Abstract
Dihydrolipoamide dehydrogenase (DLDH) is a multifunctional oxidoreductase and is well known as an essential component of four mammalian mitochondrial multienzyme complexes: pyruvate dehydrogenase, α-ketoglutarate dehydrogenase, branched chain α-keto acid dehydrogenase, and the glycine cleavage system. However, existence of extracellular DLDH in mammals, if any, has not been clearly defined. The present article reports identification and biochemical characterization of serum DLDH. Proteomic analysis of rat serum using blue native polyacrylamide gel electrophoresis (BN-PAGE) and mass spectrometry peptide sequencing led to generation of 6 tryptic peptides in one band that matched to mitochondrial DLDH, indicating the existence of DLDH in rat serum. Measurement of enzymatic activity also indicated the existence of DLDH in human and mouse serum. Further biochemical analysis of rat serum DLDH revealed that this enzyme lacked diaphorase activity and could not be detected on Western blots probed with antibodies that recognized mitochondrial DLDH. Moreover, both ammonium sulfate fractioning and gel filtration of serum samples rendered a great loss in DLDH activity, indicating that the enzyme activity of this serum protein, unlike that of mitochondrial DLDH, is very labile. When DTT was supplemented in the buffer used for gel filtration, DLDH activity was found to be largely preserved; indicating that serum DLDH is susceptible to air-implicated inactivation. Results of the present study indicate that serum DLDH differs from mitochondrial DLDH in that it is a very labile enzyme.
Keywords: blue native polyacrylamide gel electrophoresis, dihydrolipoamide dehydrogenase, mitochondria, serum
1. Introduction
Dihydrolipoamide dehydrogenase (DLDH) is a multifunctional, flavin-dependent, pyridine nucleotide disulfide oxidoreductase [1]. It is an integral component of five multi-enzyme protein complexes, including pyruvate dehydrogenase complex, α-ketoglutarate dehydrogenase complex, branched chain amino acid dehydrogenase complex, glycine cleavage system, and acetoin dehydrogenase complex [2]. Among these complexes, the first three are often collectively called 2-oxo-acid or α-keto acid dehydrogenase complexes [3]. With the exception of the acetoin dehydrogenase complex that exists only in certain bacteria such as Klebsiella pneumonia [4], all the other complexes are present in mitochondria, wherein DLDH is an extremely stable homodimer [5, 6] that catalyzes the reoxidation of acyltransferase (E2)-linked dihydrolipoamide to lipoamide using NAD+ as the electron acceptor via a disulfide relay mechanism [7, 8]. In vitro, DLDH can also catalyze NADH-dependent reduction of free lipoamide, a reaction that is usually termed the reverse reaction [7]. Moreover, DLDH has diaphorase activity that catalyzes NADH-dependent reduction of a variety of electron acceptors such as 2,6-dichlorophenolindophenol (DCPIP), nitro blue tetrazolium (NBT) [9, 10], ubiquinone [11, 12], and nitric oxide (NO) [13]. Most importantly, when dysfunctional, DLDH stimulates overproduction of reactive oxygen species and thereby causes oxidative stress [14–16]. On the other hand, a functional DLDH can serve as a protective enzyme under oxidative stress conditions [17, 18]. Additionally, DLDH also possesses metal-binding properties in certain bacteria and plants [19, 20] and has DNA binding properties that can regulate protein synthesis [21–23]. More recently, it has been reported that DLDH inhibition via RNA interference can modulate the life span of C. elegans [24] and that DLDH in this organism is also involved in mediating cellular resistance to phosphine toxicity [25]. These findings demonstrate that DLDH is truly a multifunctional oxidoreductase.
Most organisms contain only one form of DLDH. Exceptions, however, do exist. For example, there are two forms of DLDH in E. coli [26, 27]. One form plays the classical role in the aforementioned enzyme complexes [27], while the other is involved in transportation of carbohydrates [26]. The two forms of DLDH mainly differ in their molecular weight. Pseudomonas putida is the only organism that contains three forms of DLDH [28–30]. Additionally, arabidopsis mitochondria contain two forms of DLDH that are interchangeable among the different enzyme complexes [31] and the human malaria parasite Plasmodium falciparum own two distinct DLDHs [32]. In contrast, in mammals all the DLDH contained in the above mentioned mitochondrial protein complexes are reportedly encoded by a same single gene and no additional forms of this protein have been definitively identified [33]. Although several reports have described the observations of DLDH isoforms in eukaryotes, the findings are most likely due to a conformational isomerism [34, 35] or an immunological isomerism [36].
Interestingly, it was reported in 1970’s and 1980’s that DLDH existed in human serum [37–39] wherein no 2-oxo-acid dehydrogenase complexes are present. Nonetheless, whether the biochemical property of serum DLDH is identical to that of mitochondrial DLDH is unknown. Moreover, in those earlier studies, enzyme activity was the only proof that DLDH exists in serum and no other biochemical characterization of this serum protein has been reported. In our proteomic studies of rat serum proteins using blue native gel electrophoresis and mass spectrometry peptide sequencing, we found that DLDH also exists in rat serum [40]. We now report herein our further characterization of this serum enzyme. Results of the present studies indicate that serum DLDH is a labile enzyme.
2. Materials and methods
2.1. Chemicals
Ammonium sulfate [(NH4)2SO4] and dithiothreitol (DTT) were purchased from Sigma. Ammonium persulfate, bis-acrylamide, acrylamide and Coomassie brilliant blue (CBB) G-250 were from Bio-Rad laboratories (Richmond, CA). Tricine and ε-amino-N-caproic acid were purchased from MP Biochemicals. Bis-Tris was from Calbiochem (La Jolla, CA) and serva Blue G-250 was from Serva (Heidelberg, Germany). Prestained SDS-PAGE markers and native PAGE markers were from Fermentas Life Sciences (Hanover, MD) and Invitrogen (Carlsbad, CA), respectively. PD-10 columns, Q Sepharose Fast Flow, SP Sepharose Fast Flow, ECL Western blotting detection reagents, and Hybond-C nitrocellulose membrane were purchased from GE Healthcare.
2.2. Serum preparation
The present studies primarily used adult Sprague-Dawley rats from Charles River Laboratories. All animal-related experiments were conducted in adherence with the NIH Guidelines for the Care and Use of Laboratory Animals and were approved by the University of North Texas Health Science Center Animal Care and Use Committee. Where mentioned, adult C57BL/6J mice purchased from the National Institute on Aging were used. For preparation of rat and mouse sera, blood samples were collected from the tail as previously described [41]. For preparation of human serum, blood was drawn from apparent healthy adult volunteers with prior consent. All blood samples were placed at room temperature for 30 min before centrifugation at 3,000 rpm on a benchtop centrifuge. The resulting supernatants containing sera were transferred to clean microtubes, snap-frozen in liquid nitrogen, and stored at −80°C before further analysis. All protein concentrations were determined by the bicinchoninic acid assay [42] using bovine serum albumin (BSA) as the standard. Rat serum was used throughout the studies unless otherwise specified.
2.3. Isolation of mitochondria
Rat liver or kidney mitochondria were prepared as previously described [43]. Briefly, one gram of liver was homogenized in 10 ml mitochondria isolation buffer containing 70 mM sucrose, 230 mM mannitol, 15 mM MOPS (pH 7.2), and 1 mM EDTA. The homogenate was centrifuged at 700 g for 10 min and the resulting supernatant was collected for further centrifugation at 8,000 g for another 10 min. The resulting pellet containing mitochondria was washed once with the homogenization buffer and centrifuged once again at 8,000 g for 10 min. All the centrifugation steps were performed at 4°C. The isolated mitochondria were either used immediately or stored at −80°C until analysis.
2.4. Polyacrylamide gel electrophoresis, in-gel diaphorase activity staining, and Western blot assay
Nongradient blue native polyacrylamide gel electrophoresis (BN-PAGE) was performed as previously described [6, 40]. For in-gel diaphorase activity staining, gel strips were incubated in 50 ml of 50 mM potassium phosphate buffer (pH 7.0) containing 0.2 mg/ml NBT and 0.1 mg/ml NADH [45]. SDS-PAGE was performed according to Laemmli [46] using a Bio-Rad Mini-PROTEAN III electrophoresis cell. Western blot assay was performed according to Towbin et al. [47] with slight modifications [48, 49]. All gel images were captured using an EPSON PERFECTION 1670 scanner.
2.5. Spectrophotometric determination of DLDH activities
Measurement of DLDH dehydrogenase activity was performed as previously described [45] with modifications. Typically, the dehydrogenase activity was measured by DLDH-catalyzed oxidation of dihydrolipoamide in the presence of NAD+. The final mixture (1 ml) contained 100 mM potassium phosphate, pH 7.8, 1.0 mM EDTA, 3.0 mM dihydrolipoamide, and 3 mM NAD+. A solution containing all the assay components except dihydrolipoamide was used as the blank. Following 15 min of pre-incubation to eliminate any potential interference effect of serum “nothing dehydrogenase” that is known to reduce NAD+ in the absence of any added substrates [37, 50, 51], the assay was then initiated by the addition of dihydrolipoamide and the increase in absorbance at 340 nm was followed at room temperature for 5 min. An extinction coefficient of 6.22 mM−1cm−1 for NADH was used for the calculation of enzyme activity. DLDH reverse activity, catalyzing the reduction of lipoamide by NADH, was also measured spectrophotometrically as previously described [52].
2.6. Spectrophotometric assay of serum diaphorase activity
Serum diaphorase activity was measured by a method previously described for the measurement of DLDH diaphorase activity [52]. Reduction of 2,6-dichlorophenolindophenol (DCPIP) by NADH was monitored by the decrease in absorbance at 600 nm. The assay (1 ml) contained 50 mM potassium phosphate, pH 7.2, 5 mg serum protein, 1 mM EDTA, 40 μM DCPIP and 0.2 mM NADH. The reaction was initiated by the addition of NADH.
2.7. Serum DLDH activity assay using 2-p-iodophenyl-3-p-nitrophenyl-5-phenyl tetrazolium chloride (INT) as the electron acceptor
This assay was adapted from a previously published method [44] that can be used to determine a specific dehydrogenase activity when a corresponding substrate is presented. The method involves electron transfer from NADH, generated by enzyme-dependent reduction of NAD+, to INT via phenazine methosulphate (PMS) as a coupler. In this so-called INT/PMS/NAD method, the final reaction volume was 1 ml in 30 mM Tris-HCl (pH 8.0) containing 0.02% Tween-20, 50 μl serum, 0.6 mg BSA, 20 μM INT, 8 μM PMS, and 4 μM NAD+. The mixture was incubated at room temperature for 10 min followed by addition of appropriate amount of dihydrolipoamide. The reaction mixture was then further incubated at room temperature for another 10 min and the reaction was terminated by addition of 200 μl 0.2 N hydrochloric acid. The absorbance was then measured at 505 nm with blank containing no dihydrolipoamide.
2.8. Mass spectrometric identification of proteins
Protein identification was carried out at ProtTech (Norristown, PA) using the NanoLC-MS/MS peptide sequencing technology. Briefly, a given blue native gel band was destained, cleaned, and in-gel digested with sequencing grade trypsin. The resulting peptide mixture was analyzed by an LC-MS/MS system, in which a high pressure liquid chromatography with a reverse phase C18 column (inner diameter: 75 μm) was coupled on-line with an ion trap mass spectrometer. The collected mass spectrometric data were used to search the most recent non-redundant protein database using ProtTech’s proprietary software suite.
2.9. Partial purification of mitochondrial- and serum DLDHs
Isolation and purification was carried out using conventional procedures, including (NH4)2SO4 fractionation and column chromatography. For partial purification of mitochondrial DLDH, mitochondria were isolated and used for (NH4)2SO4 fractioning. For serum samples, serum was diluted 10 × by 50 mM sodium phosphate (pH 7.4) and the diluted serum sample was used for (NH4)2SO4 fractioning. The procedures for (NH4)2SO4 fractioning of mitochondria and serum were essentially identical and were adapted from previously published DLDH isolation protocols [53–55]. The procedure started with the addition of solid (NH4)2SO4 to achieve 55% saturation. After incubation with gentle stirring at 4°C for 1 h and then centrifugation at 30,000 g for 30 min, the pellet was kept for enzyme analysis while the resulting supernatant was collected and was further brought to 75% saturation by addition of solid (NH4)2SO4. This was followed by incubation and centrifugation under the same conditions to obtain the 55%–75% saturation fraction. For liver tissue, this pellet contained most of mitochondrial DLDH and was then used for further purification by column chromatography as described below.
The protein pellet obtained after 75% (NH4)2SO4 saturation was dissolved in 50 mM HEPES buffer, pH 7.4 and residue (NH4)2SO4 was removed by dialysis against the same HEPES buffer. After concentrating, the sample was applied onto an ion-exchange column (Q Sepharose Fast Flow) that was pre-equilibrated with 100 ml of 50 mM HEPES buffer (pH 7.4). The column was eluted with step-gradient NaCl (50 mM increment/step) in 50 mM HEPES buffer. Fractions containing DLDH were collected and underwent a second cycle of (NH4)2SO4 fractioning. The pellet resulting from 75% saturation was dissolved in 50 mM HEPES buffer (pH 7.4), desalted and concentrated as described above. The concentrated sample was then applied onto a cation exchange column (SP Sepharose Fast Flow) that was pre-equilibrated with 100 ml of 50 mM HEPES buffer (pH 7.4). This was followed by elution of the column with step-gradient NaCl (20 mM increment/step) in 50 mM HEPES buffer. Fractions containing DLDH were collected and underwent a third cycle of (NH4)2SO4 fractioning. The final pellet obtained after 75% saturation was dissolved in 20 mM potassium phosphate buffer (pH 7.0), desalted and concentrated. The sample was then applied onto a hydroxyapatite column pre-equilibrated with potassium phosphate buffer (pH 7.0). The column was eluted with increasing concentration of potassium phosphate buffer (pH 7.0, 40 mM per increment). Fractions containing DLDH were collected and concentrated for further analysis.
3. Results
3.1. Mass spectrometric identification of serum DLDH
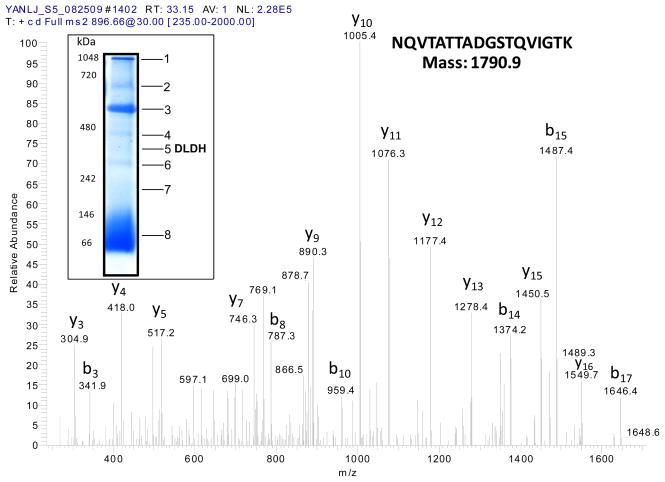
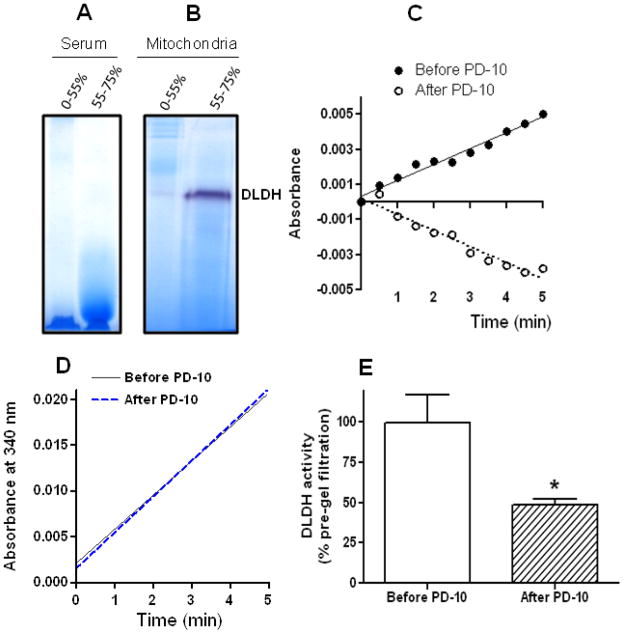
We previously noted briefly the identification of DLDH in rat serum in an unrelated study [40]. Herein we present the detailed results of this identification. In proteomic studies of rat serum proteins analyzed by BN-PAGE and mass spectrometry peptide sequencing, we found that one band contained DLDH [40]. Fig. 1 shows a mass spectrum demonstrating the identification of a serum DLDH peptide, whereby a typical blue native gel image indicating the localization of DLDH is also shown (band 5, the inset). A total of six tryptic peptides that matched to rat mitochondrial DLDH were identified (Table 1). These results provide proteomic evidence that DLDH is present in rat serum.
Fig. 1.
Mass spectrum derived from a tryptic peptide that matched to rat DLDH. The amino acid sequence of the peptide (NQVTATTADGSTQVIGTK) and the y and b ion series are shown on the spectrum. The inset shows BN-PAGE analysis of rat serum proteins and the localization of a DLDH-containing band, which was excised, destained, digested with trypsin, and analyzed by Nano-LC tandem mass spectrometry peptide sequencing technology.
Table 1.
Six peptides were generated from band 5 shown in Fig. 1 inset after trypsin digestion and mass spectrometry sequencing. Each of which matched to an internal peptide of rat DLDH. The flanking numbers on the second peptide denote, respectively, the starting and ending amino acid positions in the whole mitochondrial DLDH amino acid sequence.
| Peptide 1 | ALLNNSHYYHLAHGK 90-ALLNNSHYYHLAHGK -104 |
| Peptide 2 | ALTGGIAHLFK 133-ALTGGIAHLFK -143 |
| Peptide 3 | NQVTATTADGSTQVIGTK 160-NQVTATTADGSTQVIGTK -177 |
| Peptide 4 | LVVIGAGVIGVELGSVWQR 276-LVVIGAGVIGVELGSVWQR -294 |
| Peptide 5 | IPNIFAIGDVVAGPMLAHK 347-IPNIFAIGDVVAGPMLAHK -365 |
| Peptide 6 | SEEQLKEEGVEFK 405-SEEQLKEEGVEFK -417 |
3.2. Serum DLDH is not detectable by Western blot, but its dehydrogenase activity is
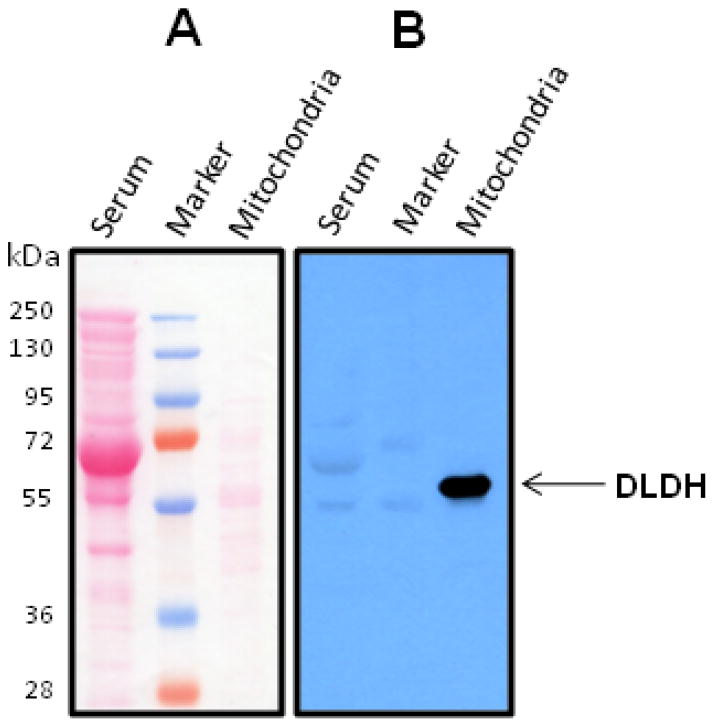
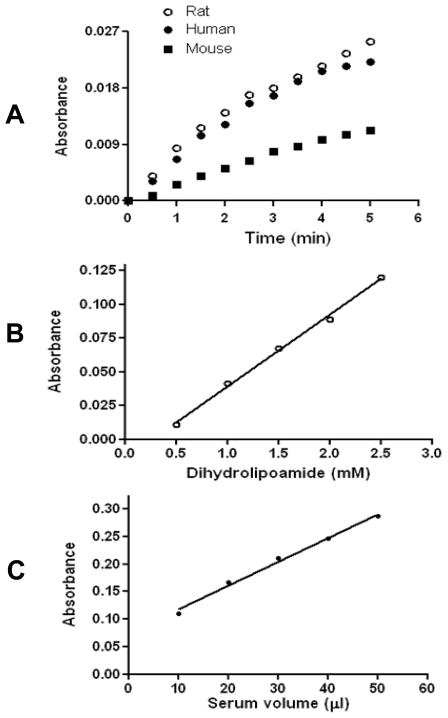
Interestingly, when we analyzed serum DLDH using Western blot assay, serum DLDH immunostaining was not detected (Fig. 2), though mitochondrial DLDH could be well-localized on the same blot (Fig. 2). The reason that the antibodies recognizing mitochondrial DLDH failed to detect serum DLDH remains unknown. When we measured serum DLDH dehydrogenase activity by spectrophotometric assay, we found that the activity could be measured under normal physiological conditions in all the three mammalian species including rat, human, and mouse (Fig. 3A), and the use of 5 mg serum protein gave a satisfactory result. Moreover, serum DLDH activity, as assayed by the INT/PMS/NAD method [44], was dependent on both substrate concentration (Fig. 3B) and serum volume (Fig. 3C). It should be noted that the dehydrogenase activity in serum was very low when compared with that in mitochondria. For example, under our experimental conditions, DLDH dehydrogenase activity in isolated mitochondria was 217 mUnit/mg mitochondrial protein (n=6), while this value in serum was 0.024 mUnit/mg serum protein (n=5). Such comparison suggests that DLDH is present in a very low abundance in serum. Nevertheless, these measurements are in agreement with previous reports that DLDH activity could be detected in human serum and the enzyme assay was generally protein concentration- and substrate concentration dependent [37, 39]. Additionally, the pronounced fall-off of the absorbance curves in Fig. 3A was likely due to product inhibition by NADH, as has been established for mitochondrial DLDH [56–59].
Fig. 2.
Western blot detection of serum and mitochondrial DLDH. A shows protein staining by Ponceau S after transfer to Hybond-C membrane (60 μg serum proteins and 10 μg mitochondrial proteins loaded, respectively); B shows Western blot detection of DLDH probed with anti-DLDH polyclonal antibodies. Arrow-indicated was the recognition of mitochondrial DLDH by the antibodies while no serum DLDH could be detected.
Fig. 3.
Serum DLDH dehydrogenase activity measured spectrophotometrically. (A) Analysis of serum samples derived from three mammalian species: rat, human, and mouse. The final assay mixture (1 ml) contained 5 mg serum protein, 3 mM dihydrolipoamide, 3 mM NAD+, 1.0 mM EDTA, 6 mg BSA, and 100 mM potassium phosphate (pH 8.0). The reaction was initiated by the addition of dihydrolipoamide and an absorbance increase at 340 nm was followed for 5 min for each sample. Serum DLDH activity shows dependence on both substrate concentration (B) and serum protein amount (C). For each point in B, 50 μl of serum was used; while for each point in C, 2.5 mM dihydrolipoamide was used. For both B and C, rat serum was used and enzyme activity was measured by the INT/PMS/NAD method [44] as described in the text with recording of absorbance at 505 nm.
3.3. Serum DLDH lacks diaphorase activity
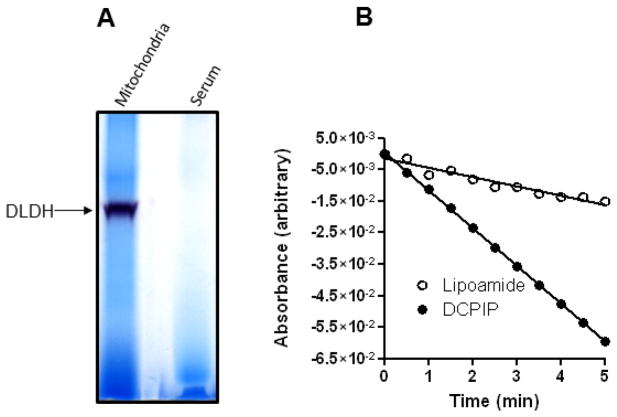
We next investigated whether serum DLDH diaphorase activity, if any, could be measured by an in-gel activity staining method that we previously described for the measurement of mitochondrial DLDH diaphorase [45]. As shown in Fig. 4A, no serum DLDH diaphorase activity could be detected while mitochondrial DLDH showed an intensive activity staining (the left lane). On the other hand, DLDH-catalyzed reverse reaction, in which lipoamide is reduced at the expense of NADH, was clearly detectable (Fig. 4B). Interestingly, there was a diaphorase activity in serum that could be detected spectrophotometrically by DCPIP in the presence of NADH (Fig. 4B). However, given the undetectable diaphorase activity in Fig. 4A, this diaphorase activity was likely contributed by other NADH-dependent enzymes in the serum [60, 61]. These results are in agreement with previous studies that serum DLDH lacks diaphorase activity [37, 39].
Fig. 4.
Analysis of serum DLDH diaphorase activity and reverse activity. (A) In-gel diaphorase activity staining of serum and mitochondrial DLDH. (B) Spectrophotometric assay of reverse activity using lipoamide (open circle) as the substrate and of diaphorase activity using DCPIP (filled circle) as the electron acceptor. NADH was used as the electron donor in both assays that contained 5 mg serum proteins.
3.4. Serum DLDH activity is lost after (NH4)2SO4 fractioning or gel filtration
When we attempted to purify serum DLDH by following its diaphorase activity determined by BN-PAGE, we found that no DLDH diaphorase activities could be detected in any of the (NH4)2SO4 fractions (Fig. 5A). This was very different from that of mitochondria (NH4)2SO4 fractionation whereby DLDH diaphorase activity could be clearly detected and majority of the activity was in the 55–75% saturation fraction (Fig. 5B). Therefore, even for concentrated and fractionated serum samples, no DLDH diaphorase activity was detectable. Moreover, no serum DLDH dehydrogenase activity could be detected spectrophotometrically in the (NH4)2SO4 fractions, indicating that serum DLDH activity was lost after (NH4)2SO4 fractioning. Likewise, no enzyme activity could be detected after serum samples underwent gel filtration on PD-10 columns (Fig. 5C). In contrast, there was no loss in mitochondrial DLDH activity after PD-10 gel filtration (Fig. 5D). To further confirm that there was indeed a loss in serum dehydrogenase activity after gel filtration, we also measured DLDH dehydrogenase activity in the gel-filtered samples by the INT/PMS/NAD method [44] in a separate experiment. Fig. 5E shows that there was more than 50% loss in enzyme activity after one-step gel filtration of serum samples, demonstrating that serum DLDH is indeed vulnerable to procedures of further isolation and purification, regardless of the methods used for measurement of enzyme activities. It should be noted that under our experimental conditions, the INT/PMS/NAD method appears to be more sensitive than the assay relying on direct measurement of NADH produced by dihydrolipoamide reduction of NAD+.
Fig. 5.
In-gel diaphorase activity staining of (A) serum and (B) liver mitochondrial DLDH following (NH4)2SO4 fractionation as well as (C) serum DLDH activity measured spectrophotometrically before and after gel filtration using PD-10 column. Note that 5 mg proteins derived from 2.5 ml of frozen serum samples were used in the activity assay both before and after gel filtration. (D) Mitochondrial DLDH activity measured in kidney mitochondrial samples before and after PD-10 gel filtration (20 μg proteins used in each assay). (E) Serum DLDH activity measured in serum samples before and after PD-10 gel filtration by the INT/PMS/NAD method in a separate experiment (p<0.05 vs. control, N = 3, Welch’s t test).
As a further comparison of the enzyme stability between serum and mitochondrial DLDH, the enzyme partially purified from rat liver mitochondria after many steps of (NH4)2SO4 fractioning and column chromatography (Fig. 6A) still remained active (Fig. 6B, control), and the activity could only be intentionally and significantly attenuated by treatment with oxidants such as peroxynitrite (Fig. 6B), a chemical that is known to inhibit enzyme activity due to its ability to induce protein carbonylation and S-nitrosylation [62, 63]. While DLDH can be inhibited by a variety of chemicals such as thiol alkylating reagents [64], heavy metals [65], valproic acid metabolites [66], diphenyleneiodonium [67], and 5-methoxyindole-2-carboxylic acid (MICA) [68–70], we chose peroxynitrite in this study because our long-term research interest is to understand the roles of DLDH oxidative modifications in disease and health [71–73]. Fig. 6B also shows that peroxynitrite might react with DLDH’s substrate dihydrolipoamide or the product NADH in the assay solution as the loss of enzyme activity in the presence of peroxynitrite alone was apparently over estimated when compared with that in the presence of GSH, a well-known peroxynitrite scavenger [74]. Nevertheless, loss of DLDH activity in the presence of GSH was still significant when compared with that of control, demonstrating an authentic action of peroxynitrite on DLDH.
Fig. 6.
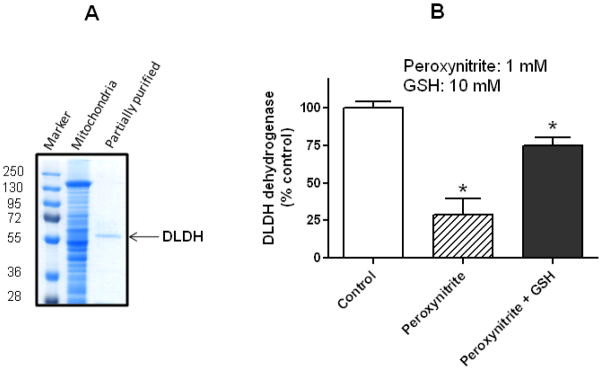
(A) Partial purification of DLDH from rat liver mitochondria assessed by SDS-PAGE, and (B) inhibition of its dehydrogenase activity by peroxynitrite. Unlike serum DLDH that lost its dehydrogenase activity after procedures of further isolation and purification, mitochondrial DLDH still remained active after the same procedures of isolation and purification, and the activity could only be purposely attenuated by incubation with peroxynitrite (1 mM). GSH (10 mM) was added after peroxynitrite incubation but before enzyme assay. *p<0.05 indicates statistical significance compared with control (Welch’s t test).
3.5. Serum DLDH is sensitive to air-inactivation
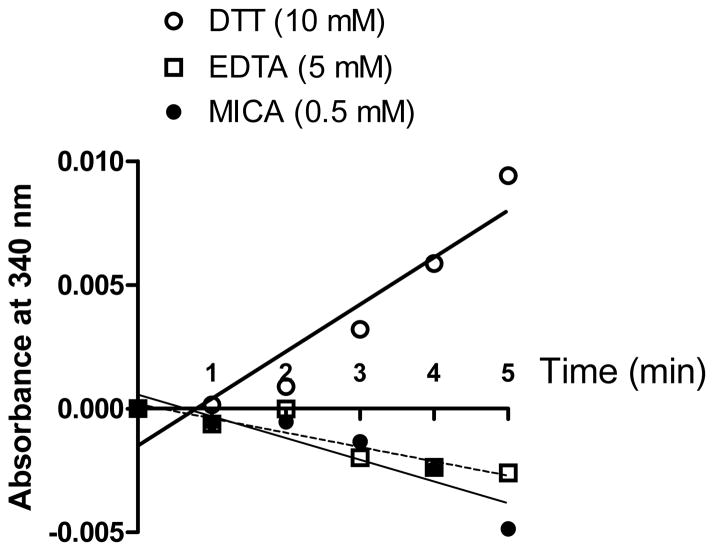
The loss of DLDH enzyme activity after gel filtration or (NH4)2SO4 precipitation is intriguing. Given the fact that xanthine dehydrogenase is very sensitive to oxygen attack and can be converted to xanthine oxidase after exposure to atmospheric oxygen during isolation and purification [75, 76], we reasoned that the loss of DLDH activity after (NH4)2SO4 precipitation and gel filtration might also be caused by atmospheric oxygen exposure. To test this possibility, we attempted to prevent oxygen exposure by supplementing DTT (final concentration: 10 mM) in both the serum and the gel filtration buffer (50 mM Tris-HCl, pH 7.4) that was used for column equilibration and elution. Based on previous findings that EDTA protects hydrogenase against air-inactivation via iron chelation [77] and that fluorocitrate, an aconitase inhibitor, protects aconitase against air-inactivation [78] via physical binding during isolation of each respective enzymes, we also tested the effects of EDTA (5 mM) and DLDH inhibitor MICA (0.5 mM) [68–70] on DLDH activity after gel filtration. Results in Fig. 7 show that while the presence of either EDTA or MICA in the equilibration and elution buffer did not prevent loss of DLDH activity after gel filtration, the presence of DTT in the same buffer largely preserved DLDH activity that, otherwise, would be lost after gel filtration, demonstrating that serum DLDH is sensitive to atmospheric oxygen inactivation. These results also indicate that, during gel filtration, binding of MICA to DLDH has no protective effect on air-inactivation of the enzyme, and that DLDH inactivation is not due to any presence of iron that might be potentially released from blood during preparation of serum samples. It should be noted that the protective effect of DTT could not last for more than 60 min after gel filtration, probably because DTT also underwent auto-oxidation by ambient oxygen (data not shown).
Fig. 7.
DLDH activity could be largely preserved by DTT (10 mM), but not by EDTA (5 mM) or its inhibitor MICA (0.5 mM) during PD-10 gel filtration of serum samples when these chemicals were supplemented in both the serum and the gel filtration buffer (50 mM Tris-HCl, pH 7.4). Shown are enzyme activities measured after PD-10 gel filtration.
4. Discussion
The major findings of the present study are that both the forward and the reverse activities of DLDH can be readily lost after (NH4)2SO4 fractioning or gel filtration of serum samples and such loss of activity is likely caused by exposure to atmospheric oxygen during isolation. Results of the present study demonstrate that serum DLDH is a very labile enzyme.
DLDH has previously been considered as a serological diagnostic in metabolic disease [37], although attempts to use this serum protein as a biomarker for Fredreich’s ataxia [38, 79, 80] proved to be controversial [39], perhaps because the serum DLDH protein was not clearly identified or well characterized. The present study provides the first peptide sequencing evidence that DLDH exists in rat serum (Fig. 1 and Table 1), and spectrophotometrically confirms previous reports that human serum contains DLDH (Fig. 3). Measurement of enzyme activity by spectrophotometric assay also indicates the existence of DLDH in mouse serum. Interestingly, our results indicate that serum DLDH is a very labile enzyme. This is in contrary to mitochondrial DLDH that is highly stable and makes it impossible to follow the enzyme’s activity during isolation and purification. Hence our attempt to purify enough of this serum enzyme for further biochemical characterization failed.
It might be argued that the reason that serum DLDH activity is undetectable after (NH4)2SO4 precipitation or native gel electrophoresis is because this enzyme has an extremely low content in the serum. However, the enzyme activity was certainly detectable in the original serum, whereby the volume of which was normalized to that after PD-10 gel filtration so that the enzyme activity could be compared on a same volume basis before and after PD-10 columns. Despite this normalization, there was a dramatic loss of activity after PD-10 gel filtration (Fig. 5, C and E) while mitochondrial DLDH treated similarly fully retained its activity (Fig. 5D). These lines of evidence led us to conclude that serum DLDH is a labile enzyme. The possibility for loss of DLDH activity could be due to the fact that serum has a reducing environment because of the existence of albumin and glutathione [81] and this reducing environment is perturbed during gel filtration, which makes DLDH vulnerable to oxygen exposure. This reasoning would be in agreement with our findings that DTT protected DLDH against air-inactivation when supplemented in serum and in the gel filtration buffer (Fig. 7), although the mechanisms underlying DLDH’s vulnerability to air-inactivation remains unknown at this point. Interestingly, in contrast to that of DTT, we did not observe any protection from EDTA supplemented in both the gel-filtration buffer and the serum, suggesting that EDTA-chelatable metals such as iron or zinc are not involved in DLDH inactivation during the gel filtration process. Nonetheless, our results do not exclude the possibility that non-EDAT chelatable metals are involved in DLDH inactivation during isolation and purification under our experimental conditions.
Another possibility for loss of DLDH activity during isolation and purification could be due to H2O2 generated by DLDH itself. This is possible because DLDH is known to be capable of producing H2O2 under appropriate conditions [15, 82–85]. Additionally, loss of DLDH activity could also be caused by conditions or disruptions favoring the formation of DLDH monomeric state [86] or 4-electron-reduced state [87] that exhibit a higher or only diaphorase activity but a lower or none dehydrogenase activity. Further, it is also known that DLDH in diluted solution is not as stable as that in mitochondria [86] and DLDH monomer actually exhibits protease properties [88]. These findings may render DLDH behavior in serum less predictable. In any case, the exact mechanisms by which DLDH loses its activity during isolation and purification needs further investigations. Nonetheless, we speculate that cysteine oxidation on DLDH could be a reason for the loss of DLDH activity as cysteine residues are redox sensitive [89] and DLDH cysteines are known to be susceptible to oxidative modifications [45, 71–73].
The origin of serum DLDH remains unknown. It has been suggested that liver could be the source of serum DLDH [37]. However, there have been no reports that DLDH isoforms exist in liver. If serum DLDH indeed originates from liver mitochondria, then it would be very difficult to explain the loss of serum DLDH activity following isolation or purification procedures. This is because mitochondrial DLDH is extremely stable, as shown in this and previous studies [6]. On the other hand, if serum DLDH results from cell destruction in tissues of origin, it would also be difficult to explain why only DLDH, other than other mitochondrial proteins, is detectable when all the visible BN-PAGE bands were sequenced [40]. All these results indicate that it is unlikely that serum samples used in our studies were contaminated by mitochondria. Additionally, mitochondrial DLDH could still remain active after heating at 75°C for 5 min [90], a property that has been repeatedly used as a key step in isolation and purification of mitochondrial DLDH [5]. In contrast, as reported in this study, serum DLDH is very labile and any further manipulation procedures would lead to severe loss of enzyme activity. Given the establishment that many mitochondrial proteins, including superoxide dismutase [91], fumerase [92], glutathione S-transferase [93], aconitase [94, 95] and plant glutathione reductase [96], have been found to also localize outside mitochondria, a phenomenon termed as dual localization of echoforms of mitochondrial proteins [92, 97–99], we speculate that serum DLDH is secreted into the circulation system and could be another mitochondrial protein that is dual-localized. Interestingly, it has been reported that mitochondrial DLDH amino acid composition possesses dual localization properties [100] and DLDH can exist extra-mitochondrially in mammalian spermatozoa [101]. Regardless, whether or not DLDH is a dual localized protein also awaits further studies.
Finally, the biological function of serum DLDH remains obscure at this time. However, we speculate that this serum protein could be involved in maintaining a proper redox balance between lipoamide and dihydrolipoamide, or between lipoic acid and dihydrolipoic acid. Both pairs of these small molecules are known to exist in the blood [102–104]. In this sense, serum DLDH is distinct from its classical role in the 2-oxo acid dehydrogenase complexes. In fact, DLDH is known to be involved in antioxidant defense systems [12, 85, 105], and can exist as a non-complexed form in mammalian tissues [106], and has also been found to be expressed in organisms that do not have 2-oxo-acid dehydrogenase complexes [107–110]. All these studies further indicate that DLDH has other biological functions.
5. Conclusion
We provided proteomic evidence that DLDH exists in serum and found that while serum DLDH also catalyzes the physiological reaction of dihydrolipoamide reoxidation using NAD+ as the electron acceptor; it differs from mitochondrial DLDH in several aspects. These include loss of activity after further procedures of isolation or purification due to its susceptibility to atmospheric oxygen inactivation, lack of diaphorase activity, and failure to be recognized by antibodies that recognize mitochondrial DLDH. These distinct properties of serum DLDH raise the question of whether this serum protein could be a form that is different from mitochondrial DLDH. Future studies will be needed to determine the biological function of this serum protein and the mechanisms underlying its susceptibility to air-implicated inactivation. Additionally, whether or not this serum enzyme could be an isoform or an echoform [92, 97, 98] of mitochondrial DLDH will also need to be investigated.
Acknowledgments
This work was supported in part by a UNTHSC-UAEM research seed grant (RI6044) and a National Institute on Aging grant (AG022550). The authors thank Dr. Drake Zhang and Ms. Kerri Jin at ProtTech for their assistance in mass spectrometry peptide sequencing.
Abbreviations
- BN-PAGE
blue native polyacrylamide gel electrophoresis
- DCPIP
2,6-dichlorophenolindophenol
- DLDH
dihydrolipoamide dehydrogenase
- INT
2-p-iodophenyl-3-p-nitrophenyl-5-phenyl tetrazolium chloride
- NADH
nicotinamide adenine dinucleotide (reduced form)
- NBT
nitro blue tetrazolium
- PMS
phenazine methosulphate
Footnotes
Conflict of interest: None declared.
References
- 1.Williams CH., Jr . Lipoamide dehydrogenase, glutathione reductase, thioredoxin reductase, and mercuric ion reductase-a family of flavoenzyme transhydrogenases. In: Muller F, editor. Chemistry and Biochemistry of Flavoenzymes. III. CRC Press; Boca Raton: 1992. pp. 121–212. [Google Scholar]
- 2.Batista AP, Kletzin A, Pereira MM. The dihydrolipoamide dehydrogenase from the crenarchaeon Acidianus ambivalens. FEMS Microbiol Lett. 2008;281:147–154. doi: 10.1111/j.1574-6968.2008.01082.x. [DOI] [PubMed] [Google Scholar]
- 3.Yeaman SJ. The 2-oxo acid dehydrogenase complexes: recent advances. Biochem J. 1989;257:625–632. doi: 10.1042/bj2570625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng HL, Deng WL, Yang YH, Chang HY. Identification and characterization of the acoD gene encoding a dihydrolipoamide dehydrogenase of the Klebsiella pneumoniae acetoin dehydrogenase system. J Biochem. 1996;119:1118–1123. doi: 10.1093/oxfordjournals.jbchem.a021357. [DOI] [PubMed] [Google Scholar]
- 5.Williams CH., Jr Studies on lipoyl dehydrogenase from Escherichia coli. J Biol Chem. 1965;240:4793–4800. [PubMed] [Google Scholar]
- 6.Yan LJ, Forster MJ. Resolving mitochondrial protein complexes using nongradient blue native polyacrylamide gel electrophoresis. Anal Biochem. 2009;389:143–149. doi: 10.1016/j.ab.2009.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carothers DJ, Pons G, Patel MS. Dihydrolipoamide dehydrogenase: functional similarities and divergent evolution of the pyridine nucleotide-disulfide oxidoreductases. Arch Biochem Biophys. 1989;268:409–425. doi: 10.1016/0003-9861(89)90309-3. [DOI] [PubMed] [Google Scholar]
- 8.de Kok A, van Berkel WJH. Lipoamide dehydrogenase. In: Patel MS, Roche TE, Harris RA, editors. Alpha-keto acid dehydrogenase complexes. Birkhauser Verlag; Berlin: 1996. pp. 53–70. [Google Scholar]
- 9.Scouten WH, McManus IR. Microbial lipoamide dehydrogenase. Purification and some characteristics of the enzyme derived from selected microorganisms. Biochim Biophys Acta. 1971;227:248–263. doi: 10.1016/0005-2744(71)90058-1. [DOI] [PubMed] [Google Scholar]
- 10.Sokatch JR, McCully V, Gebrosky J, Sokatch DJ. Isolation of a specific lipoamide dehydrogenase for a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981;148:639–646. doi: 10.1128/jb.148.2.639-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsson JM, Xia L, Eriksson LC, Bjornstedt M. Ubiquinone is reduced by lipoamide dehydrogenase and this reaction is potently stimulated by zinc. FEBS Lett. 1999;448:190–192. doi: 10.1016/s0014-5793(99)00363-4. [DOI] [PubMed] [Google Scholar]
- 12.Xia L, Bjornstedt M, Nordman T, Eriksson LC, Olsson JM. Reduction of ubiquinone by lipoamide dehydrogenase. An antioxidant regenerating pathway. Eur J Biochem. 2001;268:1486–1490. doi: 10.1046/j.1432-1327.2001.02013.x. [DOI] [PubMed] [Google Scholar]
- 13.Igamberdiev AU, Bykova NV, Ens W, Hill RD. Dihydrolipoamide dehydrogenase from porcine heart catalyzes NADH-dependent scavenging of nitric oxide. FEBS Lett. 2004;568:146–150. doi: 10.1016/j.febslet.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 14.Klivenyi P, Starkov AA, Calingasan NY, Gardian G, Browne SE, Yang L, Bubber P, Gibson GE, Patel MS, Beal MF. Mice deficient in dihydrolipoamide dehydrogenase show increased vulnerability to MPTP, malonate and 3-nitropropionic acid neurotoxicity. J Neurochem. 2004;88:1352–1360. doi: 10.1046/j.1471-4159.2003.02263.x. [DOI] [PubMed] [Google Scholar]
- 15.Ambrus A, Torocsik B, Tretter L, Ozohanics O, Adam-Vizi V. Stimulation of reactive oxygen species generation by disease-causing mutations of lipoamide dehydrogenase. Hum Mol Genet. 2011;20:2984–2995. doi: 10.1093/hmg/ddr202. [DOI] [PubMed] [Google Scholar]
- 16.Vaubel RA, Rustin P, Isaya G. Mutations in the Dimer Interface of Dihydrolipoamide Dehydrogenase Promote Site-specific Oxidative Damages in Yeast and Human Cells. J Biol Chem. 2011;286:40232–40245. doi: 10.1074/jbc.M111.274415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korotchkina LG, Yang H, Tirosh O, Packer L, Patel MS. Protection by thiols of the mitochondrial complexes from 4-hydroxy-2-nonenal. Free Radic Biol Med. 2001;30:992–999. doi: 10.1016/s0891-5849(01)00491-9. [DOI] [PubMed] [Google Scholar]
- 18.Chen HJ, Chen YM, Chang CM. Lipoyl dehydrogenase catalyzes reduction of nitrated DNA and protein adducts using dihydrolipoic acid or ubiquinol as the cofactor. Chem Biol Interact. 2002;140:199–213. doi: 10.1016/s0009-2797(02)00019-4. [DOI] [PubMed] [Google Scholar]
- 19.Siegmann A, Komarska A, Betzalel Y, Brudo I, Jindou S, Mor G, Fleminger G. The titanium binding protein of Rhodococcus ruber GIN1 (NCIMB 40340) is a cell-surface homolog of the cytosolic enzyme dihydrolipoamide dehydrogenase. J Mol Recognit. 2009;22:138–145. doi: 10.1002/jmr.919. [DOI] [PubMed] [Google Scholar]
- 20.Tan YF, O’Toole N, Taylor NL, Millar AH. Divalent metal ions in plant mitochondria and their role in interactions with proteins and oxidative stress-induced damage to respiratory function. Plant Physiol. 2010;152:747–761. doi: 10.1104/pp.109.147942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schierer T, Henderson E. A protein from Tetrahymena thermophila that specifically binds parallel-stranded G4-DNA. Biochemistry. 1994;33:2240–2246. doi: 10.1021/bi00174a034. [DOI] [PubMed] [Google Scholar]
- 22.Kee K, Niu L, Henderson E. A Tetrahymena thermophila G4-DNA binding protein with dihydrolipoamide dehydrogenase activity. Biochemistry. 1998;37:4224–4234. doi: 10.1021/bi9716377. [DOI] [PubMed] [Google Scholar]
- 23.Ajith VK, Prasad R. A novel protein that binds to dnrN-dnrO intergenic region of Streptomyces peucetius purified by DNA affinity capture has dihydrolipoamide dehydrogenase activity. Protein Expr Purif. 2009;67:132–138. doi: 10.1016/j.pep.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Butler JA, Mishur RJ, Bhaskaran S, Rea SL. A metabolic signature for long-life in the c. Elegans mit mutants. Aging Cell. 2013;12:130–138. doi: 10.1111/acel.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlipalius DI, Valmas N, Tuck AG, Jagadeesan R, Ma L, Kaur R, Goldinger A, Anderson C, Kuang J, Zuryn S, Mau YS, Cheng Q, Collins PJ, Nayak MK, Schirra HJ, Hilliard MA, Ebert PR. A core metabolic enzyme mediates resistance to phosphine gas. Science. 2012;338:807–810. doi: 10.1126/science.1224951. [DOI] [PubMed] [Google Scholar]
- 26.Richarme G, Heine HG. Galactose- and maltose-stimulated lipoamide dehydrogenase activities related to the binding-protein-dependent transport of galactose and maltose in toluenized cells of Escherichia coli. Eur J Biochem. 1986;156:399–405. doi: 10.1111/j.1432-1033.1986.tb09596.x. [DOI] [PubMed] [Google Scholar]
- 27.Stephens PE, Lewis HM, Darlison MG, Guest JR. Nucleotide sequence of the lipoamide dehydrogenase gene of Escherichia coli K12. Eur J Biochem. 1983;135:519–527. doi: 10.1111/j.1432-1033.1983.tb07683.x. [DOI] [PubMed] [Google Scholar]
- 28.Burns G, Sykes PJ, Hatter K, Sokatch JR. Isolation of a third lipoamide dehydrogenase from Pseudomonas putida. J Bacteriol. 1989;171:665–668. doi: 10.1128/jb.171.2.665-668.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burns G, Brown T, Hatter K, Sokatch JR. Sequence analysis of the lpdV gene for lipoamide dehydrogenase of branched-chain-oxoacid dehydrogenase of Pseudomonas putida. Eur J Biochem. 1989;179:61–69. doi: 10.1111/j.1432-1033.1989.tb14521.x. [DOI] [PubMed] [Google Scholar]
- 30.Palmer JA, Hatter K, Sokatch JR. Cloning and sequence analysis of the LPD-glc structural gene of Pseudomonas putida. J Bacteriol. 1991;173:3109–3116. doi: 10.1128/jb.173.10.3109-3116.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lutziger I, Oliver DJ. Characterization of two cDNAs encoding mitochondrial lipoamide dehydrogenase from Arabidopsis. Plant Physiol. 2001;127:615–623. [PMC free article] [PubMed] [Google Scholar]
- 32.McMillan PJ, Stimmler LM, Foth BJ, McFadden GI, Muller S. The human malaria parasite Plasmodium falciparum possesses two distinct dihydrolipoamide dehydrogenases. Mol Microbiol. 2005;55:27–38. doi: 10.1111/j.1365-2958.2004.04398.x. [DOI] [PubMed] [Google Scholar]
- 33.Vettakkorumakankav NN, Patel MS. Dihydrolipoamide dehydrogenase: structural and mechanistic aspects. Indian J Biochem Biophys. 1996;33:168–176. [PubMed] [Google Scholar]
- 34.Kenney WC, Zakim D, Hogue PK, Singer TP. Multiplicity and origin of isoenzymes of lipoyl dehydrogenase. Eur J Biochem. 1972;28:253–260. doi: 10.1111/j.1432-1033.1972.tb01908.x. [DOI] [PubMed] [Google Scholar]
- 35.Freudenberg W, Dietrichs D, Lebertz H, Andreesen JR. Isolation of an atypically small lipoamide dehydrogenase involved in the glycine decarboxylase complex from Eubacterium acidaminophilum. J Bacteriol. 1989;171:1346–1354. doi: 10.1128/jb.171.3.1346-1354.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carothers DJ, Raefsky-Estrin C, Pons G, Patel MS. Rat liver mitochondria contain two immunologically distinct dihydrolipoamide dehydrogenases. Arch Biochem Biophys. 1987;256:597–605. doi: 10.1016/0003-9861(87)90617-5. [DOI] [PubMed] [Google Scholar]
- 37.Pelley JW, Little GH, Linn TC, Hall FF. Lipoamide dehydrogenase in serum: a preliminary report. Clin Chem. 1976;22:275–277. [PubMed] [Google Scholar]
- 38.Melancon SB, Fontaine G, Geoffroy G, Vanasse M, Dallaire L, Potier M. Correlation between serum lipoamide dehydrogenase activity and phosphatidylcholine therapy in Friedreich’s ataxia. Can J Neurol Sci. 1980;7:413–416. doi: 10.1017/s0317167100022976. [DOI] [PubMed] [Google Scholar]
- 39.Blass JP, Hinman L, Soricelli A. Clinical assays of lipoamide dehydrogenase. Neurology. 1981;31:783–785. doi: 10.1212/wnl.31.6.783. [DOI] [PubMed] [Google Scholar]
- 40.Thangthaeng N, Sumien N, Forster MJ, Shah RA, Yan LJ. Nongradient blue native gel analysis of serum proteins and in-gel detection of serum esterase activities. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879:386–394. doi: 10.1016/j.jchromb.2010.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoff J. Methods of blood collection in the mouse. Lab Animal. 2000;29:47–53. [Google Scholar]
- 42.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 43.Navarro A, Boveris A. Rat brain and liver mitochondria develop oxidative stress and lose enzymatic activities on aging. Am J Physiol Regul Integr Comp Physiol. 2004;287:R1244–1249. doi: 10.1152/ajpregu.00226.2004. [DOI] [PubMed] [Google Scholar]
- 44.Rutkowski RB, Kaji T. Dehydrogenase activities in human and mouse serum using an ultramicro colorimetric analytical system. Int J Biochem. 1971;2:131–136. [Google Scholar]
- 45.Yan LJ, Yang SH, Shu H, Prokai L, Forster MJ. Histochemical staining and quantification of dihydrolipoamide dehydrogenase diaphorase activity using blue native PAGE. Electrophoresis. 2007;28:1036–1045. doi: 10.1002/elps.200600574. [DOI] [PubMed] [Google Scholar]
- 46.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 47.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan LJ, Sohal RS. Mitochondrial adenine nucleotide translocase is modified oxidatively during aging. Proc Natl Acad Sci USA. 1998;95:12896–12901. doi: 10.1073/pnas.95.22.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan LJ, Orr WC, Sohal RS. Identification of oxidized proteins based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunochemical detection, isoelectric focusing, and microsequencing. Anal Biochem. 1998;263:67–71. doi: 10.1006/abio.1998.2799. [DOI] [PubMed] [Google Scholar]
- 50.Mezey E, Slater KC, Holt PR. The source of the nothing dehydrogenase activity in normal serum. Clin Chim Acta. 1969;25:11–16. doi: 10.1016/0009-8981(69)90219-8. [DOI] [PubMed] [Google Scholar]
- 51.Kato S, Ishii H, Kano S, Hagihara S, Todoroki T, Nagata S, Takahashi H, Nagasaka M, Sato J, Tsuchiya M. Improved assay for alcohol dehydrogenase activity in serum by centrifugal analysis. Clin Chem. 1984;30:1817–1820. [PubMed] [Google Scholar]
- 52.Patel MS, Vettakkorumakankav NN, Liu TC. Dihydrolipoamide dehydrogenase: activity assays. Methods Enzymol. 1995;252:186–195. doi: 10.1016/0076-6879(95)52022-8. [DOI] [PubMed] [Google Scholar]
- 53.Millard SA, Kubose A, Gal EM. Brain lipoyl dehydrogenase. Purification, properties, and inhibitors. J Biol Chem. 1969;244:2511–2515. [PubMed] [Google Scholar]
- 54.Ide S, Hayakawa T, Okabe K, Koike M. Lipoamide dehydrogenase from human liver. J Biol Chem. 1967;242:54–60. [PubMed] [Google Scholar]
- 55.Koike M, Hayakawa T. Purification and properties of lipoamide dehydrogenases from pig heart alpha-keto acid dehydrogenase complexes. Methods Enzymol. 1970;18:298–307. [Google Scholar]
- 56.Massey V, Gibson QH, Veeger C. Intermediates in the catalytic action of lipoyl dehydrogenase (diaphorase) Biochem J. 1960;77:341–351. doi: 10.1042/bj0770341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reed JK. Studies on the kinetic mechanism of lipoamide dehydrogenase from rat liver mitochondria. J Biol Chem. 1973;248:4834–4839. [PubMed] [Google Scholar]
- 58.Hansen RG, Henning U. Regulation of pyruvate dehydrogenase activity in Escherichia Coli K12. Biochim Biophys Acta. 1966;122:355–358. doi: 10.1016/0926-6593(66)90076-2. [DOI] [PubMed] [Google Scholar]
- 59.Wagenknecht T, Francis N, DeRosier D. Role of excess lipoyl dehydrogenase in reconstituted alpha-ketoglutarate dehydrogenase complex of Escherichia coli. Biochem Biophys Res Commun. 1986;135:802–807. doi: 10.1016/0006-291x(86)90999-x. [DOI] [PubMed] [Google Scholar]
- 60.Friedlos F, Knox RJ. Metabolism of NAD(P)H by blood components. Relevance to bioreductively activated prodrugs in a targeted enzyme therapy system. Biochem Pharmacol. 1992;44:631–635. doi: 10.1016/0006-2952(92)90396-z. [DOI] [PubMed] [Google Scholar]
- 61.Kitamura S, Terada A, Kamio H, Sugihara K, Koga N, Ohta S. DT-diaphorase-like quinone reductase in rat plasma. Biol Pharm Bull. 1999;22:883–885. doi: 10.1248/bpb.22.883. [DOI] [PubMed] [Google Scholar]
- 62.Singh IN, Sullivan PG, Hall ED. Peroxynitrite-mediated oxidative damage to brain mitochondria: Protective effects of peroxynitrite scavengers. J Neurosci Res. 2007;85:2216–2223. doi: 10.1002/jnr.21360. [DOI] [PubMed] [Google Scholar]
- 63.Chinta SJ, Andersen JK. Nitrosylation and nitration of mitochondrial complex I in Parkinson’s disease. Free Radic Res. 2011;45:53–58. doi: 10.3109/10715762.2010.509398. [DOI] [PubMed] [Google Scholar]
- 64.Nakamura M, Yamazaki I. One-electron transfer reactions in biochemical systems. VI. Changes in electron transfer mechanism of lipoamide dehydrogenase by modification of sulfhydryl groups. Biochim Biophys Acta. 1972;267:249–257. doi: 10.1016/0005-2728(72)90113-2. [DOI] [PubMed] [Google Scholar]
- 65.Akbar SM, Jayalakshmi SK, Sharma HC, Sreeramulu K. Characterization of dihydrolipoamide dehydrogenase from the mitochondria of Helicoverpa armigera, a pest resistant to insecticides. Entomological Research. 2011;41:221–228. [Google Scholar]
- 66.Luis PB, Ruiter JP, Aires CC, Soveral G, Tavares de Almeida I, Duran M, Wanders RJ, Silva MF. Valproic acid metabolites inhibit dihydrolipoyl dehydrogenase activity leading to impaired 2-oxoglutarate-driven oxidative phosphorylation. Biochim Biophys Acta. 2007;1767:1126–1133. doi: 10.1016/j.bbabio.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 67.Stibingerova A, Velvarska H, Kynclova K, Marounkova B, Spundova M, Entlicher G. Lipoamide dehydrogenase and diaphorase catalyzed conversion of some NO donors to NO and reduction of NO scavenger 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (PTIO) Gen Physiol Biophys. 2009;28:384–390. doi: 10.4149/gpb_2009_04_384. [DOI] [PubMed] [Google Scholar]
- 68.Hanson RL, Ray PD, Walter P, Lardy HA. Mode of action of hypoglycemic agents. I. Inhibition of gluconeogenesis by quinaldic acid and 5-methoxyindole-2-carboxylic acid. J Biol Chem. 1969;244:4351–4359. [PubMed] [Google Scholar]
- 69.Bauman N, Hill CJ. Inhibition of gluconeogenesis and alpha-keto oxidation by 5-methoxyindole-2-carboxylic acid. Biochemistry. 1968;7:1322–1327. doi: 10.1021/bi00844a011. [DOI] [PubMed] [Google Scholar]
- 70.Haramaki N, Han D, Handelman GJ, Tritschler HJ, Packer L. Cytosolic and mitochondrial systems for NADH- and NADPH-dependent reduction of alpha-lipoic acid. Free Radic Biol Med. 1997;22:535–542. doi: 10.1016/s0891-5849(96)00400-5. [DOI] [PubMed] [Google Scholar]
- 71.Yan LJ, Thangthaeng N, Forster MJ. Changes in dihydrolipoamide dehydrogenase expression and activity during postnatal development and aging in the rat brain. Mech Ageing Dev. 2008;129:282–290. doi: 10.1016/j.mad.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan LJ, Liu L, Forster MJ. Reversible inactivation of dihydrolipoamide dehydrogenase by Angeli’s salt. Acta Biophysica Sinica (Sheng Wu Wu Li Hsueh Bao) 2012;28:341–350. [PMC free article] [PubMed] [Google Scholar]
- 73.Yan LJ, Sumien N, Thangthaeng N, Forster MJ. Reversible inactivation of dihydrolipoamide dehydrogenase by mitochondrial hydrogen peroxide. Free Radic Res. 2013;47:123–133. doi: 10.3109/10715762.2012.752078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bajt ML, Knight TR, Farhood A, Jaeschke H. Scavenging peroxynitrite with glutathione promotes regeneration and enhances survival during acetaminophen-induced liver injury in mice. J Pharmacol Exp Ther. 2003;307:67–73. doi: 10.1124/jpet.103.052506. [DOI] [PubMed] [Google Scholar]
- 75.Sanders SA, Massey V. The thermodynamics of xanthine oxidoreductase catalysis. Antioxid Redox Signal. 1999;1:371–379. doi: 10.1089/ars.1999.1.3-371. [DOI] [PubMed] [Google Scholar]
- 76.Nishino T, Okamoto K, Eger BT, Pai EF. Mammalian xanthine oxidoreductase - mechanism of transition from xanthine dehydrogenase to xanthine oxidase. Febs J. 2008;275:3278–3289. doi: 10.1111/j.1742-4658.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- 77.Klibanov AM, Kaplan NO, Kamen MD. Chelating agents protect hydrogenase against oxygen inactivation. Biochim Biophys Acta. 1979;547:411–416. doi: 10.1016/0005-2728(79)90021-5. [DOI] [PubMed] [Google Scholar]
- 78.Hattori Y, Hino S. Inactivation by oxygen and satbilization by fluorocitrate of aconitase from Escherichia Coli. J. Gen. Appl Microbiol. 1981;27:33–41. [Google Scholar]
- 79.Purkiss P, Baraitser M, Borud O, Chalmers RA. Biochemical and clinical studies of Friedreich’s ataxia. J Neurol Neurosurg Psychiatry. 1981;44:574–582. doi: 10.1136/jnnp.44.7.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pelley JW, Bradley CA. Lipoamide Dehydrogenase in Serum: A Potential Diagnostic Tool. Annals of the New York Academy of Sciences. 1989;573:404–404. [Google Scholar]
- 81.Chan ED, Riches DW, White CW. Redox paradox: effect of N-acetylcysteine and serum on oxidation reduction-sensitive mitogen-activated protein kinase signaling pathways. Am J Respir Cell Mol Biol. 2001;24:627–632. doi: 10.1165/ajrcmb.24.5.4280. [DOI] [PubMed] [Google Scholar]
- 82.Bando Y, Aki K. Mechanisms of generation of oxygen radicals and reductive mobilization of ferritin iron by lipoamide dehydrogenase. J Biochem (Tokyo) 1991;109:450–454. doi: 10.1093/oxfordjournals.jbchem.a123402. [DOI] [PubMed] [Google Scholar]
- 83.Kareyeva AV, Grivennikova VG, Cecchini G, Vinogradov AD. Molecular identification of the enzyme responsible for the mitochondrial NADH-supported ammonium-dependent hydrogen peroxide production. FEBS Lett. 2011;585:385–389. doi: 10.1016/j.febslet.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kareyeva AV, Grivennikova VG, Vinogradov AD. Mitochondrial hydrogen peroxide production as determined by the pyridine nucleotide pool and its redox state. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbabio.2012.03.033. [DOI] [PubMed] [Google Scholar]
- 85.Gazaryan IG, Krasnikov BF, Ashby GA, Thorneley RN, Kristal BS, Brown AM. Zinc is a potent inhibitor of thiol oxidoreductase activity and stimulates reactive oxygen species production by lipoamide dehydrogenase. J Biol Chem. 2002;277:10064–10072. doi: 10.1074/jbc.M108264200. [DOI] [PubMed] [Google Scholar]
- 86.Klyachko NL, Shchedrina VA, Efimov AV, Kazakov SV, Gazaryan IG, Kristal BS, Brown AM. pH-dependent substrate preference of pig heart lipoamide dehydrogenase varies with oligomeric state: response to mitochondrial matrix acidification. J Biol Chem. 2005;280:16106–16114. doi: 10.1074/jbc.M414285200. [DOI] [PubMed] [Google Scholar]
- 87.Argyrou A, Sun G, Palfey BA, Blanchard JS. Catalysis of diaphorase reactions by Mycobacterium tuberculosis lipoamide dehydrogenase occurs at the EH4 level. Biochemistry. 2003;42:2218–2228. doi: 10.1021/bi020654f. [DOI] [PubMed] [Google Scholar]
- 88.Babady NE, Pang YP, Elpeleg O, Isaya G. Cryptic proteolytic activity of dihydrolipoamide dehydrogenase. Proc Natl Acad Sci U S A. 2007;104:6158–6163. doi: 10.1073/pnas.0610618104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cai Z, Yan LJ. Protein oxidative modifications: Beneficial roles in disease and health. Journal of Biochemical and Pharmacological Research. 2013;1:15–26. [PMC free article] [PubMed] [Google Scholar]
- 90.Kim H, Liu TC, Patel MS. Expression of cDNA sequences encoding mature and precursor forms of human dihydrolipoamide dehydrogenase in Escherichia coli. Differences in kinetic mechanisms. J Biol Chem. 1991;266:9367–9373. [PubMed] [Google Scholar]
- 91.Miller AF. Superoxide dismutases: ancient enzymes and new insights. FEBS Lett. 2012;586:585–595. doi: 10.1016/j.febslet.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yogev O, Naamati A, Pines O. Fumarase: a paradigm of dual targeting and dual localized functions. Febs J. 2011;278:4230–4242. doi: 10.1111/j.1742-4658.2011.08359.x. [DOI] [PubMed] [Google Scholar]
- 93.Raza H. Dual localization of glutathione S-transferase in the cytosol and mitochondria: implications in oxidative stress, toxicity and disease. Febs J. 2011;278:4243–4251. doi: 10.1111/j.1742-4658.2011.08358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng L, Kennedy MC, Blondin GA, Beinert H, Zalkin H. Binding of cytosolic aconitase to the iron responsive element of porcine mitochondrial aconitase mRNA. Arch Biochem Biophys. 1992;299:356–360. doi: 10.1016/0003-9861(92)90287-7. [DOI] [PubMed] [Google Scholar]
- 95.Dupuy J, Volbeda A, Carpentier P, Darnault C, Moulis JM, Fontecilla-Camps JC. Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure. 2006;14:129–139. doi: 10.1016/j.str.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 96.Duchene AM, Giege P. Dual localized mitochondrial and nuclear proteins as gene expression regulators in plants? Front Plant Sci. 2012;3:221. doi: 10.3389/fpls.2012.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yogev O, Pines O. Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim Biophys Acta. 2011;1808:1012–1020. doi: 10.1016/j.bbamem.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 98.Ben-Menachem R, Tal M, Shadur T, Pines O. A third of the yeast mitochondrial proteome is dual localized: a question of evolution. Proteomics. 2011;11:4468–4476. doi: 10.1002/pmic.201100199. [DOI] [PubMed] [Google Scholar]
- 99.Carrie C, Small I. A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim Biophys Acta. 2013;1833:253–259. doi: 10.1016/j.bbamcr.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 100.Dinur-Mills M, Tal M, Pines O. Dual targeted mitochondrial proteins are characterized by lower MTS parameters and total net charge. PLoS One. 2008;3:e2161. doi: 10.1371/journal.pone.0002161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mitra K, Rangaraj N, Shivaji S. Novelty of the pyruvate metabolic enzyme dihydrolipoamide dehydrogenase in spermatozoa: correlation of its localization, tyrosine phosphorylation, and activity during sperm capacitation. J Biol Chem. 2005;280:25743–25753. doi: 10.1074/jbc.M500310200. [DOI] [PubMed] [Google Scholar]
- 102.Ghibu S, Richard C, Vergely C, Zeller M, Cottin Y, Rochette L. Antioxidant properties of an endogenous thiol: Alpha-lipoic acid, useful in the prevention of cardiovascular diseases. J Cardiovasc Pharmacol. 2009;54:391–398. doi: 10.1097/fjc.0b013e3181be7554. [DOI] [PubMed] [Google Scholar]
- 103.Stabler SP, Sekhar J, Allen RH, O’Neill HC, White CW. Alpha-lipoic acid induces elevated S-adenosylhomocysteine and depletes S-adenosylmethionine. Free Radic Biol Med. 2009;47:1147–1153. doi: 10.1016/j.freeradbiomed.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yadav V, Marracci GH, Munar MY, Cherala G, Stuber LE, Alvarez L, Shinto L, Koop DR, Bourdette DN. Pharmacokinetic study of lipoic acid in multiple sclerosis: comparing mice and human pharmacokinetic parameters. Mult Scler. 2010;16:387–397. doi: 10.1177/1352458509359722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295:1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 106.Matuda S, Saheki T. Intracellular distribution and biosynthesis of lipoamide dehydrogenase in rat liver. J Biochem. 1982;91:553–561. doi: 10.1093/oxfordjournals.jbchem.a133727. [DOI] [PubMed] [Google Scholar]
- 107.Danson MJ. Dihydrolipoamide dehydrogenase: a ‘new’ function for an old enzyme? Biochem Soc Trans. 1988;16:87–89. doi: 10.1042/bst0160087. [DOI] [PubMed] [Google Scholar]
- 108.Danson MJ, Conroy K, McQuattie A, Stevenson KJ. Dihydrolipoamide dehydrogenase from Trypanosoma brucei. Characterization and cellular location. Biochem J. 1987;243:661–665. doi: 10.1042/bj2430661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vettakkorumakankav N, Danson MJ, Hough DW, Stevenson KJ, Davison M, Young J. Dihydrolipoamide dehydrogenase from the halophilic archaebacterium Haloferax volcanii: characterization and N-terminal sequence. Biochem Cell Biol. 1992;70:70–75. doi: 10.1139/o92-010. [DOI] [PubMed] [Google Scholar]
- 110.Vettakkorumakankav NN, Stevenson KJ. Dihydrolipoamide dehydrogenase from Haloferax volcanii: gene cloning, complete primary structure, and comparison to other dihydrolipoamide dehydrogenases. Biochem Cell Biol. 1992;70:656–663. doi: 10.1139/o92-101. [DOI] [PubMed] [Google Scholar]