivatized neutral human milk oligosaccharides in the negative ion mode by nano-electrospray MSn (part 2: Application to isomeric mixtures) J Am Soc Mass Spec. 2002;13:1341–1348. doi: 10.1016/S1044-0305(02)00646-3. [DOI] [PubMed] [Google Scholar]
- Rohmer M, Baeumlisberger D, Stahl B, Bahr U, Karas M. Fragmentation of neutral oligosaccharides using the MALDI LTQ Orbitrap. Int J Mass Spec. 2011;305:199–208. [Google Scholar]
- Saito T, Itoh T, Adachi S. Chemical structure of three neutral trisaccharides isolated in free form from bovine colostrum. Carbohydr Res. 1987;165:43–51. doi: 10.1016/0008-6215(87)80076-9. [DOI] [PubMed] [Google Scholar]
- Saito T, Itoh T, Adachi S, Suzuki T, Usui T. The chemical structure of neutral and acidic sugar chains obtained from bovine colostrum kappa-casein. Biochim Biophys Acta. 1981;678:257–267. doi: 10.1016/0304-4165(81)90215-4. [DOI] [PubMed] [Google Scholar]
- Salvini F, Riva E, Salvatici E, Boehm G, Jelinek J, Banderali G, Giovannini M. A specific prebiotic mixture added to starting infant formula has long-lasting bifidogenic effects. J Nutr. 2011;141:1335–1339. doi: 10.3945/jn.110.136747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneir ML, Rafelson ME., Jr. Isolation and characterization of two structural isomers of N-acetylneuraminyllactose from bovine colostrum. Biochim Biophys Acta. 1966;130:1–11. [Google Scholar]
- Shen Z, Warren CD, Newburg DS. High-performance capillary electrophoresis of sialylated oligosaccharides of human milk. Anal Biochem. 2000;279:37–45. doi: 10.1006/abio.1999.4448. [DOI] [PubMed] [Google Scholar]
- Stahl B, Thurl S, Zeng J, Karas M, Hillenkamp F, Steup M, Sawatzki G. Oligosaccharides from human milk as revealed by matrix-assisted laser desorption/ionization mass spectrometry. Anal Biochem. 1994;223:218–226. doi: 10.1006/abio.1994.1577. [DOI] [PubMed] [Google Scholar]
- Stiasny K, Allison SL, Marchler-Bauer A, Kunz C, Heinz FX. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J Virol. 1996;70:8142–8147. doi: 10.1128/jvi.70.11.8142-8147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadasu U, Tadao S, Kenzi O, Keiichi S. Structural determination of three neutral oligosaccharides in bovine (Holstein-Friesian) colostrum, including the novel trisaccharide; GalNAc αl-3Gal β1–4Glc. Biochim Biophys Acta. 1991;1073:225–229. doi: 10.1016/0304-4165(91)90207-w. [DOI] [PubMed] [Google Scholar]
- Tao N, DePeters EJ, Freeman S, German JB, Grimm R, Lebrilla CB. Bovine milk glycome. J Dairy Sci. 2008;91:3768–3778. doi: 10.3168/jds.2008-1305. [DOI] [PubMed] [Google Scholar]
- Tao N, DePeters EJ, German JB, Grimm R, Lebrilla CB. Variations in bovine milk oligosaccharides during early and middle lactation stages analyzed by high-performance liquid chromatography-chip/mass spectrometry. J Dairy Sci. 2009;92:2991–3001. doi: 10.3168/jds.2008-1642. [DOI] [PubMed] [Google Scholar]
- Urashima T, Saito T, Nakamura T, Messer M. Oligosaccharides of milk and colostrum in non-human mammals. Glycoconj J. 2001;18:357–371. doi: 10.1023/a:1014881913541. [DOI] [PubMed] [Google Scholar]
- Urashima T, Saito T, Ohmisya K, Shimazaki K. Structural determination of three neutral oligosaccharides in bovine (Holstein-Friesian) colostrum, including the novel trisaccharide; GalNAc alpha 1–3Gal beta 1–4Glc. Biochim Biophys Acta. 1991;1073:225–229. doi: 10.1016/0304-4165(91)90207-w. [DOI] [PubMed] [Google Scholar]
- Veh RW, Michalski J-C, Corfield AP, Sander-Wewer M, Gies D, Schauer R. New chromatographic system for the rapid analysis and preparation of colostrum sialyloligosaccharides. J Chromatogr A. 1981;212:313–322. doi: 10.1016/s0021-9673(01)84044-9. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe S, Hua S, Rincon G, Islas-Trejo A, German JB, Lebrilla CB, Medrano JF. Transcriptome profiling of bovine milk oligosaccharide metabolism genes using RNA-sequencing. PLoS One. 2011;6:e18895. doi: 10.1371/journal.pone.0018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Grimm R, German JB, Lebrilla CB. Annotation and structural analysis of sialylated human milk oligosaccharides. J Proteome Res. 2010;10:856–868. doi: 10.1021/pr101006u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. 2010;9:4138–4151. doi: 10.1021/pr100362f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhrer M, Koeleman CAM, Hokke CH, Deelder AM. Mass spectrometry of proton adducts of fucosylated N-glycans: Fucose transfer between antennae gives rise to misleading fragments. Rapid Commun Mass Spectrom. 2006;20:1747–1754. doi: 10.1002/rcm.2509. [DOI] [PubMed] [Google Scholar]
- Xie Y, Tseng K, Lebrilla CB, Hedrick JL. Targeted use of exoglycosidase digestion for the structural elucidation of neutral O-linked oligosaccharides. J Am Soc Mass Spec. 2001;12:877–884. doi: 10.1016/S1044-0305(01)00267-7. [DOI] [PubMed] [Google Scholar]
- Yang H, Yu Y, Song F, Liu S. Structural characterization of neutral oligosaccharides by laser-enhanced in-source decay of MALDI-FTICR MS. J Am Soc Mass Spec. 2011;22:845–855. doi: 10.1007/s13361-011-0085-0. [DOI] [PubMed] [Google Scholar]
- Zivkovic AM, Barile D. Bovine milk as a source of functional oligosaccharides for improving human health. Adv Nutr. 2011;2:284–289. doi: 10.3945/an.111.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.