Abstract
The crystal structure for cce_0566 (171 aa, 19.4 kDa), a DUF269 annotated protein from the diazotrophic cyanobacterium Cyanothece sp. ATCC 51142, was determined to 1.60 Å resolution. Cce_0566 is a homodimer with each molecule composed of eight α-helices folded on one side of a three strand anti-parallel β-sheet. Hydrophobic interactions between the side chains of largely conserved residues on the surface of each β-sheet hold the dimer together. The fold observed for cce_0566 may be unique to proteins in the DUF269 family, hence, the protein may also have a function unique to nitrogen fixation. A solvent accessible cleft containing conserved charged residues near the dimer interface could represent the active site or ligand-binding surface for the protein’s biological function.
Keywords: Nitrogen fixation, Nitrogenase, Cyanobacteria, Diazotroph, DUF269
1. Introduction
The conversion of one molecule of dinitrogen (N2) to two molecules of bioavailable ammonia (NH3) is a key component of the global biogeochemical nitrogen cycle [1]. Excluding made–made nitrogen fixation via the industrial Haber–Bosch reaction [2], 99% of the total N2 fixed per year occurs via biological processes performed exclusively by diazotrophic microorganisms [3]. The enzyme responsible for catalyzing the chemically difficult reduction of the N2 triple bond under ambient conditions is a multimeric metal-bound complex, nitrogenase [1, 3, 4]. Four different types of nitrogenases have been identified that differ by the metal composition at the active site [5]. Despite identification of the proteins that compose the nitrogenase complex, the mechanism of enzymatic nitrogen fixation is still the subject of much research [4]. Indeed, interest in fully understanding the biological nitrogen fixation process may be at an all time high because a byproduct of nitrogen fixation is dihydrogen (H2) [6], an appealing source of renewable and green energy [7].
The only living organisms capable of fixing nitrogen are a select number of bacterial and archaea genera and include species of cyanobacteria, phototrophic organisms that were the progenitors of chloroplasts in plants and algae [8, 9]. Through the release of the photosynthesis byproduct, oxygen, cyanobacteria radically altered the composition of early earth’s reducing atmosphere into an oxidizing atmosphere 2.3 billion years ago [10, 11]. However, nitrogen fixation is biochemically intolerant of oxygen because nitrogenase is rapidly and permanently inactivated by it [12–14]. To aid in the transition from an anerobic to aerobic environment some cyanobacteria evolved metabolic and regulatory processes to perform both photosynthesis and nitrogen fixation [15]. In the marine diazotrophic cyanobacterium Cyanothece sp. ATCC 51142 this was achieved by the temporal separation of photosynthesis and nitrogen fixation into daytime and nighttime activities, respectively, making cyanobacteria the simplest known organisms to display circadian rhythms [16, 17].
The genome of Cyanothece 51142 was recently sequenced and deposited into the GenBank database (Accession Nos. CP000806–CP000811) [18]. Present in the conserved syntenic nif cluster of 34 genes is cce_0566 that falls into a family of proteins with a “Domain of Unknown Function” annotated DUF269 (PF03270). To date, the DUF269 gene has only been observed in nitrogen-fixing species [19] suggesting that it plays a role, yet undefined, in the biosynthesis, assembly, transport, and insertion of components of the nitrogenase complex. The cce_0566 gene is located between nifX (cce_0565) and a protein that falls into the DUF683 family (cce_0567) [20, 21]. To obtain clues as to the biochemical function of proteins in the DUF269 family of conserved proteins [22, 23] the crystal structure for cce_0566 was determined to a resolution of 1.60 Å.
2. Materials and methods
2.1. Cloning, expression, and purification
The cce_0566 gene from Cyanothece sp. ATCC 51142, minus the 18-residue, N-terminal signal sequence (MFIDNGNALIVIVIMTTT), was synthesized between NdeI and BamHI restriction endonuclease sites (BIOS&T, Montreal, Canada) and inserted into the expression vector pET28b (Novagen, Madison, WI). The 19.3 kDa gene product contained a 21-residue tag (MGSSHHHHHHSSGLVPRGSHM) prior to the first native residue (T19) that included a poly-histidine stretch to assist protein purification by metal-affinity chromatography. The recombinant plasmid was transformed into Escherichia coli BL21(DE3) cells (Novagen, Madison, WI) using a heat shock method. Nitrogen-15 substituted protein was prepared using standard minimal media protocols [20] with 15NH4Cl and isopropyl β-d-1-thiogalactopyranoside induction at an OD600 of ~0.8 at 25 °C. Expressed protein from 750 mL of growth media was processed sequentially in two steps involving Ni–NTA (Qiagen, Valencia, CA) affinity chromatography followed by size exclusion chromatography on a Superdex75 HiLoad 26/60 column (Amersham Pharmacia Biotech, Piscataway, NJ) [21] that simultaneously exchanged cce_0566 into the buffer used for the crystallization, CD, and NMR experiments (500 mM NaCl, 20 mM TrisHCl, 1.0 mM dithiothreitol, pH 7.1).
2.2. Nuclear magnetic resonance spectroscopy
An overall rotational correlation time (τc) for an 15N-labeled cce_0566 sample (~1 mM) was rapidly estimated from backbone amide 15N T1/T1ρ ratios measured using a modified 1H–15N HSQC experiment to record a 15N-edited one-dimensional spectrum [24]. Data was collected at 25 °C on a Varian Inova-600 spectrometer equipped with a triple resonance cyroprobe and pulse field gradients.
2.3. Circular dichroism spectroscopy
Circular dichroism data was collected on an Aviv Model 410 spectropolarimeter (Lakewood, NJ) calibrated with an aqueous solution of ammonium d-(+)camphorsulfonate using a 13 µM cce_0566 sample in NMR buffer and in a quartz cell of 0.1 cm path length. A thermal denaturation curve was obtained by recording and plotting the ellipticity at 215 nm in 2.0 °C intervals from 5 to 80 °C. Steady-state wavelength spectra for cce_0566 were recorded in 0.5 nm increments between 200 and 250 nm at 25 °C. Each wavelength spectrum was the result of averaging two consecutive scans with a bandwidth of 1.0 nm and a time constant of 1.0 s. Steady-state wavelength spectra were processed by subtracting a blank spectrum from the protein spectrum and then automatically line smoothing the data using Aviv software.
2.4. Crystallization, structure determination and refinement
Crystallization conditions for 15N-labeled cce_0566 were identified using screens from Hampton Research (Aliso Viejo, CA) and the hanging-drop, vapor-diffusion method set-up at room temperature (~22 °C). Crystals began appearing 24–48 h later under one condition that was then optimized by adjusting the protein concentration and crystallization protocols. The data used to determine the structure for cce_0566 were obtained from plate-like crystals grown by microbatch under paraffin oil [25]. Crystals were harvested approximately two weeks after mixing 1.5 µL of protein (~2 mg/mL) with 1.5 µL of reservoir buffer containing 0.2 M ammonium acetate, 0.1 M sodium acetate trihydrate, and 30% (w/v) polyethylene glycol 4000, pH 4.6. Crystals were directly mounted in nylon CryoLoops (Hampton Research), flash-frozen in liquid nitrogen, stored under liquid nitrogen, and shipped to the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory for X-ray data collection.
2.5. X-ray data collection and processing
Native X-ray diffraction data were collected at the X29A beamline with an ADSC Q315r CCD detector. A solution was determined by molecular replacement using MOLREP [26] from the CCP4 suite [27] and the crystal structure from a related protein, B7JA91_ACIF2 (YP_002425942.1) from Acidithiobacillus ferrooxidans (PDB ID 3G7P). The final model was generated after numerous iterative rounds of refinement in REFMAC (v 5.5.0109) [28]. A final check on the stereochemical quality of the final model was assessed using the program MolProbity [29] and PROCHECK [30] and any conflicts addressed. The data collection and structure refinement statistics are given in Supplementary Table S1 and the coordinates deposited in the Protein Data Bank (PDB ID 3NJ2). The amino acid sequence for the coordinates of cce_0566 deposited in the RCSBPDB is numbered sequentially, G1–L174, beginning with the 21-residue, non-native tag. Here, the protein is numbered following the order in the native sequence, and hence, the first native residue, T22 in the RCSB-PDB, is labeled T20.
3. Results and discussion
3.1. Crystal structure
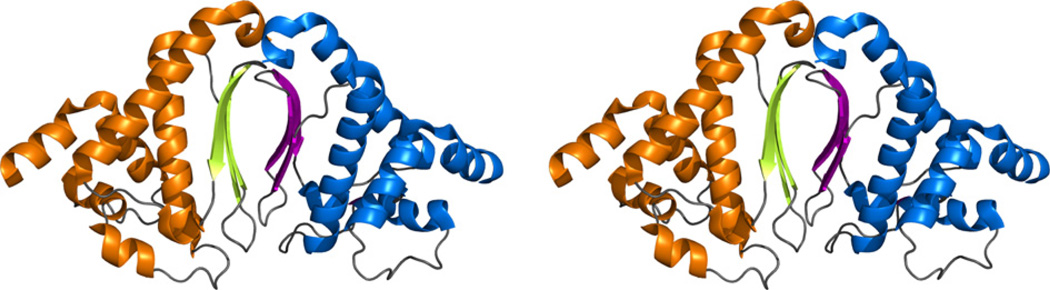
The first indication that DUF269 formed a dimer in solution was its elution time off a size exclusion column consistent with an ~36 kDa protein (data not shown). This was corroborated by an estimated rotational correlation time (τc) of 18.1 ± 2.7 ns (25 °C) that was more consistent with an ~36 kDa dimer than an ~18 kDa monomer [31]. Further evidence for dimer formation was the observation of broad line shapes, untypical for a < 20 kDa protein, for most of the amide cross peaks in the 1H–15N HSQC spectrum of cce_0566 (data not shown). Consequently, the dimer observed in the asymmetric unit of the crystal structure of cce_0566, shown in Fig. 1, likely represents the species present in solution. The buried surface area due to the formation of such a dimer, 1505 Å2, is 8.3% of the solvent-accessible surface and adequate to stabilize this species [32]. There is some asymmetry between the two molecules in the dimer, as molecule A and B in the asymmetric unit have a backbone RMSD of 0.73 Å and an all atom RMSD of 1.73 Å over residues S21–L171 (superposition server SuperPose, v 1.0) [33]. The variations are most prominent in the loops and turns in the structure as the backbone and all atom RMSDs reduce to 0.51 and 1.36 Å, respectively, when only the α-helical and β-strand regions are used for the alignment.
Fig. 1.
(A) Stereo-view cartoon representation of the two molecules that form the asymmetric unit in the crystal structure of cce_0566 (3NJ2). The dimer is the biological unit. In molecule A the helices and β-strands are colored orange and lime, respectively, and in molecule B they are colored blue and magenta.
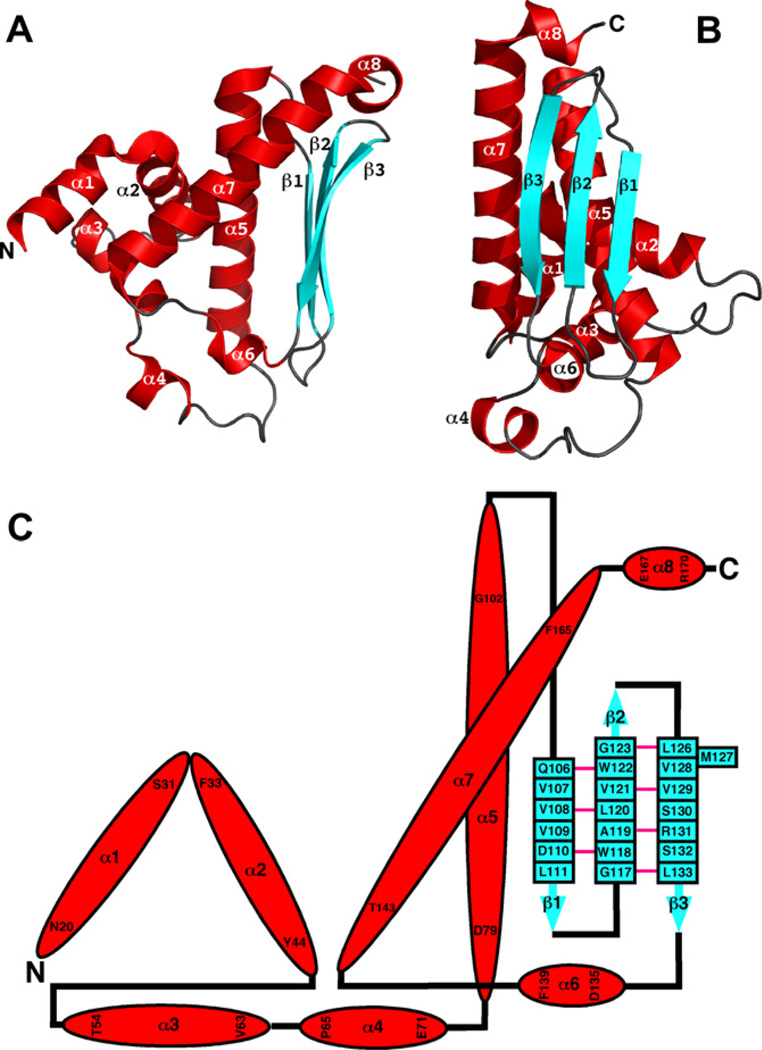
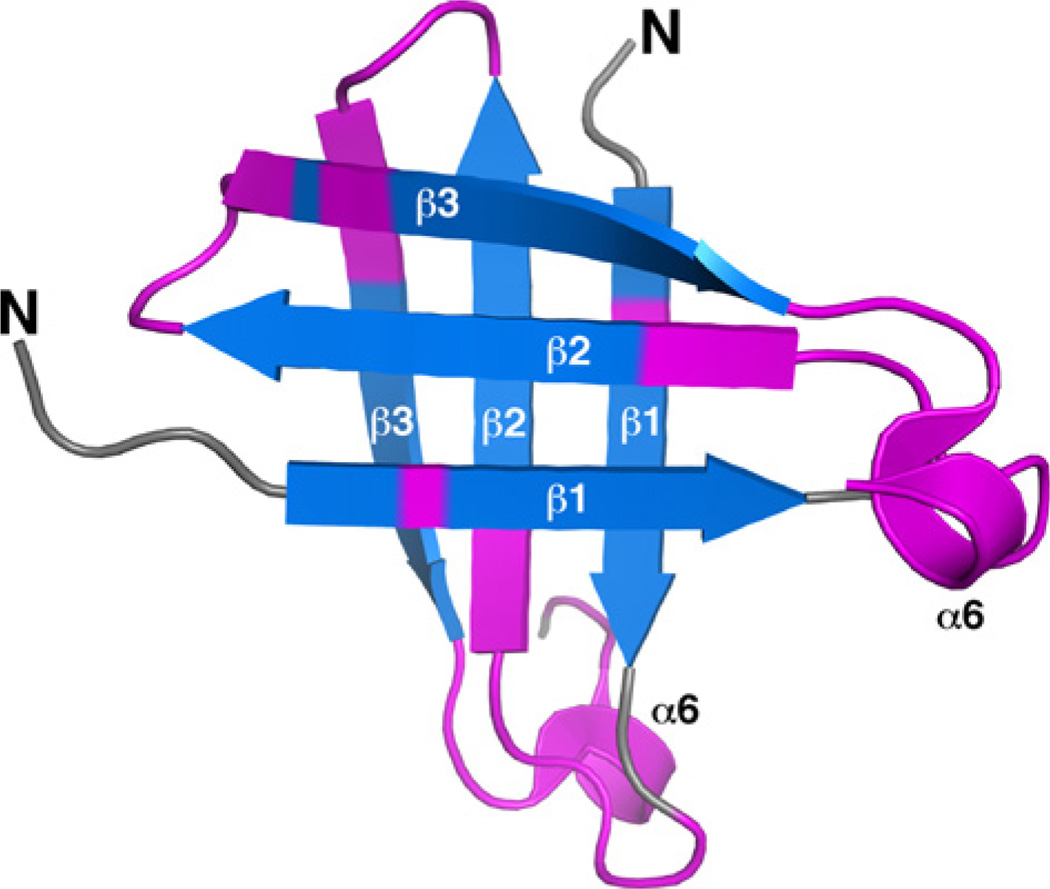
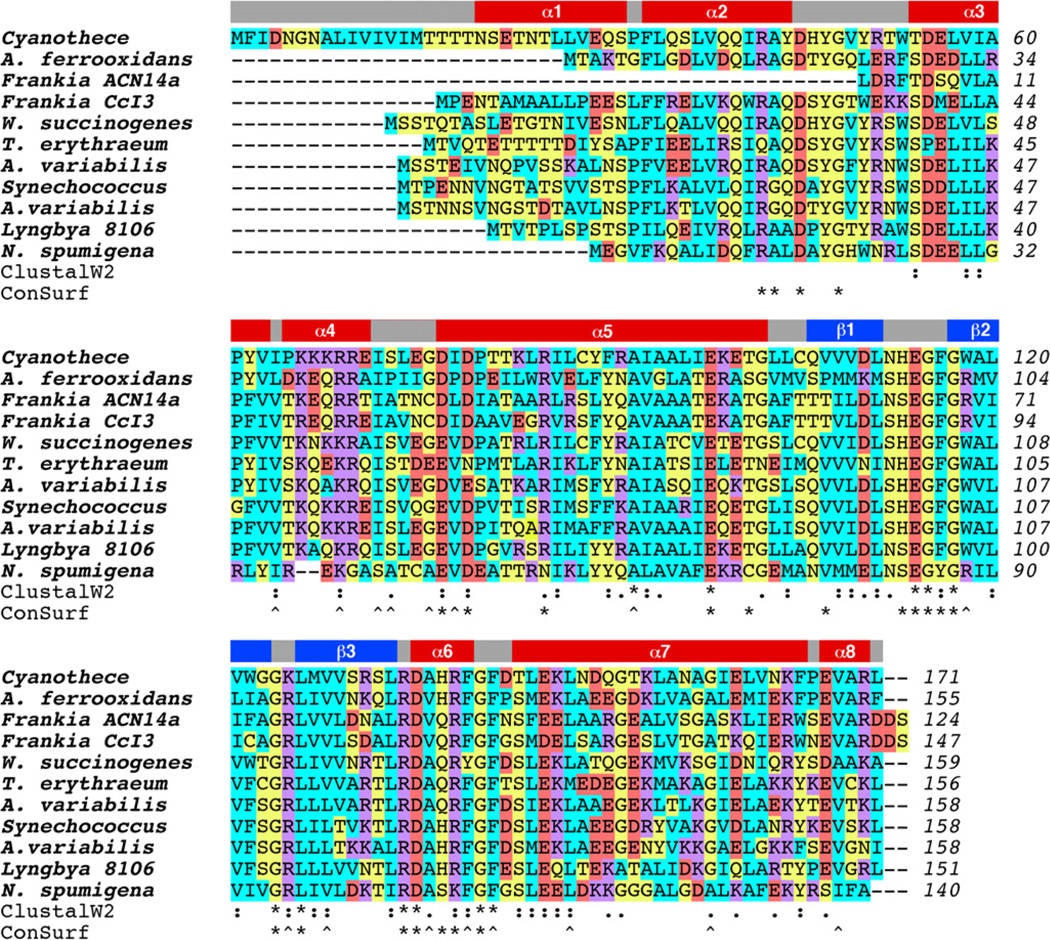
With dimensions of approximately 72 × 42 × 32 Å, the cce_0566 dimer shown in Fig. 1 has a roughly elliptical shape. Fig. 2A and B are cartoon representations of a single cce_0566 molecule shown in two different orientations with the representation on the right (B) rotated ~90° relative to the orientation on the left (A). Fig. 2C is a secondary structure diagram that details the residues in each secondary structure element and approximates the relative orientation of the secondary structure elements to each other. Each monomer is composed of eight α-helices folded on one side of a three strand anti-parallel β-sheet. The first three helices form the narrow ends of the ellipse while the two longest helices, α5 and α 7, cross over each other to form the framework for the wide central part of the eclipse. As highlighted in Fig. 3, the strands of each β-sheet cross over at a ~90° angle to form a “checkered” lattice at the dimer interface. Most of the side chains of the residues in the β-sheet are hydrophobic and hydrophobic contacts between the two β-sheets likely play a significant role in holding the dimer together. Indeed, this feature may be universal in all DUF269 proteins as sequence alignment of the most dissimilar proteins in the DUF269 family, shown in Fig. 4, illustrates that these β-strands are composed largely of conserved hydrophobic residues.
Fig. 2.
(A + B) Cartoon representation of a single molecule of the cce_0566 dimer shown in two orientations. Each molecule contains eight α-helices (red) and three β-strands (cyan). (C) Secondary structure diagram for cce_0566. The α-helices and β-strands are drawn to scale but not the connecting loops and turns. The α-helices are colored red with the residue number for the first and last residue shown. The β-strands are colored cyan with each residue labeled. A solid pink link between β-sheet residues indicates dual hydrogen bonds between two residues in an antiparallel β-sheet.
Fig. 3.
View of the “checkered” lattice formed by the β-sheets at the dimer interface. The β-sheet in the back is positioned as in Fig. 2B. Highly conserved residues, as identified by ConSurf [34] in Fig. 4, are colored magenta.
Fig. 4.
Color-coded ClustalW2 sequence alignment for Cyanothece 51142 cce_0566, B7JA91_ACIF2 (Acidithiobacillus ferrooxidans strain ATCC 23270), plus nine other proteins identified as most dissimilar by Pmaf in the DUF269 protein family (PF03270): Q2TT24_9ACTO Frankia sp. ACN14a; Q2J4G4_FRASC Frankia sp. CcI3; Q7MRG2_WOLSU Wolinella succinogenes; Q9KJL5_TRIEI Trichodesmium erythraeum IMS101; Q44490_ANAVA Anabaena variabilis; Q2JTK4_SYNJA Synechococcus sp. JA-3–3Ab; Q3M646_ANAVT Anabaena variabilis ATCC 29413; ZP_01620760 Lyngbya sp. PCC 8106; ZP_01628420 Nodularia spumigena CCY9414. The amino acids are color-coded as follows: cyan = hydrophobic (A, V, F, P, M, I, L, W); yellow = hydrophilic (S, T, Y, H, C, N, G, Q); red = acidic (D, E); magenta = basic (R, K). On top of the alignment are the elements of secondary structure observed in the crystal structure of cce_0566. Below the alignment is shown the position of residues that are highly conserved following ClustalW2 [42] (* = identical, : = conserved substitutions, . = semi-conserved substitutions) and ConSurf [34] (scores of 9 = asterisk, 8 = caret) analysis. The ClustalW2 analysis used the amino acid sequence of cce_0566 and the ten other proteins listed in Fig. 5 while the ConSurf analysis used the complete RCSB-PDB and SWISS-PROT databases.
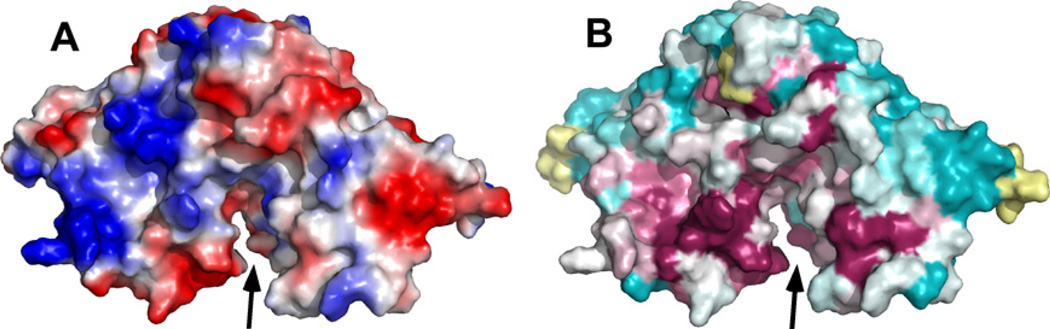
Fig. 5 shows the solvent accessible surface on the cce_0566 dimer labeled by electrostatic surface potential (Fig. 5A) and conserved regions identified by ConSurf analysis [34] (Fig. 5B). While there are many large, well defined regions of positive and negative charge on the protein’s surface, they do not correspond to highly conserved regions, suggesting that much of the surface electrostatic potential is not biologically significant. On the other hand, the solvent accessible surface rendering shows a cleft (solid black arrow) composed of highly conserved residues near the dimer interface. Furthermore, some of conserved surface in this cleft is charged. The sequence alignment in Fig. 4 indicates that these conserved regions are principally α6 plus adjacent residues and the turn between β1 and β2, regions that are physically near each other (Figs. 2 and 3). Included in this conserved region are the charged residues E114, K125, R134, D135, and R138. Such conservation of charged residues in a solvent accessible cleft may represent the active site or ligand-binding surface of cce_0566.
Fig. 5.
(A) PyMOL generated maps of the electrostatic potentials [43] at the solvent-accessible surface of the cce_0566 dimer. The orientation in (A) is similar to the orientation shown in Fig. 1. (B) ConSurf generated maps [34] of the conserved residues on the solvent-accessible surface of the cce_0566 dimer in the same orientation as shown in Fig. 5A. Highly conserved residues are colored magenta and pink, poorly conserved residues are colored cyan, and residues with insufficient information are colored yellow. Black arrow indicates the cleft opening at the dimer interface. Only one face of the molecule is shown because, upon rotating the molecule 180° about the y-axis, the electrostatic potential and ConSurf surfaces are mirror images of the shown face due to symmetry.
3.2. Circular dichroism profile and thermal stability
Fig. S1A is the steady-state CD spectrum for cce_0566 collected at 25 °C. The spectrum is dominated by a double minimum at approximately 224 and 208 nm and an extrapolated maximum around 195 nm, features characteristic of α-helical secondary structure [35, 36]. Such an observation is consistent with the amount of helical structure (57%) observed in the crystal structure of the protein. Note that the double minimum is skewed and more intense around 208 nm, likely due to the contribution of other elements of secondary structure to the CD steady-state spectrum.
The thermal stability of cce_0566 was measured by monitoring the ellipicity at a specific wavelength as a function of temperature [37]. As shown in Fig. S1B, a gradual increase in ellipticity at 215 nm is observed for cce_0566 up to ~60 °C followed by a rapid increase in ellipticity that plateaus slightly above ~70 °C. Visual inspection of the sample after heating to 80 °C showed evidence for precipitation, indicating the heat induced protein unfolding was irreversible. Consequently, the CD data for cce_0566 may not be analyzed thermodynamically [37], however, a quantitative estimation of the Tm for this transition may still be obtained by assuming a two-state model and taking a first derivative of the curve in Fig. S1B [36]. The maximum of this first derivative, shown in Fig. S1C, is 70 °C.
3.3. Biological context
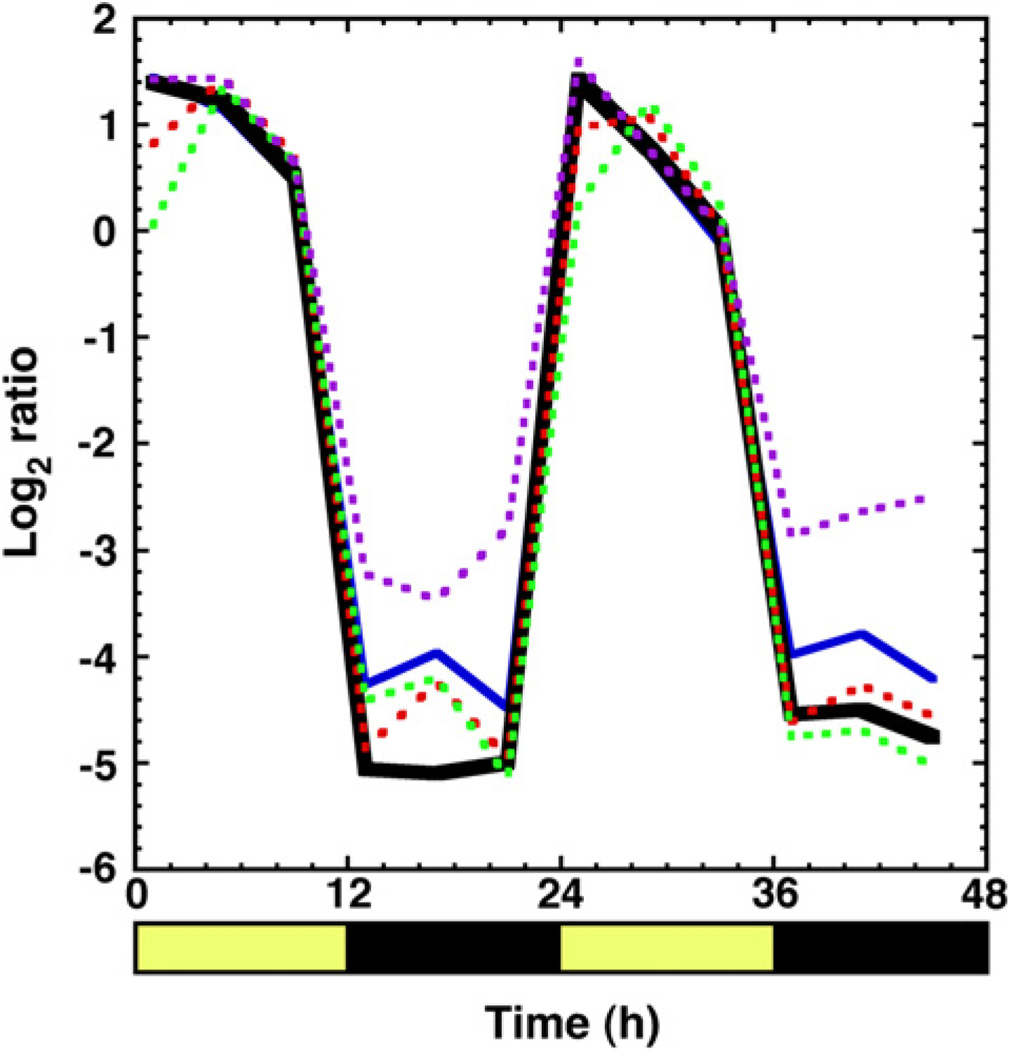
The primary reason that cce_0566 is believed to play a role in nitrogen fixation is circumstantial evidence: the gene is located in an operon of genes involved in the biochemical process. Further circumstantial evidence that cce_0566 may play a biochemical role in nitrogen fixation comes from transcriptomics microarray data for Cyanothece 51142 cultures grown as previously described [38] for 48 h in alternating 12 h periods of light and dark [39]. Fig. 6 shows the results of global transcriptomic data (European Bioinformatics Institute AssayExpress database – accession numbers AMEXP-864 and E-TABM-337) for cce_0566, cce_0567 21], and three essential nitrogen fixation genes, nifB, nifH, and nifN 40]. The expression profile for cce_0566 oscillates in sync with its neighbor, cce_0567, and other known, essential nif genes. Hence, the transcriptomics evidence places cce_0566 one step closer to a potential biological function, from gene location in the nitrogen fixation operon to physical expression at the appropriate time in the cell cycle. Indeed, in Cyanothece 51142 it was observed that all 34 genes in the nitrogen fixation transcriptional regulon exhibited strong co-regulation in their expression [39]. It is possible that the cce_0566 gene product is now non-essential and it is only expressed because it still happens to exist within the essential genes in the nitrogen fixation transcriptional regulon. However, the functions of 16 of the 34 genes in the nitrogen fixation transcriptional regulon, including cce_0566 (DUF269), are not known. If unknown function translates into non-essential in the nif operon, it seems illogical that the cell would waste resources expressing cce_0566 along with so many other non-essential genes.
Fig. 6.
Expression profile for cce_0566 (solid black), cce_0567 (solid blue) and three genes essential for the formation of a functional nitrogen fixation complex (dashed: nifB (red), nifH (green), and nifN (purple)) plotted as the log2 ratios of transcript abundance relative to the pooled sample control. Dark and light cycles are indicated with black and yellow bars, respectively, below the x-axis with the time displayed in hours [39].
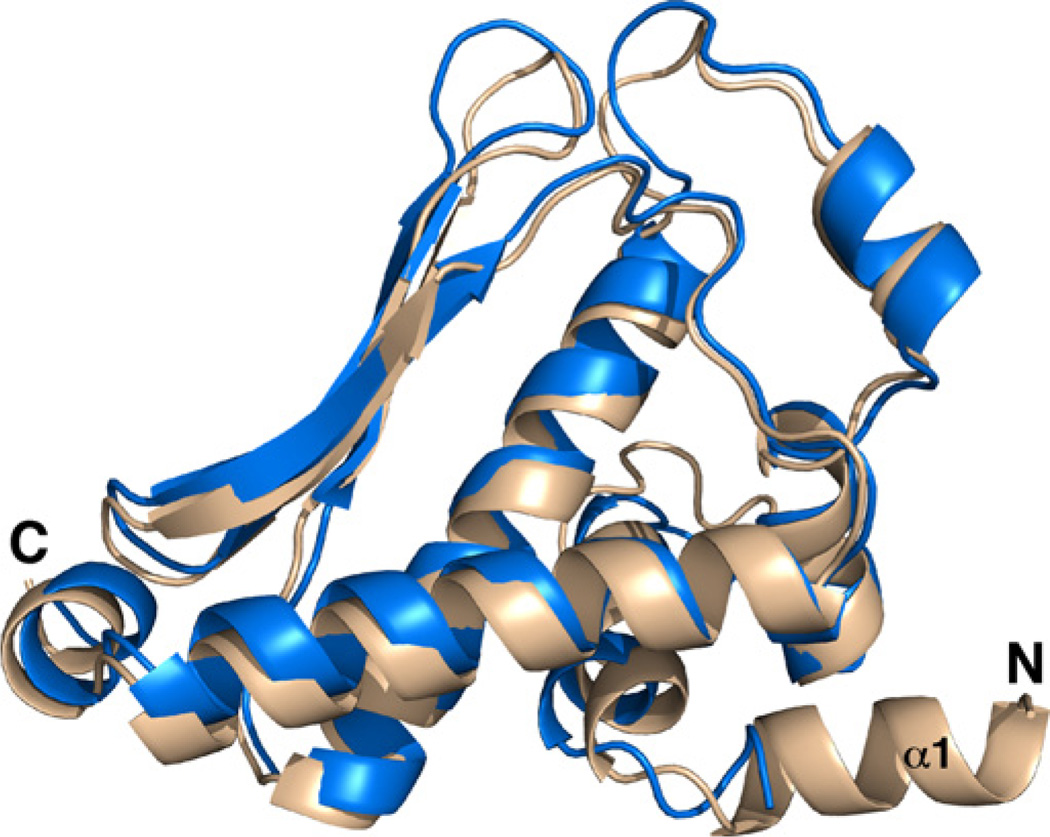
To identify a possible biological function for cce_0566 and the other proteins in the DUF269 family, the RCSB Protein Data Bank was searched for structures with similarities to cce_0566 using the DALI search engine [41]. The best match, with a Z-score of 22.4, was the structure used as the molecular replacement model to determine the structure for cce_0566, the DUF269 annotated protein from A. ferrooxidans (3G7P). The amino acid sequences of the two proteins are 41% identical and 61% conserved. The two structures are similar with a backbone RMSD of 2.47 Å as shown in their superposition illustrated in Fig. 7 (SuperPose, v 1.0) [33]. Indeed, the major difference is an additional α-helix, α1, at the N-terminus of cce_0566. Aside from the DUF269 protein from A. ferrooxidans, the best Z-scores above 4.0 identified by the search were three dissimilar proteins: Eco RI endonuclease (4.6), F-actin-capping protein subunit alpha-1 (4.4), and prolyl-tRNA synthetase (4.1). Hence, the fold adopted by the DUF269 proteins appears to be unique to this family of proteins. Given that the DUF269 gene is only found in the nitrogen fixation transcriptional regulon present in a select number of diazotrophic genera, the DUF269 gene product may have a specialized function in nitrogen fixation, and hence, warrant further biochemical studies to identify its precise biological role in the nitrogen fixation process.
Fig. 7.
Superposition of two DUF269 structures from Cyanothece 51142 (wheat, 3NJ2) and A. ferrooxidans (blue, 3G7P), cce_0566 and B7JA91_ACIF2, respectively, using the program SuperPose [33]. The additional α-helix at the N-terminus present in the structure of cce_0566 is labeled (α1).
Supplementary Material
Acknowledgements
This work was initiated as part of a Membrane Biology EMSL Scientific Grand Challenge project at the W.R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by U.S. Department of Energy’s Office of Biological and Environmental Research (BER) program located at Pacific Northwest National Laboratory (PNNL). Battelle operates PNNL for the U.S. Department of Energy. The PNNL Laboratory Directed Research Development (LDRD) program assisted completion of this research. The assistance of the X29A beam line scientists at the National Synchrotron Light Source at Brookhaven National Laboratory is appreciated. Support for beamline X29A at the National Synchrotron Light Source comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.febslet.2012.01.037.
References
- 1.Howard JB, Rees DC. Structural basis of biological nitrogen fixation. Chem. Rev. 1996;96:2965–2982. doi: 10.1021/cr9500545. [DOI] [PubMed] [Google Scholar]
- 2.Haber F. The production of ammonia from nitrogen and hydrogen. Naturwissenschaften. 1922;10:1041. [Google Scholar]
- 3.Igarashi RY, Seefeldt LC. Nitrogen fixation: the mechanism of the Mo-dependent nitrogenase. Crit. Rev. Biochem. Mol. Biol. 2003;38:351–384. doi: 10.1080/10409230391036766. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman BM, Dean DR, Seefeldt LC. Climbing nitrogenase: Toward a mechanism of enzymatic nitrogen fixation. Acc. Chem. Res. 2009;42:609–619. doi: 10.1021/ar8002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y, Bian S-M, Zhou H-N, Huang J-F. Diversity of nitrogenase systems in diazotrophs. J. Integr. Plant Biol. 2006;48:745–755. [Google Scholar]
- 6.Simpson FB, Burris RH. A nitrogen pressure of 50 atmospheres does not prevent evolution of hydrogen by nitrogenase. Science. 1984;224:1095–1097. doi: 10.1126/science.6585956. [DOI] [PubMed] [Google Scholar]
- 7.Sakurai H, Masukawa H. Promoting R & D in photobiological hydrogen production utilitzing mariculture-raised cyanobacteria. Mar. Biotechnol. 2007;9:128–145. doi: 10.1007/s10126-006-6073-x. [DOI] [PubMed] [Google Scholar]
- 8.Grey MW. The evolutionary origins of organelles. Trends Gen. 1993;5:294–299. doi: 10.1016/0168-9525(89)90111-x. [DOI] [PubMed] [Google Scholar]
- 9.Cavalier-Smith T. Chloroplast evolution: secondary symbiogenesis and multiple-losses. Curr. Biol. 2002;12:R62–R64. doi: 10.1016/s0960-9822(01)00675-3. [DOI] [PubMed] [Google Scholar]
- 10.Kasting JF. The rise of atmospheric oxygen. Science. 2001;293:819–820. doi: 10.1126/science.1063811. [DOI] [PubMed] [Google Scholar]
- 11.Berman-Frank I, Lundgren P, Falkowski P. Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res. Microbiol. 2003;154:157–164. doi: 10.1016/S0923-2508(03)00029-9. [DOI] [PubMed] [Google Scholar]
- 12.Fay P. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 1992;56:340–373. doi: 10.1128/mr.56.2.340-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Postgate JR. Nitrogen Fixation. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- 14.Zehr JP, Methe B, Foster R. New nitrogen-fixing microorganisms from the oceans: biological aspects and global implications. In: Wang Y-P, Lin M, Tian Z-X, Elmerich C, Newton WE, editors. Biological Nitrogen Fixation. Netherlands: Sustainable Agriculture and the Environment; 2005. pp. 361–365. [Google Scholar]
- 15.Bandyopadhyay A, Stockel J, Min H, Sherman LA, Pakrasi HB. High rates of photobiological H2 production by a cyanobacterium under aerobic conditions. Nature Commun. 2010;1:139. doi: 10.1038/ncomms1139. [DOI] [PubMed] [Google Scholar]
- 16.Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. USA. 1993;90:5672–5681. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golden SS, Canales SR. Cyanobacterial circadian clocks–timing is everything. Nat. Rev. Microbiol. 2003;1:191–199. doi: 10.1038/nrmicro774. [DOI] [PubMed] [Google Scholar]
- 18.Welsh EA, et al. The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle. Proc. Natl. Acad. Sci. USA. 2008;105:15094–15099. doi: 10.1073/pnas.0805418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamoto S, Yamanishi Y, Shira S, Kawashima S, Tonomura K, Kanehisa M. Prediction of nitrogen metabolism-related genes in Anabaena by kernel-based network analysis. Proteomics. 2007;7:900–909. doi: 10.1002/pmic.200600862. [DOI] [PubMed] [Google Scholar]
- 20.Buchko GW, Sofia HJ. Backbone 1H, 13C, and 15N NMR assignments for the Cyanothece 51142 protein cce_0567: a protein associated with nitrogen fixation in the DUF683 family. Biomol. NMR Assign. 2008;2:25–28. doi: 10.1007/s12104-007-9075-3. [DOI] [PubMed] [Google Scholar]
- 21.Buchko GW, Robinson H, Addlagatta A. Structural characterization of the protein cce_0567 from Cyanothece 51142, a metalloprotein associated with nitrogen fixation in the DUF683 family. Biochim. Biophy. Acta. 2009;1794:627–633. doi: 10.1016/j.bbapap.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskowski RA, Watson JD, Thornton JM. From protein structure to biochemical function? J. Struct. Func. Gen. 2003;4:167–177. doi: 10.1023/a:1026127927612. [DOI] [PubMed] [Google Scholar]
- 23.Bateman A, Coggill P, Finn RD. DUFs: families in search of a function. Acta Cryst. 2010;F66:1148–1152. doi: 10.1107/S1744309110001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szyperski T, Yeh D, Sukumaran D, Moseley H, Montelione G. Reduced-dimensionality NMR spectroscopy for high-throughput protein resonance assignment. Proc. Natl. Acad. Sci. USA. 2002;99:8009–8014. doi: 10.1073/pnas.122224599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chayen NE, Shaw-Stewart PD, Maeder DL, Blow DM. An automated system for micro-batch protein crystallization and screening. J. Appl. Cryst. 1990;23:297–302. [Google Scholar]
- 26.Vagin AA, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 27.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Cryst. 2011;D67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murshudova GN, Vagin AA, Dodson EJ. Refinement of macaromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Lovell SC, Davis IW, Arendall WB, III, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Cα geometry: Φ, Ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 31.Rossi P, et al. A microscale protein NMR sample screening pipeline. J. Biomol. NMR. 2010:46. doi: 10.1007/s10858-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krissinel E, Henrick K. Inference of macromolecular assembles from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Maiti R, van Domselaar GH, Zhang H, Wishart DS. SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 2004;23:W590–W594. doi: 10.1093/nar/gkh477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landau M, Mayrose I, Rosengerg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holzwarth GM, Doty P. The ultraviolet circular dichroism of polypeptides. J. Amer. Chem. Soc. 1965;87:218–228. doi: 10.1021/ja01080a015. [DOI] [PubMed] [Google Scholar]
- 36.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Prot. 2006;6:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karantzeni I, Ruiz C, Liu C-C, LiCata VJ. Comparative thermal denaturation of Thermus aquaticus and Escherichia coli type 1 DNA polymerases. Biochem. J. 2003;374:785–792. doi: 10.1042/BJ20030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reddy KJ, Haskell JB, Sherman DM, Sherman LA. Unicellular, aerobic nitrogen-fixing cyanobacteria of the genus Cyanothece . J. Bact. 1993;175:1284–1292. doi: 10.1128/jb.175.5.1284-1292.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stöckel J, Welsch EA, Liberton M, Kunnvakkam R, Aurora R, Pakrasi HB. Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proc. Natl. Acad. Sci. USA. 2008;105:6156–6161. doi: 10.1073/pnas.0711068105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curatti L, Hernandez JA, Igarashi RY, Soboh B, Zhao D, Rubio LM. In vitro synthesis of the iron-molybdenum cofactor of nitrogenase from iron, sulfur, molybdenum and homocitrate using purified proteins. Proc. Natl. Acad. Sci. USA. 2007;104:17626–17631. doi: 10.1073/pnas.0703050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larkin MA, et al. ClustalW2 and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 43.DeLano WL. The PyMOL molecular graphics system. Palo Alto CA USA: DeLano Scientific LLC; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.