Low-frequency HIV variants are increasingly recognized as a key factor that increases the risk of HIV treatment failure. This article will provide a review of HIV minority variants, including their demonstrated clinical impact and areas of controversy.
Keywords: HIV-1 drug resistance, minority variants, treatment failure
Abstract
Technologic advances in human immunodeficiency virus type 1 (HIV-1) sequencing have revolutionized the study of antiretroviral drug resistance and are increasingly moving from the laboratory to clinical practice. These techniques are able to detect HIV-1 drug resistance mutations present at low frequencies not detectable by current HIV-1 genotyping assays. For a number of commonly used antiretroviral medications, such as nonnucleoside reverse transcriptase inhibitors, the detection of these drug-resistant minority variants significantly increases the risk of treatment failure. The level of evidence, however, is insufficient to determine the impact of HIV-1 minority variants for several other classes of antiretroviral medications. Clinicians should be aware of the novel technologies that are moving into routine clinical use and the clinical implications of HIV-1 minority variants. Additional studies are needed to determine the optimal platform for clinical application of these new technologies and to provide guidance to clinicians on the type and frequency of clinically important HIV-1 minority variants.
Genotypic tests for human immunodeficiency virus type 1 (HIV-1) drug resistance employ polymerase chain reaction (PCR) amplification and population sequencing techniques that detect resistance-associated mutations present at ≥15%–25% of the viral population [1, 2]. These assays do not reliably detect the presence of low-frequency resistance mutations present as minority variants within the population of HIV-1 in an infected individual. A number of studies have now shown that such low-frequency mutations, also known as minority variants, can have significant clinical implications on the risk of combination antiretroviral treatment (cART) failure. However, there remains much confusion over which HIV-1 minority variants are clinically significant and how their presence should affect clinical practice.
Minority HIV-1 drug resistance mutations found in treatment-naive patients originate from 1 of 2 sources: transmitted drug resistance or de novo generation as part of natural viral diversification. Compared to wild-type HIV-1, those viruses harboring resistance mutations generally have lower fitness. In the absence of drug-selective pressure, the frequency of such transmitted HIV-1 drug resistance mutations is likely to decay and at a certain time would no longer be detectable by current genotyping assays that rely on population sequencing [3, 4]. HIV-1 minority variants can also arise due to the underlying diversity of the viral population. This remarkable diversity stems from a high replication rate and the error-prone reverse transcriptase enzyme. It is estimated that up to 5 mutations may arise with each replication cycle [5]. The daily production of more than a billion new virions in a typical chronically infected patient implies that the virus undergoes 10–100 million rounds of replication daily, resulting in the rapid generation of viral progeny carrying every possible mutation throughout the viral genome [6, 7]. Because of this underlying diversity, it is estimated that drug resistance mutations are likely to be present during chronic infection even in the absence of drug exposure, with the frequency of the mutations dependent on their fitness costs [8]. This situation allows HIV-1 drug resistance to emerge rapidly in patients who are on antiretroviral therapy that is not adequately suppressive or during episodes of treatment interruption.
Minority HIV-1 drug resistance mutations can be detected by a number of ultrasensitive assays. The characteristics of the most commonly used assays are compared in Table 1. These assays can generally be categorized as point-mutation assays (eg, allele-specific PCR [ASPCR] and oligonucleotide ligation assay [OLA]) or deep-sequencing techniques. Although ASPCR is a highly sensitive assay with a limit of detection of much less than 1% of the viral population, this technique is limited by the select number of resistance mutations that can be interrogated concurrently, because the detection of each mutation requires a separate PCR reaction [9–11]. The OLA uses labeled probes that preferentially bind to either the wild-type sequence or a sequence with the mutation of interest [12]. This assay is relatively inexpensive and does not require costly equipment. However, like other point-mutation assays, the number of mutations it evaluates concurrently is limited and OLA is not quantitative. Recent advances in high-throughput sequencing have revolutionized HIV-1 sequencing and the study of HIV-1 minority variants. Unlike point-mutation assays, deep sequencing confers the benefit of evaluating an entire region of HIV-1 (eg, HIV-1 reverse transcriptase or the third variable [V3] loop of HIV-1 envelope) and all mutations contained in that region. The most commonly used next-generation sequencing platforms are a pyrosequencing system developed by Roche/454 and a sequencing-by-synthesis system developed by Illumina. The general principle behind both of these technologies lies in the clonal amplification of millions of individual fragments of HIV-1 DNA that can then be sequenced in parallel. The Roche/454 system was the first to enter the market and has been the most popular platform for deep sequencing HIV-1 to date. As a whole, the deep sequencing field remains quite fluid due to the continual development and evolution of these and other next-generation sequencing platforms [13].
Table 1.
Characteristics of Conventional Genotypic, Phenotypic, and Ultrasensitive Drug Resistance Assays
| Assay | Advantages | Disadvantages | Currently in Clinical Use |
|---|---|---|---|
| Conventional genotypic (Sanger DNA sequencing) |
|
|
Yes |
| Phenotypic |
|
|
Yes |
| Ultrasensitive techniques | |||
| Allele-specific polymerase chain reaction |
|
|
No |
| Deep sequencingb |
|
|
Yesc |
| Oligonucleotide ligation assay |
|
|
No |
Adapted from [104].
a Test sensitivity and limits of detection vary by testing site and patient-specific variables (eg, viral load, sample volume).
b Refers to the Roche/454, Illumina, and other next-generation sequencing platforms.
c The Roche/454 deep-sequencing system is currently used as part of a coreceptor tropism test developed by Quest Diagnostics (Madison, New Jersey).
In this review, we explore the evidence behind 3 scenarios where HIV-1 minority variants have been shown to affect the risk of virologic failure and discuss the antiretroviral medications (ARVs) for which the impact of minority variants is still controversial.
NONNUCLEOSIDE REVERSE TRANSCRIPTASE INHIBITOR RESISTANCE AFTER SINGLE-DOSE NEVIRAPINE EXPOSURE
The use of peripartum single-dose nevirapine (sdNVP) is an inexpensive and effective means of reducing the risk of mother-to-child transmission of HIV-1 [14]. This strategy was rapidly adopted in many developing countries, especially in Africa. However, the use of sdNVP has also been associated with the emergence of nonnucleoside reverse transcriptase inhibitor (NNRTI) resistance, detected by standard genotyping, in a significant subset of women and infants exposed to sdNVP [15–17]. Even more concerning was the finding that with the use of more sensitive assays, the majority of women and children who had received sdNVP harbored NNRTI resistance mutations, often at frequencies below the detection of standard genotyping [18–25]. The proportion of NNRTI-resistant minority variants decays over time, but they could still be detected in both women and infants a year or more after sdNVP exposure [23–27]. Three factors help explain these findings: (1) high-level resistance to the most commonly used NNRTIs can be conferred by a single mutation; (2) a long elimination half-life resulting in prolonged exposure to subtherapeutic drug levels [28]; and (3) the relatively small negative impact on viral replication kinetics (fitness) conferred by these NNRTI resistance mutations.
But do these minority variants increase the risk of treatment failure? The OCTANE Trial 1 was performed to determine the efficacy of a nevirapine-based regimen compared to a regimen containing ritonavir-boosted lopinavir (LPV/r) for women previously exposed to sdNVP. The study found that women who had received sdNVP are more likely to reach the primary endpoint (virologic failure or death) when assigned to nevirapine-based antiretroviral therapy than when assigned to a regimen containing LPV/r (26% vs 8%) [29]. Interestingly, this outcome was observed even in participants in whom nevirapine resistance was not detectable in pretreatment samples by standard genotyping. A subsequent analysis of this trial revealed that women without NNRTI resistance by standard genotyping, but harboring K103N or Y181C detected by an ultrasensitive ASPCR assay, had >3 times the risk of virologic failure or death compared to women without these mutations [30]. This association between the presence of NNRTI-resistant minority variants and treatment failure has also been detected in several other studies of women and children exposed to sdNVP [31–34]. These results have led to several clinical trials evaluating the use of a nucleoside reverse transcriptase inhibitor (NRTI) regimen concurrent with sdNVP to prevent HIV-1 from developing resistance as the nevirapine is metabolized. These studies have shown that treatment with a short “tail” of NRTIs significantly decreases, but does not eliminate the emergence of NNRTI resistance mutations detectable by either conventional genotyping [35, 36] or by ultrasensitive techniques [37–42].
NNRTI RESISTANCE IN cART-NAIVE and -EXPERIENCED INDIVIDUALS
The effect of drug-resistant minority variants in cART-naive patients has been studied most rigorously for patients on an NNRTI-based first-line regimen. Interpretation of these studies has been challenging due to heterogeneity in the patient population and outcomes (ie, definition of virologic failure). To facilitate the interpretation of these studies, a pooled analysis was performed of 10 studies involving 985 participants [43–52]. This pooled analysis included only individuals with no detectable NNRTI and NRTI resistance by standard genotyping and standardized the definition of virologic failure across studies. Overall, 14% of participants were found to harbor either an NNRTI or NRTI minority variant, but this estimate varied depending on the sensitivity of the assay and the mutations tested. However, the finding was consistent with prior studies showing that ultrasensitive assays significantly increase the detection of HIV-1 drug resistance mutations in treatment-naive individuals [53, 54]. The pooled analysis also revealed that the presence of a minority HIV-1 drug resistance mutation at baseline is associated with more than twice the risk of virologic failure [55]. The increased risk of virologic failure was most evident early after therapy initiation (Figure 1) and was mediated primarily by NNRTI-resistant minority variants (hazard ratio, 2.6). To evaluate whether a threshold existed for the minority variant effect, analyses were performed categorizing participants by either the minority variant percentage or minority variant copy numbers, which takes into account both the minority variant percentage and viral load for each individual. The effect of the minority variants was dose-dependent and was detectable even after controlling for medication adherence and other potential confounding factors [55, 56]. In addition, an increased risk of treatment failure was detected even at very low minority variant frequencies (<0.5% and 10–99 mutant copies/mL).
Figure 1.
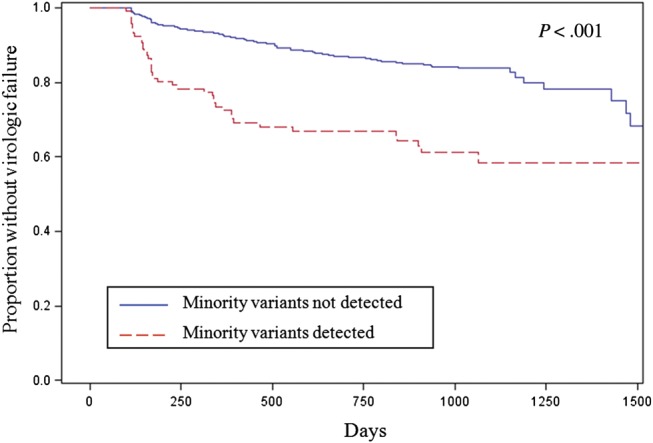
Kaplan-Meier curves for the proportion of patients without virologic failure on a first-line nonnucleoside reverse transcriptase inhibitor (NNRTI)–based combination antiretroviral therapy regimen by the presence of human immunodeficiency virus type 1 nucleoside reverse transcriptase inhibitor and NNRTI-resistant minority variants. Adapted from [55].
Studies in treatment-naive African patients have also shown an association between NNRTI minority variant detection and an increased risk of treatment failure [31, 57]. However, a recently reported analysis of OCTANE Trial 2 of treatment-naive African women did not detect a significant association between NNRTI minority variant detection and risk of treatment failure [58]. The reason behind this discrepant finding is unclear.
Ultrasensitive techniques may also be useful for detecting drug-resistant minority variants in NNRTI-experienced patients [59–62]. As in treatment-naive patients, presence of minority variants increases the risk of treatment failure [63, 64]. In addition, the use of ultrasensitive drug resistance assays in NNRTI-experienced patients frequently reveals the presence of minority variants associated with reduced efficacy of etravirine [65, 66].
HIV-1 TROPISM
HIV-1 requires the use of either CCR5 or CXCR4 as a coreceptor for cellular entry. Maraviroc, a CCR5 antagonist that blocks HIV-1 entry, is approved by the US Food and Drug Administration for treatment of HIV-1 infection. Resistance to the CCR5 antagonists occurs either by adaptation of HIV-1 to use the drug-bound receptor or through the use of the CXCR4 coreceptor. Approximately 10%–15% of treatment-naive individuals and 50% of treatment-experienced individuals harbor virus that can use CXCR4 [67, 68]. To avoid prescribing maraviroc to patients harboring CXCR4-using variants, coreceptor usage of a patient's virus must first be determined by a genotypic or phenotypic tropism test. Historically, the phenotypic assay (Trofile, Monogram Biosciences) has been the preferred assay in the United States whereas the genotypic assay (by population sequencing of the V3 region of the HIV-1 envelope) has been used predominantly outside the United States. Despite tropism testing prior to the use of CCR5 antagonists, virologic failure is frequently accompanied by evidence of CXCR4-using virus. Using both cloning and deep sequencing, a number of studies have shown that virologic failure of a CCR5 antagonist can be caused by the outgrowth of a preexisting minority population of CXCR4-using HIV-1 not detected by current tropism assays [69–73]. In a retrospective analysis of 2 phase 3 clinic trials of maraviroc in treatment-experienced patients, the use of 454 deep sequencing was found to be a potentially better predictor of maraviroc response than the original Trofile phenotypic test [74]. Ultradeep sequencing also performed as well as an improved version of the Trofile assay in predicting the response to maraviroc [75]. A limitation of that analysis is that all the patients who received maraviroc were prescreened by the original Trofile assay. Thus, it was not possible to estimate the true sensitivity and specificity of deep sequencing for predicting treatment response to maraviroc.
A genotypic tropism assay that includes initial population sequencing with deep sequencing of samples that show only R5-tropic virus by population sequencing is now available commercially in the United States (HIV-1 Coreceptor Tropism with Reflex to Ultradeep Sequencing, Quest Diagnostics). This Reflex assay was compared to both population sequencing alone and the enhanced sensitivity Trofile test in the retrospective analysis of 327 treatment-experienced patients who received maraviroc in the MOTIVATE and A4001029 studies [76]. The assay was found to have nearly identical ability to predict week 8 and 24 viral load declines and similar positive and negative predictive values when compared to the standard phenotypic assay. Population sequencing alone had significantly lower positive and negative predictive values compared to either the Reflex or phenotypic assay. These results represent the first commercial application of HIV-1 deep sequencing for the detection of HIV-1 minority variants. The use of deep sequencing as part of a genotypic tropism test has additional advantages as it is currently more cost-effective and faster to perform than phenotypic tropism testing.
CONTROVERSIES
Nucleoside Reverse Transcriptase Inhibitors
In the pooled analysis of treatment-naive patients initiating an NNRTI-based regimen, those with NRTI-resistant minority variants were found to have 1.6 times the risk of treatment failure compared to those without. However, only a subset of individuals was tested for NRTI minority variants, which were mainly limited to M184V and K65R [55]. Other studies evaluating the importance of NRTI-resistant minority variants have reported conflicting results. Several small studies have shown possible associations between NRTI-resistant minority variants and virologic failure [77–79], while others have detected no association [80–82]. There is evidence though, that low-frequency NRTI mutations can be detected after virologic rebound/failure [62, 83]. Additional studies evaluating the impact of NRTI-resistant minority variants would benefit from deep sequencing to assess the full spectrum of NRTI resistance mutations as there are currently ≥60 NRTI resistance mutations listed in the Stanford HIV Drug Resistance Database.
Integrase Inhibitors
Resistance to the integrase strand-transfer inhibitors (INSTI) raltegravir and elvitegravir share a number of characteristics with NNRTI resistance that may increase the likelihood that drug-resistant minority variants could contribute to the risk of virologic failure. First, these INSTIs have relatively low barriers to resistance and single amino acid changes are sufficient to confer a substantial decrease in antiretroviral activity. Integrase inhibitor resistance is usually selected more rapidly during in vitro passage experiments than is resistance to NRTIs or protease inhibitors (PIs). As with NNRTIs, clinical failure of raltegravir and elvitegravir is frequently accompanied by genotypic evidence of INSTI resistance [84]. One report documents a case of virologic failure and emergence of raltegravir resistance in a patient with preexisting raltegravir-resistant minority variants [85]. The proportion of patients with low-frequency drug resistance mutations is also an important component in determining to what degree these minority variants contribute to the risk of treatment failure. In one study of 32 patients, 81% were found to have Q148R minority variants before exposure to integrase inhibitors [86]. In a retrospective case-control analysis of the BENCHMRK-2 study, 46% of those with treatment failure arm as compared to 31% with treatment success were found to harbor at least 1 primary or secondary minority raltegravir resistance mutation prior to raltegravir treatment [87]. However, neither study showed a significant association between the presence of raltegravir-resistant minority variants and an increased risk of treatment failure. In addition, a number of other studies have not detected either significant pretreatment raltegravir-resistant minority variants or an increased risk of virologic failure [88–91]. However, those studies have been relatively small and larger studies are needed. The investigational INSTI dolutegravir appears to have a higher genetic barrier to resistance than raltegravir and elvitegravir. Its activity is less likely to be affected by minority variants containing single INSTI resistance mutations.
Protease Inhibitors
The use of more sensitive genotyping methods has significantly increased the number of PI resistance mutations detectable in pre- [47, 52, 77, 92, 93] and posttreatment failure samples [62, 94–96]. However, evidence has not yet emerged for a significant association between low-frequency PI resistance mutations and a significantly increased risk of treatment failure [52, 77]. A number of factors may contribute to this lack of association. Unlike most NNRTIs and integrase inhibitors, the majority of ritonavir-boosted PIs have a high barrier to resistance as they require multiple mutations to confer significant resistance [97, 98]. The appropriate combination of mutations is unlikely to arise de novo in any significant proportion in the absence of drug selective pressure. Although a single resistance mutation (I50L) confers high-level resistance to atazanavir, the significant reduction in viral fitness associated with this mutation may minimize the frequency and hence the impact of minority variants carrying this mutation [99]. It is possible that drug-resistant minority variants could play a role in PI failure in patients with transmitted resistance or in those who have previously failed PI therapy with resistance mutations that have decayed in frequency over time. However, unless multiple linked mutations are detected, it can be difficult to distinguish between transmitted drug resistance and those generated de novo. Because of the number of PIs in clinical use and the variety of mutations implicated in resistance to these drugs, it will be challenging to perform a pooled analysis (as was done for the NNRTIs) to assess the clinical significance of PI-resistant minority variants.
CONCLUSIONS
The detection of low-frequency variants of HIV-1 with altered drug susceptibility has been shown to be clinically significant in 3 settings: (1) detection of NNRTI-resistant minority variants after exposure to single-dose nevirapine; (2) detection of NNRTI-resistant minority variants prior to the initiation of a first-line NNRTI-based regimen; and (3) detection of CXCR4-using variants prior to treatment with CCR5 antagonists. In addition, there is evidence that ultrasensitive techniques could be useful after virologic failure of either NNRTI- or PI-based regimens to detect resistance mutations that emerged during treatment failure, but have decayed after drug discontinuation due to reduced fitness of the resistant virus in the absence of drug. Whether the efficacy of drugs in other classes are also affected by the presence of minority variants remains controversial. Uncertainty also exists as to the best method for detecting minority variants in a clinical setting, whether a threshold exists for what constitutes a significant frequency or number of resistant variants, and the importance of evaluating resistance mutations in the HIV-1 DNA reservoir.
At present, testing for HIV-1 minority variants is not available to clinicians for patient management with the exception of testing for coreceptor usage. However, efforts are ongoing worldwide to validate and implement deep sequencing for routine HIV-1 drug resistance genotyping [57, 100–103]. When compared to current HIV-1 genotyping assays, deep sequencing not only has improved sensitivity for detecting low-frequency resistance mutations, but will likely prove to be a lower-cost method of HIV-1 resistance testing for centers with a high demand for HIV-1 genotyping. However, the increased sensitivity of these novel assays for minority variant detection is also coupled with a heightened risk of detecting resistance artifacts generated in the laboratory during the amplification or sequencing steps. It is important to define conservative limits of detection for these ultrasensitive assays and to rigorously validate sequence analysis software that are to be used in clinical practice. In addition, the challenge for both clinicians and researchers alike will be in defining appropriate resistance interpretation algorithms for low-frequency variants detected by these ultrasensitive assays.
Additional research is also needed to provide clarity on the importance of minority variants outside of NNRTIs and CCR5 antagonists, and to define the frequency or copy number of minority variants that should prompt selection of an alternative regimen. For the time being, clinicians should be aware of these novel technologies and the clinical implications of HIV-1 minority variants, especially for those who have received sdNVP or who are initiating a CCR5 antagonist.
Notes
Financial support. This work was supported by the National Institutes of Health (AI100699 to J. Z. L. and AI068636).
Potential conflicts of interest. J. Z. L. has served as a consultant for Therapy Edge and has received speaking honoraria from Quest Diagnostics. D. R. K. has served as a consultant to and/or has received research grant support from Abbott, Avexa, Bristol-Myers Squibb, Boehringer-Ingelheim, Gilead, GlaxoSmithKline, Merck, Roche, Tobira, Vertex, ViroStatistics, and ViiV Healthcare; and has received speaking honoraria from Gilead and Roche.
Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Larder BA, Kohli A, Kellam P, Kemp SD, Kronick M, Henfrey RD. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature. 1993;365:671–3. doi: 10.1038/365671a0. [DOI] [PubMed] [Google Scholar]
- 2.Church JD, Jones D, Flys T, et al. Sensitivity of the ViroSeq HIV-1 genotyping system for detection of the K103N resistance mutation in HIV-1 subtypes A, C, and D. J Mol Diagn. 2006;8:430–2. doi: 10.2353/jmoldx.2006.050148. quiz 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain V, Sucupira MC, Bacchetti P, et al. Differential persistence of transmitted HIV-1 drug resistance mutation classes. J Infect Dis. 2011;203:1174–81. doi: 10.1093/infdis/jiq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yanik EL, Napravnik S, Hurt CB, et al. Prevalence of transmitted antiretroviral drug resistance differs between acutely and chronically HIV-infected patients. J Acquir Immune Defic Syndr. 2012;61:258–62. doi: 10.1097/QAI.0b013e3182618f05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Preston BD, Poiesz BJ, Loeb LA. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–71. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 6.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–6. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 7.Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–9. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 8.Gadhamsetty S, Dixit NM. Estimating frequencies of minority nevirapine-resistant strains in chronically HIV-1-infected individuals naive to nevirapine by using stochastic simulations and a mathematical model. J Virol. 2010;84:10230–40. doi: 10.1128/JVI.01010-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paredes R, Marconi VC, Campbell TB, Kuritzkes DR. Systematic evaluation of allele-specific real-time PCR for the detection of minor HIV-1 variants with pol and env resistance mutations. J Virol Methods. 2007;146:136–46. doi: 10.1016/j.jviromet.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson JA, Li JF, Wei X, et al. Simple PCR assays improve the sensitivity of HIV-1 subtype B drug resistance testing and allow linking of resistance mutations. PLoS One. 2007;2:e638. doi: 10.1371/journal.pone.0000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boltz VF, Maldarelli F, Martinson N, et al. Optimization of allele-specific PCR using patient-specific HIV consensus sequences for primer design. J Virol Methods. 2010;164:122–6. doi: 10.1016/j.jviromet.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelstein RE, Nickerson DA, Tobe VO, Manns-Arcuino LA, Frenkel LM. Oligonucleotide ligation assay for detecting mutations in the human immunodeficiency virus type 1 pol gene that are associated with resistance to zidovudine, didanosine, and lamivudine. J Clin Microbiol. 1998;36:569–72. doi: 10.1128/jcm.36.2.569-572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Archer J, Weber J, Henry K, et al. Use of four next-generation sequencing platforms to determine HIV-1 coreceptor tropism. PLoS One. 2012;7:e49602. doi: 10.1371/journal.pone.0049602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guay LA, Musoke P, Fleming T, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet. 1999;354:795–802. doi: 10.1016/S0140-6736(99)80008-7. [DOI] [PubMed] [Google Scholar]
- 15.Eshleman SH, Mracna M, Guay LA, et al. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012) AIDS. 2001;15:1951–7. doi: 10.1097/00002030-200110190-00006. [DOI] [PubMed] [Google Scholar]
- 16.Eshleman SH, Guay LA, Mwatha A, et al. Characterization of nevirapine resistance mutations in women with subtype A vs. D HIV-1 6-8 weeks after single-dose nevirapine (HIVNET 012) J Acquir Immune Defic Syndr. 2004;35:126–30. doi: 10.1097/00126334-200402010-00004. [DOI] [PubMed] [Google Scholar]
- 17.Jackson JB, Becker-Pergola G, Guay LA, et al. Identification of the K103N resistance mutation in Ugandan women receiving nevirapine to prevent HIV-1 vertical transmission. AIDS. 2000;14:F111–5. doi: 10.1097/00002030-200007280-00001. [DOI] [PubMed] [Google Scholar]
- 18.Johnson JA, Li JF, Morris L, et al. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis. 2005;192:16–23. doi: 10.1086/430741. [DOI] [PubMed] [Google Scholar]
- 19.Flys TS, Chen S, Jones DC, et al. Quantitative analysis of HIV-1 variants with the K103N resistance mutation after single-dose nevirapine in women with HIV-1 subtypes A, C, and D. J Acquir Immune Defic Syndr. 2006;42:610–3. doi: 10.1097/01.qai.0000221686.67810.20. [DOI] [PubMed] [Google Scholar]
- 20.Hauser A, Mugenyi K, Kabasinguzi R, et al. Detection and quantification of minor human immunodeficiency virus type 1 variants harboring K103N and Y181C resistance mutations in subtype A and D isolates by allele-specific real-time PCR. Antimicrob Agents Chemother. 2009;53:2965–73. doi: 10.1128/AAC.01672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner TA, Kress CM, Beck I, et al. Detection of HIV-1 drug resistance in women following administration of a single dose of nevirapine: comparison of plasma RNA to cellular DNA by consensus sequencing and by oligonucleotide ligation assay. J Clin Microbiol. 2010;48:1555–61. doi: 10.1128/JCM.02062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilger D, Hauser A, Kuecherer C, et al. Minor drug-resistant HIV type-1 variants in breast milk and plasma of HIV type-1-infected Ugandan women after nevirapine single-dose prophylaxis. Antivir Ther. 2011;16:109–13. doi: 10.3851/IMP1698. [DOI] [PubMed] [Google Scholar]
- 23.Loubser S, Balfe P, Sherman G, Hammer S, Kuhn L, Morris L. Decay of K103N mutants in cellular DNA and plasma RNA after single-dose nevirapine to reduce mother-to-child HIV transmission. AIDS. 2006;20:995–1002. doi: 10.1097/01.aids.0000222071.60620.1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer S, Boltz V, Martinson N, et al. Persistence of nevirapine-resistant HIV-1 in women after single-dose nevirapine therapy for prevention of maternal-to-fetal HIV-1 transmission. Proc Natl Acad Sci U S A. 2006;103:7094–9. doi: 10.1073/pnas.0602033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt GM, Coovadia A, Abrams EJ, et al. HIV-1 drug resistance at antiretroviral treatment initiation in children previously exposed to single-dose nevirapine. AIDS. 2011;25:1461–9. doi: 10.1097/QAD.0b013e3283492180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flys T, Nissley DV, Claasen CW, et al. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J Infect Dis. 2005;192:24–9. doi: 10.1086/430742. [DOI] [PubMed] [Google Scholar]
- 27.Hauser A, Mugenyi K, Kabasinguzi R, Kuecherer C, Harms G, Kunz A. Emergence and persistence of minor drug-resistant HIV-1 variants in Ugandan women after nevirapine single-dose prophylaxis. PLoS One. 2011;6:e20357. doi: 10.1371/journal.pone.0020357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muro E, Droste JA, Hofstede HT, Bosch M, Dolmans W, Burger DM. Nevirapine plasma concentrations are still detectable after more than 2 weeks in the majority of women receiving single-dose nevirapine: implications for intervention studies. J Acquir Immune Defic Syndr. 2005;39:419–21. doi: 10.1097/01.qai.0000167154.37357.f9. [DOI] [PubMed] [Google Scholar]
- 29.Lockman S, Hughes MD, McIntyre J, et al. Antiretroviral therapies in women after single-dose nevirapine exposure. N Engl J Med. 2010;363:1499–509. doi: 10.1056/NEJMoa0906626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boltz VF, Zheng Y, Lockman S, et al. Role of low-frequency HIV-1 variants in failure of nevirapine-containing antiviral therapy in women previously exposed to single-dose nevirapine. Proc Natl Acad Sci U S A. 2011;108:9202–7. doi: 10.1073/pnas.1105688108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coovadia A, Hunt G, Abrams EJ, et al. Persistent minority K103N mutations among women exposed to single-dose nevirapine and virologic response to nonnucleoside reverse-transcriptase inhibitor-based therapy. Clin Infect Dis. 2009;48:462–72. doi: 10.1086/596486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rowley CF, Boutwell CL, Lee EJ, et al. Ultrasensitive detection of minor drug-resistant variants for HIV after nevirapine exposure using allele-specific PCR: clinical significance. AIDS Res Hum Retroviruses. 2010;26:293–300. doi: 10.1089/aid.2009.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacLeod IJ, Rowley CF, Thior I, et al. Minor resistant variants in nevirapine-exposed infants may predict virologic failure on nevirapine-containing ART. J Clin Virol. 2010;48:162–7. doi: 10.1016/j.jcv.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehman DA, Wamalwa DC, McCoy CO, et al. Low-frequency nevirapine resistance at multiple sites may predict treatment failure in infants on nevirapine-based treatment. J Acquir Immune Defic Syndr. 2012;60:225–33. doi: 10.1097/QAI.0b013e3182515730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McIntyre JA, Hopley M, Moodley D, et al. Efficacy of short-course AZT plus 3TC to reduce nevirapine resistance in the prevention of mother-to-child HIV transmission: a randomized clinical trial. PLoS Med. 2009;6:e1000172. doi: 10.1371/journal.pmed.1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chi BH, Sinkala M, Mbewe F, et al. Single-dose tenofovir and emtricitabine for reduction of viral resistance to non-nucleoside reverse transcriptase inhibitor drugs in women given intrapartum nevirapine for perinatal HIV prevention: an open-label randomised trial. Lancet. 2007;370:1698–705. doi: 10.1016/S0140-6736(07)61605-5. [DOI] [PubMed] [Google Scholar]
- 37.Chi BH, Ellis GM, Chintu N, et al. Intrapartum tenofovir and emtricitabine reduces low-concentration drug resistance selected by single-dose nevirapine for perinatal HIV prevention. AIDS Res Hum Retroviruses. 2009;25:1099–106. doi: 10.1089/aid.2009.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lallemant M, Ngo-Giang-Huong N, Jourdain G, et al. Efficacy and safety of 1-month postpartum zidovudine-didanosine to prevent HIV-resistance mutations after intrapartum single-dose nevirapine. Clin Infect Dis. 2010;50:898–908. doi: 10.1086/650745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer S, Boltz VF, Chow JY, et al. Short-course Combivir after single-dose nevirapine reduces but does not eliminate the emergence of nevirapine resistance in women. Antivir Ther. 2012;17:327–36. doi: 10.3851/IMP1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hauser A, Sewangi J, Mbezi P, et al. Emergence of minor drug-resistant HIV-1 variants after triple antiretroviral prophylaxis for prevention of vertical HIV-1 transmission. PLoS One. 2012;7:e32055. doi: 10.1371/journal.pone.0032055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Micek MA, Blanco AJ, Carlsson J, et al. Effects of short-course zidovudine on the selection of nevirapine-resistant HIV-1 in women taking single-dose nevirapine. J Infect Dis. 2012;205:1811–5. doi: 10.1093/infdis/jis282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Dyke RB, Ngo-Giang-Huong N, Shapiro DE, et al. A comparison of 3 regimens to prevent nevirapine resistance mutations in HIV-infected pregnant women receiving a single intrapartum dose of nevirapine. Clin Infect Dis. 2012;54:285–93. doi: 10.1093/cid/cir798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balduin M, Oette M, Daumer MP, Hoffmann D, Pfister HJ, Kaiser R. Prevalence of minor variants of HIV strains at reverse transcriptase position 103 in therapy-naive patients and their impact on the virological failure. J Clin Virol. 2009;45:34–8. doi: 10.1016/j.jcv.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Geretti AM, Fox ZV, Booth CL, et al. Low-frequency K103N strengthens the impact of transmitted drug resistance on virologic responses to first-line efavirenz or nevirapine-based highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;52:569–73. doi: 10.1097/QAI.0b013e3181ba11e8. [DOI] [PubMed] [Google Scholar]
- 45.Goodman D, Zhou Y, Margot NA, et al. Low level of the K103N HIV-1 above a threshold is associated with virological failure in treatment-naive individuals undergoing efavirenz-containing therapy. AIDS. 2011;25:325–33. doi: 10.1097/QAD.0b013e3283427dcb. [DOI] [PubMed] [Google Scholar]
- 46.Jakobsen MR, Tolstrup M, Sogaard OS, et al. Transmission of HIV-1 drug-resistant variants: prevalence and effect on treatment outcome. Clin Infect Dis. 2010;50:566–73. doi: 10.1086/650001. [DOI] [PubMed] [Google Scholar]
- 47.Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008;5:e158. doi: 10.1371/journal.pmed.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metzner KJ, Giulieri SG, Knoepfel SA, et al. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin Infect Dis. 2009;48:239–47. doi: 10.1086/595703. [DOI] [PubMed] [Google Scholar]
- 49.Metzner KJ, Rauch P, Braun P, et al. Prevalence of key resistance mutations K65R, K103N, and M184V as minority HIV-1 variants in chronically HIV-1 infected, treatment-naive patients. J Clin Virol. 2011;50:156–61. doi: 10.1016/j.jcv.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Paredes R, Lalama CM, Ribaudo HJ, et al. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis. 2010;201:662–71. doi: 10.1086/650543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peuchant O, Thiebaut R, Capdepont S, et al. Transmission of HIV-1 minority-resistant variants and response to first-line antiretroviral therapy. AIDS. 2008;22:1417–23. doi: 10.1097/QAD.0b013e3283034953. [DOI] [PubMed] [Google Scholar]
- 52.Simen BB, Simons JF, Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199:693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 53.Buckton AJ, Harris RJ, Pillay D, Cane PA. HIV type-1 drug resistance in treatment-naive patients monitored using minority species assays: a systematic review and meta-analysis. Antivir Ther. 2010;16:9–16. doi: 10.3851/IMP1687. [DOI] [PubMed] [Google Scholar]
- 54.Buckton AJ, Prabhu D, Motamed C, et al. Increased detection of the HIV-1 reverse transcriptase M184V mutation using mutation-specific minority assays in a UK surveillance study suggests evidence of unrecognized transmitted drug resistance. HIV Med. 2010;12:250–4. doi: 10.1111/j.1468-1293.2010.00882.x. [DOI] [PubMed] [Google Scholar]
- 55.Li JZ, Paredes R, Ribaudo HJ, et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA. 2011;305:1327–35. doi: 10.1001/jama.2011.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li JZ, Paredes R, Ribaudo HJ, et al. Relationship between minority nonnucleoside reverse transcriptase inhibitor resistance mutations, adherence, and the risk of virologic failure. AIDS. 2012;26:185–92. doi: 10.1097/QAD.0b013e32834e9d7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Papathanasopoulos M, Ketseoglou I, Travers S, et al. Antiretroviral drug-resistant minority variants are significantly associated with first-line treatment failure in antiretroviral drug-naive patients. International Workshop on HIV and Hepatitis Virus Drug Resistance and Curative Strategies, Sitges, Spain, 5–9 June 2012. Abstract 25. [Google Scholar]
- 58.Boltz VF, Bao Y, Lockman S, et al. The risk of virologic failure associated with low frequency nevirapine-resistant variants in women initiating nevirapine-containing ART varies depending on the history of exposure to sd-Nevirapine: OCTANE/ACTG 5208. 19th Conference on Retroviruses and Opportunistic Infections, Seattle, WA, 5–8 March 2012. Abstract 105. [Google Scholar]
- 59.Garcia-Gonzalez C, Garcia-Bujalance S, Ruiz-Carrascoso G, et al. Detection and quantification of the K103N mutation in HIV reverse transcriptase by pyrosequencing. Diagn Microbiol Infect Dis. 2012;72:90–6. doi: 10.1016/j.diagmicrobio.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 60.Delobel P, Saliou A, Nicot F, et al. Minor HIV-1 variants with the K103N resistance mutation during intermittent efavirenz-containing antiretroviral therapy and virological failure. PLoS One. 2011;6:e21655. doi: 10.1371/journal.pone.0021655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hare CB, Mellors J, Krambrink A, et al. Detection of nonnucleoside reverse-transcriptase inhibitor-resistant HIV-1 after discontinuation of virologically suppressive antiretroviral therapy. Clin Infect Dis. 2008;47:421–4. doi: 10.1086/589867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le T, Chiarella J, Simen BB, et al. Low-abundance HIV drug-resistant viral variants in treatment-experienced persons correlate with historical antiretroviral use. PLoS One. 2009;4:e6079. doi: 10.1371/journal.pone.0006079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Halvas EK, Wiegand A, Boltz VF, et al. Low frequency nonnucleoside reverse-transcriptase inhibitor-resistant variants contribute to failure of efavirenz-containing regimens in treatment- experienced patients. J Infect Dis. 2010;201:672–80. doi: 10.1086/650542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lecossier D, Shulman NS, Morand-Joubert L, et al. Detection of minority populations of HIV-1 expressing the K103N resistance mutation in patients failing nevirapine. J Acquir Immune Defic Syndr. 2005;38:37–42. doi: 10.1097/00126334-200501010-00007. [DOI] [PubMed] [Google Scholar]
- 65.Varghese V, Shahriar R, Rhee SY, et al. Minority variants associated with transmitted and acquired HIV-1 nonnucleoside reverse transcriptase inhibitor resistance: implications for the use of second-generation nonnucleoside reverse transcriptase inhibitors. J Acquir Immune Defic Syndr. 2009;52:309–15. doi: 10.1097/QAI.0b013e3181bca669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Codoner FM, Pou C, Thielen A, et al. Added value of deep sequencing relative to population sequencing in heavily pre-treated HIV-1-infected subjects. PLoS One. 2011;6:e19461. doi: 10.1371/journal.pone.0019461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moyle GJ, Wildfire A, Mandalia S, et al. Epidemiology and predictive factors for chemokine receptor use in HIV-1 infection. J Infect Dis. 2005;191:866–72. doi: 10.1086/428096. [DOI] [PubMed] [Google Scholar]
- 68.Raymond S, Saliou A, Nicot F, et al. Frequency of CXCR4-using viruses in primary HIV-1 infections using ultra-deep pyrosequencing. AIDS. 2011;25:1668–70. doi: 10.1097/QAD.0b013e3283498305. [DOI] [PubMed] [Google Scholar]
- 69.Westby M, Lewis M, Whitcomb J, et al. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol. 2006;80:4909–20. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Archer J, Braverman MS, Taillon BE, et al. Detection of low-frequency pretherapy chemokine (CXC motif) receptor 4 (CXCR4)-using HIV-1 with ultra-deep pyrosequencing. AIDS. 2009;23:1209–18. doi: 10.1097/QAD.0b013e32832b4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsibris AM, Korber B, Arnaout R, et al. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One. 2009;4:e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Archer J, Rambaut A, Taillon BE, Harrigan PR, Lewis M, Robertson DL. The evolutionary analysis of emerging low frequency HIV-1 CXCR4 using variants through time—an ultra-deep approach. PLoS Comput Biol. 2010;6:e1001022. doi: 10.1371/journal.pcbi.1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baatz F, Struck D, Lemaire M, et al. Rescue of HIV-1 long-time archived X4 strains to escape maraviroc. Antiviral Res. 2011;92:488–92. doi: 10.1016/j.antiviral.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 74.Swenson LC, Mo T, Dong WW, et al. Deep sequencing to infer HIV-1 co-receptor usage: application to three clinical trials of maraviroc in treatment-experienced patients. J Infect Dis. 2011;203:237–45. doi: 10.1093/infdis/jiq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swenson LC, Mo T, Dong WW, et al. Deep V3 sequencing for HIV type 1 tropism in treatment-naive patients: a reanalysis of the MERIT trial of maraviroc. Clin Infect Dis. 2011;53:732–42. doi: 10.1093/cid/cir493. [DOI] [PubMed] [Google Scholar]
- 76.Kagan RM, Johnson EP, Siaw M, et al. A Genotypic Test for HIV-1 tropism combining Sanger sequencing with Ultradeep sequencing predicts virologic response in treatment-experienced patients. PLoS One. 2012;7:e46334. doi: 10.1371/journal.pone.0046334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lataillade M, Chiarella J, Yang R, et al. Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naive subjects in the CASTLE study. PLoS One. 2010;5:e10952. doi: 10.1371/journal.pone.0010952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vignoles M, Barboni G, Agosti MR, et al. Evaluation of minority populations of HIV type-1 with K103N and M184V drug resistance mutations among children in Argentina. Antivir Ther. 2009;14:1175–81. doi: 10.3851/IMP1461. [DOI] [PubMed] [Google Scholar]
- 79.Bansal V, Metzner KJ, Niederost B, et al. Minority K65R variants and early failure of antiretroviral therapy in HIV-1-infected Eritrean immigrant. Emerg Infect Dis. 2011;17:1966–8. doi: 10.3201/eid1710.110592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Metzner KJ, Rauch P, von Wyl V, et al. Efficient suppression of minority drug-resistant HIV type 1 (HIV-1) variants present at primary HIV-1 infection by ritonavir-boosted protease inhibitor-containing antiretroviral therapy. J Infect Dis. 2010;201:1063–71. doi: 10.1086/651136. [DOI] [PubMed] [Google Scholar]
- 81.Gianella S, Delport W, Pacold ME, et al. Detection of minority resistance during early HIV-1 infection: natural variation and spurious detection rather than transmission and evolution of multiple viral variants. J Virol. 2011;85:8359–67. doi: 10.1128/JVI.02582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stekler JD, Ellis GM, Carlsson J, et al. Prevalence and impact of minority variant drug resistance mutations in primary HIV-1 infection. PLoS One. 2011;6:e28952. doi: 10.1371/journal.pone.0028952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.D'Aquila RT, Geretti AM, Horton JH, et al. Tenofovir (TDF)-selected or abacavir (ABC)-selected low-frequency HIV type 1 subpopulations during failure with persistent viremia as detected by ultradeep pyrosequencing. AIDS Res Hum Retroviruses. 2011;27:201–9. doi: 10.1089/aid.2010.0077. [DOI] [PubMed] [Google Scholar]
- 84.Blanco JL, Varghese V, Rhee SY, Gatell JM, Shafer RW. HIV-1 integrase inhibitor resistance and its clinical implications. J Infect Dis. 2011;203:1204–14. doi: 10.1093/infdis/jir025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Codoner FM, Pou C, Thielen A, et al. Dynamic escape of pre-existing raltegravir-resistant HIV-1 from raltegravir selection pressure. Antiviral Res. 2010;88:281–6. doi: 10.1016/j.antiviral.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 86.Charpentier C, Laureillard D, Piketty C, et al. High frequency of integrase Q148R minority variants in HIV-infected patients naive of integrase inhibitors. AIDS. 2010;24:867–73. doi: 10.1097/QAD.0b013e3283367796. [DOI] [PubMed] [Google Scholar]
- 87.Liu J, Miller MD, Danovich RM, et al. Analysis of low-frequency mutations associated with drug resistance to raltegravir before antiretroviral treatment. Antimicrob Agents Chemother. 2011;55:1114–9. doi: 10.1128/AAC.01492-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ceccherini-Silberstein F, Van Baelen K, Armenia D, et al. Secondary integrase resistance mutations found in HIV-1 minority quasispecies in integrase therapy-naive patients have little or no effect on susceptibility to integrase inhibitors. Antimicrob Agents Chemother. 2010;54:3938–48. doi: 10.1128/AAC.01720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mukherjee R, Jensen ST, Male F, et al. Switching between raltegravir resistance pathways analyzed by deep sequencing. AIDS. 2011;25:1951–9. doi: 10.1097/QAD.0b013e32834b34de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Armenia D, Vandenbroucke I, Fabeni L, et al. Study of genotypic and phenotypic HIV-1 dynamics of integrase mutations during raltegravir treatment: a refined analysis by ultra-deep 454 pyrosequencing. J Infect Dis. 2012;205:557–67. doi: 10.1093/infdis/jir821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nguyen H, Charpentier C, Nguyen N, et al. Longitudinal analysis of integrase N155H variants in heavily treated patients failing raltegravir-based regimens. HIV Med. 2013;14:85–91. doi: 10.1111/j.1468-1293.2012.01039.x. [DOI] [PubMed] [Google Scholar]
- 92.Ross LL, Weinberg WG, DeJesus E, et al. Impact of low abundance HIV variants on response to ritonavir-boosted atazanavir or fosamprenavir given once daily with tenofovir/emtricitabine in antiretroviral-naive HIV-infected patients. AIDS Res Hum Retroviruses. 2010;26:407–17. doi: 10.1089/aid.2009.0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Metzner KJ, Rauch P, Walter H, et al. Detection of minor populations of drug-resistant HIV-1 in acute seroconverters. AIDS. 2005;19:1819–25. doi: 10.1097/01.aids.0000189878.97480.ed. [DOI] [PubMed] [Google Scholar]
- 94.McKinnon JE, Delgado R, Pulido F, Shao W, Arribas JR, Mellors JW. Single genome sequencing of HIV-1 gag and protease resistance mutations at virologic failure during the OK04 trial of simplified versus standard maintenance therapy. Antivir Ther. 2011;16:725–32. doi: 10.3851/IMP1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fisher R, van Zyl GU, Travers SA, et al. Deep sequencing reveals minor protease resistance mutations in patients failing a protease inhibitor regimen. J Virol. 2012;86:6231–7. doi: 10.1128/JVI.06541-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lataillade M, Chiarella J, Yang R, et al. Virologic failures on initial boosted-PI regimen infrequently possess low-level variants with major PI resistance mutations by ultra-deep sequencing. PLoS One. 2012;7:e30118. doi: 10.1371/journal.pone.0030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van Maarseveen N, Boucher C. Geretti AM, editor. Resistance to protease inhibitors. 2006 Antiretroviral resistance in clinical practice. London: Mediscript. [PubMed] [Google Scholar]
- 98.Tang MW, Shafer RW. HIV-1 antiretroviral resistance: scientific principles and clinical applications. Drugs. 2012;72:e1–25. doi: 10.2165/11633630-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Colonno R, Rose R, McLaren C, Thiry A, Parkin N, Friborg J. Identification of I50L as the signature atazanavir (ATV)-resistance mutation in treatment-naive HIV-1-infected patients receiving ATV-containing regimens. J Infect Dis. 2004;189:1802–10. doi: 10.1086/386291. [DOI] [PubMed] [Google Scholar]
- 100.Stelzl E, Proll J, Bizon B, et al. Human immunodeficiency virus type 1 drug resistance testing: evaluation of a new ultra-deep sequencing-based protocol and comparison with the TRUGENE HIV-1 Genotyping Kit. J Virol Methods. 2011;178:94–7. doi: 10.1016/j.jviromet.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 101.Dudley DM, Chin EN, Bimber BN, et al. Low-cost ultra-wide genotyping using Roche/454 pyrosequencing for surveillance of HIV drug resistance. PLoS One. 2012;7:e36494. doi: 10.1371/journal.pone.0036494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.De Wolf H, Van Marck H, Mostmans W, et al. HIV-1 nucleotide mixture detection in the virco((R))TYPE HIV-1 genotyping assay: a comparison between Sanger sequencing and 454 pyrosequencing. J Virol Methods. 2011;175:129–32. doi: 10.1016/j.jviromet.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 103.Avidor B, Girshengorn S, Matus N, et al. Evaluation of a Benchtop HIV ultradeep pyrosequencing drug-resistance assay in the clinical laboratory. J Clin Microbiol. 2013;51:880–6. doi: 10.1128/JCM.02652-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li JZ. Novel HIV-1 resistance and tropism testing. 2012. Cases on the Web. San Francisco, CA: IAS-USA, Available at: https://www.iasusa.org/cow . [Google Scholar]


