Abstract
During the past ten years, dramatic advances have been made in unraveling the biological bases of age-related macular degeneration (AMD), the most common cause of irreversible blindness in western populations. In that timeframe, two distinct lines of evidence emerged which implicated chronic local inflammation and activation of the complement cascade in AMD pathogenesis. First, a number of complement system proteins, complement activators, and complement regulatory proteins were identified as molecular constituents of drusen, the hallmark extracellular deposits associated with early AMD. Subsequently, genetic studies revealed highly significant statistical associations between AMD and variants of several complement pathway-associated genes including: Complement factor H (CFH), complement factor H-related 1 and 3 (CFHR1 and CFHR3), complement factor B (CFB), complement component 2 (C2), and complement component 3 (C3).
In this article, we revisit our original hypothesis that chronic local inflammatory and immune-mediated events at the level of Bruch’s membrane play critical roles in drusen biogenesis and, by extension, in the pathobiology of AMD. Secondly, we report the results of a new screening for additional AMD-associated polymorphisms in a battery of 63 complement-related genes. Third, we identify and characterize the local complement system in the RPE-choroid complex -- thus adding a new dimension of biological complexity to the role of the complement system in ocular aging and AMD. Finally, we evaluate the most salient, recent evidence that bears directly on the role of complement in AMD pathogenesis and progression. Collectively, these recent findings strongly re-affirm the importance of the complement system in AMD. They lay the groundwork for further studies that may lead to the identification of a transcriptional disease signature of AMD, and hasten the development of new therapeutic approaches that will restore the complement-modulating activity that appears to be compromised in genetically susceptible individuals.
1. The Complement System and AMD: A Brief Overview
1.1. The Complement Cascade
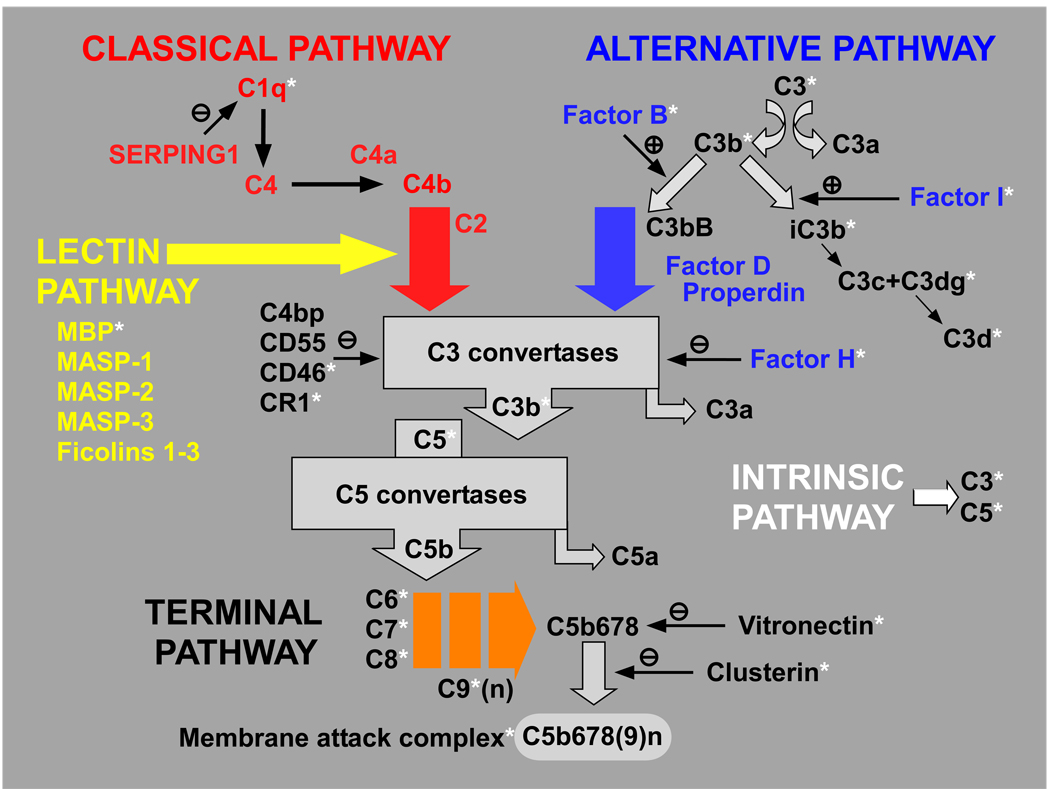
As a component of the innate immune system, the complement system provides for opsonization and lysis of microorganisms, removal of foreign particles and dead cells, recruitment and activation of inflammatory cells, regulation of antibody production, and elimination of immune complexes (Markiewski and Lambris, 2007). The complement cascade’s four activation pathways converge upon a common terminal pathway that culminates in the formation of the cytolytic membrane attack complex (MAC) (see Figure 1). Typically, binding of circulating C1q to antigen-antibody complexes activates the classical pathway. The lectin pathway is activated by mannan-binding lectin following recognition of, and binding to, molecular patterns on pathogen surfaces. The recently characterized intrinsic pathway is activated by proteases that cleave C3 and C5 directly. Unlike the other three pathways, the alternative pathway is in a continuous low-level state of activation (‘tickover’) characterized by the spontaneous hydrolysis of C3 into C3a and C3b fragments. C3b binds complement factor B (CFB) and, once bound, factor B is cleaved by complement factor D (CFD) into Ba and Bb, thereby forming the active C3 convertase (C3bBb). The convertase cleaves additional C3 molecules, generating more C3a and C3b, thereby promoting further amplification of the cascade. In addition to the 30 or more complement components and fragments, there are numerous soluble and membrane-bound regulatory proteins that modulate the complement system. Activation surfaces on host cells and tissues not protected by these regulatory molecules are rapidly coated with C3b, and become targets of attack by the membrane attack complex (MAC) formed by the sequential assembly of the terminal components C5b, C6, C7, C8, and up to 16 molecules of C9. The MAC generates a pore in the cell’s lipid bilayer, thereby promoting cell lysis. Host cells and tissues that are inadequately protected from complement attack are subject to bystander cell lysis.
Figure 1.
Schematic of the complement system. The complement cascade consists of 4 activation pathways, including the recently characterized intrinsic pathway, all of which converge upon a terminal pathway that results in the assembly of the membrane attack complex (MAC). Multiple complement regulatory proteins act at different levels to modulate the system. The complement components and regulatory molecues that are localized in drusen are highlighted by an asterisk.
1.2. Complement-related Molecules in Drusen
In the mid-1990s, evidence of complement in one class of sub-RPE deposits (basal laminar deposits) began to emerge(van der Schaft et al., 1993) and, in the ensuing years, it became evident that a large number of complement activators, complement components and complement regulatory proteins are molecular constituents of drusen, the hallmark extracellular deposits associated with early stage AMD (Anderson et al., 2002; Anderson et al., 2004; Crabb et al., 2002; Hageman and Mullins, 1999; Hageman et al., 1999a; Johnson et al., 2002; Johnson et al., 2001; Johnson et al., 2000; Johnson et al., 2003; Johnson et al., 2006; Laine et al., 2007; Mullins et al., 2001; Mullins and Hageman, 1997; Mullins et al., 2000; Sakaguchi et al., 2002; van der Schaft et al., 1993; Zhou et al., 2006; Zhou et al., 2009). A list of these complement-related molecules in drusen is provided in Table 1 below, and are indicated by an asterisk in Figure 1.
Table 1.
Complement-related Molecules in Drusen
Age-related drusen accumulate between the basal surface of the retinal pigmented epithelium (RPE) and a unique, stratified extracellular matrix known as Bruch’s membrane (BM) (Hogan, 1971). Identification of this compositional profile formed the basis for a new paradigm of AMD pathogenesis in which drusen are regarded as byproducts of chronic, local inflammatory events at the level of BM. According to this “inflammation” model, local inflammation accompanied by complement activation, bystander cell lysis, and immune responsiveness are important facets of AMD pathogenesis and progression (Anderson et al., 2002; Hageman et al., 2001; Johnson et al., 2001).
1.3. Variants of Complement-related Genes Associated with AMD
Definitive support for the “inflammation” model of AMD pathogenesis emerged within the past few years from multiple genetic studies which revealed highly significant associations between AMD and sequence variants of several complement pathway-associated genes including: Complement factor H (CFH) (Edwards et al., 2005; Hageman et al., 2005; Haines et al., 2005; Klein et al., 2005), complement factor B (CFB) and complement component 2 (C2) (Gold et al., 2006; Jakobsdottir et al., 2008). Subsequently, two non-synonymous polymorphisms in the C3 gene were also reported to be associated with AMD (Despriet et al., 2009; Maller et al., 2007; Park et al., 2009a; Yates et al., 2007), a result that further reinforced the conclusion that the complement system---and the interactive CFH, CFB, and C3 triad in particular---is a defining feature of AMD genetic susceptibility. Combined analyses of the CFH and CFB/C2 variants indicated that they can account for nearly 75% of all AMD cases in the European and North American populations (Gold et al., 2006). The total population attributable risk for the variants of the C3 gene was estimated to be 14.5%, a figure that is apparently independent of the risk attributable to the CFH variants (Despriet et al., 2009). Other studies yielded evidence for a protective CFH haplotype tagged, in part, by a deletion of two additional members of the CFH gene family (CFHR1 and CFHR3) (Hageman et al., 2006; Hughes et al., 2006). In two cohorts, deletion homozygotes represented 4.9% and 6.7% of controls versus 1.2% and 0.94% of cases respectively (Hageman et al., 2006). CFHR1 and CFHR3 possess significant amino acid sequence homology, and also share binding properties with CFH. However, unlike CFH that regulates the C3 convertase, CFHR1 appears to act downstream by modulating the activity of the C5 convertase and inhibiting formation of the MAC. As such, it has been proposed that the protective effect conferred by deletion of CFHR1/CFHR3 in AMD is mediated by removal of the C5a blockade and disinhibition of MAC formation (Heinen et al., 2009).
Most recently, an inverse relationship between AMD and a non-coding variant in intron 6 of the classical pathway gene SERPING1 (C1 inhibitor) was reported in two cohorts(Ennis et al., 2008); but this protective effect was not replicated in other independent studies (Allikmets et al., 2009; Park et al., 2009b). Lastly, AMD-associated variations in a region in close proximity to another alternative pathway gene (CFI) on chromosome 4 have been reported (Ennis et al., 2009; Fagerness et al., 2009). In order to determine whether variants in other complement-related genes also contribute to AMD susceptibility, we screened for single nucleotide polymorphisms (SNPs) in a battery of 63 complement-related genes, plus several non-complement related genes/loci previously linked to AMD (ARMS2, PLEKHA1, PRSS11/HTRA1) (See supplemental Table 1). New SNPs were ‘discovered’ from a pool of DNA samples derived from 478 study participants with an established history of AMD and 300 controls. For this analysis, we employed a multiplexed SNP enrichment technology (Mismatch Repair Detection; ParAllele Biosciences/Affymetrix), an approach that enriches for variants from pooled samples. The SNP discovery phase was skewed through the use of DNA from individuals with AMD, based upon the rationale that the library of identified SNPs should be enriched in those with AMD.
DNA samples derived from 1,162 total case and control subjects were used for genotyping all complement gene-associated SNPs identified in the SNP discovery phase, as well as additional SNPs chosen from public databases. In choosing additional SNPs, we applied a higher SNP density in the genic regions defined as 5Kb upstream from the start of transcription to 5Kb downstream from the end of transcription. In these regions, we utilized an average density of 1 SNP per 10Kb. In the non-genic regions of clusters of complement-related genes, we employed an average of 1 SNP per 20 kb. Selection included intronic SNPs, variants from the regulatory regions (mainly promoters) and cSNPs included in open reading frames. Data obtained by direct screening were used to validate the information extracted from databases. We concentrated on the overall sequence variation of functionally important regions of candidate complement pathway-associated genes, not only on a few polymorphisms, and a previously described algorithm for tag selection. Positive controls included CEPH members of the HapMap trios and the nomenclature used for these samples was the Coriell sample name. The panel also contained a limited number of X-chromosome probes from two regions.
All study subjects underwent clinical examination and stereoscopic color fundus photography, and were graded according to the Rotterdam Grading System and/or according to the classification established by the AREDS study. Subjects were graded as ‘unaffected controls’ only when photographs showed clear fundi at >60 years of age or fewer than 5 small hard drusen (grade 1 by AREDS classification or grade 0 by Rotterdam classification). Detailed descriptions of this cohort have been published previously(Allikmets et al., 2009; Gold et al., 2006; Hageman et al., 2005).
Genotype associations with ‘discovered’ and other established complement gene-associated SNPs were assessed using SAS® and the results were sorted by genotype p-value. Genotypes with SNPs showing significant association with AMD were then ranked by order of statistical significance based on logistic likelihood (genotype p value) (Table 2).
Table 2.
AMD-associated Genes Identified by Mismatch Repair Detection
| Gene | SNP | Genotype P Value |
|---|---|---|
| ARMS2 | rs3750847 | 0.00000000000000000 |
| ARMS2 | rs10490924 | 0.00000000000000002 |
| CFH | rs3753395 | 0.00000000000000018 |
| CFH | rs1410996 | 0.00000000000000034 |
| CFH | rs393955 | 0.00000000000005833 |
| CFH | rs403846 | 0.00000000000005833 |
| CFH | rs1329421 | 0.00000000000016925 |
| CFH | rs10801554 | 0.00000000000016925 |
| CFH | rs12144939 | 0.00000000604366378 |
| HTRA1 | rs2253755 | 0.00000016209469941 |
| CFH | rs800292 | 0.00000039291447774 |
| PLEKHA1 | rs6585827 | 0.00000837318772810 |
| PLEKHA1 | rs10887150 | 0.00001151791573703 |
| CFH | rs12124794 | 0.00000697204343841 |
| CFH | rs2284664 | 0.00001520716964024 |
| CFH | rs16840422 | 0.00002131200064255 |
| F13B | rs5997 | 0.00002484721176248 |
| F13B | rs6003 | 0.00003907893662798 |
| F13B | rs6428380 | 0.00004105218136048 |
| F13B | MRD_3922 | 0.00004105218136073 |
| F13B | rs1794006 | 0.00006131751806055 |
| CFH | rs6695321 | 0.00051708256369126 |
| F13B | rs10801586 | 0.00024294166371323 |
| HTRA1 | rs760336 | 0.00089646179932454 |
| ARMS2 | rs10490923 | 0.00635524863142015 |
| PLEKHA1 | rs2421018 | 0.00738987158600264 |
| PLEKHA1 | rs1045216 | 0.00914700496886938 |
| HTRA1 | rs4237540 | 0.01062131492381840 |
| C3 | MRD_4273 | 0.01013214997745320 |
| FCN1 | rs10117466 | 0.00928927952072993 |
| FCN1 | rs10120023 | 0.01474171750688250 |
| FCN1 | MRD_4502 | 0.01827326920746700 |
| PLEKHA1 | rs10082476 | 0.01652667604649510 |
| C9 | rs476569 | 0.02226012069559650 |
| PLEKHA1 | rs10399971 | 0.02881915184734270 |
| C2 | rs9332739 | 0.02877688379436240 |
| F13B | rs2990510 | 0.01308948584736170 |
This analysis confirmed previously published associations of AMD with variations/SNPs in the CFH, ARMS2, PLEKHA1 and HTRA1 genes/loci, thus providing confidence in the validity of results obtained using this approach. Out of 595 verified new SNPs, 48 SNPs associated with AMD at a q-value <0.50 (the minimum false discovery rate for which the test is statistically significant). Twenty-seven of those SNPs passed the Bonferroni level of adjustment. These include markers located in the ARMS2, CFH, HTRA1, PLEKHA1, F13B, and C9 genes. In addition, there were additional markers associated with AMD that did not pass the Bonferroni cut-off, but had a low false discovery rate [q-value < 0.20). These were located in the CFH, HTRA1, PLEKHA1, C3, FCN1, F13B, and C2 genes.
FCN1 is one of 3 collagen-like ficolin genes involved in the activation of the lectin pathway. Two of the 3 FCN1 SNPs identified (rs10117466 and rs10120023) are located in the promoter region, and are part of a larger haplotype block that has been associated previously with rheumatoid arthritis (Hummelshoj et al., 2008; Vander Cruyssen et al., 2007). F13B is part of the regulation of complement activation (RCA) gene cluster on chromosome 1q32 along with CFH, the CFH related genes, and a number of other complement genes. The protein encoded by F13B is structurally similar to CFH in that it also contains a series of SCR domains. As such, it is thought to have a close evolutionary connection to CFH. As part of the coagulation cascade, activated F13B functions as a transglutaminase that catalyzes the formation of fibrin crosslinks and promotes the stabilization of fibrin clots. One of the six significant F13B SNPs identified (rs6003) has been linked to neovascular AMD in a previous study (Zhang et al., 2008). Finally, we found new SNPs in the C3 (MRD_4273), C9 (rs476569), and FCN1 (rs10117466, rs10120023, MRD_4502) genes that showed borderline associations with AMD (i.e. p-values < 0.05; q-values between 0.20 and 0.50).
These results implicate new SNPs in C3 as well as in several other complement-related genes (F13B, FCN1, C9) in AMD susceptibility. Replication of the latter findings in other cohorts will be required before their associations with AMD may be regarded as definitive. None of the newly-identified SNPs in any other complement-related genes examined made a statistically significant contribution to AMD susceptibility, over and above that which is conferred by the CFH, CFB/C2, and C3 gene variants.
For a further discussion of the molecular genetics of AMD, including a consideration of the ARMS2/HTRA1 locus on chromosome 10q26, the reader is referred to the following excellent articles (Bergeron-Sawitzke et al., 2009; DeWan et al., 2007; Donoso et al., 2006; Edwards, 2008; Lotery and Trump, 2007; Montezuma et al., 2007).
1.4 Structure and Function of the CFH Y402H Risk Variant
The CFH gene contains 20 short consensus repeat (SCR) domains, each of which encodes a functional peptide domain of approximately 60 amino acids. A single nucleotide polymorphism (T1277C) in SCR7 results in an amino acid substitution of histidine for tyrosine at position 402 in a CFH domain that contains binding sites for C-reactive protein (CRP), heparin, and streptococcal M6 protein (Giannakis et al., 2003). At least five groups have reported that the binding properties of recombinant CFH fragments that included SCR7, and/or full length CFH purified from the serum of individuals who are homozygous for the CFH402H variant, show strongly reduced affinity for CRP (Laine et al., 2007; Ormsby et al., 2008; Sjoberg et al., 2007; Skerka et al., 2007; Yu et al., 2007).
CRP exists in both monomeric and pentameric forms, but their respective biological functions are incompletely understood. Both forms bind to damaged cells, and recruit soluble CFH to the cell surface where the CRP-CFH complex binds to phosphocholine and blocks the complement cascade at the level of the C3 convertase (Mihlan et al., 2009). However, only the monomeric form of CRP binds to immobilized CFH, in a calcium independent manner (Mihlan et al., 2009). By contrast, CFH interacts with monomeric CRP, but apparently not with the pentameric form in the liquid phase at physiological concentrations (Hakobyan et al., 2008). Two groups have also reported that the Streptomyces M6 protein shows reduced affinity for purified CFH402H (Ormsby et al., 2008; Yu et al., 2007). The effect of the Y402H substitution on heparin binding is more problematic, with some finding higher affinity (Herbert et al., 2006; Skerka et al., 2007), others concluding that changes depend upon the type of heparin preparation (Clark et al., 2006), and still others finding no significant difference (Ormsby et al., 2008; Yu et al., 2007).
At the cellular level, a reduction in the binding of CFH402H “risk” variant to the surfaces of both cultured human RPE (ARPE-19) and endothelial cells (HUVEC’s) cells has been reported, an effect also demonstrable in the alternately spliced, truncated form of CFH that consists of SCR’s1-7 and a unique 8th exon (Skerka et al., 2007). This reduction was not attributable to differential binding to C3b. Finally, other investigators found an increased interaction of the CFH402H variant with necrotic Jurkat T cells, fibromodulin, and DNA (Gershov et al., 2000; Sjoberg et al., 2007).
Despite these differences in binding properties, there are no detectable differences in either CFH or CRP immunolabeling in drusen between CFH402HH and CFH402YY homozygotes. However, CFH402HH homozygotes do show elevated levels of CRP in the choroid that is apparently systemic in origin, since there is no evidence for CRP transcription in either the RPE, neural retina, or choroid in vivo (Johnson et al., 2006). These results suggest that CRP is most likely extravasated from the choroidal capillaries in response to local signals generated by the RPE-choroid. Beyond that, however, a more precise explanation for the elevated levels of CRP in the choroid in CFH402HH individuals is not immediately obvious.
Like other members of the pentraxin family, CRP consists of five subunits arranged around a central core. It is the prototypical acute phase reactant (proteins, the levels of which rise dramatically in response to acute inflammation), and has been shown to be a serum biomarker for inflammatory conditions, including cardiovascular disease and AMD (Seddon et al., 2004). CRP has a multiplicity of functions that may be pro-inflammatory or anti-inflammatory depending upon context (Black et al., 2004). It also binds numerous ligands, including complement component C1q, an interaction that promotes formation of the C3 convertase and activation of the classical pathway. On the other hand, by virtue of its binding to CFH, CRP can inhibit the formation of C5 convertases and the subsequent assembly of the MAC (C5b-9). Furthermore, in endothelial cells, CRP upregulates the expression of the complement inhibitors DAF (CD55), MCP (CD46), and CD59 thus protecting host cells from complement attack (Li et al., 2004). If, as we have proposed, drusen and other sub-RPE deposits are biomarkers of chronic local inflammation at the RPE-choroid interface (Anderson et al., 2002; Hageman et al., 2001; Johnson et al., 2001), then the recruitment of circulating CRP to the choroid is most likely a indication of cellular injury that may be exacerbated by the reduced binding of CRP to CFH402HH, diminished complement regulatory activity, and heightened complement activation in predisposed ‘risk’ individuals.
2. Complement-related Gene Expression in the RPE, Choroid, and Neural Retina
Most complement components and many circulating complement regulatory proteins are synthesized primarily by liver hepatocytes and then released into the bloodstream. As such, most tissues and organs depend upon the circulation as a primary source for many complement-related proteins. However, in some tissues with limited access to circulating plasma proteins such as the brain, an extrahepatic system of complement biosynthesis also exists (Gasque et al., 1995; Johnson et al., 1992; Walker and McGeer, 1992). In the brain, local complement expression may be required to compensate for the restricted entry of bloodborne proteins normally excluded by the blood-brain barrier. The access of circulating proteins to the neural retina is similarly constrained by its counterpart, the blood-retinal barrier [see (Cunha-Vaz, 2004) for review].
In view of the similar barrier properties present in the neural retina and brain, and of the recognized importance of the alternative pathway of complement in AMD pathogenesis, we sought to characterize the profile of local complement-related gene expression in the human neural retina, the apposed RPE, and the adjacent choroid, in order to enhance our understanding of the role of complement in ocular aging and AMD.
2.1 Quantitative PCR Analyses
Complement-related transcription was quantified by real-time PCR in the RPE-choroid tissue complex, in isolated tissue samples of RPE, choroid, neural retina, and in differentiated primary cultures of human fetal RPE cells grown under conditions described previously (Hu and Bok, 2001). Detailed human donor information is listed in Supplemental Table 2. A number of other non-ocular tissue samples and cultured cell types were also included for cross-comparison and as controls including whole blood, peripheral blood leukocytes, vein, placenta, lung, kidney, fetal liver and adult liver, human umbilical vein endothelial cells (HUVECs), and primary human fibroblasts. The analysis was carried out using the SYBR Green method (Woo et al., 1999) utilizing the reaction conditions and parameters described in Radeke et al. (Radeke et al., 2007). The complement-related genes of interest, and the sequences of the corresponding PCR primers, are listed in Supplemental Table 3.
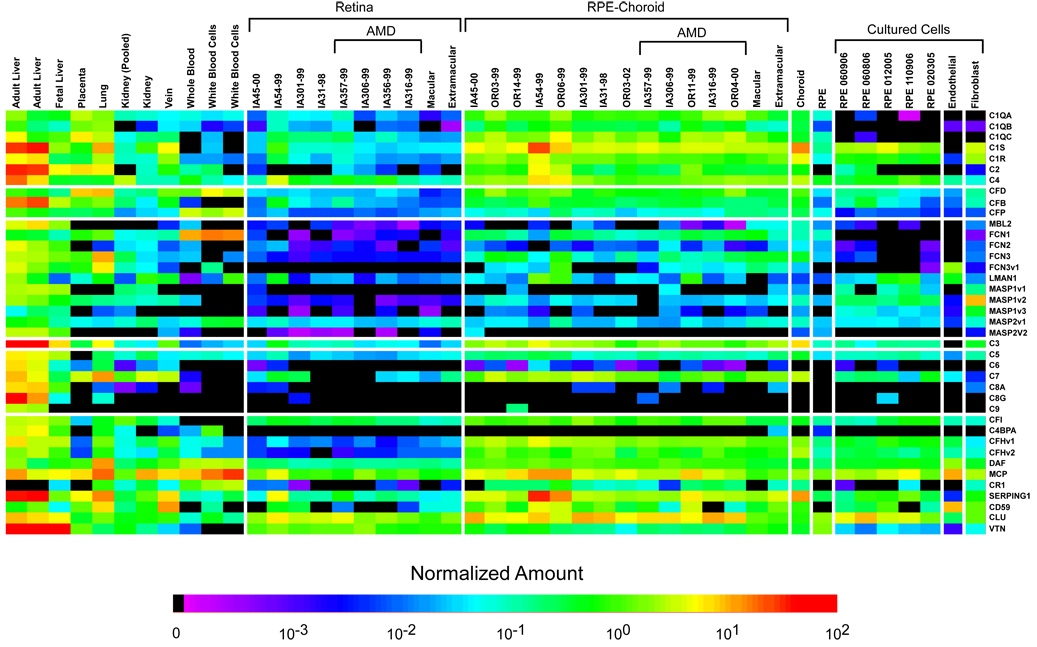
Quantitative analyses of the neural retina and RPE-choroid complement gene expression profiles have the potential to be complicated by the presence of circulating blood cells trapped within the microvasculature. However, analyses showed that levels of hemoglobin transcripts were extremely low in all of the neural retina-RPE-choroid tissue specimens examined (<0.5% of that obtained in whole blood), thereby indicating that the contamination attributable to blood cells in these tissue specimens was minor and did not contribute significantly to apparent levels of complement-associated gene transcription. A summary of the complement-related gene expression data for all of tissues and cultured cells examined is illustrated in heat map format in Figure 2, and the corresponding quantitative data are included in Supplemental Table 4.
Figure 2.
Complement expression profiles of cells and tissues used in this study. The normalized expression values, as determined by real-time quantitative PCR, for all of the genes analyzed, are depicted graphically in the form of a pseudocolored heatmap. The numerical values can be found in Supplemental Table 3.
2.2. Classical Pathway Gene Transcription
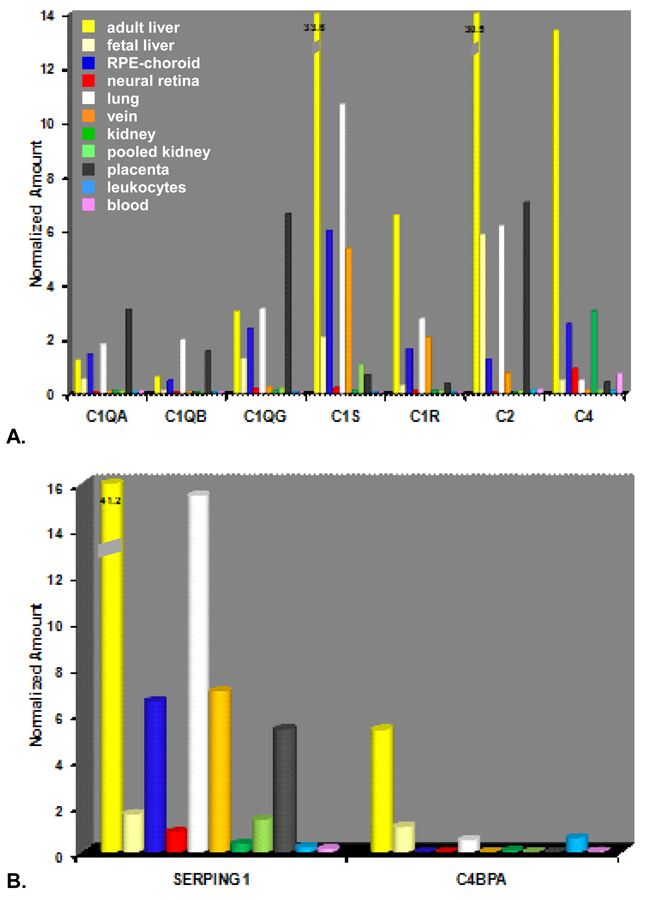
The expression levels of classical pathway-related transcripts in the adult liver, RPE-choroid, and several other tissues including lung, placenta, and vein were relatively robust (Figs. 3A-B). In contrast, relative expression levels in other tissues including the neural retina and kidney were considerably lower. As anticipated, transcript levels of most classical pathway components in adult liver exceeded those in the other cells/tissues examined by a wide margin. That was not the case for the three C1Q genes. In the RPE-choroid, relative expression levels of C1QA (1.42±0.21; n=13), C1QB (0.45±0.07; n=13), and C1QG (2.37±0.34; n=13) were roughly equivalent to those in adult liver (Figure 3A). Similar C1Q transcript levels were also detected in lung and placenta. Their corresponding signals in neural retina were extremely low. In adult liver, the serine protease C1S gave the most abundant signal followed closely by C2, and then by C4, C1R, C1QG, C1QA, and C1QB in descending order. The expression ratios of adult liver relative to RPE-choroid were highest for C2 (24:1), intermediate for C4 (5:1) and the serine proteases C1R (4:1) and C1S (6:1), and lowest for the three C1Q transcripts (1:1). The rank order of classical pathway expression in the RPE-choroid (C1S>C4>C1QG > C1R>C1QA >C2>C1QB) (Figure 3A) was similar, but not identical, to that in adult liver. Analysis of the two classical pathway regulatory genes, SERPING1 and C4BPA, showed widespread expression of SERPING1 transcripts in adult liver as well as in the other tissues/cells examined including RPE-choroid and neural retina; whereas C4BPA was readily detectable only in liver, lung, and in one of two leukocyte donor samples (Figure 3B). Expression levels of SERPING1 in adult liver (41.2) were about six fold higher than in the RPE-choroid (6.55±2.6; n=13).
Figure 3.
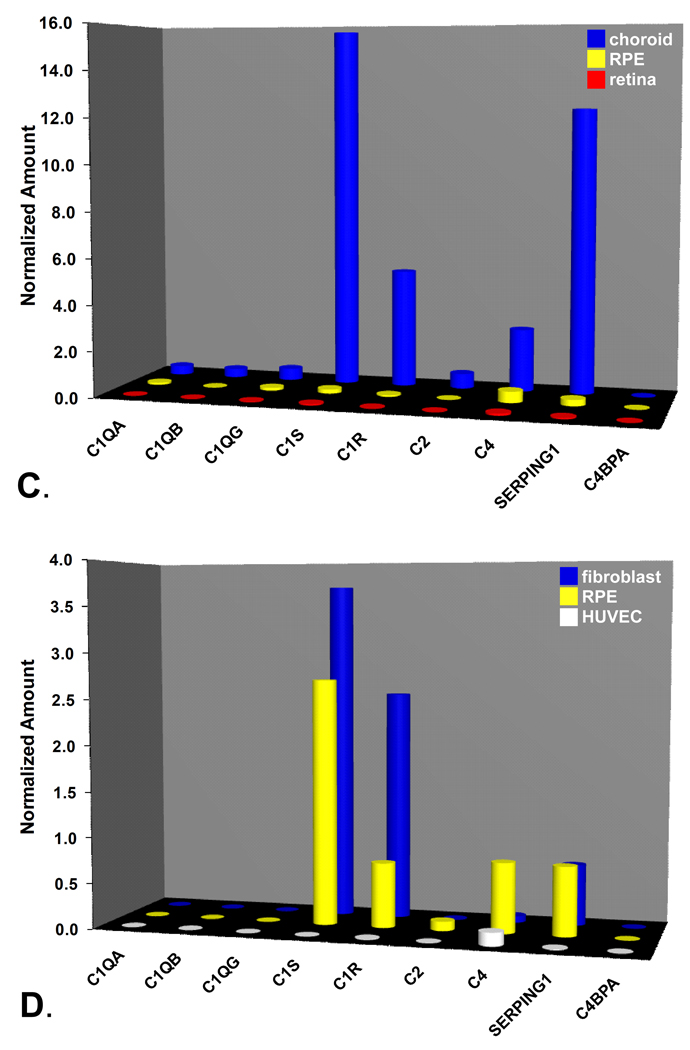
Real-time quantitative PCR analysis of gene expression in classical pathway-related genes. A-B. Expression profiles of classical pathway components (A) and regulators (B) in the RPE-choroid (dark blue), neural retina (red), and various tissue samples including: adult liver (yellow), fetal liver (pale yellow), lung (white), vein (orange),kidney (green), pooled kidney (light green), placenta (black), leukocytes (light blue) and whole blood (pink). Note off-scale expression levels of C1S (33.6) C2 (30.5), and SERPING1 (41.2) in adult liver. C. Expression profiles of classical pathway components and regulators (SERPING1, C4BPA) in microdissected tissue isolates. Choroid (blue), RPE (yellow), neural retina (red). D. Expression analysis of classical pathway components and regulators in cultured human cells. Fibroblasts (blue), RPE (yellow), HUVECs (white).
A separate comparative analysis of isolated preparations of RPE, choroid, and neural retina pooled from five donor eyes revealed that cells in the choroid are the main source(s) of classical pathway gene expression (Figure 3C). Transcription levels of the two serine proteases (C1S, C1R), as well as C4, were particularly high in the choroid relative to the other ocular tissue compartments. Considerably lower levels of C1S, C1R and C4 transcripts were detected in the isolated RPE, and signals in the neural retina were at or close to background levels. Cells in the choroid also appeared to be the predominant source of most SERPING1 transcription (Figure 3C).
Cultured human fetal RPE cells (n=5) showed expression levels of C1S (2.70±0.52), C1R (0.71±0.08), C4 (0.77±0.15), and SERPING1 (0.87±0.66) that were higher than those observed in the pooled native human RPE sample (Figure 3D). Similar levels of C1S, C1R, C4, and SERPING1 were detected in the cultured fibroblasts as well. Little to no expression of the three C1Q subunits was evident in either cultured RPE or fibroblasts. In HUVEC’s, expression of transcripts for most classical pathway-related genes was extremely low, with detectable levels evident only for C4 (Figure 3D).
2.3. Lectin Pathway Gene Transcription
Transcription levels of most lectin pathway component genes were low in nearly all of the various tissues/cells examined, except adult liver. However, FCN1 (ficolin1) transcripts were extremely high in leukocytes, and levels of transcripts for both ficolin 2 variants (FCN2ν1 and FCN2ν2) were higher in lung relative to liver by a factor of two. In general, lectin pathway component expression levels in the RPE, choroid, and neural retina were very low (Fig. 2; Supplemental Table 4). Endothelial (HUVEC) cell cultures showed significant expression of LMAN1 and FCN3ν1 (Fig. 2).
2.4. Alternative Pathway Gene Transcription
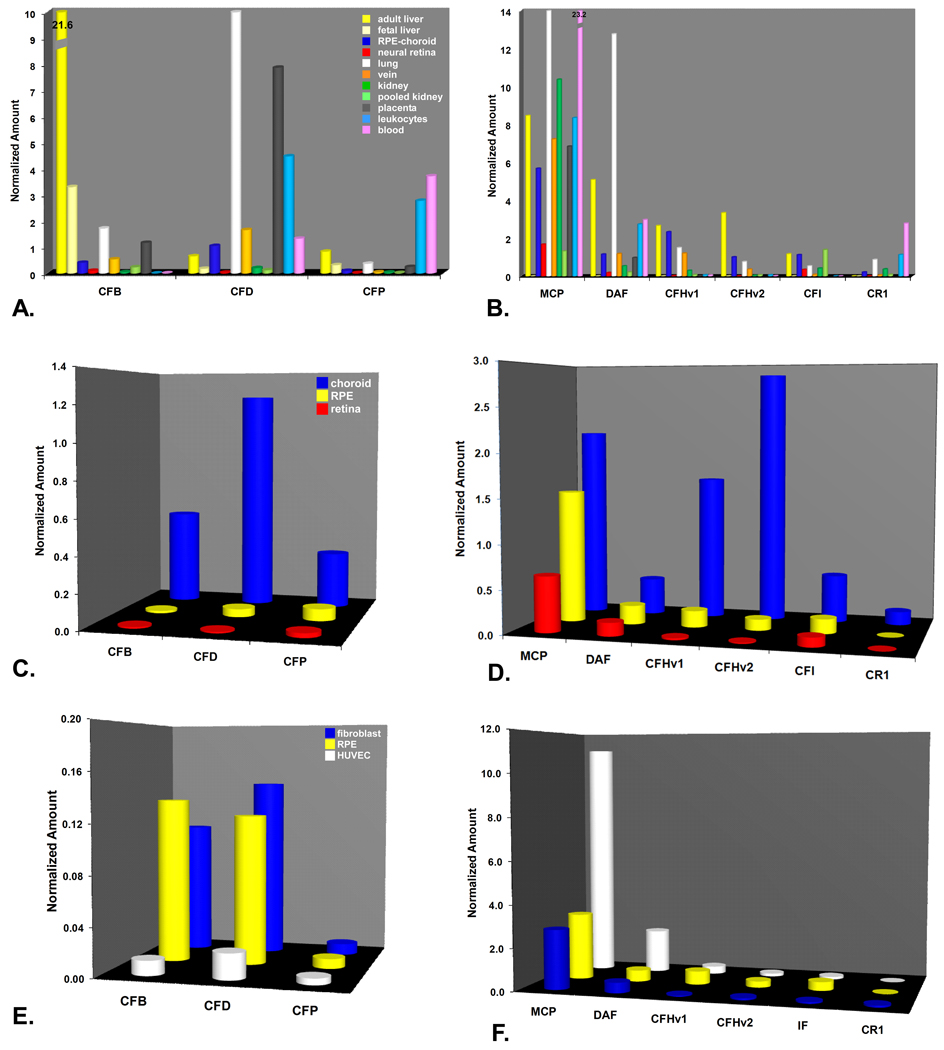
For the alternative pathway-related genes, differential expression in adult liver relative to the other cells and tissues was not as striking as it was for the classical pathway-related genes (Fig. 4A-B). Levels of CFB transcripts were considerably higher in adult liver (21.6) than in all other tissues examined including RPE-choroid (0.41±0.07; n=13) and neural retina (0.10±0.06; n=8). However, CFD showed much higher relative levels in leukocytes, lung, and placenta. CFD levels were roughly comparable to adult liver (0.70) in both RPE-choroid (1.05±0.17; n=13) and vein (Figure 4A). Expression of properdin (CFP), the alternative pathway convertase stabilizer, was relatively low in all tissues/cells examined except in whole blood and peripheral blood leukocytes.
Figure 4.
Real-time quantitative PCR analysis of gene expression in alternative pathway-related genes. A-B. Expression profiles of alternative pathway components (A) and regulators (B) in the same set of tissues shown in Figs. 1A-B. Note off-scale expression levels of CFB (21.6) in adult liver and MCP (23.2) in leukocytes. C-D. Expression profiles of alternative pathway components (C) and regulators (D) in tissue isolates. Choroid (blue), RPE (yellow), retina (red). E-F. Expression levels of alternative pathway components (E) and regulators (F) in cultured human fibroblasts (blue), RPE (yellow), and HUVECs (white). Note the relatively low levels of alternative pathway component expression in the cultured cells (E) relative to the intact choroids (C).
Expression of the alternative pathway inhibitors was also widespread in the various tissues/cells examined, but somewhat less prevalent in adult liver. Measurable levels of MCP (5.7±1.00; n=13), DAF (1.16±0.06; n=13), CFH variants 1 (2.33±0.25; n=13) and 2 (1.01±0.13; n=13), and CFI (1.12±0.10; n=13) transcripts were detected in the RPE-choroid as well as in nearly all of the other tissues/cells examined (Figure 4B). The rank order of expression in the RPE-choroid was MCP>CFHν1>IF>DAF>CFHν2>CR1. In other tissues, levels of MCP and DAF were highest in lung and in peripheral leukocytes. Measurable levels of CR1 (complement receptor 1) were detected only in whole blood, leukocytes, kidney, and lung. In the neural retina, only low levels of MCP, DAF and CFI were detected; CFH and CR1 were at or below the level of detection.
Analysis of neural retina, RPE, and choroid isolates indicated that choroidal cells are the predominant local source of most alternative pathway components and regulators (Figs. 4C-D). The expression ratios of CFHν1 and CFHν2 in the choroid relative to the RPE were 9:1 and 24:1 respectively, and much higher in relation to neural retina. MCP, DAF, and CFI expression levels were also higher in the choroid than in either RPE or neural retina.
In cultured cells, alternative pathway component expression (CFB, CFD, CFP) was relatively low (Fig. 4E); in the RPE, the levels in culture were comparable to those in situ (Fig. 4C). CFP was at or close to background levels in all three cell types (Fig. 4E). Fibroblasts had levels of CFD, CFB, and CFP that were very similar to those in cultured RPE, whereas HUVECs showed only background levels of all three (Fig. 4E). Levels of the two membrane-associated inhibitors (MCP and DAF) were high in all three cell types, and particularly in the HUVECs compared to CFH and IF. Cultured RPE cells showed highest levels of transcripts for MCP, followed by CFHν1, DAF, CFI, CFHν2, and CR1 (Fig. 4F).
2.5. Complement C3 and the Terminal Pathway Genes
A number of tissues including lung, RPE-choroid (2.06±0.30; n=13), and vein showed significant expression of the shared complement component C3. C3 levels in RPE-choroid were ~5% of those measured in adult liver (data not shown). These findings are consistent with previous reports, most notably in the kidney (Tang et al., 1999), showing that at least 10% of the circulating pool of C3 cannot be attributed to hepatic synthesis. C3 expression in the neural retina was 16 fold lower than in the RPE-choroid (0.13±0.06; n=8).
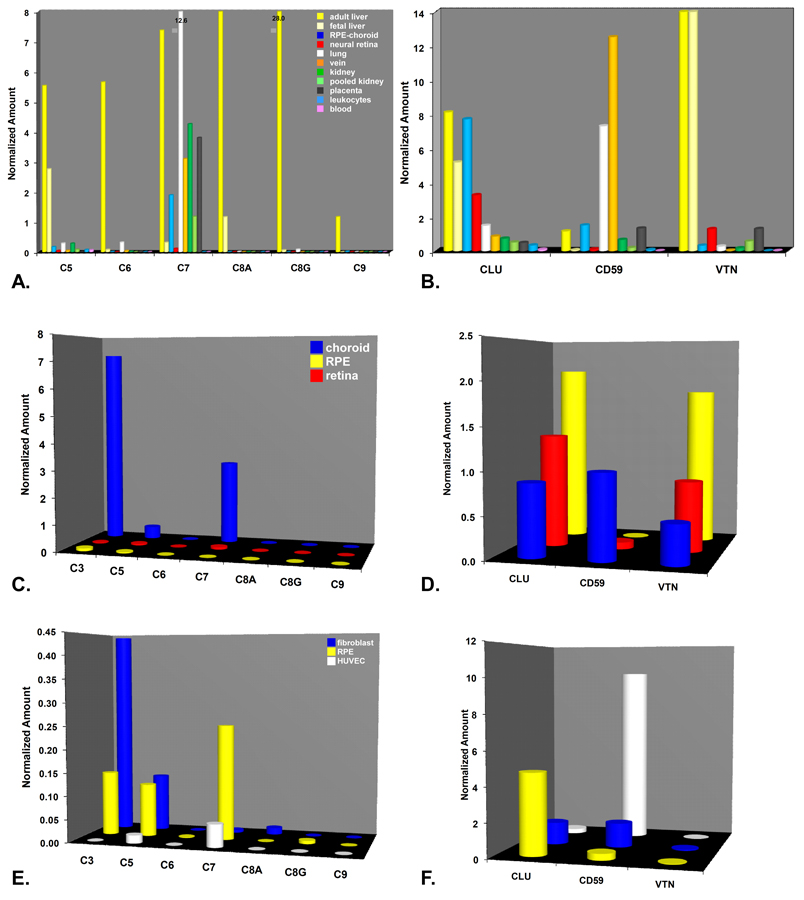
Expression of most terminal pathway components (C5-C9) was also consistently higher in adult liver than in most of the other tissues examined including the RPE-choroid and neural retina (Figs 5A). However, this was not the case for C7, which was most highly expressed in lung (12.6) (Figure 5A). Robust expression levels of C7 were also detected in the RPE-choroid (1.89±0.30; n=13), with relatively low levels in neural retina (0.18± 0.09; n=5) and only background levels in fetal liver and peripheral leukocytes. Expression ratios of C7 in the adult liver relative to other tissues examined ranged from approximately ~2:1 in vein, kidney, and placenta to ~4:1 in RPE-choroid (Figure 5A).
Figure 5.
Real-time quantitative PCR analysis of gene expression in terminal pathway-related genes. A-B. Expression profile of terminal pathway components (A) and regulators (B) using the same set of tissues/cells described above in Figs. 2A-B. A. Expression of most terminal pathway components was low in the RPE-choroid and neural retina, with the exception of C7 where levels were 1.9±0.3 (N=13) and 0.18±0.09 (N=5) respectively. Note the off-scale expression levels of C7 in lung (12.6) and C8G in adult liver (28.0). B. Expression profiles of the three terminal regulators (CLU, CD59, VTN). CLU expression levels were relatively high in both in the RPE-choroid (7.72±0.68; N=13) and neural retina (3.28±0.57; N=8) relative to adult liver. Levels of CD59 in the RPE-choroid (1.6±0.47; N=12) were also comparable to those in adult liver. In contrast, VTN expression in retina and RPE-choroid was over 10 fold lower than in adult liver, with much higher levels in the neural retina (1.29±0.15; N=12) than in the RPE-choroid (0.19+0.05* N=13). C-D. C3 and terminal pathway-related expression profiles in isolated choroids (blue), RPE (yellow), and neural retina (red). C. In the isolated choroid, robust expression of C3, C7, and, to a lesser extent, C5 was evident. In contrast, there was little or no evidence of C3 or terminal pathway component expression in either isolated neural retina or RPE. D. Expression levels of CLU and VTN expression were highest in the isolated RPE, and somewhat lower in neural retina and choroid. Levels of CD59 were highest in the choroid and extremely low in the neural retina and RPE. E-F. Terminal pathway-related expression in cultured human fibroblasts blue), RPE cells (yellow), and HUVECs (white). E. As in the isolated tissues (C), expression in the cultured cells was limited to C3, C5, and C7; overall expression levels in the cultured cells were considerably lower than in the isolated tissues. F. CD59 expression levels were highest in HUVECs, whereas levels of CLU were highest in the RPE cells.
Expression of the three terminal pathway inhibitors [CLU (clusterin; apolipoprotein J), CD59, and VTN (vitronectin)] was widespread in the various cells/tissues examined (Fig. 5B). Levels of CLU, a secreted glycoprotein, were particularly high in the RPE-choroid (7.72+ 0.68; n=13) and in neural retina (3.28 +0.57; n=8) relative to adult liver (~8.0) (Figure 5B). CD59, a membrane protein that inhibits formation of the MAC on autologous cells, was most highly expressed in non-hepatic tissues such as lung and vein, with much lower levels (4–8 fold) in RPE-choroid (1.64±0.47; n=13) and neural retina (0.22±0.12; n=4). As expected, levels of VTN (vitronectin), a circulating plasma protein and acute phase reactant, were most abundant in liver (~14). However, in this case, VTN levels in the neural retina (1.29±0.15; n=8) exceeded those in the RPE-choroid (0.32±0.05; n=13) by a factor of four.
In the isolated tissues, there was little or no detectable expression of transcripts for C3 or C5-C9 in either the RPE or neural retina (Figure 5C). In the choroid, however, there were significant levels of C3, C7 and, to a lesser extent, C5 transcripts. Levels of the three terminal pathway inhibitors (CLU, CD59, VTN) were similar in the choroid (Fig. 5D). The RPE and neural retina had comparable levels of both CLU and VTN, but little detectable CD59 (Fig. 5D). The rank order for both CLU and VTN expression was RPE>neural retina>choroid; the ranking for CD59 was the reverse, with predominant expression in the vascular rich choroid, lower levels in the neural retina, and only background levels in the RPE (Figure 5D).
C3, C5, and C7 signals were also detected, but at lower levels, in the three cultured cell types (Fig. 5E). For the three inhibitors, HUVECs showed significantly higher relative expression levels of CD59 compared to cultured RPE and fibroblasts (Fig. 5F). Conversely, RPE cells showed much higher levels of CLU transcripts relative to the other cell types (Fig. 5F). VTN expression levels were uniformly low in all three cell types.
3. Localization of Complement-related Proteins in Neural Retina, RPE, and Choroid
For those complement-related genes that showed relatively high transcription levels in the neural retina, RPE, and/or choroid, we localized some of the corresponding proteins to specific cells and extracellular compartments within these tissues. Some of the most salient results are illustrated in Figure 6–Figure 7.
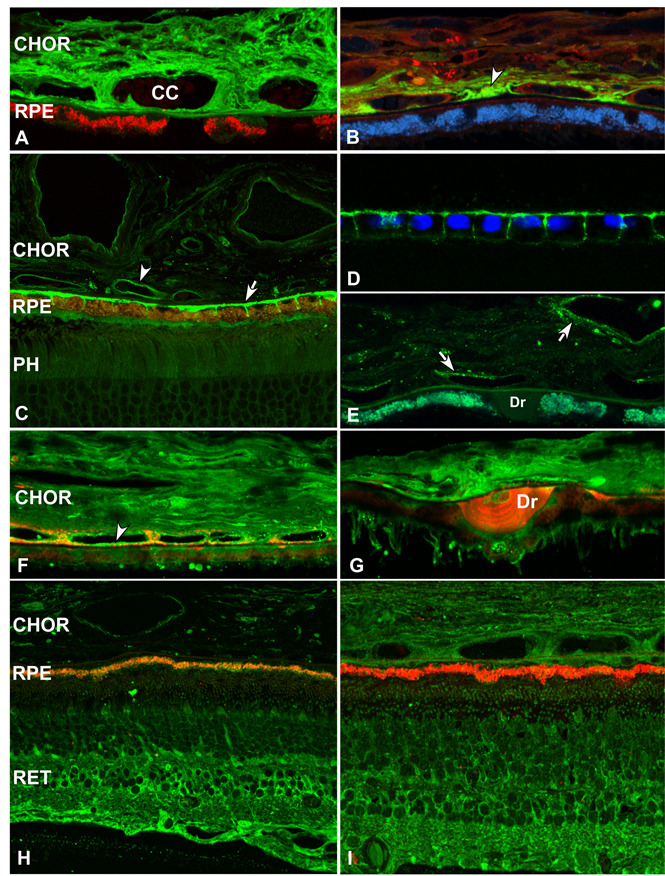
Figure 6.
Localization of alternative pathway components and inhibitors in human RPE, choroid (CHOR), and retina (RET) by confocal immunofluorescence microscopy. A. Albumin immunoreactivity is distributed throughout the choroid (CY2; green). Lipofuscin autofluorescence is present in the RPE cytoplasm (CY3; red) B. Factor H is localized to choroidal capillaries and the intercapillary pillars (arrowhead) (CY2; green). Lipofuscin autofluorescence (CY5; blue), C-reactive protein (CY3; red). C-D. MCP (CD46) is localized to choroidal vessel walls (arrowhead) and to the basolateral surface of the RPE (arrow). Photoreceptor layer of the retina (PH) (CY2; green). The polarized basolateral distribution of MCP is preserved in cultured human fetal RPE cells (shown in D) (CY2; green). RPE cell nuclei are stained by the DNA binding dye Hoescht 33258 (violet) (D). E. DAF (CD55) immunoreactivity is associated with choroidal vessel walls, but absent in the RPE and in drusen arrows) (CY2; green). F-G. Factor B/C5b-9 co-localization [anti-Factor B (CY2, green); anti-C5b-9 (CY3, red)]. Diffuse anti-Factor B labeling is present throughout the choroid; choroidal capillary vessel walls are heavily labeled (arrowhead). Factor B immunofluorescence can be localized to the cores of some drusen (Dr). H. Factor I localization (CY2, green). Lipofuscin autofluorescence is visualized on the CY3 channel (red). Most Factor I immunoreactivity is concentrated in the inner retina (RET) and, to a lesser extent, in the choroid. I. Factor D adipsin) localization (CY2, green). Lipofuscin autofluorescence (CY3, red). Diffuse Factor D immunoreactivity is present throughout the retina and choroid.
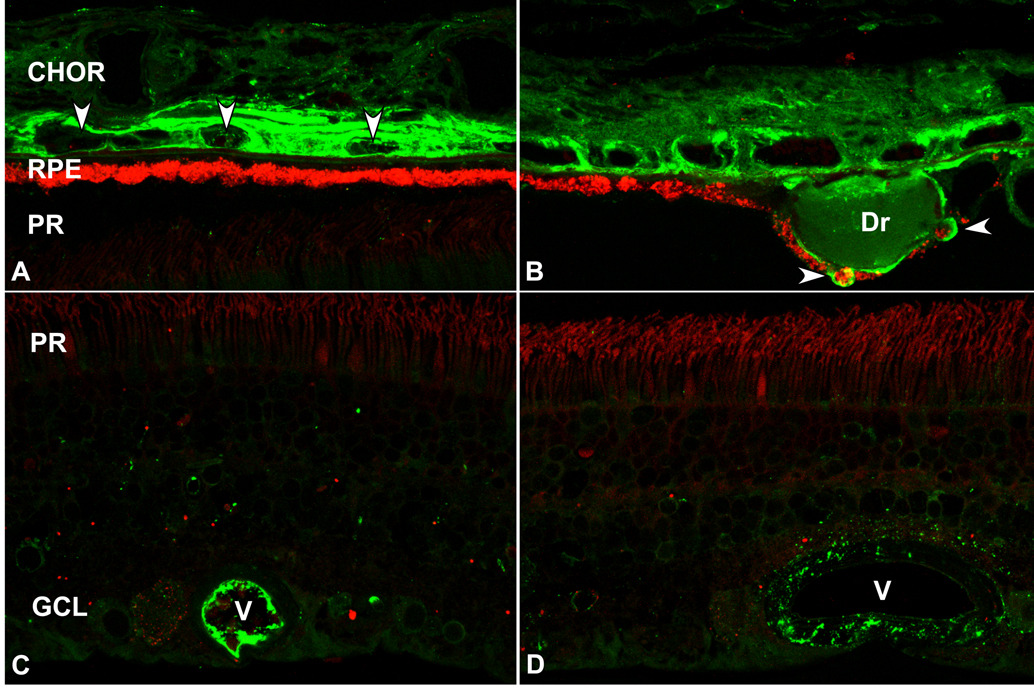
Figure 7.
Localization of complement C3 in human RPE, choroid (CHOR), and neural retina by confocal immunofluorescence microscopy. A-B. C3 immunoreactivity is concentrated in the RPE-choroid, but absent in the photoreceptor layer (PR) of the neural retina (CY2, green). Lipofuscin autofluorescence (CY3, red). A. Intense C3 immunofluorescence is associated with the microvasculature in the choroid. Arrowheads indicate cross-sections of choroidal capillaries. B. In those eyes with drusen (Dr), C3 immunoreactivity is also present in the extracellular space between the RPE and Bruch’s membrane (i.e. the sub-RPE space) and in the cytoplasm of some RPE cells (arrowheads) overlying drusen. C-D. Localization of C3 in the retina (CY2, green). C3 immunoreactivity is confined to the lumens and walls of blood vessels (V) in the inner retina.
Abundant plasma proteins such as albumin (Rs= 3.5 nm; 66 kDa) are localized throughout the choroid (Fig. 6A). In contrast, immunolabeling for factor H (142 kDa), a larger elongated molecule synthesized by both the RPE and resident choroidal cells, is confined primarily to the choriocapillaris and intercapillary pillars (Fig. 6B). MCP immunoreactivity is concentrated primarily along the basolateral surface of the RPE cells and, to a lesser extent, on the luminal surfaces of choroidal endothelial cells (Fig. 6C). This polarized, basolateral distribution is preserved in cultured fetal human RPE cells (Fig. 6D). DAF immunoreactivity is limited to the inner aspect of choroidal capillaries (Fig. 6E). CFB (90 kDa) immunolabeling is found throughout the choroid and is also associated with the luminal surfaces of choroidal endothelial cells (Figs. 6F-G). In contrast, CFI labeling is highest in the inner retina and relatively low in the choroid (Fig. 6H). CFD labeling appears to be diffusely distributed throughout the neural retina and choroid (Fig. 6I).
C3 (Rs=5.3 nm; 187 kDa) is an acute phase reactant and is among the most highly expressed complement-related genes in the choroid (Fig. 5C), relative to adult liver. This finding is consistent with previous reports, most notably in the kidney (Tang et al., 1999), showing that at least 10% of the circulating pool of C3 cannot be attributed to hepatic synthesis. In normal adult RPE-choroid, most C3 immunoreactivity is restricted to the choriocapillaris, a narrow zone of microvasculature immediately adjacent to Bruch’s membrane (Fig. 7A). In those with drusen deposits, however, C3 immunoreactivity is also present in the sub-RPE space and in the cytoplasm of some RPE cells overlying drusen (Fig. 7B). In the neural retina, C3 immunoreactivity is associated with the retinal vessels (Fig. 7C). In some larger vessels, punctuate labeling is located in a narrow zone surrounding the vessel walls (Fig. 7D).
4. Functional Implications of the Local Complement System in the RPE-Choroid
The results from the qPCR analyses indicate that cells in the human RPE-choroid complex express a virtually complete set of transcripts for complement components and regulatory molecules associated with both the classical (Fig. 2–Fig. 3) and alternative branches of the complement cascade (Fig. 2,Fig. 4). In contrast, we found little evidence for gene expression of lectin pathway components or terminal pathway components (Fig. 2,Fig. 5), with the exception of C5 and C7, thereby indicating the RPE-choroid depends upon the circulation as the source of most lectin pathway components and terminal pathway components that are required for assembly of the MAC. Further comparisons between isolated preparations of RPE, choroid, and neural retina reveal that the predominant cellular source(s) for most classical and alternative pathway gene expression, including the shared C3 component, resides in the choroid rather than in the RPE or neural retina. In nearly all cases, transcript levels of classical and alternative pathway components (Fig. 3C,Fig. 4C), as well as their respective regulatory molecules (Fig. 3C,Fig. 4D) were substantially higher in the choroid than in either isolated RPE or neural retina. In short, the results indicate that cells in the choroid have the capacity to synthesize an extensive array of classical and alternative activation pathway components and regulatory molecules, most of which are apparently not produced by cells in the RPE or neural retina.
The choroid is a heterogeneous tissue that houses a well-developed network of sympathetic and parasympathetic nerves, an arteriovenous system, an extensive capillary network, a pigmented stroma containing melanocytes and fibroblasts, and several types of plasma-derived cells including macrophages and mast cells. A number of cell types represented in the intact choroid have been shown to synthesize one or more complement components in vitro (Guc et al., 1993; Tenner and Volkin, 1986). Further study will be required to identify the resident cell type(s) in the choroid that express specific complement pathway components and/or their respective regulatory molecules.
4.1. CFH and MCP are the Predominant Cell Surface-associated Alternative Pathway Inhibitors in the RPE-Choroid Complex
An exception to the generalization that the choroid is the predominant source of complement regulatory proteins is MCP, an alternative pathway regulator which showed robust levels of transcription in the RPE, neural retina, and choroid (Fig. 4D), as well as an immunoreactivity pattern consistent with its known role as a membrane bound inhibitor of complement activation (Figs. 6C-D). MCP codes for a transmembrane protein that down-regulates complement activation by serving as a cofactor for the cleavage and subsequent inactivation of surface-bound C3b and C4b by the serine protease CFI. It is one of three related alternative pathway regulators, including DAF and CR1, which provide protection to host cell surfaces by accelerating the decay of the C3 and C5 convertases. It is expressed by a number of cell types including endothelial and epithelial cells, with four predominant isoforms derived through alternative splicing of a single gene. In the eye, MCP has been localized to RPE cells overlying drusen (Johnson et al., 2001) and, more recently, to the basolateral surface of RPE cells in situ (Fig. 6C) (McLaughlin et al., 2003; Vogt et al., 2006) and in vitro (Fig. 6D). Evidence of MCP and DAF immunoreactivity has also been reported in the anterior segment of the eye (Bora et al., 1993). Prior results, together with those reported here, strongly suggest that CFH and MCP are the two predominant cell surface-associated alternative pathway inhibitors in the RPE-choroid complex
4.2. Local Expression of C1r, C1s, and C1 inhibitor (SERPING1)
The relatively robust expression levels of several other complement-related components in the RPE-choroid suggests that they may fulfill important functions at that location. Although hepatocytes synthesize C1r and C1s in culture, they do not express C1q (reviewed by Morgan and Gasque, 1997) and, therefore, are incapable of assembling the C1 macromolecular complex. This finding is consistent with our results which show robust local expression of the C1S and C1R mRNAs in the RPE/choroid, but much lower levels of C1QA and C1QB, that are required for assembly of the C1 complex (Fig. 3C). Numerous cell types represented in the intact RPE/choroid including epithelial cells, fibroblasts and monocytic cells have been shown to synthesize C1q, C1r, and C1s in vitro (Tenner and Volkin, 1986; Gulati et al, 1993).
In addition to its inhibitory role in classical pathway activation, SERPING1, also known as C1 inhibitor, has other biological activities that are anti-inflammatory in nature (Davis et al., 2007). By virtue of its non-covalent interaction with C3b, it is regarded, along with Factor H, to be a regulator of the alternative pathway (Jiang et al., 2001). Although an association between AMD and several non-coding SNPs in the SERPING1 gene has been reported (Ennis et al., 2008b), this result has not been confirmed in other large case control studies (Park et al., 2009b).
In contrast to the high levels of SERPING1 expression, transcripts for C4BPA, the major fluid phase inhibitor of the classical pathway convertase, were not detected in the RPE-choroid. C4BP is a large multimeric 570 kDa protein consisting of seven alpha subunits and one beta subunit; in plasma, it is almost exclusively associated with the phosphatidylserine binding protein, protein S. In a mechanism analogous to that of CFH, C4BP binds to the classical C3 convertase (C4bC2b) and recruits CFI to inactivate the convertase through proteolysis of C4b. Therefore, in the absence of local C4BP synthesis or extravasation from the choroidal capillaries, regulation of the complement cascade in the RPE-choroid at the C3 level rests more heavily on CFH. Since the CFH402H variant associated with AMD is thought to have altered complement inhibitory activity (Laine et al., 2007), the RPE-choroid and other tissues with little or no local C4BP synthesis may be inherently more susceptible to complement attack.
4.3. Local Expression of Terminal Pathway Component C7
Unlike the other terminal pathway components (C5,C6,C8,C9), the liver is not the predominant site of C7 biosynthesis, and local C7 production is therefore regarded as a limiting factor for generation of the MAC on activating surfaces (Wurzner, 2000; Wurzner et al., 1994). Enhanced local synthesis of C7 in the RPE-choroid, as well as in other tissues, may be advantageous, particularly at those locations that are highly vulnerable to inflammation and/or microbial invasion. A membrane-bound form of C7 (mC7)has been identified recently on the surfaces of endothelial cells in various tissues, and it has been proposed that mC7 may attenuate inflammation by acting as a trap for the assembly of the MAC(Bossi et al., 2009). Unlike the soluble C5b-9 terminal complex, the mC7-associated terminal complex does not promote cell permeability or interleukin-8 expression.
4.4. Local Expression of Alternative Pathway Component Factor D (Adipsin)
The liver is also not the primary biosynthetic source of the alternative pathway component CFD (Factor D), a serine protease aptly named adipsin since adipocytes are the primary cellular source (Choy, Rosen et al, 1992; White, Damm et al, 1992). Local levels of CFD mRNA are consistently high in the RPE-choroid (Figs. 4A, C), as they are in some other tissues including the kidney glomerulus (Song, Zhou et al, 1998). CFD is essential for alternative pathway activation in humans and, as shown recently by Abrera-Abeleda et al (2007), targeted deletion of the CFD gene in mice results in the formation of deposits containing C3 and IgM within the renal mesangium, and in the development of immune complex glomerulonephritis. Thus, local synthesis of CFD appears to be an essential requirement for normal kidney homeostasis, and the same could be true in the RPE/Choroid. By analogy, it may be predicted that, in the presence of a suitable inflammatory triggering event, the CFD(−/−) mouse may also express an ocular phenotype that has similarities to AMD(Rohrer et al., 2007).
5. Similarities between the Local Complement Systems in the RPE-Choroid and Kidney
The overall complement expression profile in the RPE-choroid is quite similar to the one identified previously in the kidney [reviewed by (Zhou et al., 2001)]. In both tissues, expression appears to be limited primarily to activation pathway components and a subset of complement inhibitors, with limited or no expression of terminal pathway components. The functional implications of this shared tissue-specific expression pattern strengthens the general concept that extrahepatic complement expression imparts an extra level of protection to those tissues/organs which are particularly vulnerable to disease (Laufer et al., 2001).
The glomerulus and the RPE-choroid share a number of structural attributes and functional properties that may also help to explain the similarities between their local complement systems. Glomerular epithelial cells (podocytes), like the RPE cells, are separated from an adjacent fenestrated capillary network by a basement membrane that acts as a structural scaffold, as well as a charge and filtration barrier. Molecular traffic through the glomerulus is mediated by the glomerular epithelium, just as the traffic into and out of the neural retina is mediated, in part, by the RPE. In the choroid, the fenestrated capillaries permit the extravasation of at least some plasma proteins throughout the choroid. Experimental studies in primates indicate that circulating proteins at least as large horseradish peroxidase (Rs =3.0 nm; 40 kDa) extravasate from the choroidal capillaries and diffuse across Bruch’s membrane down to the apicolateral surface of the RPE where they encounter occluding junctional complexes (Peyman, 1972). In the glomerulus, extravasation out of the capillaries and into the urinary space is limited by the glomerular basement membrane and specialized junctional complexes (i.e. slit diaphragms) between podocytes.
In those afflicted with dense deposit disease (DDD; previously glomerulonephritis type II, or MPGN II), subepithelial deposits containing C3 complement form beneath the glomerular basement membrane. Similarly, in those afflicted with AMD, complement-containing subepithelial deposits form in the sub-RPE space. In addition, a substantial number of individuals with DDD show evidence of drusen deposits indistinguishable in appearance and molecular composition from those in AMD(Mullins et al., 2001). Approximately 70% of those with DDD harbor the CFH402H ‘risk’ haplotype, the common variant of the CFH gene that is strongly associated with drusen and development of AMD (Hageman et al., 2005). It is highly likely that further insights into pathogenesis and progression of AMD will be gleaned from studies of patients with DDD and other kidney diseases, as well as experimental animal models of glomerulonephritis. In that regard, there is at least one published report of a 56 year old patient with DDD who had a partial CFH plasma deficiency caused by a novel CFH mutation (Cys431Tyr), and went on to develop neovascular AMD at age 60 (Montes et al., 2008). The entire spectrum of disease phenotypes caused by variants in the CFH gene has been addressed in recent reviews (Boon et al., 2009; de Cordoba and de Jorge, 2008; Jozsi and Zipfel, 2008; Zipfel et al., 2007).
6. Potential Role(s) of the Local Complement System in Aging and AMD
The identification of a local complement system in the RPE-choroid adds a new dimension, and imparts an extra level of biological complexity, to the role of the alternative pathway in the context of aging and AMD. The expression of complement pathway components by resident cells in the choroid constitutes a supplemental system for complement activation/regulation that may be mediated through either the alternative or classical pathways. In contrast, complement component expression in the RPE is significantly lower in comparison to the choroid (except for C4 and CFI), and is virtually undetectable in the neural retina where access to circulating complement components is normally constrained by the blood-retinal barrier. At least under normal conditions, therefore, complement-related gene expression in both neural retina and RPE is limited to a subset of inhibitors associated with the alternative (MCP, DAF, CFHν1/ν2, CFI) and terminal (CLU, VTN) pathways.
It has been proposed that extrahepatic complement systems facilitate a more rapid response to microbial invasion in tissues/organs that are either highly vulnerable, or not adequately exposed, to a source of complement components in the circulation (Laufer et al., 2001). It is widely recognized that the choroid, iris, and ciliary body -- defined collectively as the uvea -- are highly susceptible to local inflammation that can arise from numerous infectious agents and/or systemic diseases [reviewed by (Jha et al., 2007)]. Thus, it is significant that individuals with AMD who are homozygous for the CFH Y402H ‘risk’ variant have elevated levels of the pro-inflammatory, acute phase reactant C-reactive protein in the choroid (Johnson et al., 2006). Furthermore, an association has recently been reported between patients with one form of uveitis (multifocal choroiditis) and the major alleles/haplotypes in the CFH locus that are associated with AMD (Ferrara et al., 2008). Such studies suggest that the RPE-choroid may be the target of choice for the development of new therapeutic agents to treat AMD, uveitis, and other posterior segment diseases with an inflammatory component.
In theory, extrahepatic expression of the ‘risk’ variants of complement components and/or complement regulatory molecules may pose substantial additional risk to vulnerable tissues, such as the macula, by providing a supplemental local biosynthetic source(s) for the dysfunctional proteins. For example, biosynthesis of the ‘risk’ variant(s) of CFH and/or C3 proteins could result in a localized over/under accumulation in the RPE-choroid that could be harmful over time.
It is also possible that local complement expression may be inducible in response to aging, injury, inflammation, or other ocular diseases as has been reported recently in the neural retina of glaucomatous eyes (Khalyfa et al., 2007; Kuehn et al., 2006; Stasi et al., 2006). Dysregulation of the local complement system in the RPE-choroid could enhance susceptibility to AMD, uveitis and other ocular diseases with an inflammatory component. Thus far, we have detected no statistically significant age-related or AMD-related changes in the local expression of complement-related genes in the RPE-choroid at the transcriptional level. However, results from two recent studies indicate that circulating levels of several complement proteins and activation peptides including Ba, Bb, C5a, C3, C3d, iC3b, CFH, and CFD are correlated with AMD overall, and with specific AMD genotypes (Reynolds et al., 2009; Scholl et al., 2008). Interestingly, circulating levels of CFH were inversely related to incidence of AMD and risk-conferring AMD genotypes. The respective biosynthetic contributions of the RPE-choroid and the liver to these changes in circulating complement component/fragment levels remain to be assessed.
The local synthesis of proteins involved in immune function is an important determinant in many other diseases and injuries that possess an inflammatory component including Alzheimer’s disease, glomerulonephritis, rheumatoid arthritis, systemic lupus erythematosus, inflammatory bowel disease, and ischemia reperfusion injury [see (Bonifati and Kishore, 2007; Laufer et al., 2001) for reviews]. In view of these already established disease associations, further studies that focus on the impact(s) of aging, AMD, uveitis, and other ocular inflammatory diseases on gene expression in the RPE, choroid, and neural retina should prove to be a highly productive avenue for further investigation.
7. Future Directions
7.1. A Transcriptional Signature of AMD
It is highly likely that AMD, as well as many other diseases, possesses a characteristic transcriptional disease signature that is indicative of its pathogenic mechanisms (Kittleson and Hare, 2005; Klee, 2008). In the case of AMD, however, its transcriptional signature may be masked to varying degrees by different phenotypic manifestations, disease stage and duration, concurrent age-related changes, and multiple genetic linkages that have not as yet been characterized completely. In order to compensate for this inherent variability, a sufficiently large sample size is essential to draw meaningful conclusions with respect to the disease process.
Accordingly, we have recently completed a whole genome microarray analysis on the macular and extramacular RPE, choroid, and retina from over 90 well-characterized human donors, with and without AMD, ranging in age from 9 to 101 years of age. Approximately 50% of these donors had a clinical diagnosis of AMD, and those eyes have been sub-classified using an established AMD grading system. In addition, all donors have been genotyped with respect to the known genetic variants associated with AMD. Tissue specimens from the retina and RPE/choroid are also available from most donors, and preliminary morphological data has already been obtained. A representative subset of these donors is currently being evaluated for transcriptional evidence of microbial infection. A preliminary statistical evaluation of the data, utilizing class comparison and time course analyses, indicates that there is a large subset of genes associated with aging (~1500; false detection rate < 0.001), and smaller subsets associated with AMD (~130, p < 0.0001) and/or geographic location (i.e. macular verses extramacula) (~450; p < 0.0001). This kind of large scale gene expression profiling has the potential to facilitate the discovery of the critical pathways involved in AMD, and to identify subsets of genes that are differentially expressed as a function of aging, AMD phenotype, AMD genotype, and retinal location.
7.2. Unresolved Issues in AMD Pathobiology
Over the past decade, a definitive body of evidence has emerged that implicates the complement system in the pathogenesis and progression of AMD. Pathobiologic investigations have led to the identification of numerous complement proteins in drusen, and genetic studies have led to the discovery of variants in several complement genes that confer significant risk for, or protection from, the development of AMD late in life. Thus, it is now apparent that dysregulation of the complement cascade, and of the alternative pathway in particular, is a critical early predisposing step in the development of AMD, regardless of ocular phenotype.
For reasons not yet completely understood, the phenotypic profile of alternative pathway dysregulation is most often manifested in the aging macula and, much less frequently, in the glomerulus as aHUS or DDD. The evidence accumulated thus far also suggests that, in those individuals who carry at least one CFH Y402H “risk” allele, quantitative changes or functional alteration(s) in the biological activity(ies) of the CFH protein are primarily responsible for dysregulation of the alternative pathway.
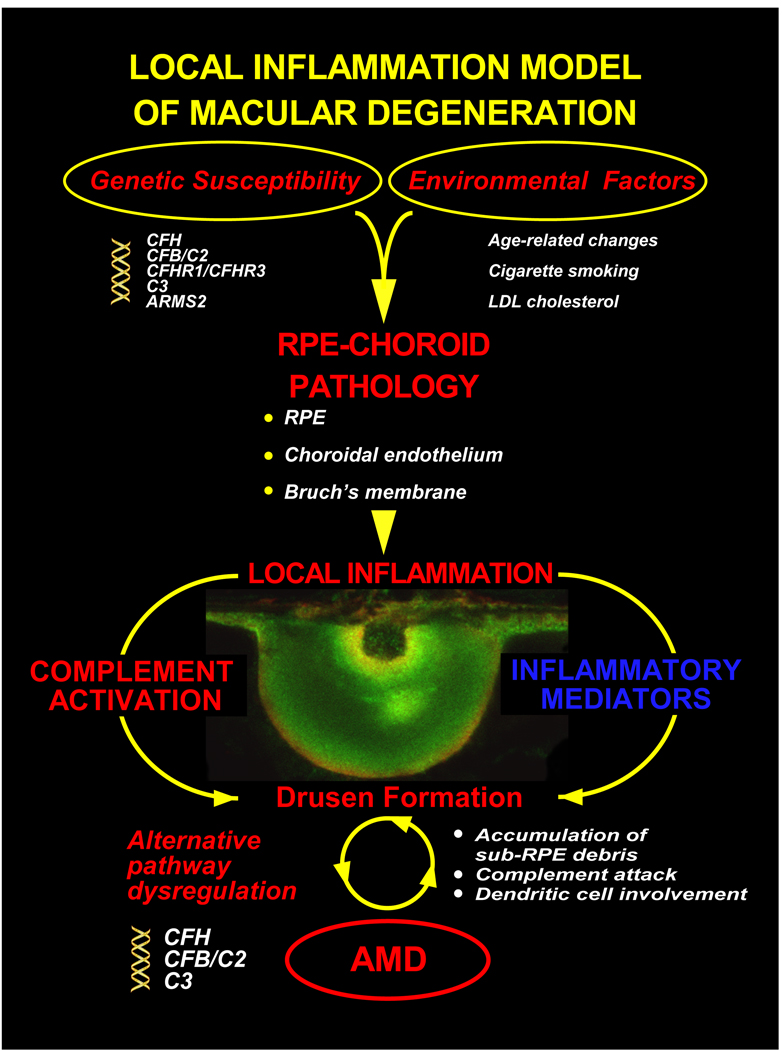
Great strides have been made during the past ten years in identifying the genetic and environmental factors that give rise to AMD, as well as the ensuing cellular events that characterize the disease process. A schematic summarizing our current understanding of these factors and events is illustrated in Figure 8. It remains unclear whether, and how, other inflammation-mediated pathways, including the cytokine system, contribute to the development and progression of early AMD. Also unclarified is the role of complement in the manifestation of the more advanced forms of AMD, particularly geographic atrophy and choroidal neovascularization (CNV). Recent immunohistochemical evidence indicates that complement-related proteins are also present in the BLDs often associated with CNV (Lommatzsch et al., 2008; Lommatzsch et al., 2007; Wolf-Schnurrbusch et al., 2009).
Figure 8.
Inflammation model of macular degeneration (updated from Anderson et al, 2002). According to the model, AMD may triggered by one or more environmental risk factors (see text) that occurs against the background of a genetic susceptibility profile conferred by variants in the CFH, CFB/C2, and/or C3 gene triad (The potential role of the ARMS2 locus in the inflammatory process, if any, remains to be clarified). This confluence of environmental and genetic risk factors gives rise to pathological changes in the RPE-choroid late in life which generates a chronic, local inflammatory response that includes complement activation and other inflammation-mediated events characterized, in part, by alterations in Bruch’s membrane, drusen formation and the accumulation of other sub-RPE deposits, bystander cell lysis, and dendritic cell involvement. Over time, these processes/events result in photoreceptor degeneration and the loss of central vision that defines the clinical entity of AMD.
Among the important specific issues that have yet to be fully resolved are the following:
What cellular event(s) leads to complement activation at the RPE-choroid interface? Is complement activation primarily systemic, or is the local system also engaged?
What are the respective roles of acute phase proteins and pro-inflammatory cytokines in RPE-choroidal pathology, and when do they come into play?
What is the temporal relationship between local inflammation, complement activation, dendritic cell involvement, and drusen biogenesis at the RPE-choroid interface?
What is the significance of the coagulation cascade in AMD, and of various coagulation and fibrinolytic proteins found in drusen including thrombin, fibrin/fibrinogen, and plasmin/plasminogen(Anderson et al., 2002; Mullins et al., 2000)? Is coagulation associated with activation of complement via the intrinsic pathway?
Does the presence of HLA-DR in drusen, and the invasion of drusen by dendritic cells, signify an ongoing process of antigen presentation to the immune system(Anderson et al., 2002; Hageman and Mullins, 2007)? If so, what are the antigens involved?
To what extent does the adaptive immune system contribute to the AMD disease process?
How does one explain the unique susceptibility of the macula to degeneration in the context of established immune-mediated pathways?
Although the initiating event(s) that gives rise to RPE-choroid pathology in AMD remains elusive, a plethora of different insults have been proposed as causative. These include microbial infection, cigarette smoking, photooxidative stress, phagocytic overload, lipofuscin toxicity, buildup of advanced glycation endproducts, alterations of iron and/or lipid homeostasis, beta amyloid toxicity, RPE atrophy, RPE autophagy, immune complex formation, and choroidal ischemia. Narrowing this list to the most critical element(s) is among the greatest remaining future challenges in AMD research. Irrespective of the precise triggering event(s) that provokes RPE-choroidal pathology, it is clear that a major downstream consequence is the deposition and sequestration of cellular and acellular debris in the sub-RPE space. Failure to dispose of the entrapped debris may be sufficient in itself to trigger a “seeding event”, generate a local pro-inflammatory signal, and activate the complement system in an attempt to eliminate the debris. For those who lack sufficient alternative pathway complement modulating activity, this would most likely result in sustained complement attack, bystander injury to neighboring cells, continued formation of drusen and other sub-RPE deposits, photoreceptor degeneration and, eventually, concomitant loss of vision.
7.3. Complement Modulation Therapy for the Treatment of AMD
Our current understanding of the role of the complement system in the AMD disease process has now been refined to the point where new therapeutic approaches can be envisioned that are designed to restore the complement-modulating activity that is deficient in genetically susceptible individuals. In its simplest form, this approach would augment the native form of CFH in ‘risk’ individuals with a supplemental source of ‘protective’ CFH that would modulate the ongoing complement attack, attenuate the local inflammation, and thereby delay the onset and slow the progression of the disease. Various delivery systems including gene transfer, cell-based therapies, organ (liver) transplantation, systemic or intraocular injections can be envisioned. In theory, the introduction of ‘protective’ CFH, or molecules with similar biological activity, should slow or arrest the progression of AMD in those individuals who carry one or two risk CFH alleles by reasserting control over the alternative pathway. If this augmentation concept is proven to be correct, we can anticipate the introduction of new complement-modulation therapeutics in the near future that will drastically reduce, or even eliminate, the devastating loss of vision that accompanies AMD.
Supplementary Material
Acknowledgments
This study was supported by NIH R24 EY017404 (GSH, LVJ, DHA), EY00331 (DB), EY014799 (LVJ), the Dolly Green Endowed Chair in Ophthalmology at UCLA (DB), the Foundation Fighting Blindness (CBR), the Ruth and Milton Steinbach Fund (CBR), and an unrestricted grant to the University of Utah Department of Ophthalmology and Visual Sciences from Research to Prevent Blindness.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: GSH has a financial interest in Optherion, Inc., New Haven, CT
Bibliography
- Allikmets R, Dean M, Hageman GS, Baird PN, Klaver CC, Bergen AA, Weber BH. The SERPING1 gene and age-related macular degeneration. Lancet. 2009;374:875–876. doi: 10.1016/S0140-6736(09)61618-4. author reply 876-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DH, Mullins RF, Hageman GS, Johnson LV. A role for local inflammation in the formation of drusen in the aging eye. Am J Ophthalmol. 2002;134:411–431. doi: 10.1016/s0002-9394(02)01624-0. [DOI] [PubMed] [Google Scholar]
- Anderson DH, Talaga KC, Rivest AJ, Barron E, Hageman GS, Johnson LV. Characterization of beta amyloid assemblies in drusen: the deposits associated with aging and age-related macular degeneration. Exp Eye Res. 2004;78:243–256. doi: 10.1016/j.exer.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Bergeron-Sawitzke J, Gold B, Olsh A, Schlotterbeck S, Lemon K, Visvanathan K, Allikmets R, Dean M. Multilocus analysis of age-related macular degeneration. Eur J Hum Genet. 2009 doi: 10.1038/ejhg.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black S, Kushner I, Samols D. C-reactive Protein. J Biol Chem. 2004;279:48487–48490. doi: 10.1074/jbc.R400025200. [DOI] [PubMed] [Google Scholar]
- Bonifati DM, Kishore U. Role of complement in neurodegeneration and neuroinflammation. Mol Immunol. 2007;44:999–1010. doi: 10.1016/j.molimm.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Boon CJ, van de Kar NC, Klevering BJ, Keunen JE, Cremers FP, Klaver CC, Hoyng CB, Daha MR, den Hollander AI. The spectrum of phenotypes caused by variants in the CFH gene. Mol Immunol. 2009;46:1573–1594. doi: 10.1016/j.molimm.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Bora NS, Gobleman CL, Atkinson JP, Pepose JS, Kaplan HJ. Differential expression of the complement regulatory proteins in the human eye. Invest Ophthalmol Vis Sci. 1993;34:3579–3584. [PubMed] [Google Scholar]
- Bossix F, Rizzi L, Bulla R, Debeus A, Tripodo C, Picotti P, Betto E, Macor P, Pucillo C, Wurzner R, Tedesco F. C7 is expressed on endothelial cells as a trap for the assembling terminal complement complex and may exert anti-inflammatory function. Blood. 2009;113:3640–3648. doi: 10.1182/blood-2008-03-146472. [DOI] [PubMed] [Google Scholar]
- Clark SJ, Higman VA, Mulloy B, Perkins SJ, Lea SM, Sim RB, Day AJ. His-384 allotypic variant of factor H associated with age-related macular degeneration has different heparin binding properties from the non-disease-associated form. J Biol Chem. 2006;281:24713–24720. doi: 10.1074/jbc.M605083200. [DOI] [PubMed] [Google Scholar]
- Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, Kamei M, Hasan A, Yan L, Rayborn ME, Salomon RG, Hollyfield JG. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci U S A. 2002;99:14682–14687. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Vaz JG. The blood-retinal barriers system. Basic concepts and clinical evaluation. Exp Eye Res. 2004;78:715–721. doi: 10.1016/s0014-4835(03)00213-6. [DOI] [PubMed] [Google Scholar]
- Curcio CA, Millican CL, Bailey T, Kruth HS. Accumulation of cholesterol with age in human Bruch's membrane. Invest Ophthalmol Vis Sci. 2001;42:265–274. [PubMed] [Google Scholar]
- Curcio CA, Presley JB, Millican CL, Medeiros NE. Basal deposits and drusen in eyes with age-related maculopathy: evidence for solid lipid particles. Exp Eye Res. 2005;80:761–775. doi: 10.1016/j.exer.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Davis AE, 3rd, Cai S, Liu D. C1 inhibitor: biologic activities that are independent of protease inhibition. Immunobiology. 2007;212:313–323. doi: 10.1016/j.imbio.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cordoba SR, de Jorge EG. Translational mini-review series on complement factor H: genetics and disease associations of human complement factor H. Clin Exp Immunol. 2008;151:1–13. doi: 10.1111/j.1365-2249.2007.03552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentchev T, Milam AH, Lee VM, Trojanowski JQ, Dunaief JL. Amyloid-beta is found in drusen from some age-related macular degeneration retinas, but not in drusen from normal retinas. Mol Vis. 2003;9:184–190. [PubMed] [Google Scholar]
- Despriet DD, van Duijn CM, Oostra BA, Uitterlinden AG, Hofman A, Wright AF, ten Brink JB, Bakker A, de Jong PT, Vingerling JR, Bergen AA, Klaver CC. Complement component C3 and risk of age-related macular degeneration. Ophthalmology. 2009;116:474–480. doi: 10.1016/j.ophtha.2008.09.055. e2. [DOI] [PubMed] [Google Scholar]
- DeWan A, Bracken MB, Hoh J. Two genetic pathways for age-related macular degeneration. Curr Opin Genet Dev. 2007;17:228–233. doi: 10.1016/j.gde.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Donoso LA, Kim D, Frost A, Callahan A, Hageman G. The Role of Inflammation in the Pathogenesis of Age-related Macular Degeneration. Surv Ophthalmol. 2006;51:137–152. doi: 10.1016/j.survophthal.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AO. Genetics of age-related macular degeneration. Adv Exp Med Biol. 2008;613:211–219. doi: 10.1007/978-0-387-74904-4_24. [DOI] [PubMed] [Google Scholar]
- Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- Ennis S, Gibson J, Cree AJ, Collins A, Lotery AJ. Support for the involvement of complement factor I in age-related macular degeneration. Eur J Hum Genet. 2009 doi: 10.1038/ejhg.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis S, Jomary C, Mullins R, Cree A, Chen X, Macleod A, Jones S, Collins A, Stone E, Lotery A. Association between the SERPING1 gene and age-related macular degeneration: a two-stage case-control study. Lancet. 2008;372:1828–1834. doi: 10.1016/S0140-6736(08)61348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerness JA, Maller JB, Neale BM, Reynolds RC, Daly MJ, Seddon JM. Variation near complement factor I is associated with risk of advanced AMD. Eur J Hum Genet. 2009;17:100–104. doi: 10.1038/ejhg.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhrai-Rad H, Zheng J, Willis TD, Wong K, Suyenaga K, Moorhead M, Eberle J, Thorstenson YR, Jones T, Davis RW, Namsaraev E, Faham M. SNP discovery in pooled samples with mismatch repair detection. Genome Res. 2004;14:1404–1412. doi: 10.1101/gr.2373904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara DC, Merriam JE, Freund KB, Spaide RF, Takahashi BS, Zhitomirsky I, Fine HF, Yannuzzi LA, Allikmets R. Analysis of major alleles associated with age-related macular degeneration in patients with multifocal choroiditis: strong association with complement factor h. Arch Ophthalmol. 2008;126:1562–1566. doi: 10.1001/archopht.126.11.1562. [DOI] [PubMed] [Google Scholar]
- Gasque P, Fontaine M, Morgan BP. Complement expression in human brain. Biosynthesis of terminal pathway components and regulators in human glial cells and cell lines. J Immunol. 1995;154:4726–4733. [PubMed] [Google Scholar]
- Gershov D, Kim S, Brot N, Elkon KB. C-Reactive protein binds to apoptotic cells, protects the cells from assembly of the terminal complement components, and sustains an antiinflammatory innate immune response: implications for systemic autoimmunity. J Exp Med. 2000;192:1353–1364. doi: 10.1084/jem.192.9.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakis E, Jokiranta TS, Male DA, Ranganathan S, Ormsby RJ, Fischetti VA, Mold C, Gordon DL. A common site within factor H SCR 7 responsible for binding heparin, C-reactive protein and streptococcal M protein. Eur J Immunol. 2003;33:962–969. doi: 10.1002/eji.200323541. [DOI] [PubMed] [Google Scholar]
- Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs K, Cramer K, Neel J, Bergeron J, Barile GR, Smith RT, Hageman GS, Dean M, Allikmets R. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guc D, Gulati P, Lemercier C, Lappin D, Birnie GD, Whaley K. Expression of the components and regulatory proteins of the alternative complement pathway and the membrane attack complex in normal and diseased synovium. Rheumatol Int. 1993;13:139–146. doi: 10.1007/BF00301260. [DOI] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman GS, Hancox LS, Taiber AJ, Gehrs KM, Anderson DH, Johnson LV, Radeke MJ, Kavanagh D, Richards A, Atkinson J, Meri S, Bergeron J, Zernant J, Merriam J, Gold B, Allikmets R, Dean M. Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: characterization, ethnic distribution and evolutionary implications. Ann Med. 2006;38:592–604. [PMC free article] [PubMed] [Google Scholar]
- Hageman GS, Luthert PJ, Victor Chong NH, Johnson LV, Anderson DH, Mullins RF. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the rpe-bruch's membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res. 2001;20:705–732. doi: 10.1016/s1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- Hageman GS, Mullins RF. Molecular composition of drusen as related to substructural phenotype. Mol Vis. 1999;5:28. [PubMed] [Google Scholar]
- Hageman GS, Mullins RF. Association of major histocompatibility class II antigens with core subdomaims present within human ocular drusen. In: Zierhut M, editor. Antigen-presenting Cells and the Eye. New York/London: Taylor and Francis Group; 2007. pp. 209–216. [Google Scholar]
- Hageman GS, Mullins RF, Russell SR, Johnson LV, Anderson DH. Vitronectin is a constituent of ocular drusen and the vitronectin gene is expressed in human retinal pigmented epithelial cells. Faseb Journal. 1999a;13:477–484. doi: 10.1096/fasebj.13.3.477. [DOI] [PubMed] [Google Scholar]
- Hageman GS, Mullins RF, Russell SR, Johnson LV, Anderson DH. Vitronectin is a constituent of ocular drusen and the vitronectin gene is expressed in human retinal pigmented epithelial cells. Faseb J. 1999b;13:477–484. doi: 10.1096/fasebj.13.3.477. [DOI] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- Hakobyan S, Harris CL, van den Berg CW, Fernandez-Alonso MC, de Jorge EG, de Cordoba SR, Rivas G, Mangione P, Pepys MB, Morgan BP. Complement factor H binds to denatured rather than to native pentameric C-reactive protein. J Biol Chem. 2008;283:30451–30460. doi: 10.1074/jbc.M803648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen S, Hartmann A, Lauer N, Wiehl U, Dahse HM, Schirmer S, Gropp K, Enghardt T, Wallich R, Halbich S, Mihlan M, Schlotzer-Schrehardt U, Zipfel PF, Skerka C. Factor H related protein 1 (CFHR-1) inhibits complement C5 convertase activity and terminal complex formation. Blood. 2009 doi: 10.1182/blood-2009-02-205641. [DOI] [PubMed] [Google Scholar]
- Herbert AP, Soares DC, Pangburn MK, Barlow PN. Disease-associated sequence variations in factor H: a structural biology approach. Adv Exp Med Biol. 2006;586:313–327. doi: 10.1007/0-387-34134-X_21. [DOI] [PubMed] [Google Scholar]
- Hogan MJ, Alvarado JA, Weddell JE. Histology of the Human Eye. Philadelphia: W.B. Saunders Company; 1971. [Google Scholar]
- Hu J, Bok D. A cell culture medium that supports the differentiation of human retinal pigment epithelium into functionally polarized monolayers. Mol Vis. 2001;7:14–19. [PubMed] [Google Scholar]
- Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38:1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- Hummelshoj T, Munthe-Fog L, Madsen HO, Garred P. Functional SNPs in the human ficolin (FCN) genes reveal distinct geographical patterns. Mol Immunol. 2008;45:2508–2520. doi: 10.1016/j.molimm.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Murata T, Hangai M, Nagai R, Horiuchi S, Lopez PF, Hinton DR, Ryan SJ. Advanced glycation end products in age-related macular degeneration. Archives of Ophthalmology. 1998;116:1629–1632. doi: 10.1001/archopht.116.12.1629. [DOI] [PubMed] [Google Scholar]
- Jakobsdottir J, Conley YP, Weeks DE, Ferrell RE, Gorin MB. C2 and CFB genes in age-related maculopathy and joint action with CFH and LOC387715 genes. PLoS ONE. 2008;3:e2199. doi: 10.1371/journal.pone.0002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha P, Bora PS, Bora NS. The role of complement system in ocular diseases including uveitis and macular degeneration. Mol Immunol. 2007;44:3901–3908. doi: 10.1016/j.molimm.2007.06.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wagner E, Zhang H, Frank MM. Complement 1 inhibitor is a regulator of the alternative complement pathway. J Exp Med. 2001;194:1609–1616. doi: 10.1084/jem.194.11.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LV, Leitner WP, Rivest AJ, Staples MK, Radeke MJ, Anderson DH. The Alzheimer's A beta -peptide is deposited at sites of complement activation in pathologic deposits associated with aging and age-related macular degeneration. Proc Natl Acad Sci U S A. 2002;99:11830–11835. doi: 10.1073/pnas.192203399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LV, Leitner WP, Staples MK, Anderson DH. Complement Activation and Inflammatory Processes in Drusen Formation and Age Related Macular Degeneration. Experimental Eye Research. 2001;73:887–896. doi: 10.1006/exer.2001.1094. [DOI] [PubMed] [Google Scholar]
- Johnson LV, Ozaki S, Staples MK, Erickson PA, Anderson DH. A potential role for immune complex pathogenesis in drusen formation. Experimental Eye Research. 2000;70:441–449. doi: 10.1006/exer.1999.0798. [DOI] [PubMed] [Google Scholar]
- Johnson LV, Talaga KC, Rivest AJ, Staples MK, Radeke MJ, Barron E, Anderson DH. Characterization of Beta Amyloid-Containing Vesicles within Drusen. 2003 [Google Scholar]
- Johnson PT, Betts KE, Radeke MJ, Hageman GS, Anderson DH, Johnson LV. Individuals homozygous for the age-related macular degeneration risk-conferring variant of complement factor H have elevated levels of CRP in the choroid. Proc Natl Acad Sci U S A. 2006;103:17456–17461. doi: 10.1073/pnas.0606234103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Lampert-Etchells M, Pasinetti GM, Rozovsky I, Finch CE. Complement mRNA in the mammalian brain: responses to Alzheimer's disease and experimental brain lesioning. Neurobiology of Aging. 1992;13:641–648. doi: 10.1016/0197-4580(92)90086-d. [DOI] [PubMed] [Google Scholar]
- Jozsi M, Zipfel PF. Factor H family proteins and human diseases. Trends Immunol. 2008;29:380–387. doi: 10.1016/j.it.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Khalyfa A, Chlon T, Qiang H, Agarwal N, Cooper NG. Microarray reveals complement components are regulated in the serum-deprived rat retinal ganglion cell line. Mol Vis. 2007;13:293–308. [PMC free article] [PubMed] [Google Scholar]
- Kittleson MM, Hare JM. Molecular signature analysis: using the myocardial transcriptome as a biomarker in cardiovascular disease. Trends Cardiovasc Med. 2005;15:130–138. doi: 10.1016/j.tcm.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Klee EW. Data mining for biomarker development: a review of tissue specificity analysis. Clin Lab Med. 2008;28:127–143. doi: 10.1016/j.cll.2007.10.009. viii. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn MH, Kim CY, Ostojic J, Bellin M, Alward WL, Stone EM, Sakaguchi DS, Grozdanic SD, Kwon YH. Retinal synthesis and deposition of complement components induced by ocular hypertension. Exp Eye Res. 2006;83:620–628. doi: 10.1016/j.exer.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Laine M, Jarva H, Seitsonen S, Haapasalo K, Lehtinen MJ, Lindeman N, Anderson DH, Johnson PT, Jarvela I, Jokiranta TS, Hageman GS, Immonen I, Meri S. Y402H polymorphism of complement factor H affects binding affinity to C-reactive protein. J Immunol. 2007;178:3831–3836. doi: 10.4049/jimmunol.178.6.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer J, Katz Y, Passwell JH. Extrahepatic synthesis of complement proteins in inflammation. Mol Immunol. 2001;38:221–229. doi: 10.1016/s0161-5890(01)00044-x. [DOI] [PubMed] [Google Scholar]
- Li CM, Clark ME, Rudolf M, Curcio CA. Distribution and composition of esterified and unesterified cholesterol in extra-macular drusen. Exp Eye Res. 2007;85:192–201. doi: 10.1016/j.exer.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Li SH, Szmitko PE, Weisel RD, Wang CH, Fedak PW, Li RK, Mickle DA, Verma S. C-reactive protein upregulates complement-inhibitory factors in endothelial cells. Circulation. 2004;109:833–836. doi: 10.1161/01.CIR.0000117087.27524.0E. [DOI] [PubMed] [Google Scholar]
- Loeffler K, Edward D, Tso M. Immunoreactivity against tau, amyloid precursor protein, and beta-amyloid in the human retina. Investigative Ophthalmology & Visual Science. 1995;36:24–31. [PubMed] [Google Scholar]
- Lommatzsch A, Hermans P, Muller KD, Bornfeld N, Bird AC, Pauleikhoff D. Are low inflammatory reactions involved in exudative age-related macular degeneration? Morphological and immunhistochemical analysis of AMD associated with basal deposits. Graefes Arch Clin Exp Ophthalmol. 2008;246:803–810. doi: 10.1007/s00417-007-0749-4. [DOI] [PubMed] [Google Scholar]
- Lommatzsch A, Hermans P, Weber B, Pauleikhoff D. Complement factor H variant Y402H and basal laminar deposits in exudative age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2007;245:1713–1716. doi: 10.1007/s00417-007-0649-7. [DOI] [PubMed] [Google Scholar]
- Lotery A, Trump D. Progress in defining the molecular biology of age related macular degeneration. Hum Genet. 2007;122:219–236. doi: 10.1007/s00439-007-0406-3. [DOI] [PubMed] [Google Scholar]
- Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39:1200–1201. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- Markiewski MM, Lambris JD. The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am J Pathol. 2007;171:715–727. doi: 10.2353/ajpath.2007.070166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin BJ, Fan W, Zheng JJ, Cai H, Del Priore LV, Bora NS, Kaplan HJ. Novel role for a complement regulatory protein (CD46) in retinal pigment epithelial adhesion. Invest Ophthalmol Vis Sci. 2003;44:3669–3674. doi: 10.1167/iovs.02-0813. [DOI] [PubMed] [Google Scholar]
- Mihlan M, Stippa S, Jozsi M, Zipfel PF. Monomeric CRP contributes to complement control in fluid phase and on cellular surfaces and increases phagocytosis by recruiting factor H. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.103. [DOI] [PubMed] [Google Scholar]
- Montes T, Goicoechea de Jorge E, Ramos R, Goma M, Pujol O, Sanchez-Corral P, Rodriguez de Cordoba S. Genetic deficiency of complement factor H in a patient with age-related macular degeneration and membranoproliferative glomerulonephritis. Mol Immunol. 2008;45:2897–2904. doi: 10.1016/j.molimm.2008.01.027. [DOI] [PubMed] [Google Scholar]
- Montezuma SR, Sobrin L, Seddon JM. Review of genetics in age related macular degeneration. Semin Ophthalmol. 2007;22:229–240. doi: 10.1080/08820530701745140. [DOI] [PubMed] [Google Scholar]
- Mullins RF, Aptsiauri N, Hageman GS. Structure and composition of drusen associated with glomerulonephritis: implications for the role of complement activation in drusen biogenesis. Eye. 2001;15:390–395. doi: 10.1038/eye.2001.142. [DOI] [PubMed] [Google Scholar]
- Mullins RF, Hageman GS. Histochemical comparison of ocular "drusen" in monkey and human. In: MH LaVail JG, Anderson RE, editors. Degenerative Retinal Diseases. New York, NY: Plenum Press; 1997. pp. 1–10. [Google Scholar]
- Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. Faseb J. 2000;14:835–846. [PubMed] [Google Scholar]
- Ormsby RJ, Ranganathan S, Tong JC, Griggs KM, Dimasi DP, Hewitt AW, Burdon KP, Craig JE, Hoh J, Gordon DL. Functional and structural implications of the complement factor H Y402H polymorphism associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:1763–1770. doi: 10.1167/iovs.07-1297. [DOI] [PubMed] [Google Scholar]
- Park KH, Fridley BL, Ryu E, Tosakulwong N, Edwards AO. Complement component 3 (C3) haplotypes and risk of advanced age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009a;50:3386–3393. doi: 10.1167/iovs.08-3231. [DOI] [PubMed] [Google Scholar]
- Park KH, Ryu E, Tosakulwong N, Wu Y, Edwards AO. Common variation in the SERPING1 gene is not associated with age-related macular degeneration in two independent groups of subjects. Mol Vis. 2009b;15:200–207. [PMC free article] [PubMed] [Google Scholar]
- Peyman G, Bok D. Peroxidase diffusion in the normal and laser-coagulated primate retina. Invest Ophthalmol. 1972;11:35–45. [PubMed] [Google Scholar]
- Radeke MJ, Peterson KE, Johnson LV, Anderson DH. Disease susceptibility of the human macula: differential gene transcription in the retinal pigmented epithelium/choroid. Exp Eye Res. 2007;85:366–380. doi: 10.1016/j.exer.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Reynolds R, Hartnett ME, Atkinson JP, Giclas PC, Rosner B, Seddon JM. Plasma Complement Components and Activation Fragments are Associated with Age-Related Macular Degeneration Genotypes and Phenotypes. Invest Ophthalmol Vis Sci. 2009 doi: 10.1167/iovs.09-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer B, Guo Y, Kunchithapautham K, Gilkeson GS. Eliminating complement factor D reduces photoreceptor susceptibility to light-induced damage. Invest Ophthalmol Vis Sci. 2007;48:5282–5289. doi: 10.1167/iovs.07-0282. [DOI] [PubMed] [Google Scholar]
- Sakaguchi H, Miyagi M, Shadrach KG, Rayborn ME, Crabb JW, Hollyfield JG. Clusterin is present in drusen in age-related macular degeneration. Exp Eye Res. 2002;74:547–549. doi: 10.1006/exer.2002.1186. [DOI] [PubMed] [Google Scholar]
- Scholl HP, Charbel Issa P, Walier M, Janzer S, Pollok-Kopp B, Borncke F, Fritsche LG, Chong NV, Fimmers R, Wienker T, Holz FG, Weber BH, Oppermann M. Systemic complement activation in age-related macular degeneration. PLoS One. 2008;3:e2593. doi: 10.1371/journal.pone.0002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddon JM, Gensler G, Milton RC, Klein ML, Rifai N. Association between C-reactive protein and age-related macular degeneration. Jama. 2004;291:704–710. doi: 10.1001/jama.291.6.704. [DOI] [PubMed] [Google Scholar]
- Sjoberg AP, Trouw LA, Clark SJ, Sjolander J, Heinegard D, Sim RB, Day AJ, Blom AM. The factor H variant associated with age-related macular degeneration (His-384) and the non-disease-associated form bind differentially to C-reactive protein, fibromodulin, DNA, and necrotic cells. J Biol Chem. 2007;282:10894–10900. doi: 10.1074/jbc.M610256200. [DOI] [PubMed] [Google Scholar]
- Skerka C, Lauer N, Weinberger AA, Keilhauer CN, Suhnel J, Smith R, Schlotzer-Schrehardt U, Fritsche L, Heinen S, Hartmann A, Weber BH, Zipfel PF. Defective complement control of factor H (Y402H) and FHL-1 in age-related macular degeneration. Mol Immunol. 2007;44:3398–3406. doi: 10.1016/j.molimm.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Stasi K, Nagel D, Yang X, Wang RF, Ren L, Podos SM, Mittag T, Danias J. Complement component 1Q (C1Q) upregulation in retina of murine, primate, and human glaucomatous eyes. Invest Ophthalmol Vis Sci. 2006;47:1024–1029. doi: 10.1167/iovs.05-0830. [DOI] [PubMed] [Google Scholar]
- Tang S, Zhou W, Sheerin NS, Vaughan RW, Sacks SH. Contribution of renal secreted complement C3 to the circulating pool in humans. J Immunol. 1999;162:4336–4341. [PubMed] [Google Scholar]
- Tenner AJ, Volkin DB. Complement subcomponent C1q secreted by cultured human monocytes has subunit structure identical with that of serum C1q. Biochem J. 1986;233:451–458. doi: 10.1042/bj2330451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Schaft T, Mooy C, de Bruijn W, de Jong P. Early stages of age-related macular degeneration: an immunofluorescence and electron microscopy study. British Journal of Ophthalmology. 1993;77:657–661. doi: 10.1136/bjo.77.10.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Cruyssen B, Nuytinck L, Boullart L, Elewaut D, Waegeman W, Van Thielen M, De Meester E, Lebeer K, Rossau R, De Keyser F. Polymorphisms in the ficolin 1 gene (FCN1) are associated with susceptibility to the development of rheumatoid arthritis. Rheumatology (Oxford) 2007;46:1792–1795. doi: 10.1093/rheumatology/kem266. [DOI] [PubMed] [Google Scholar]
- Vogt SD, Barnum SR, Curcio CA, Read RW. Distribution of complement anaphylatoxin receptors and membrane-bound regulators in normal human retina. Exp Eye Res. 2006;83:834–840. doi: 10.1016/j.exer.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Walker DG, McGeer PL. Complement gene expression in human brain: comparison between normal and Alzheimer disease cases. Brain Research. Molecular Brain Research. 1992;14:109–116. doi: 10.1016/0169-328x(92)90017-6. [DOI] [PubMed] [Google Scholar]
- Wolf-Schnurrbusch UE, Stuck AK, Hess R, Wolf S, Enzmann V. Complement Factor P in choroidal neovascular membranes of patients with age-related macular degeneration. Retina. 2009;29:966–973. doi: 10.1097/IAE.0b013e3181a2f40f. [DOI] [PubMed] [Google Scholar]
- Woo TH, Patel BK, Cinco M, Smythe LD, Norris MA, Symonds ML, Dohnt MF, Piispanen J. Identification of Leptospira biflexa by real-time homogeneous detection of rapid cycle PCR product. J Microbiol Methods. 1999;35:23–30. doi: 10.1016/s0167-7012(98)00095-5. [DOI] [PubMed] [Google Scholar]
- Wurzner R. Modulation of complement membrane attack by local C7 synthesis. Clin Exp Immunol. 2000;121:8–10. doi: 10.1046/j.1365-2249.2000.01263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzner R, Joysey VC, Lachmann PJ. Complement component C7. Assessment of in vivo synthesis after liver transplantation reveals that hepatocytes do not synthesize the majority of human C7. J Immunol. 1994;152:4624–4629. [PubMed] [Google Scholar]
- Yamada Y, Ishibashi K, Ishibashi K, Bhutto IA, Tian J, Lutty GA, Handa JT. The expression of advanced glycation endproduct receptors in rpe cells associated with basal deposits in human maculas. Exp Eye Res. 2006;82:840–848. doi: 10.1016/j.exer.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR, Sepp T, Matharu BK, Khan JC, Thurlby DA, Shahid H, Clayton DG, Hayward C, Morgan J, Wright AF, Armbrecht AM, Dhillon B, Deary IJ, Redmond E, Bird AC, Moore AT. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- Yu J, Wiita P, Kawaguchi R, Honda J, Jorgensen A, Zhang K, Fischetti VA, Sun H. Biochemical analysis of a common human polymorphism associated with age-related macular degeneration. Biochemistry. 2007;46:8451–8461. doi: 10.1021/bi700459a. [DOI] [PubMed] [Google Scholar]
- Zhang H, Morrison MA, Dewan A, Adams S, Andreoli M, Huynh N, Regan M, Brown A, Miller JW, Kim IK, Hoh J, Deangelis MM. The NEI/NCBI dbGAP database: genotypes and haplotypes that may specifically predispose to risk of neovascular age-related macular degeneration. BMC Med Genet. 2008;9:51. doi: 10.1186/1471-2350-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jang YP, Kim SR, Sparrow JR. Complement activation by photooxidation products of A2E, a lipofuscin constituent of the retinal pigment epithelium. Proc Natl Acad Sci U S A. 2006;103:16182–16187. doi: 10.1073/pnas.0604255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Kim SR, Westlund BS, Sparrow JR. Complement activation by bisretinoid constituents of RPE lipofuscin. Invest Ophthalmol Vis Sci. 2009;50:1392–1399. doi: 10.1167/iovs.08-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Marsh JE, Sacks SH. Intrarenal synthesis of complement. Kidney Int. 2001;59:1227–1235. doi: 10.1046/j.1523-1755.2001.0590041227.x. [DOI] [PubMed] [Google Scholar]
- Zipfel PF, Edey M, Heinen S, Jozsi M, Richter H, Misselwitz J, Hoppe B, Routledge D, Strain L, Hughes AE, Goodship JA, Licht C, Goodship TH, Skerka C. Deletion of complement factor H-related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet. 2007;3:e41. doi: 10.1371/journal.pgen.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.