Summary
Bacterial chemoreceptors cluster into exquisitively sensitive, tunable, highly ordered, polar arrays. While these arrays serve as paradigms of cell signalling in general, it remains unclear what conformational changes transduce signals from the periplasmic tips, where attractants and repellents bind, to the cytoplasmic signalling domains. Conflicting reports support and contest the hypothesis that activation causes large changes in the packing arrangement of the arrays, up to and including their complete disassembly. Using electron cryotomography, here we show that in Caulobacter crescentus, chemoreceptor arrays in cells grown in different media and immediately after exposure to the attractant galactose all exhibit the same 12 nm hexagonal packing arrangement, array size and other structural parameters. ΔcheB and ΔcheR mutants mimicking attractant- or repellent-bound states prior to adaptation also show the same lattice structure. We conclude that signal transduction and amplification must be accomplished through only small, nanoscale conformational changes.
Introduction
Most motile bacteria move towards favourable environments through a process known as chemotaxis (Wadhams and Armitage, 2004). With only a small set of proteins, the cells are able to sense attractant and repellent concentrations (Duke and Bray, 1999), compare the current conditions with the recent past, and continuously adapt to their changing circumstances (Li and Hazelbauer, 2005; Endres and Wingreen, 2006). Attractants and repellents are sensed by transmembrane receptors known as methyl-accepting chemotaxis proteins (MCPs). Together with the histidine kinase CheA and the linker protein CheW, these receptors form large clusters at the cell poles (Maddock and Shapiro, 1993; Lybarger and Maddock, 2001; Boukhvalova et al., 2002). Receptors bind nutrients and toxins at their periplasmic tips either directly or via periplasmic binding proteins (PBPs) (Tam and Saier, 1993). The binding state is communicated through the membrane and down the receptor length, typically across one or more HAMP (histidine kinases, adenyl cyclases, MCPs and some phosphatases) domain(s) and along the coiled-coil cytoplasmic signalling domain, where the activity of the histidine kinase bound at distal tip of the receptors is ultimately modulated (Hazelbauer et al., 2008). Activated CheA autophosphorylates and transfers the phosphoryl group to the response regulator CheY, which in turn diffuses through the cell, binds to the flagellar motor and changes the direction of motor rotation (Stock et al., 2000). While repellents lead to an increased tumbling frequency due to elevated levels of CheY-P, attractants reduce tumbling by decreasing levels of CheY-P (Borkovich et al., 1989).
Chemotactic bacteria are also able to adapt to current conditions by regulating the methylation state of the receptors. In the case of Escherichia coli, receptor methylation is regulated by two additional enzymes, the methylesterase CheB (Stock and Koshland, 1978) and the methyltransferase CheR (Springer and Koshland, 1977). Constitutive receptor methylation by CheR shifts receptors towards a more CheA-activating state; receptor demethylation by CheB is dependent on phosphorylation by CheA and shifts receptors away from a CheA-activating state. The regulation of receptor methylation is slow, however, compared with stimulant binding: in the case of E. coli, for example, attractant binding occurs in milliseconds (Jiang et al., 2010), but adaptation requires a minute or more (Parkinson and Revello, 1978).
Although a wealth of high-resolution information on isolated chemotaxis proteins is available, details about their arrangement in the cluster and the structural changes that accompany activation and drive signal transduction are still elusive. Several results suggest that small, piston-like shifts of the receptors' transmembrane helix TM2 communicate the binding state through the membrane (reviewed in Falke and Hazelbauer, 2001; Hazelbauer et al., 2008; Falke and Erbse, 2009). Inside the cytoplasm, small structural changes such as rotation and/or modulated packing stabilities of the HAMP bundle and the signalling helices are thought to carry the signal to the kinase (Manson, 2010; Parkinson, 2010). The cooperative behaviour of the receptors (Li and Weis, 2000; Sourjik and Berg, 2002) suggests, however, that larger structural alterations such as changes in the receptor packing might also be involved. Indeed, immunofluorescence studies using antibodies raised against the cytoplasmic portion of MCPs showed that while chemoreceptors clustered together at the poles in adapted cells and cells treated with repellents, addition of attractant resulted in more diffuse staining (Lamanna et al., 2005; Borrock et al., 2008; Wu et al., 2011). This same delocalization was also observed in a study using receptors that were labelled directly with GFP (Wu et al., 2011), and receptor methylation (equivalent to shifting the arrays towards a repellent-bound state) appeared to stabilize GFP-tagged clusters (Shiomi et al., 2005). FRET experiments suggested that receptors move away from each other upon attractant addition and closer together upon repellent exposure (Vaknin and Berg, 2007; Besschetnova et al., 2008), but a tilting, bending or rotation of receptor homodimers was noted as another possible explanation of the observations (Vaknin and Berg, 2007). In contrast to these studies, no significant effects on the polar receptor localization were seen upon attractant binding using a GFP-tagged Tar receptor (Homma et al., 2004). Furthermore, by indirectly monitoring receptor clusters with the fluorescently labelled phosphatase CheZ, no change in receptor localization was observed upon attractant exposure (Liberman et al., 2004). While it is unclear how to reconcile all these results, they all share the resolution limitations of light microscopy. Unfortunately, not even the super-resolution PALM light microscopy technique was able to resolve the receptor arrangement in the arrays (Greenfield et al., 2009). The issues that arise from the introduction and use of tags all further complicate these results.
Electron cryotomography (ECT) is an emerging technique that allows intact cells to be visualized directly in a near-native, ‘frozen-hydrated’ state without tags in 3-D to ‘macromolecular’ (~4 nm) resolution. In our initial cryotomographic studies of chemoreceptor arrays in adapted states (Briegel et al., 2008; 2009), we showed that chemoreceptors consistently form 12 nm hexagonal arrays. Also using ECT, the Subramanian group confirmed the basic array architecture in Caulobacter crescentus (Khursigara et al., 2008a), but argued that the arrays were only partially ordered. Later, Subramanian et al. reported that the density of the receptors in the array increases under the nutrient-limiting conditions of a minimal medium, and argued that changes in lateral packing density modulate the array's responses (Khursigara et al., 2011). Subramanian et al. also reported that in cells overexpressing chemoreceptors, receptor activation resulted in a large outward movement of certain HAMP helices (Khursigara et al., 2008b).
To investigate the effects of receptor activation on the array packing in vivo, we chose the Gram-negative bacterium C. crescentus as a model system. This organism has a characteristic, dimorphic life cycle: motile swarmer cells mature into sessile cells by releasing their single polar flagellum and developing a stalk, which enables the cells to attach to surfaces (Wagner and Brun, 2007). Once sessile, cells undergo multiple rounds of cell division, each time releasing a motile daughter cell. Since the chemotactic swarmer cells only possess a single flagellum, they swim up or down gradients by modulating forward swimming paths (equivalent to ‘runs’) and short reversals in swimming direction (equivalent to ‘tumbles’) through a switch in the direction of flagellar rotation (Alley et al., 1991). Both chemotaxis and flagellar genes are cell cycle regulated and are only expressed in the swarmer cells (Jensen et al., 2002). C. crescentus contains 18 predicted mcp, 4 cheW, 2 cheA, 3 cheR and 2 cheB genes (Briegel et al., 2009; Ulrich and Zhulin, 2010). Based on the observed receptor length of 31 nm in the arrays (measured from the centre of the inner membrane to the centre of the CheA/CheW base plate), only 9 of the 18 receptors are predicted to form the polar receptor arrays, all of them belonging to the MCP class 36H, and containing an extra linker region as well as a second HAMP domain (Briegel et al., 2009). The chemoreceptors localize to the flagellated pole at the convex side of the swarmer cells in hexagonal arrays with the same architecture seen in other bacteria (Briegel et al., 2008; 2009). The adaptation system of C. crescentus is similar to that of E. coli and also relies on the regulation of receptor methylation by CheR and CheB (Shaw et al., 1983; Gomes and Shapiro, 1984; Ely et al., 1986).
Here, we applied ECT to intact, wild-type C. crescentus cells in different growth media and with or without recent exposure to an attractant, as well as mutants that mimic either the non-adapted attractant- or repellent-bound state. Our data show clearly that there are no major structural changes in receptor packing in different activation states, so signal transduction and amplification must be accomplished through only small, nanoscale conformational changes. We discuss possible explanations for the conflicting results of previous studies.
Results
The hexagonal arrangement of chemoreceptor arrays in C. crescentus is independent of the growth media
Previous ECT studies have already shown that when grown in rich media (peptone yeast extract, PYE), C. crescentus swarmer cells exhibit hexagonally packed chemoreceptor arrays at the convex side of the flagellated cell pole (Briegel et al., 2008; 2009; Khursigara et al., 2008a). Khursigara et al. also reported that the density of receptors in E. coli arrays increased when cells were grown in minimal media (Khursigara et al., 2011). To test whether the composition of the growth media influences the packing arrangement of chemoreceptors in C. crescentus, wild-type cells (CB15N) were grown in M2G minimal medium (Ely, 1991). Tomograms of these cells revealed the typical swarmer cell morphology with an S-layer, outer membrane, peptidoglycan layer and inner membrane surrounding a densely packed cytoplasm.
As before, chemoreceptor arrays were found inserted into the inner membrane at the convex side of the flagellated cell pole. The long, pillar-like densities of the chemoreceptors extended into the cytoplasm and terminated at the ‘base plate’ parallel to the inner membrane formed by CheA and CheW. The distance from the centre of the inner membrane to the centre of the base plate was 31 nm, as seen in arrays of cells grown in PYE. The receptors also formed an ~12 nm, honeycomb-like hexagonal lattice just above the base plate (Fig. 1A–C, Movie S1), and the estimated number of receptors (monomers) in the four arrays analysed was 600, 1100, 2000 and 3500. Except for a wider range in the observed array size, the packing arrangement and receptor length measured here are identical to the values reported in swarmer cells grown in PYE (Briegel et al., 2008; 2009). Thus we conclude that receptor packing in C. crescentus is the same in either rich or minimal growth media.
Figure 1.
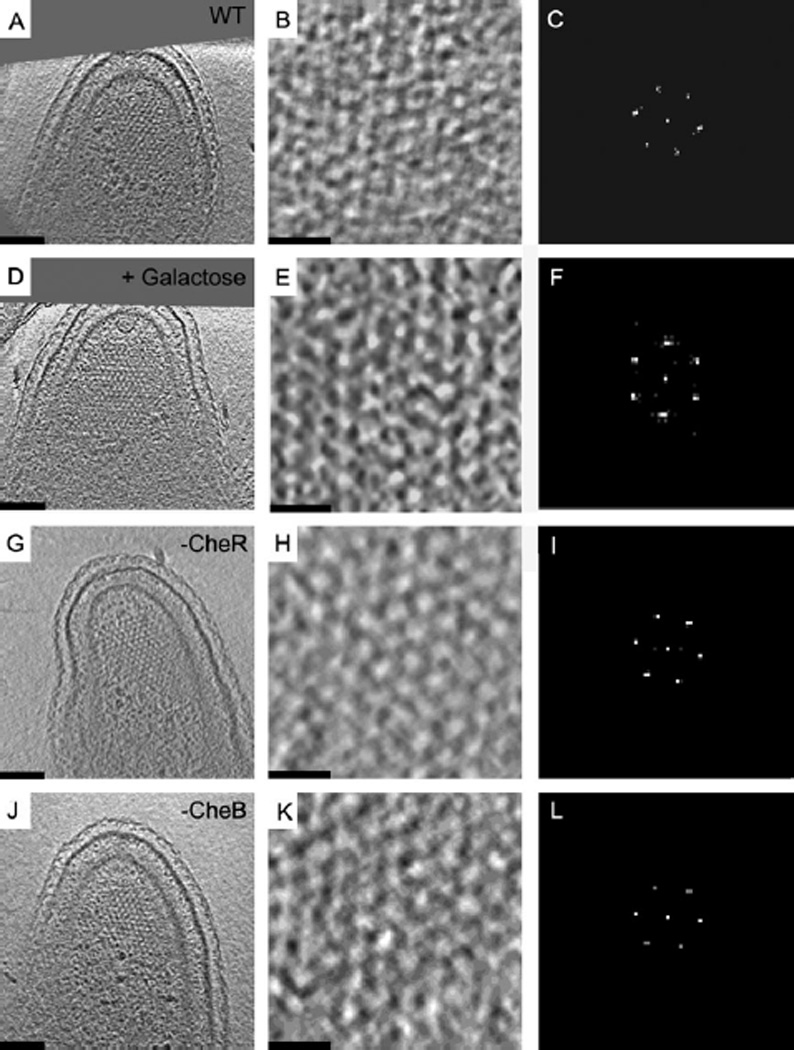
Twelve-nanometre hexagonal arrangement of receptors is preserved upon activation. Tomographic slices (left column), enlarged to highlight the organization of the chemoreceptor arrays (middle column), and corresponding power spectra (right column) revealing 12 nm hexagonal order seen in a wild-type swarmer cell grown in M2G minimal media (A–C), a wild-type cell frozen ~7 s after exposure to 10 mM galactose (D–F), a ΔcheR mutant cell mimicking a fully attractant-bound state prior to adaptation (G–I), and a ΔcheB mutant cell mimicking a fully repellent-bound state prior to adaptation (J–L). Bars (A, D, G, J): 100 nm, Bars (B, E, H, K): 25 nm. Power-spectra not to scale.
Galactose is a chemoattractant for C. crescentus
Glucose, maltose, galactose, xylose, ribose, alanine, proline and glutamine were listed in a review and mentioned in a paper as chemoattractants to C. crescentus, but the experiments were not described (Shapiro, 1985; Ely et al., 1986). To confirm that galactose is an attractant, C. crescentus was grown in the presence of a galactose gradient on swim plates (see Experimental procedures). The swim colonies show a clear bias towards the galactose source (Fig. 2A), confirming that galactose is a chemoattractant.
Figure 2.
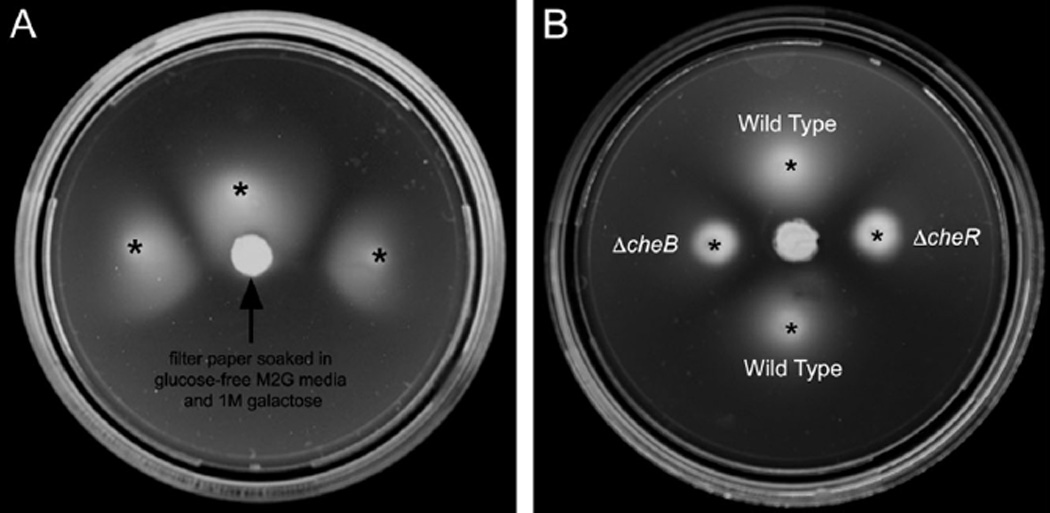
Swim assays. A. Wild-type C. crescentus was stabbed at the indicated locations (asterisks) into 0.126% agar M2G media plates without glucose. A filter paper soaked with glucose-free M2G media supplemented with 1 M galactose was placed in the centre of the plate. After 72 h at room temperature, the swim colonies showed a clear bias towards the galactose source, confirming that galactose is an attractant for this organism. B. Cultures of C. crescentus wild-type, ΔcheR and ΔcheB cells were spotted onto 0.25% agar plates made with glucose-free M2G media. A filter paper soaked with glucose-free M2G media supplemented with 1 M galactose was placed in the centre of the plate. After 7-day incubation at room temperature, both the ΔcheR and the ΔcheB mutants failed to form a swarm on the semisolid plate and did not exhibit any bias towards the galactose source, confirming their loss of chemotaxis.
Galactose exposure does not alter the hexagonal packing of the chemoreceptors
To investigate the structural effects of attractant binding, synchronized swarmer cells were resuspended in pre-warmed minimal medium (30°C) for 5 min, and then 10 mM galactose was added to the cells for 7, 10 and 110 s before vitrification. The high concentration of galactose and short incubation times were chosen to ensure high receptor occupation but prevent adaptation. The chemoreceptor arrays in the resulting cryotomograms were indistinguishable from the adapted arrays observed previously (Fig. 1D–F), exhibiting the same ~12 nm hexagonal packing right above the CheA/CheW base plate where the lattice was most obvious. The number of receptors in these arrays ranged from 1500 to 3500 (1500, 1900, 2000, 2200, 2400 and 3500), which is not significantly different from arrays grown in minimal medium as determined be the Mann–Whitney rank-sum test (P < 0.05, U = 8, where n1,n2 = 4,6).
ΔcheB and ΔcheR mutants exhibit constant kinase-‘on’ and ‘off’ states
While the previous experiment showed that chemoreceptor arrays do not undergo large structural changes upon addition of the attractant galactose, at present we do not know which MCP in the C. crescentus genome is responsible for galactose sensing or whether that type of receptor is abundant in the array. If those MCPs are rare, for instance, their impact on the overall array architecture may have been small. Additionally, while the adaptation time of E. coli to high concentrations of attractants such as 10 µM aspartate or serine ranged between 4 and 7 min (Parkinson and Revello, 1978), we cannot rule out the possibility that the adaptation time in C. crescentus is much faster, and that the arrays were already re-adapted in the 7–110 s that elapsed before freezing. To eliminate these uncertainties, mutants locked into kinase-‘on’ and ‘off’ states without the ability to adapt were analysed. Like in E. coli, deletion of the methylesterase CheB in C. crescentus prevents demethylation of the MCPs, trapping the cells in a fully methylated state that activates CheA (Ely et al., 1986). Deletion of the methyltransferase CheR has the opposite effect, preventing receptor methylation and locking the cells into a state in which CheA is inactive (Ely et al., 1986). For our analysis, we generated two mutant strains: a ΔcheB strain (MT292) with in-frame deletions of both chromosomal cheB copies (cheB1 and cheB2), and a ΔcheR strain (MT293) with in-frame deletions of all three cheRs present in the genome (cheR1, cheR2 and cheR3). Cell growth, cell cycle progression and cell morphology were similar to wild type in both cases. As expected, both strains were unable to perform chemotaxis, as both mutants lack the ability to compare current environmental conditions to the recent past (Fig. 2B). To confirm the effect of the mutations, we analysed swimming behaviour in light microscope movies. While ΔcheB cells reversed swimming direction unusually frequently, the trajectories of ΔcheR cells were generally very smooth, as expected for kinase-on and -off states respectively (Fig. 3, Movies S2, S3 and S4).
Figure 3.
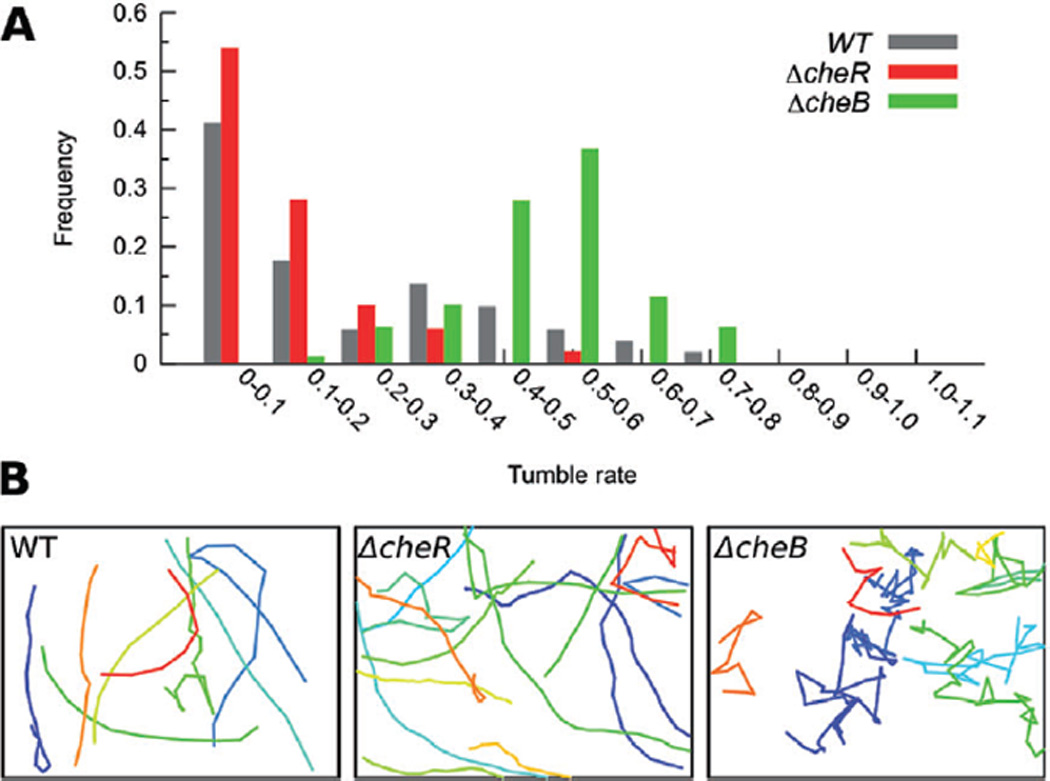
Swimming behaviour of the ΔcheB and ΔcheR mutants. A histogram of tumble rate (defined as a change in direction by more than 45°) is shown for the wild-type and mutant strains (A) above sample trajectories of different cells (colours) in movies of swimming cells (B). The frequent changes in swimming direction (‘tumbles’) observed in the ΔcheB mutant mimic the behaviour of cells sensing repellent prior to adaptation, whereas the smooth trajectories of the ΔcheR strain mimic cells sensing attractant prior to adaptation. Trajectories from 80 ΔcheB, 61 ΔcheR and 72 wild-type cells were analysed for the histogram; the trajectories in (B) represent a subset of these.
ΔcheB and ΔcheR strains exhibit similarly ordered arrays
To visualize the receptor packing in the genetically locked kinase-on and -off states, ΔcheB and ΔcheR strains were grown overnight in PYE media at 30°C and plunge-frozen for ECT as before. Cryotomograms of 10 ΔcheB and 7 ΔcheR swarmer cells were recorded, and all exhibited chemoreceptor arrays. In four of these tomograms (two of each mutant), the orientation of the arrays with respect to the beam prevented clear visualization of their packing arrangement (the oft-cited ‘missing wedge’ effect caused by the restricted tilt range (typically ± 60°) of the specimen holder). In the other 13 cells, the chemoreceptors were clearly packed in the same ~12 nm hexagonal lattice, measured right above the CheA/CheW base plate where the lattice was most obvious (Fig. 1G–L, Movies S5 and S6), and the distance between the base plate and the inner membrane was 31 nm (measured from the centre of the inner membrane to the centre of the CheA/CheW base plate), just as in adapted chemoreceptor arrays grown in either PYE or M2G minimal media or recently exposed to galactose. The estimated number of receptors ranged from 2100 to 4800 (2100, 2700, 2800, 4000 and 4800) for ΔcheB and 3000–4800 (3000, 3000, 3100, 4700 and 4800) for ΔcheR. Although average size gave the impression that these arrays were larger than wild-type arrays grown in minimal medium, this difference was not significant as determined by the Mann–Whitney rank-sum test (P < 0.05, U = 3 for both, and n1,n2 = 4,5 for both).
The best cryotomogram of each mutant was selected for subvolume averaging. Side views of the averages from the ΔcheB (Fig. 4A) and ΔcheR strains (Fig. 4B) again reveal their similarity to each other and to the published wild-type average (Briegel et al., 2008). In both averages, the cytoplasmic receptor tips are clearly discernable, indicating a well-organized array. About 18 nm from the base plate towards the inner membrane, however, the receptor densities disappear from both averages, just as we have observed previously in adapted wild-type cells (Briegel et al., 2008), showing that the cytoplasmic region near the inner membrane of the receptors is dynamic and does not retain the ordered hexagonal packing of the cytoplasmic tips. In the well-ordered region, the receptor densities emerge at a slight angle to the base plate (not perpendicular), in agreement with the expected trimer-of-dimers packing (Kim et al., 1999; Briegel et al., 2008).
Figure 4.
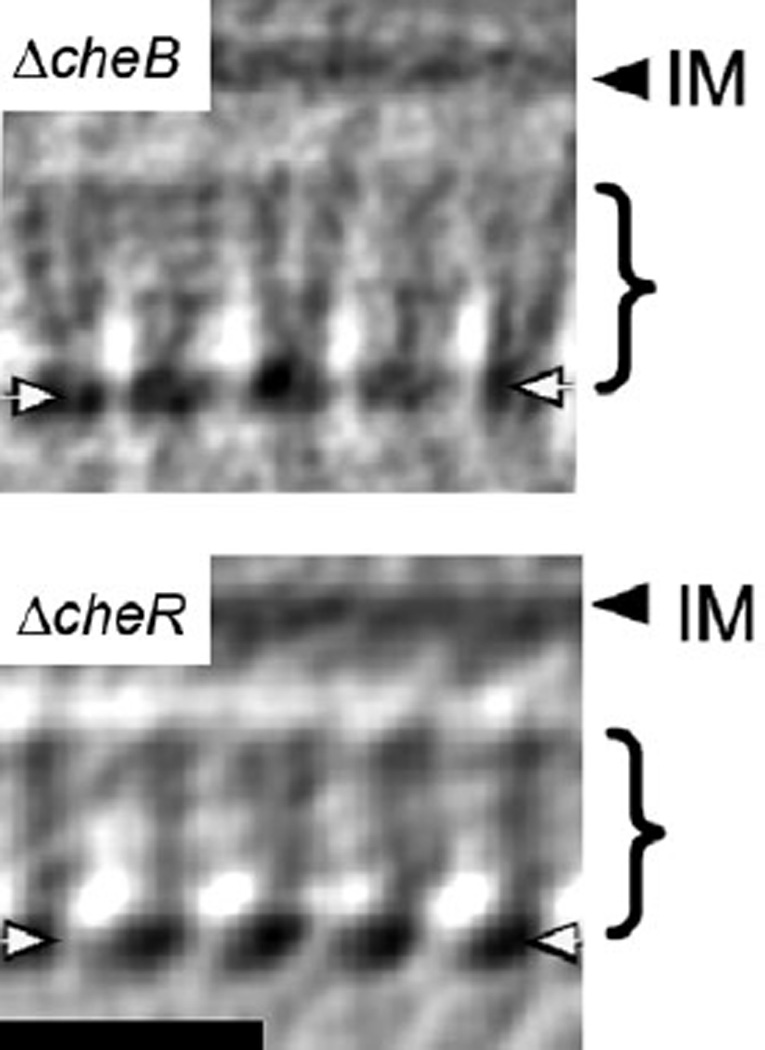
Side views of subvolume averages of the mutant strains. Receptors are seen as a series of regularly spaced densities in the CheA/CheW base plate (white arrows) and extending towards the inner membrane for ~18 nm (brackets). The absence of densities in the remaining ~13 nm towards the inner membrane (IM), as well as in the periplasmic region, indicates a lack of regular order. This region presumably includes the two HAMP and linker regions characteristic of C. crescentus MCPs as well as the transmembrane and periplasmic domains. Receptor densities emerge at an acute angle to the base plate, as expected for trimers-of-dimers. Note that the quality of both averages is not identical due to slight quality differences in the tomograms and the different number of subvolumes averaged (100 ΔcheB and 50 ΔcheR subvolumes). Bar: 25 nm. Braces indicate the ordered region of the receptors.
Discussion
Bacterial chemotaxis has served as a paradigm for cell signalling in general. Despite its importance, however, the structural changes involved in signal transduction are not yet understood. Recent advances in imaging technologies, such as the development of ECT, provide us with the tools necessary to investigate chemoreceptor arrays in a near-native state of the cells at ‘macromolecular’ resolution. We previously discovered that chemoreceptors are packed in 12 nm hexagonal arrays in a wide range of bacterial species (Briegel et al., 2008; 2009; Khursigara et al., 2008a). In the current study, we tested the hypothesis that chemoreceptor arrays become disordered upon activation. Our results show that there are no major changes in receptor arrangement or array size upon activation.
Our results agree well with both in vivo FRAP studies (Schulmeister et al., 2008) and in vitro biochemical experiments (Gegner et al., 1992; Erbse and Falke, 2009). These studies showed that the receptors in the arrays do not exchange during the generation time of cells, but instead form an ultrastable core together with CheA and CheW, and that the stability of this complex is not significantly altered by either repellents or attractants. Our results also agree with two fluorescence light microscopy studies which showed that GFP-tagged chemoreceptors and CheZ phosphatases (which localize to chemoreceptor arrays) remain polarly localized upon attractant binding (Homma et al., 2004; Liberman et al., 2004).
Our results conflict, however, with other fluorescence studies. As an example, different immunofluorescent staining patterns were observed for MCPs in the attractant- or repellent-bound state. The diffuse staining pattern seen when attractant was added gave rise to the idea that attractants disrupt the chemoreceptor array packing (Lamanna et al., 2005; Borrock et al., 2008; Wu et al., 2011). In light of the results presented here, we speculate that the diffuse staining pattern did not reflect array disassembly, but instead a small conformational change that blocked antibody binding. GFP fusions may therefore be more instructive, but unfortunately these results have been conflicting as well: in one case GFP-tagged receptors remained localized upon activation (Homma et al., 2004), but in another, diffuse staining was observed (Wu et al., 2011). At present, we cannot explain this difference. We note, however, that protein localization patterns are often disrupted by the addition of tags and differed in many cases in C. crescentus, for instance, when the tag was fused to the opposite terminus (Werner et al., 2009). Given the tight packing of the receptors in the array, one wonders how the GFP tags would influence the structure.
In light of our results, we also favour the interpretation that small conformational changes such as tilting, bending or rotation of the homodimers (rather than array disassembly) could have led to the FRET signals observed (Vaknin and Berg, 2007; Besschetnova et al., 2008). Such shifts would have to be subtle, however, since severe tilting or bending would presumably pull the base plate closer to the inner membrane, and no change in that distance was detected here upon activation. Changes in FRET efficiencies can occur, however, simply because the dipole moments of the fluorophores rotate with respect to each other (Lakowicz, 1999). In the FRET experiments under discussion, fluorophores were fused to the C-termini of receptors, near the HAMP domain in space. In subvolume averages, receptor densities are seen from the CheA/W base plate towards the inner membrane for only ~18 nm, but not the remaining ~13 nm to the centre of the inner membrane or in the periplasm, indicating that the arrays are not well ordered in those layers. We estimate that the disordered region includes the two HAMP domains and linker region typical for C. crescentus MCPs (Briegel et al., 2009), the transmembrane regions and the periplasmic domains. If large conformational changes in the receptors occur during activation, they are limited to these regions, and could explain the FRET results.
The lack of density in the HAMP region observed in our averages disagrees with results described in a different ECT study (Khursigara et al., 2008b), where densities were seen near the membrane and interpreted as HAMP domains undergoing conformational changes. These differences may have arisen due to differences in receptor environments: while the arrays in our study were imaged under physiological conditions and normal expression levels, the apparently ordered HAMP regions reported in Khursigara et al. (2008b) were studied in cells strongly overexpressing the Tsr receptor. In such samples, the receptors pack much more tightly together, tail-to-tail in a ‘zippered’ arrangement in the likely absence of CheA and CheW. Thus it remains unclear what conformational changes occur in HAMP domains in natural arrays upon activation.
We further showed that, at least in C. crescentus, chemoreceptor array packing is not affected by growth in minimal media. This result also conflicts with another ECT study where it was claimed that in E. coli, receptors pack more tightly in minimal media (Khursigara et al., 2011). While it is possible that E. coli and C. crescentus arrays react differently, this seems unlikely, given the strong homologies of chemotactic systems throughout Bacteria (Briegel et al., 2009; Wuichet and Zhulin, 2010). Our interpretation is that Khursigara et al.'s attempt to locate receptor tips within the CheA/W base plate manually in ‘side’ views (where the base plate is parallel to the electron beam) was unsuccessful due to smearing from the missing wedge, as evidenced by the presence of vertical rows of presumed receptor tips in Fig. 3B of that paper. In fact, if one calculates the power spectrum of this manual segmentation (their Fig. 3B), no hexagonal order at all is apparent. Our study relied on ‘top’ views of the arrays where the hexagonal lattice was unobscured by the missing wedge, and clearly showed that the 12 nm packing is maintained regardless of growth media or activation state.
We conclude that chemoreceptors form stable scaffolds that mediate chemotactic signals through small structural changes that do not involve array disassembly. This seems reasonable since the signal communicated through the membrane seems to be just a small, piston-like shift of transmembrane helix TM2 (Falke and Hazelbauer, 2001; Hazelbauer et al., 2008; Falke and Erbse, 2009), and the arrays appear to switch cooperatively between activated and inactivated states rapidly (Li and Weis, 2000; Sourjik and Berg, 2002).
Experimental procedures
Construction of ΔcheB and ΔcheR mutant strains
To generate plasmids for in-frame deletion of the C. crescentus cheB and cheR homologues, the upstream and downstream flanking regions of each target gene, including its first and last 12 codons, were PCR-amplified. After treatment with suitable restriction enzymes, the two fragments of each gene were combined and ligated into pNPTS138. All plasmids were verified by DNA sequencing. Gene replacement was achieved by a two-step procedure, using sucrose counter-selection to identify clones that have undergone double homologous recombination (Stephens et al., 1996). Strain MT292 was generated by consecutive deletion of cheB1 and cheB2 with the help of plasmids pMT941 and pMT942. Strain MT293 resulted from consecutive deletion of cheR1, cheR2 and cheR3 using pMT940, pMT943 and pMT944.
Strains and plasmids
Strains and plasmids used in this study are listed in Tables 1 and 2 respectively.
Table 1.
Strains used in this study.
| Strain | Characteristic | References |
|---|---|---|
| CB15N | Wild-type strain | Evinger and Agabian (1977) |
| MT292 | CB15N ΔCC0436 (cheB1) ΔCC0597 (cheB2) | This study |
| MT293 | CB15N ΔCC0435 (cheR1) ΔCC0598 (cheR2) ΔCC3472 (cheR3) | This study |
Table 2.
Plasmids used in this study.
| Plasmids | Characteristic | References |
|---|---|---|
| pNPTS138 | sacB-containing suicide vector for double homologous recombination, KanR | M.R.K. Alley (unpublished) |
| pMT940 | pNPTS138 derivative for generating an in-frame deletion in CC0435 (cheR1) | This study |
| pMT941 | pNPTS138 derivative for generating an in-frame deletion in CC0436 (cheB1) | This study |
| pMT942 | pNPTS138 derivative for generating an in-frame deletion in CC0597 (cheB2) | This study |
| pMT943 | pNPTS138 derivative for generating an in-frame deletion in CC0598 (cheR2) | This study |
| pMT944 | pNPTS138 derivative for generating an in-frame deletion in CC3472 (cheR3) | This study |
Primers
Capital letters indicate bases that are not complementary to the target sequences. Bold letters indicate restriction sites.
| CC0597-1 | ATAAGCTTcgaccattacggtctggtcgtcgacga |
| CC0597-2 | TTAAGCTAGCcacgggatgggctgtcgccgcgac |
| CC0597-3 | TTAAGCTAGCccctatatagcgtcaagagcccggagcg |
| CC0597-4 | TAGAATTCctgctgcgaagctcattcttggcgc |
| CC0436-1 | ATAAGCTTttcgatgatctgcgcgccaatgtcc |
| CC0436-2 | TTAAGCTAGCgtcgtcgacgacaagaacgcgaatcttg |
| CC0436-3 | TTAAGCTAGCcagtccatcctcgatttggcctccgc |
| CC0436-4 | TAGAATTCcggagtccggcagcaggaagtgattc |
| CC0435-1 | ATAAGCTTgatcctcgacgtcgacgccacgatc |
| CC0435-2 | TTAAGCTAGCgcggcggtcagaagagggtgcggtg |
| CC0435-3 | TTAAGCTAGCtgcggcttgaccgcctaccggaagg |
| CC0435-4 | TAGAATTCcatgtgctgggtgatgacggtcggc |
| CC0598-1 | ATAAGCTTgtggtggtggcgtcctccacaggag |
| CC0598-2 | TAGGATCCgagctcggcgaaaaacagaaggttctgagg |
| CC0598-3 | TAGGATCCgacgccagcgccgcgccgcac |
| CC0598-4 | TAGAATTCttcgtgatcctcgctcgtggcagtg |
| CC3472-1 | ATAAGCTTtgggtgcaggcggcctatctgaacg |
| CC3472-2 | TTAAGCTAGCcgccagcaggtccatgtcctcgggc |
| CC3472-3 | TTAAGCTAGCctgcgcgatccgtccatccgcgcg |
| CC3472-4 | TAGAATTCgaggccatcaccaacgcccagaagc |
Specimen preparation for ECT experiments
Caulobacter wild type (CB15N) and chemotaxis mutant strains were grown overnight at 30°C in PYE or minimal media (M2G) with glucose as the carbon source (Ely, 1991). One millilitre of cell suspension was concentrated for 5 min at 1500 rcf and resuspended in 30–50 µl of supernatant. Alternatively, to enrich swarmer cells for the attractant exposure experiments, cells were synchronized according to published protocols (Tsay and Alley, 2001). Immediately before plunge freezing, 20 µl of concentrated cell suspension was mixed with colloidal gold solution that was previously treated with BSA to avoid particle aggregation (Iancu et al., 2007). Four microlitres of the gold-and-cell solution was applied to a glow discharged, R2/2 copper/Rhodium Quantifoil™ grid (Quantifoil Micro Tools, Jena, Germany) and blotted and plunged into liquid ethane or a liquid ethane/propane mixture using a Vitrobot (FEI Company, Hillsboro, OR) (Iancu et al., 2007; Tivol et al., 2008). For the attractant experiments, the cells were resuspended in pre-warmed M2G medium after synchronization and incubated for 5 min to allow recovery from the cold temperatures (4°C) necessary during the synchronization procedure. Cells were then mixed with the colloidal gold pellet, 3 µl were applied to the EM grid, and 1 µl of 40 mM Galactose in M2G medium was added right before blotting and freeing. The time from attractant addition until complete vitrification was 7, 10 or 110 s. Images were collected using a FEI Polara™ (FEI Company, Hillsboro, OR, USA), 300 kV FEG transmission electron microscope equipped with a Gatan energy filter (slit width 20 eV) on a lens-coupled 4 × 4 k Ultracam (Gatan, Pleasanton, CA). Tilt series from −60° to 60° with an increment of 1° were recorded semi-automatically at 8–12 µm underfocus using UCSF-Tomo or Leginon (Zheng et al., 2007; Suloway et al., 2009). A cumulative dose of 200 e−/Å2 or less was used for each tilt series.
Image processing
Three-dimensional reconstructions were calculated using the IMOD software package (Mastronarde, 2005). To enhance the signal-to-noise ratio of the well-ordered parts of the chemoreceptor arrays, subvolume averaging was applied using the PEET software package (Nicastro et al., 2006). Segmentations were done manually using the Amira software package (Mercury Computer Systems). The number of receptors in the arrays was calculated as described previously (Briegel et al., 2008).
Light microscopy
Motion analysis was performed using a Nikon Eclipse 90i microscope, equipped with a Coolsnap HQ2 camera (Photometrics, Tuscon, AZ) and operated using the Metamorph software (Molecular Devices, Chicago, IL). Cell positions in individual frames were manually determined using IMOD (Mastronarde, 1997), and tracked with custom-written python scripts (available on request). A bacterial ‘tumble’ was defined as a change in trajectory greater than 45° over three successive time points.
Swim assays
Galactose assay
Wild-type cultures were stabbed about halfway between the centre of the plate and the outer rim into minimal media swim plates (M2G media without glucose containing 0.126% agar). A sterile filter paper ~7 mm in diameter was soaked in glucose-free M2G media containing 1 M galactose and placed in the centre of the swim plate. The plates were incubated for 72 h at room temperature until photographed (Parkinson, 2007).
Wild-type and mutant assay
Wild-type and mutant strains were spotted onto glucose-free M2G plates about halfway between the centre of the plate and the outer rim (M2G media without glucose containing 0.25% agar). A sterile filter paper ~7 mm in diameter was soaked in glucose-free M2G media containing 1 M galactose and placed in the centre of the swim plate. The plates were incubated at room temperature and incubated for 7 days until photographed (Parkinson, 2007).
Supplementary Material
Acknowledgements
This work was supported in part by the Howard Hughes Medical Institute and by gifts to Caltech from the Gordon and Betty Moore Foundation.
References
- Alley MRK, Gomes SL, Alexander W, Shapiro L. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus. Genetics. 1991;129:333–342. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besschetnova TY, Montefusco DJ, Asinas AE, Shrout AL, Antommattei FM, Weis RM. Receptor density balances signal stimulation and attenuation in membrane-assembled complexes of bacterial chemotaxis signaling proteins. Proc Natl Acad Sci USA. 2008;105:12289–12294. doi: 10.1073/pnas.0802868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Kaplan N, Hess JF, Simon MI. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci USA. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrock MJ, Kolonko EM, Kiessling LL. Chemical probes of bacterial signal transduction reveal that repellents stabilize and attractants destabilize the chemoreceptor array. ASC Chem Biol. 2008;3:101–109. doi: 10.1021/cb700211s. [DOI] [PubMed] [Google Scholar]
- Boukhvalova MS, Dahlquist FW, Stewart RC. CheW binding interactions with CheA and Tar: importance for chemotaxis signaling in E. coli. J Biol Chem. 2002;277:22251–22259. doi: 10.1074/jbc.M110908200. [DOI] [PubMed] [Google Scholar]
- Briegel A, Ding HJ, Li Z, Werner J, Gitai Z, Dias DP, et al. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, et al. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci USA. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke TAJ, Bray D. Heightened sensitivity of a lattice of membrane receptors. Proc Natl Acad Sci USA. 1999;96:10104–10108. doi: 10.1073/pnas.96.18.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- Ely B, Gerardot CJ, Fleming DL, Gomes SL, Frederikse P, Shapiro L. General nonchemotactic mutants of C. crescentus. Genetics. 1986;114:717–730. doi: 10.1093/genetics/114.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres RG, Wingreen NS. Precise adaptation in bacterial chemotaxis through ‘assistance neighborhoods’. Proc Natl Acad Sci USA. 2006;103:13040–13044. doi: 10.1073/pnas.0603101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbse AH, Falke JJ. The core signaling proteins of bacterial chemotaxis assemble to form an ultrastable complex. Biochemistry. 2009;48:6975–6987. doi: 10.1021/bi900641c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke JJ, Erbse AH. The piston rises again. Structure. 2009;17:1149–1151. doi: 10.1016/j.str.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegner JA, Graham DR, Roth AF, Dahlquist FW. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- Gomes SL, Shapiro L. Differential expression and positioning of chemotaxis methylation proteins in Caulobacter. J Mol Biol. 1984;178:551–568. doi: 10.1016/0022-2836(84)90238-9. [DOI] [PubMed] [Google Scholar]
- Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J. Self-organization of the Escherichia coli chemotaxis network imaged with super resolution light microscopy. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000137. e1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M, Shiomi D, Homma M, Kawagishi I. Attractant binding alters arrangement of chemoreceptor dimers within its cluster at the cell pole. Proc Natl Acad Sci USA. 2004;101:3462–3467. doi: 10.1073/pnas.0306660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu CV, Tivol WF, Schooler JB, Dias DP, Henderson GP, Murphy GE, et al. Electron cryotomography sample preparation using the Vitrobot. Nat Protoc. 2007;1:2813–2819. doi: 10.1038/nprot.2006.432. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Wang SC, Shapiro L. Dynamic localisation of proteins and DNA during a bacterial cell cycle. Nat Rev. 2002;3:167–176. doi: 10.1038/nrm758. [DOI] [PubMed] [Google Scholar]
- Jiang L, Ouyang Q, Tu Y. Quantitative modeling of Escherichia coli chemotactic motion in environments varying in space and time. PLOS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000735. e1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursigara CM, Wu X, Subramaniam S. Chemoreceptors in Caulobacter crescentus: trimers of receptor dimers in a partially ordered hexagonally packed array. J Bacteriol. 2008a;190:6805–6810. doi: 10.1128/JB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursigara CM, Wu X, Zhang P, Lefman J, Subramaniam S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc Natl Acad Sci USA. 2008b;105:16555–16560. doi: 10.1073/pnas.0806401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursigara CM, Lan G, Neumann S, Wu X, Ravindran S, Borgia MJ, et al. Lateral density of receptor arrays in the membrane plane influences sensitivity of the E. coli chemotaxis response. EMBO J. 2011;30:1719–1729. doi: 10.1038/emboj.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- Lakowicz JR. Principles of Fluorescence Spectroscopy. New York: Kluwer Academic/Plenum; 1999. [Google Scholar]
- Lamanna AC, Ordal GW, Kiessling LL. Large increases in attractant concentration disrupt the polar localization of bacterial chemoreceptors. Mol Microbiol. 2005;57:774–785. doi: 10.1111/j.1365-2958.2005.04728.x. [DOI] [PubMed] [Google Scholar]
- Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. Adaptational assistance in clusters of bacterial chemoreceptors. Mol Microbiol. 2005;56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- Liberman L, Berg HC, Sourjik V. Effect of chemoreceptor modification on assembly and activity of the receptor–kinase complex in Escherichia coli. J Bacteriol. 2004;186:6643–6646. doi: 10.1128/JB.186.19.6643-6646.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybarger SR, Maddock J. Polarity in action: asymmetric protein localization in bacteria. J Bacteriol. 2001;183:3261–3267. doi: 10.1128/JB.183.11.3261-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Manson MD. Bacterial chemoreceptors as membrane-spanning allosteric enzymes. In: Spiro S, Dixon R, editors. Sensory Mechanisms in Bacteria: Molecular Aspects of Signal Recognition. Norfolk: Caister Academic Press; 2010. pp. 107–149. [Google Scholar]
- Mastronarde DA. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz CL, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Parkinson JS. A ‘bucket of light’ for viewing bacterial colonies in soft agar. Methods Enzymol. 2007;423:432–435. doi: 10.1016/S0076-6879(07)23020-4. [DOI] [PubMed] [Google Scholar]
- Parkinson JS. Signaling mechanisms of HEMP domains in chemoreceptors and sensor kinases. Annu Rev Microbiol. 2010;64:101–122. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Revello PT. Sensory adaptation mutants of E. coli. Cell. 1978;15:1221–1230. doi: 10.1016/0092-8674(78)90048-x. [DOI] [PubMed] [Google Scholar]
- Schulmeister S, Ruttorf M, Thiem S, Kentner D, Lebiedz D, Sourjik V. Protein exchange dynamics at chemoreceptor clusters in E. coli. Proc Natl Acad Sci USA. 2008;105:6403–6408. doi: 10.1073/pnas.0710611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L. Generation of polarity during Caulobacter cell differentiation. Annu Rev Cell Biol. 1985;1:173–207. doi: 10.1146/annurev.cb.01.110185.001133. [DOI] [PubMed] [Google Scholar]
- Shaw P, Gomes SL, Sweeney K, Ely B, Shapiro L. Methylation involved in chemotaxis is regulated during Caulobacter differentiation. Proc Natl Acad Sci USA. 1983;80:5261–5265. [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Banno S, Homma M, Kawagishi I. Stabilization of polar localization of a chemoreceptor via its covalent modifications and its communication with a different chemoreceptor. J Bacteriol. 2005;187:7647–7654. doi: 10.1128/JB.187.22.7647-7654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Berg H. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer WR, Koshland DE. Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci USA. 1977;74:533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C, Reseinauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci USA. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–251. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Stock JB, Koshland DE. A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci USA. 1978;75:3659–3663. doi: 10.1073/pnas.75.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, et al. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol. 2009;167:11–18. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam R, Saier MHJ. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivol W, Briegel A, Jensen GJ. An improved cryogen for plunge freezing. Microsc Microanal. 2008;14:375–379. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay JW, Alley MRK. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J Bacteriol. 2001;183:5001–5007. doi: 10.1128/JB.183.17.5001-5007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich LE, Zhulin IB. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 2010;38:D401–D407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaknin A, Berg HC. Physical responses of bacterial chemoreceptors. J Mol Biol. 2007;366:1416–1423. doi: 10.1016/j.jmb.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- Wagner JK, Brun YV. Out on a limb: how the Caulobacter stalk can boost the study of bacterial shape. Mol Microbiol. 2007;64:28–33. doi: 10.1111/j.1365-2958.2007.05633.x. [DOI] [PubMed] [Google Scholar]
- Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci USA. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Walukietwicz HE, Glekas GD, Ordal GW, Rao CV. Attractant binding induces distinct structural changes to the polar and lateral signaling clusters in Bacillus subtilis chemotaxis. J Biol Chem. 2011;286:2587–2595. doi: 10.1074/jbc.M110.188664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010;3 doi: 10.1126/scisignal.2000724. ra. 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng QS, Keszthelyi B, Branlund E, Lyle JM, Braunfeld MB, Sedat JW, Agard DA. UCSF tomography: an integrated software suite for real-time electron microscopic tomographic data collection, alignment and reconstruction. J Struct Biol. 2007;157:138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


