Abstract
Therapeutic ultrasound is widely employed in clinical applications but its mechanism of action remains unclear. Here we report prompt fluidization of a cell and dramatic acceleration of its remodeling dynamics when exposed to low intensity ultrasound. These physical changes are caused by very small strains (10−5) at ultrasonic frequencies (106 Hz), but are closely analogous to those caused by relatively large strains (10−1) at physiological frequencies (100 Hz). Moreover, these changes are reminiscent of rejuvenation and aging phenomena that are well-established in certain soft inert materials. As such, we suggest cytoskeletal fluidization together with resulting acceleration of cytoskeletal remodeling events as a mechanism contributing to the salutary effects of low intensity therapeutic ultrasound.
Introduction
Low intensity pulsed ultrasound (LIPUS) is a non-invasive therapeutic tool that is widely used for clinical applications including physiotherapy, drug delivery, bone fracture healing, and thrombolysis.1–5 In vivo and in vitro studies indicate that LIPUS facilitates wound repair, microvascular remodeling, blood flow restoration, and angiogenesis and also activates mechanosensitive signaling pathways.6–11 Despite its wide applicability, the physical mechanisms responsible for the beneficial effects of LIPUS are not understood. In particular, LIPUS must ultimately affect individual cells, but its direct physical effects on cells have never been investigated.
Most of the biological processes associated with LIPUS stimulation, such as wound repair, microvascular remodeling, and angiogenesis, necessarily entail structural remodeling on the cytoskeletal level. Such structural remodeling not only is typically mediated by events at the levels of signaling or energy metabolism12–14 but also can be mediated mechanically by direct application of physical forces such as shear or tensile stress.15–17 For example, strains of 10−1 at physiological frequencies of 100 Hz effectively fluidize the cytoskeleton and accelerate microscale dynamics in a manner analogous to the rejuvenation of soft glassy materials.16,18 In contrast, during LIPUS agitation, as described below, the levels of oscillatory strain are on the order of 10−5 at the frequency of 106 Hz, but the physical consequences of such ultrasonic stimulation on the cytoskeletal structure remain unknown.
In this paper we demonstrate that although oscillatory strains are quite small during LIPUS agitation, they are large enough nonetheless to promote prompt and nearly complete fluidization of the cytoskeleton. This fluidization response is reversible in the sense that cessation of the exposure leads to a slow cytoskeletal resolidification. The fluidization response is accompanied by a dramatic but reversible acceleration in the rate of cytoskeletal remodeling. These results highlight the potential of low intensity ultrasound to perturb cytoskeletal dynamics and demonstrate for the first time a direct and prompt mechanical response of cellular structure to LIPUS.
Methods
Experimental setup
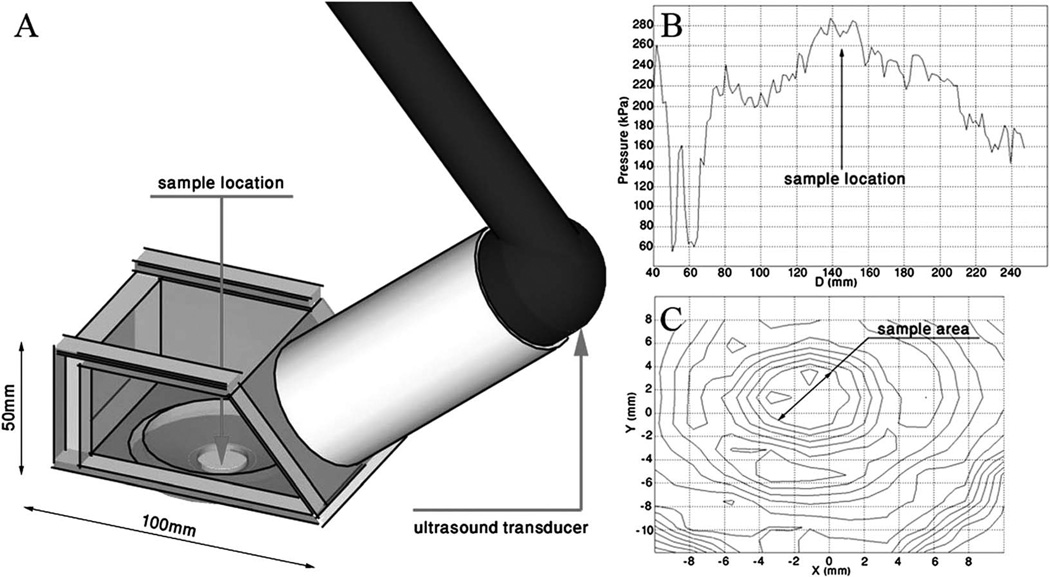
Our experiments are conducted in an exposure chamber mounted on an inverted microscope, allowing us to monitor changes in cell dynamics in real time (Fig. 1A). The acoustic agitation is applied using a clinical ultrasound device (Sonicator 730—Mettler Electronics, Anaheim, CA) and the acoustic field propagates at an angle of 45° to the sample, and was calibrated to ensure a stable uniform field across the sample (Fig. 1B and C). We apply two levels of acoustic pressure commensurate with intensity levels utilized in clinical therapies: 1 W cm−2 and 2 W cm−2 at the transducer, which we refer to as moderate and higher exposure respectively. The actual acoustic pressure as measured at the sample location using a needle hydrophone (HNR-500, Onda, Sunnyvale, CA) is 170 kPa and 290 kPa which correspond to moderate and higher exposure respectively. A 1 MHz pulsed signal with a 20% duty cycle is used to minimize sample heating; the maximum temperature rise during 30 s irradiation is less than 1 K.
Fig. 1.
(A) Scheme of the experimental setup comprised an irradiation chamber, a circular unfocused ultrasound transducer, and an adaptor filled with degassed water, connecting the irradiation chamber and the transducer. The adaptor length is designed to locate the sample in the far-field region providing stable pressure values. (B) Axial pressure as a function of distance from the transducer; the far-field was determined to start at a distance of ~140 mm. (C) Contour plot of the radial pressure distribution at the far field region (43.3 kPa per contour). The sample area is confined to 0.8 cm2 at the center of the beam where an approximately homogeneous field is measured. Measurements of acoustic pressure are performed using an HNR-0500 needle hydrophone (Onda Corp., Sunnyvale, CA).
Cell culture
To study the changes in cell dynamics mediated by LIPUS we used Human Airway Smooth Muscle (HASM) cells; the mechanical properties and responses of these cells to physical stimuli have been well established previously.13,16,17,19,20 HASM cells21 are cultured on plastic flasks in Ham’s F-12 medium supplemented with 10% fetal bovine serum, 100 U ml−1 penicillin, 100 mg ml−1 streptomycin, 200 mg ml−1 amphotericin B, 12 mM NaOH, 1.7 mM CaCl2, 2 mM l-glutamine, and 25 mM HEPES. Cells in passages 4–6 are plated in serum-free medium on type I collagen-coated (0.1 mg ml−1) polyacrylamide gel overnight before being tested.
Live fluorescent imaging of actin fibers
For imaging of actin fibers, HASM cells were transfected for 48 h with an adenoviral vector encoding enhanced yellow fluorescent protein (YFP)-tagged actin.22 Then the cells were plated sparsely on a polyacrylamide gel substrate. The fluorescent images of the cell actin network were obtained before US application, at the end of 30 s irradiation and every 30 s during the 5 min relaxation period.
Traction force microscopy
We measure cell contractility using traction force microscopy.23 Cells were prepared following a protocol previously established. 17 Briefly, we use a substrate comprising an elastic polyacrylamide gel with a shear modulus of 4 kPa, and embedded 0.2 µm diameter fluorescent microspheres (Molecular Probes, Eugene, OR) in the gel. We image the cell using phase contrast microscopy and the microspheres using fluorescence microscopy. We then determine the displacement field of the gel by comparing the positions of the microspheres with the cell present to those obtained for the relaxed gel when the cell is removed. The traction field is computed from the displacement field using Fourier transform traction microscopy (FTTM),17,23,24 and traction maps are obtained at each time point and integrated to yield the net contractile moment, T;23 this quantifies the cell contractility. Both cell shape and cell area are found to vary only marginally during the experiment.
Spontaneous bead motions
To probe the dynamics of CSK remodeling, we measured spontaneous movement of an individual ferromagnetic bead (4.5 µm) coated with an Arg-Gly-Asp (RGD) peptide which forms focal adhesions and becomes tightly anchored to the CSK via integrin receptors.25,26 We reason that such a firmly attached bead cannot move unless the structure to which it is attached somehow reorganizes. If so, then bead motion as quantified by mean square displacement (MSD) reports the rate of ongoing structural rearrangements over time.13,26 As a control, we used beads coated with acetylated low-density lipoprotein (acLDL), which promotes non-specific binding to the cell membrane and is not associated with CSK rearrangement.20,25,26 For MSD measurements ferromagnetic beads are attached to a confluent monolayer of HASM cells which are grown on collagen-coated polyacrylamide gel. MSD profiles are measured by tracking the centre position of each bead during spontaneous bead motion. The MSD of the bead is calculated for each 30 s interval as: MSD (Δt) = 〈(x(t + Δt) − (x(t))2)〉, where Δt is the time lag, x is the bead coordinate, and brackets indicate an average over time t. For each experiment, data represent the following conditions: 30 s of baseline followed by 30 s of US exposure and continuous relaxation during the following 180 s after excitation removal. The distribution of the MSD(Δt) from bead to bead was approximately log-normal.13 Accordingly, data are reported as the median of the bead population. Each MSD curve is the result of an average over ~300 beads over a likewise number of cells.
Results and discussion
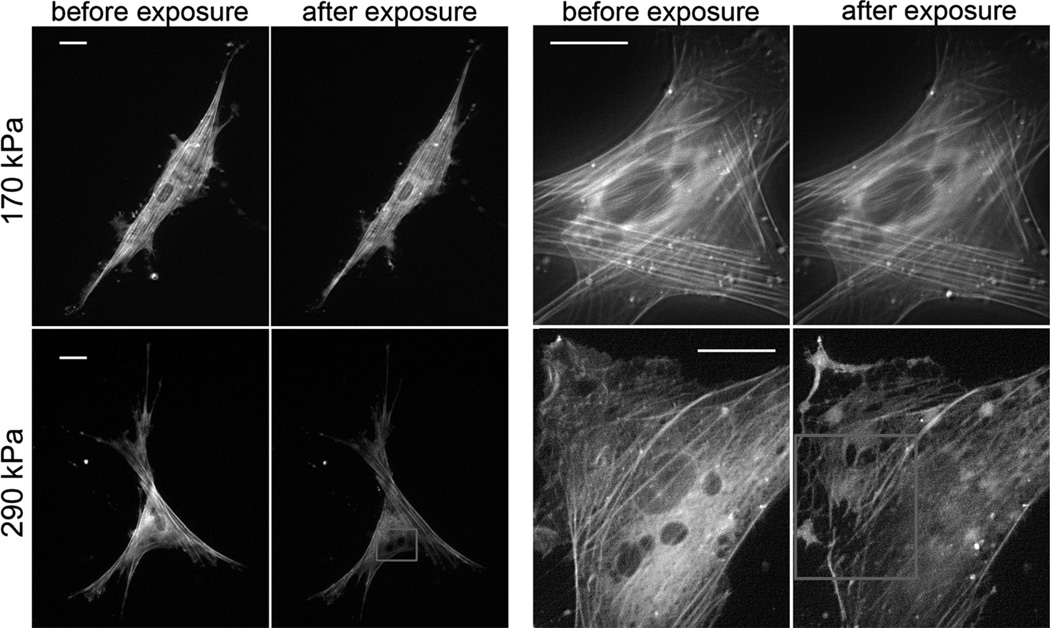
To characterize dynamic changes in cell structure before, during, and after LIPUS exposure, we visualize the actin network of human airway smooth muscle (HASM) cells using YFP-live staining. At moderate acoustic intensities, we observe no structural changes as shown by the comparison of before and after microscopy images (Fig. 2, top panel and Movie S1†). In contrast, when subjected to a higher acoustic intensity, the actin network is progressively disrupted and disassembles within 3 min following exposure (Fig. 2, bottom panel and Movies S2 and S3†). Quantification of overall cell fluorescence demonstrates the same trend in reduction of global actin content of the cell following exposure to higher levels of acoustic intensity (Fig. S1†).
Fig. 2.
Live YFP-actin network staining of cells before and 5 min after exposure to 170 kPa (top panel) and 290 kPa (bottom panel) acoustic pressures. Regions with perturbed actin structure (bottom panel—left) and disrupted individual fibers (bottom panel—right) are marked by squares in 10× and 40× magnification images correspondingly. Scale bar 10 µm.
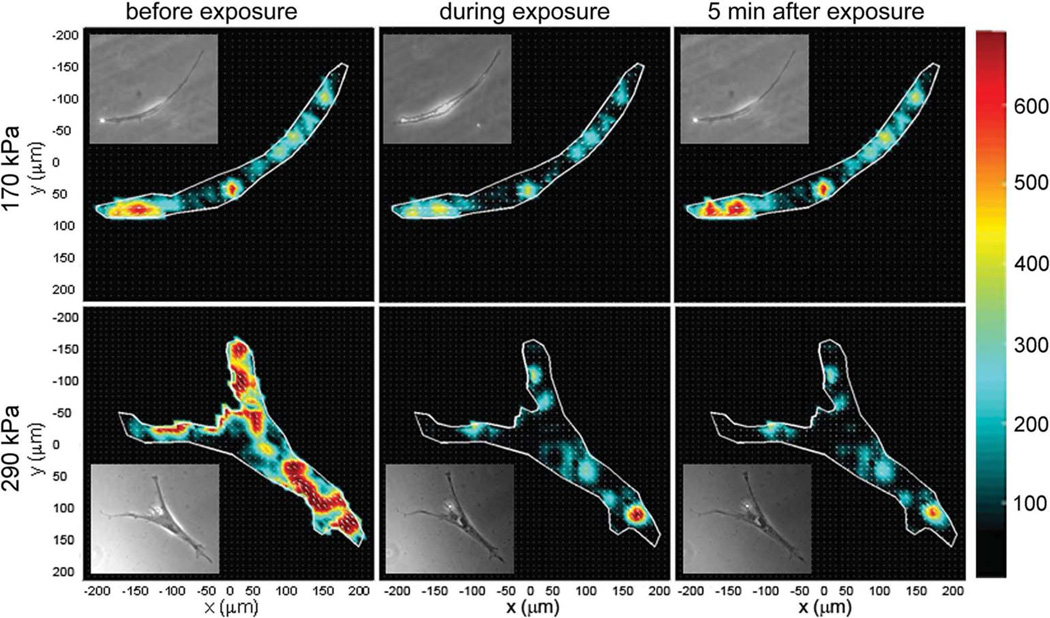
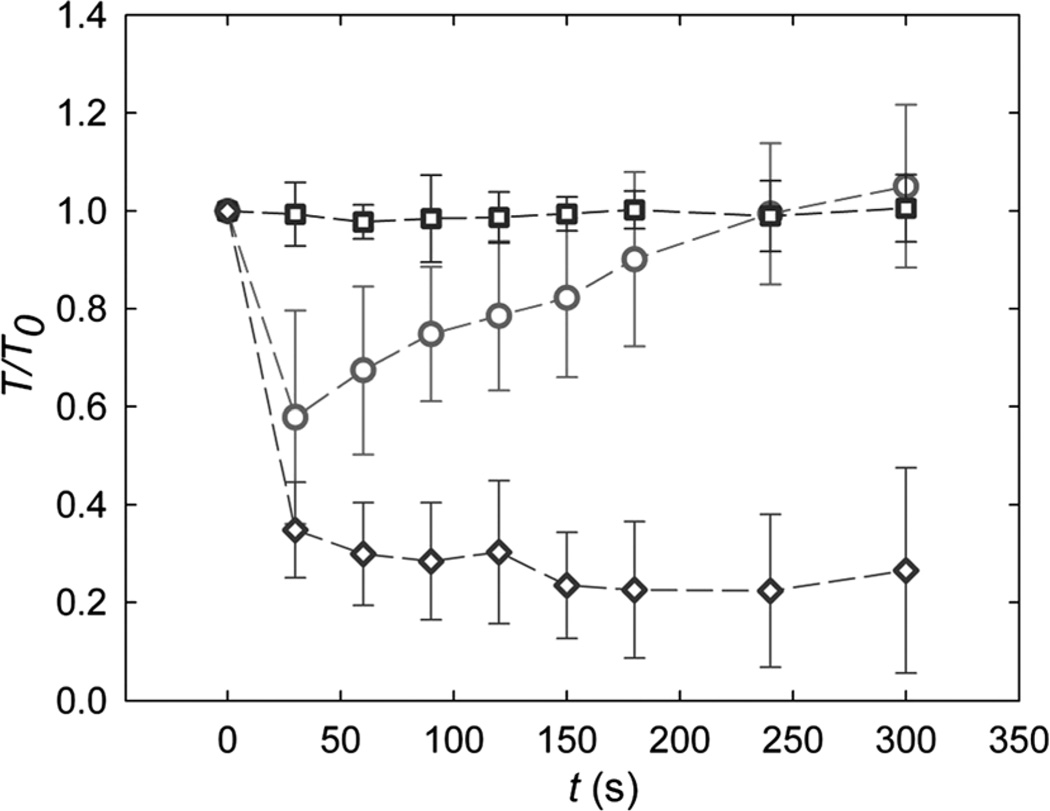
To quantify the global effects of LIPUS on the cytoskeleton, we measure the changes in cell contractility. Cell traction maps are obtained at each time point (Fig. 3) and integrated to yield the net contractile moment, T.23 When no LIPUS is applied, the contractility remains constant and T is independent of time (Fig. 4, squares). When a moderate acoustic pressure of 170 kPa is applied for 30 s, however, T drops promptly to 50% of its initial value, and then following exposure gradually recovers to its initial value over about 200 s (Fig. 4, circles). In contrast, when the acoustic pressure is increased to 290 kPa, T exhibits the same immediate drop but does not recover over the experimental times (Fig. 4, diamonds). Interestingly, the rapid decrease and slow recovery of T following exposure to the moderate intensity ultrasound is reminiscent of the cellular fluidization induced by a single transient strain of 10%.17
Fig. 3.
Representative cell traction maps before, during and 5 min after exposure to 170 kPa (top panel) and 290 kPa (bottom panel) acoustic pressure. Inset: the corresponding phase contrast images of the cell.
Fig. 4.
Average and standard deviation of cell contractile moment, T, normalized with respect to the initial value before exposure to LIPUS: during time control (squares) and when exposed to moderate (170 kPa— circles) and higher (290 kPa—diamonds) acoustic pressure.
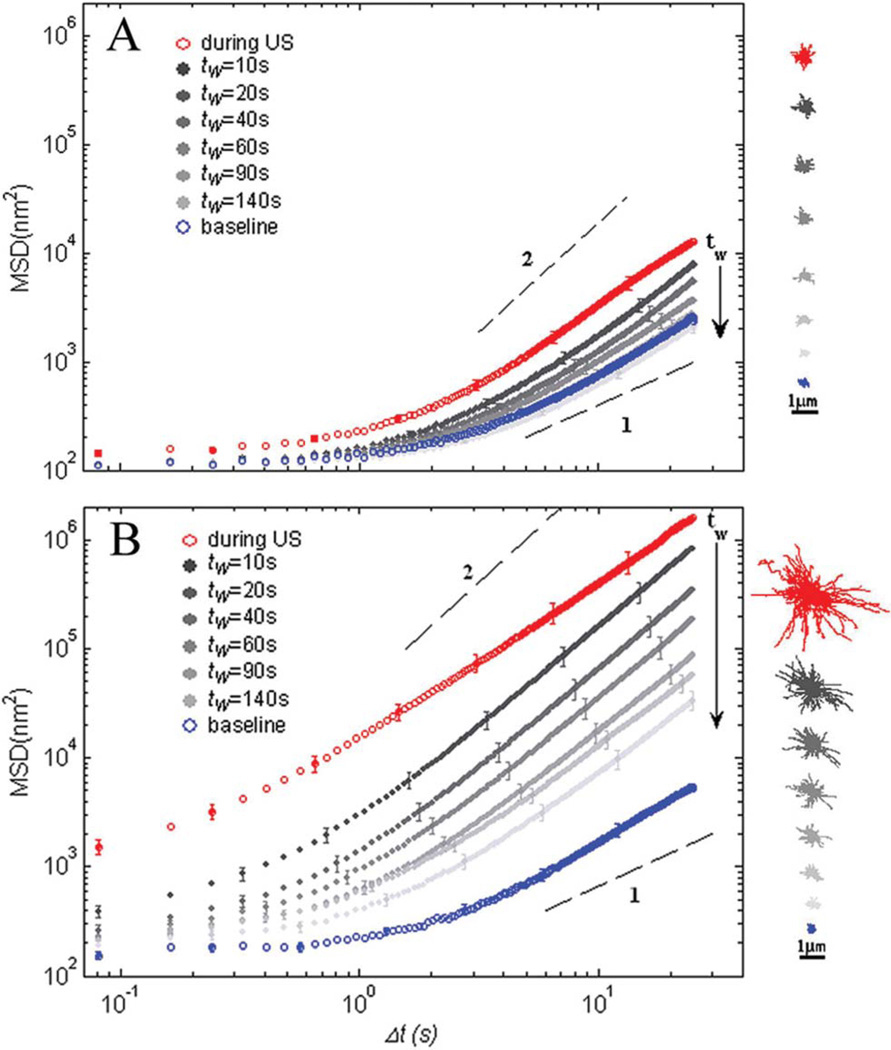
To determine the effects of LIPUS on the rate of cytoskeletal remodeling, we measure the mean square displacement (MSD) of beads tightly attached to the cytoskeleton as a function of time lag Δt.13,16 When no LIPUS is applied, the MSD(Δt) exhibits subdiffusive behavior for small Δt and superdiffusive behavior for large Δt, as previously reported13,27 (Fig. 5A and B, bottom blue symbols). During exposure to moderate intensity, the MSD increases significantly as shown by the topmost red symbols in Fig. 5A. Upon removal of the ultrasound, the MSD gradually relaxes back to its baseline position over ~180 s (Fig. 5A, closed symbols). Exposure to the higher intensity ultrasound leads to a significantly different behavior: the MSD curve shifts by three orders of magnitude during the exposure (Fig. 5B, topmost red symbols), but no longer relaxes to the baseline; instead it only exhibits partial relaxation (Fig. 5B). Such behavior during both agitation regimes is consistent with the traction data presented in Fig. 4 and further supports the notion that LIPUIS induces cytoskeleton fluidization.
Fig. 5.
MSD of beads tightly bound to cell cytoskeleton as a function of time lag after application of moderate, 170 kPa (A) or higher, 290 kPa (B) acoustic pressures. For both irradiation regimes the MSD measurements were taken before exposure to ultrasound (blue symbols), during exposure (red symbols), and after exposure removal (gray symbols). n = 280 and 320 beads for A and B correspondingly, and the error bars represent the standard error of MSD for each experiment. Waiting time, tw, indicates consequent time intervals after ultrasound removal. The dashed lines show diffusion exponents of 1 and 2. Insets: representative trajectories of 30 tracers, during each time interval.
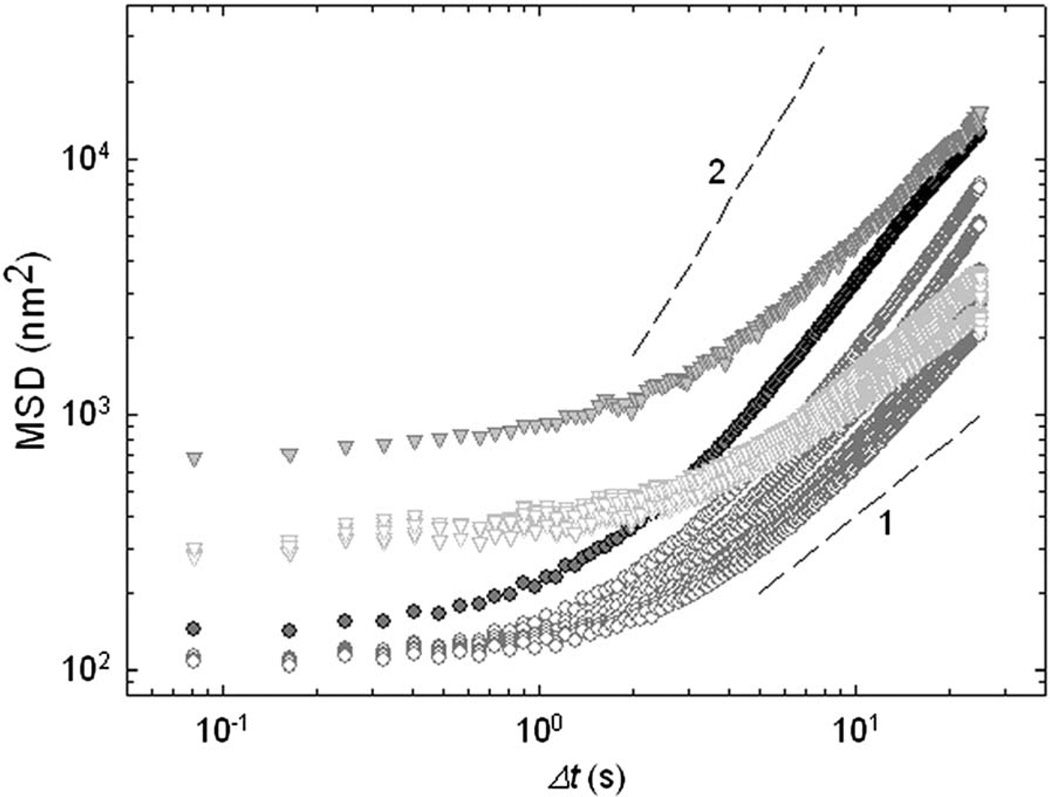
As a control, we use beads coated with acetylated low-density lipoprotein (acLDL) which bind to cell membrane but not to cytoskeleton.19,20,25 In this case the MSD–Δt relationship is impacted only during application of the ultrasound, but exhibits immediate recovery upon cessation (Fig. 6—triangles). Immediate recovery seen in acLDL beads versus gradual relaxation seen in RGD beads (Fig. 6—triangles vs. circles) suggests that the locus of acoustically driven fluidization is the cytoskeleton.
Fig. 6.
MSD measurements with different attachment specificity. Ferromagnetic beads were coated with acetylated low-density lipoprotein (acLDL) to the cell membrane (triangles) or with RGD to cell cytoskeleton (circles). Topmost closed symbols represent the measurements during acoustic excitation. The dashed lines show diffusion exponents of 1 and 2.
Taken together, cellular structure as probed by traction, MSDs, and live-actin stainings are irreversibly perturbed after exposure to higher acoustic pressure (Fig. 4, diamonds; Fig. 5B). This suggests that the cytoskeleton is fluidized by the higher intensity ultrasound but remains disrupted when irradiation stops because of the catastrophic disruption of actin fibers (Fig. 2, bottom panel and Movies S2 and S3†). In response to lower intensity ultrasound, in contrast, cellular responses exhibit gradual but complete recovery (Fig. 4, circles and 5(A)). Such mechanical responses are consistent with those observed upon application of a transient strain at physiological amplitude, and suggest prompt fluidization and subsequent re-solidification of the cytoskeleton.16,17,28
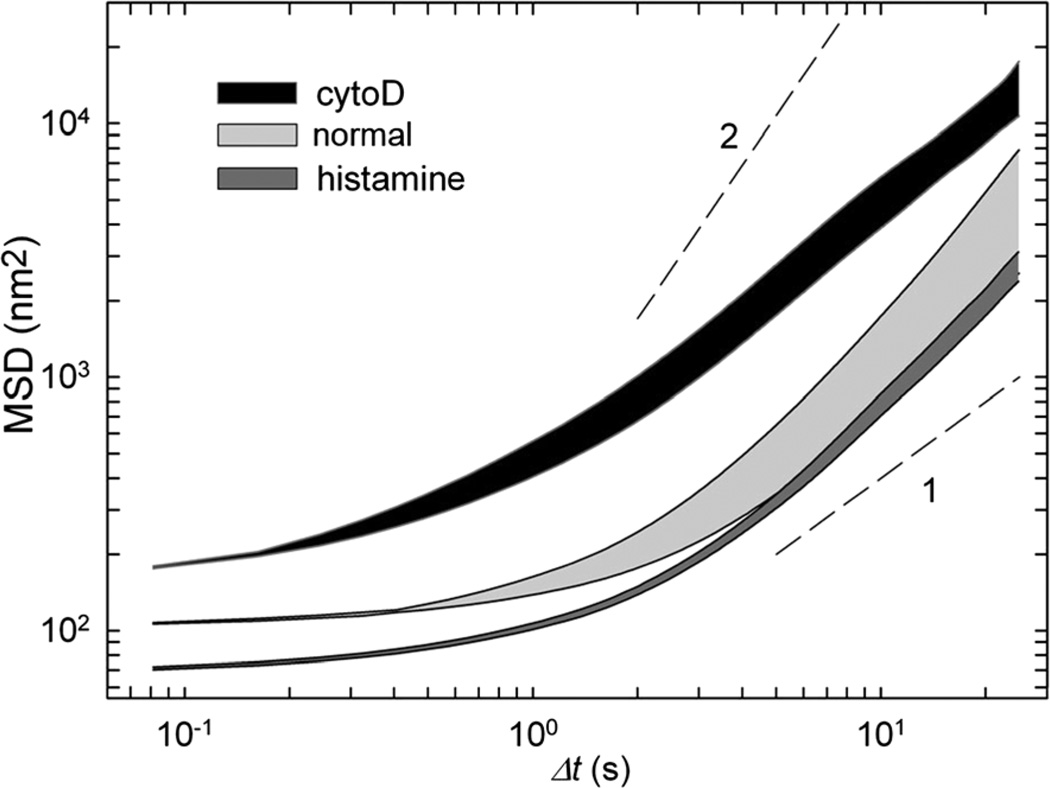
To further characterize this mechanical response, we pretreated cells with cytochalasin D and histamine (Fig. 7). Actin disruption by cytochalasin D shifts the baseline MSD values upward while stiffening the cytoskeleton using histamine shifts it downward.13 In both cases, acoustic stimulation causes a prompt increase in MSD, although the effect of acoustic agitation is reduced when cells are pre-treated with histamine. Such a response of pharmacologically pre-treated cells suggests that acoustic agitation acts as a mechanical stimulation and remodels the cytoskeleton with respect to its initial prestressed state. Histamine-activated cells are stiffer with a higher density of actin–myosin bonds; in such cells the same acoustic pressure would supply less energy per single bond resulting in a smaller increase in remodeling rates. Conversely, cells pretreated with cytochalasin D have lower density of actin–myosin bonds and would attain higher energy per bond resulting in a larger increase in the rate of structural rearrangement.
Fig. 7.
MSD intervals represented by the area restricted between the baseline MSD values (bottom line) and values obtained immediately after exposure to 170 kPa ultrasound (top line). Three min after exposure to ultrasound, all MSD curves returned to the baseline position. Each area represents different conditions: untreated cells (light gray), cells treated with histamine (dark gray) and cells pretreated with cytochalasin D (black). The dashed lines show diffusion exponents of 1 and 2.
To rationalize the origin of these structural rearrangements, we estimate the local strain induced by LIPUS. For an acoustic plane wave, the maximum displacement is given by Pac/ρcω, where Pac is the acoustic pressure amplitude, ρ and c are the density and speed of sound in the medium, respectively, and ω is the angular frequency. For the moderate acoustic intensity, the oscillatory displacement amplitude corresponds to about 30 nm. Since the wavelength is 1.75 mm, the maximum oscillatory strain is on the order of 10−5; were they applied at physiological frequencies, such strains would produce no discernible effect.16,18 We further calculated the energy supplied during exposure to moderate ultrasound for 30 s which is defined by AtDc/ρc, where α is the cell absorption coefficient,29 l and A are the cell length and area, respectively, and tDc is the duty cycle and is in the order of 10−13 J or ~2.4 × 108 kBT. Interestingly, in cell stretching experiments an equivalent amount of energy is sufficient to reach the transition point to the inelastic regime and was estimated to cause ~10% of bond rupture in 10 µm cells.30 Correspondingly, for HASM cells which are five times bigger in area and assuming the same bond mesh size, the same energy injected during moderate intensity agitation would result in ~2% of bond rupture. Overall, this estimated level of bond rupture further supports that LIPUS is capable of delivering enough energy per bond to drive profound changes in cell mechanics including inelastic effects, as shown in the high intensity measurements (Fig. 4—diamonds and 5B). Because a comparable amount of energy absorbed by a cell will raise the temperature only by 10−4 K, this rules out temperature changes as a driving force for cellular remodeling14 and emphasizes instead the importance of direct mechanical agitation during ultrasound stimulation. In this connection, forces originating from ultrasound oscillations have been shown to remodel inert polymer systems by altering the shape of the potential energy landscape and biasing chemical pathways to yield atypical molecular conformation.31–33 By analogy, acoustic oscillations can drive the cytoskeleton through structural remodeling by disrupting weak nonspecific bonds, altering protein conformations, and remodeling initial structural integrity.18,30,34 Alternatively, a model of intramembrane cavitation was presented recently which predicts periodic expansion and contraction of the intramembrane spaces that can stretch and compress the cytoskeleton and transmembrane proteins.35
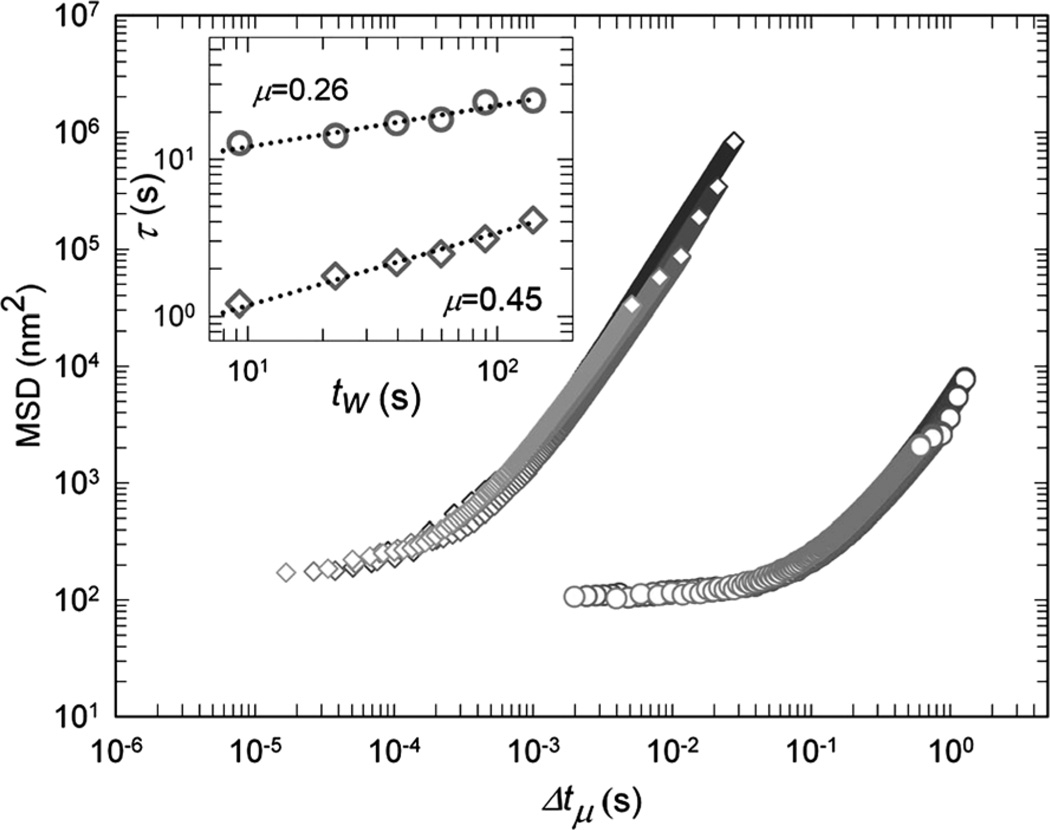
Phenomenologically, acoustically driven cytoskeleton remodeling is highly reminiscent of deformation-driven rejuvenation of soft glassy systems and living cells in particular.16,36 The subsequent recovery, commonly observed in glassy systems, has a well-defined characteristic behavior and has been interpreted as a form of physical rejuvenation followed by aging.16,37 To explore the analogy with rejuvenation and aging of cells exposed to LIPUS, we investigated the time evolution of the MSD during the recovery following fluidization. A characteristic feature of all systems undergoing rejuvenation is the self-similarity in the relaxation dynamics, which allows the data to be scaled onto a master curve. To scale the MSD curves here, we characterize the progressive slowing of the rearrangement kinetics with time by defining a waiting time, tw, which is the elapsed time from cessation of exposure to LIPUS. We measure the time τ required for each bead to diffuse an arbitrary MSD value, which we take to be d2 = 103 nm2; we find that τ increases with tw as a power law (Fig. 8, inset), where μ = 0.26. After rescaling the time axis using , all data for the moderate intensity LIPUS collapse onto a master curve (Fig. 8, diamonds), confirming that relaxation kinetics at each waiting time are self-similar. Interestingly, however, the MSD following the higher-intensity LIPUS also exhibits a similar scaling behavior, albeit with a different value of μ = 0.45 (Fig. 8, circles). In both cases, the decay in remodeling rate is slower than any exponential process. Such a behavior is consistent with the aging dynamics of soft glassy systems subjected to mechanical stimulation.37–39 Nevertheless, living cells are active matter, and respond actively to mechanical stimulation. However active response such as activation of contractile regime and cell reinforcement will follow on a longer time scale.40
Fig. 8.
All MSD curves at different waiting times, tw, are collapsed onto one master curve by horizontal shifting (170 kPa and 290 kPa, circles and diamonds correspondingly). The amount of shift for each curve defines a characteristic time τ which is the Δt at which MSD(Δt, tw) crosses an arbitrary MSD value of 103 nm2. Inset: τ increases with tw as a power law (), while μ varies according to the applied acoustic pressure; 0.26 and 0.45 for 170 kPa (circles) and 290 kPa (diamonds), respectively.
Conclusions
The physical mechanism responsible for the effects of LIPUS is poorly understood and thereby limits the extent of its applications. We report here that LIPUS acts as a mechanical stimulus in a manner similar to a transient physiological stretch. Short exposure to LIPUS drives the cytoskeleton through fluidization followed by slow recovery, which is interpreted as a form of physical rejuvenation followed by aging. Accordingly, here we propose that LIPUS acts as a direct mechanical stimulus mediating its beneficial therapeutic effects through accelerated cytoskeletal remodeling that is associated with physical rejuvenation.
Supplementary Material
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c2sm07246g
References
- 1.Mitragotri S. Nat. Rev. Drug Discovery. 2005;4:255–260. doi: 10.1038/nrd1662. [DOI] [PubMed] [Google Scholar]
- 2.Robertson VJ. Phys. Ther. Sport. 2002;3:124–133. [Google Scholar]
- 3.Rapoport NY, Christensen DA, Fain HD, Barrows L, Gao Z. Ultrasonics. 2004;42:943–950. doi: 10.1016/j.ultras.2004.01.087. [DOI] [PubMed] [Google Scholar]
- 4.Hadjiargyrou M, McLeod K, Ryaby JP, Rubin C. Clin. Orthop. Relat. Res. 1998;355:S216. doi: 10.1097/00003086-199810001-00022. [DOI] [PubMed] [Google Scholar]
- 5.Daffertshofer M, Fatar M. Eur. J. Ultrasound. 2002;16:121–130. doi: 10.1016/s0929-8266(02)00049-6. [DOI] [PubMed] [Google Scholar]
- 6.Khanna A, Nelmes RT, Gougoulias N, Maffulli N, Gray J. Br. Med. Bull. 2009;89:169–182. doi: 10.1093/bmb/ldn040. [DOI] [PubMed] [Google Scholar]
- 7.Kimmel E. CRC Crit. Rev. Bioeng. 2006;34:105. [Google Scholar]
- 8.Hart J. J. Wound Care. 1998;7:25. doi: 10.12968/jowc.1998.7.1.25. [DOI] [PubMed] [Google Scholar]
- 9.Song J, Cottler PS, Klibanov AL, Kaul S, Price RJ. Am. J. Physiol.: Heart Circ. Physiol. 2004;287:H2754. doi: 10.1152/ajpheart.00144.2004. [DOI] [PubMed] [Google Scholar]
- 10.Barzelai S, Sharabani-Yosef O, Holbova R, Castel D, Walden R, Engelberg S, Scheinowitz M. Ultrasound Med. Biol. 2006;32:139–145. doi: 10.1016/j.ultrasmedbio.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Mizrahi N, Seliktar D, Kimmel E. Ultrasound Med. Biol. 2007;33:1818–1829. doi: 10.1016/j.ultrasmedbio.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Massiera G, Van Citters KM, Biancaniello PL, Crocker JC. Biophys. J. 2007;93:3703–3713. doi: 10.1529/biophysj.107.111641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bursac P, Lenormand G, Fabry B, Oliver M, Weitz DA, Viasnoff V, Butler JP, Fredberg JJ. Nat. Mater. 2005;4:557–561. doi: 10.1038/nmat1404. [DOI] [PubMed] [Google Scholar]
- 14.Sunyer R, Ritort F, Farre R, Navajas D. Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 2009;79 doi: 10.1103/PhysRevE.79.051920. 051920. [DOI] [PubMed] [Google Scholar]
- 15.Dangaria JH, Butler PJ. Am. J. Physiol.: Cell Physiol. 2007;293:C1568–C1575. doi: 10.1152/ajpcell.00193.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trepat X, Deng L, An SS, Navajas D, Tschumperlin DJ, Gerthoffer WT, Butler JP, Fredberg JJ. Nature. 2007;447:592–595. doi: 10.1038/nature05824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnan R, Park CY, Lin YC, Mead J, Jaspers RT, Trepat X, Lenormand G, Tambe D, Smolensky AV, Knoll AH, Butler JP, Fredberg JJ. PLoS One. 2009;4:e5486. doi: 10.1371/journal.pone.0005486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver M, Kovats T, Mijailovich S, Butler JP, Fredberg JJ, Lenormand G. Phys. Rev. Lett. 2010;105:158102. doi: 10.1103/PhysRevLett.105.158102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabry B, Maksym GN, Butler JP, Glogauer M, Navajas D, Fredberg JJ. Phys. Rev. Lett. 2001;87:148102. doi: 10.1103/PhysRevLett.87.148102. [DOI] [PubMed] [Google Scholar]
- 20.Puig-de-Morales M, Millet E, Fabry B, Navajas D, Wang N, Butler JP, Fredberg JJ. Am. J. Physiol.: Cell Physiol. 2004;287:C643–C654. doi: 10.1152/ajpcell.00070.2004. [DOI] [PubMed] [Google Scholar]
- 21.Panettieri RA, Murray RK, DePalo LR, Yadvish PA, Kotlikoff MI. Am. J. Physiol. 1989;256:C329–C335. doi: 10.1152/ajpcell.1989.256.2.C329. [DOI] [PubMed] [Google Scholar]
- 22.Hu S, Chen J, Fabry B, Numaguchi Y, Gouldstone A, Ingber DE, Fredberg JJ, Butler JP, Wang N. Am. J. Physiol.: Cell Physiol. 2003;285:C1082–C1090. doi: 10.1152/ajpcell.00159.2003. [DOI] [PubMed] [Google Scholar]
- 23.Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Am. J. Physiol.: Cell Physiol. 2002;282:C595–C605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 24.Wang JHC, Lin JS. Biomech. Model. Mechanobiol. 2007;6:361–371. doi: 10.1007/s10237-006-0068-4. [DOI] [PubMed] [Google Scholar]
- 25.Wang N, Butler JP, Ingber DE. Science. 1993;260:1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 26.An SS, Fabry B, Mellema M, Bursac P, Gerthoffer WT, Kayyali US, Gaestel M, Shore SA, Fredberg JJ. J. Appl. Physiol. 2004;96:1701–1713. doi: 10.1152/japplphysiol.01129.2003. [DOI] [PubMed] [Google Scholar]
- 27.Hoffman BD, Massiera G, Van Citters KM, Crocker JC. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10259–10264. doi: 10.1073/pnas.0510348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C, Krishnan R, Zhou EH, Ramachandran A, Tambe D, Rajendran K, Adam RM, Deng LH, Fredberg JJ. PLoS One. 2010;5:e12035. doi: 10.1371/journal.pone.0012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo L, Molnar J, Ding H, Lv X, Spengler G. Diagn. Pathol. 2006;1:35. doi: 10.1186/1746-1596-1-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernández P, Ott A. Phys. Rev. Lett. 2008;100:238102. doi: 10.1103/PhysRevLett.100.238102. [DOI] [PubMed] [Google Scholar]
- 31.Hickenboth CR, Moore JS, White SR, Sottos NR, Baudry J, Wilson SR. Nature. 2007;446:423–427. doi: 10.1038/nature05681. [DOI] [PubMed] [Google Scholar]
- 32.Piermattei A, Karthikeyan S, Sijbesma RP. Nat. Chem. 2009;1:133–137. doi: 10.1038/nchem.167. [DOI] [PubMed] [Google Scholar]
- 33.Naota T, Koori H. J. Am. Chem. Soc. 2005;127:9324–9325. doi: 10.1021/ja050809h. [DOI] [PubMed] [Google Scholar]
- 34.Wolff L, Fernandez P, Kroy K. New J. Phys. 2010;12:053024. [Google Scholar]
- 35.Krasovitski B, Frenkel V, Shoham S, Kimmel E. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3258. doi: 10.1073/pnas.1015771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sollich P, Lequeux F, Hébraud P, Cates ME. Phys. Rev. Lett. 1997;78:2020–2023. [Google Scholar]
- 37.Viasnoff V, Lequeux F. Phys. Rev. Lett. 2002;89:065701. doi: 10.1103/PhysRevLett.89.065701. [DOI] [PubMed] [Google Scholar]
- 38.Sollich P. Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top. 1998;58:738–759. [Google Scholar]
- 39.Cloitre M, Borrega R, Leibler L. Phys. Rev. Lett. 2000;85:4819–4822. doi: 10.1103/PhysRevLett.85.4819. [DOI] [PubMed] [Google Scholar]
- 40.Thoumine O, Ott A. J. Cell Sci. 1997;110(Pt 17):2109–2116. doi: 10.1242/jcs.110.17.2109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.