Abstract
We have identified from the mutualistic grass endophyte Epichloë festucae a non-ribosomal peptide synthetase gene (sidN) encoding a siderophore synthetase. The enzymatic product of SidN is shown to be a novel extracellular siderophore designated as epichloënin A, related to ferrirubin from the ferrichrome family. Targeted gene disruption of sidN eliminated biosynthesis of epichloënin A in vitro and in planta. During iron-depleted axenic growth, ΔsidN mutants accumulated the pathway intermediate N5-trans-anhydromevalonyl-N5-hydroxyornithine (trans-AMHO), displayed sensitivity to oxidative stress and showed deficiencies in both polarized hyphal growth and sporulation. Infection of Lolium perenne (perennial ryegrass) with ΔsidN mutants resulted in perturbations of the endophyte-grass symbioses. Deviations from the characteristic tightly regulated synchronous growth of the fungus with its plant partner were observed and infected plants were stunted. Analysis of these plants by light and transmission electron microscopy revealed abnormalities in the distribution and localization of ΔsidN mutant hyphae as well as deformities in hyphal ultrastructure. We hypothesize that lack of epichloënin A alters iron homeostasis of the symbiotum, changing it from mutually beneficial to antagonistic. Iron itself or epichloënin A may serve as an important molecular/cellular signal for controlling fungal growth and hence the symbiotic interaction.
Author Summary
Perennial ryegrass is a cool-season grass that is agriculturally important as forage, especially in Australasia. Essential to the longevity and maintenance of healthy pastures are epichloae fungi, such as Epichloë/Neotyphodium spp. that live inside these grasses in a symbiotic manner. These fungi produce bioactive compounds that can protect its host grass from various abiotic and biotic stresses. They depend on resource supply by their host, including nutrients that are taken up by the roots of the grass host. We have found that iron, an indispensable element for growth, is acquired by Epichloë festucae from its grass host via the secretion of the extracellular siderophore epichloënin A. Without iron-mediated siderophore uptake to capture host iron, the symbiotic relationship becomes disturbed and the growth habit of the fungus is no longer restricted. Our results indicate that secretion of epichloënin A enables the symbiotic fungus to compete for plant iron while avoiding fungal over-growth inside the host plant.
Introduction
Iron is an essential nutrient for almost all organisms (except for some lactobacilli [1]) due to its central role in vital cellular reactions. The redox properties of iron confer a catalytic function essential for fundamental metabolic pathways such as DNA synthesis, respiration and photosynthesis [2], [3]. Even though iron is a highly abundant metal, in aerobic environments bioavailability is low because ferric iron forms insoluble oxides through reaction with oxygen. Iron concentrations in tissues must also be carefully regulated since free iron is deleterious given that it has the potential to catalyze the production of cytotoxic reactive oxygen species (ROS) through the Haber-Weiss/Fenton reactions [4]. Controlling iron homeostasis is therefore an essential function and organisms have developed complex strategies for iron uptake, utilization, transport and storage [5]. The uptake of iron is presumably the principal regulatory point of iron homeostasis and multiple high- and low-affinity iron uptake systems have evolved [6].
In mammals and plants excess iron is tightly sequestered by high-affinity binding proteins [7], [8], and deliberately withdrawing iron is a documented defense strategy employed by animal hosts against invading bacterial pathogens in order to limit their growth [9], [10]. For microbes to acquire limited iron from animal or plant hosts, high affinity iron uptake systems are required. One such mechanism is siderophore-mediated iron uptake. Under iron starvation, most fungi and bacteria synthesize siderophores, low molecular weight (Mr<1,500) iron-chelating agents, to solubilize ferric iron and control their intracellular iron levels [11]–[13]. The functional role of siderophores may be extracellular – secreted as iron-free siderophores to chelate iron for cellular iron uptake; and/or intracellular – located within the cell for intracellular iron storage. A number of roles have been attributed to fungal siderophores and these include functions such as virulence factors and asexual/sexual determinants [14]–[21]. Another major type of high affinity iron acquisition in fungi is reductive iron assimilation (RIA) which is based on a ferroxidation/permeation uptake [6], [22], [23]. Some fungi are capable of utilizing both of these high affinity uptake systems; examples are Ustilago maydis, Aspergillus fumigatus and Fusarium graminearum [15], [16], [24]–[28]. Nevertheless, characterization studies of both siderophore and RIA uptake systems in the same fungal species through respective gene deletion studies have shown that only one of these two systems is essential for mammalian or plant disease by a given fungal pathogen and which one is indispensable thus appears to be fungal species dependent and possibly related to their lifestyles. Examples of fungi demonstrated to require the siderophore system for virulence are A. fumigatus, Histoplasma capsulatum and F. graminearum [16], [19], [29], whereas RIA is essential for the virulence of the basidomycetes U. maydis, Candida albicans and Cryptococcus neoformans [15], [30], [31].
Nearly all fungi produce hydroxamate-type siderophores [32], which are typically composed of three hydroxamate groups linked by peptide or ester bonds to form an octahedral complex. Siderophores are classified into four major structural types: ferrichromes, fusarinines, coprogens and rhodotorulic acid [12]. Their formation by the linkage of hydroxamate groups or additional amino acids in the case of ferrichrome-type siderophores is catalyzed by non-ribosomal peptide synthetases (NRPS) [28]. NRPSs are multifunctional enzymes that synthesize peptides by a thio-template mechanism and are modular in structure. A typical module consists of an adenylation (A) domain for substrate specificity, a peptidyl carrier domain (T), which binds a 4′-phosphopantetheine cofactor, and a condensation (C) domain for bond formation [33]. NRPS genes functionally identified to encode fungal siderophore synthetases with an extracellular role are sid2 and fer3 of U. maydis and sib1 of Schizosacccharomyces pombe, involved in the synthesis of ferrichrome-type siderophores [28], [34], [35], sidD from A. fumigatus involved in triacetylfusarinine C (TAFC) production [36] as well as NPS6 of Alternaria brassicicola, F. graminearum, and Cochliobolus heterostrophus involved in the respective production of Nα-dimethylcoprogen, TAFC and coprogens [19]. NPS6 deletion studies have demonstrated that these extracellular siderophores play a role in fungal virulence to plants [19]. Recently, another NPS6 ortholog responsible for the production of coprogens was identified from the plant pathogenic fungus Magnaporthe grisea, but loss of the corresponding gene did not affect virulence in rice [37], whereas the gene encoding the NRPS responsible for the synthesis of the intracellular siderophore, ferricrocin, was required for full virulence of M. grisea [20]. Fungi that form mutualistic relationships with plants such as the widespread symbioses of mycorrhizal fungi with terrestrial plant communities also produce siderophores [38]–[40]. In contrast to fungal pathogens, siderophore production by mycorrhizal fungi has been postulated to aid their hosts by improving solubilization of insoluble iron oxides resulting in a positive effect on the iron nutrition of the host plant [41].
The epichloae fungi of family Clavicipitaceae (comprising genera Epichloë and Neotyphodium) form symbioses with temperate grasses of the subfamily Poöideae [42]. These Poöideae-epichloae associations comprise an evolutionary continuum from mutualistic to antagonistic, with the nature of this relationship determined by the importance of vertical (via host seeds) versus horizontal (ascospore mediated) transmission of the fungus [42]. During colonisation of grass leaves, the endosymbiont's growth is tightly regulated and synchronized with the growth of its host plant [43]. Fungal hyphae are confined to the intercellular spaces of leaf sheaths and blades where the endophyte induces no symptoms [44]. Epichloid symbioses can be mutually beneficial with the plant providing the endophyte with nutrients, and the endophyte conferring biotic and abiotic advantages to the host plant [44]–[48]. Improved herbivore resistance of infected plants is linked to the production of fungal secondary metabolites (alkaloids), and it appears that the host plant plays a key role in the regulation of some of these metabolites [49], [50]. In a study to investigate the distribution and diversity of NRPS gene families within the epichloae, two NRPS fragments, NRPS2 and NRPS9, were isolated with proposed functions identified as siderophore-like [51]. Our unpublished data shows that NRPS9 encodes sidC, the siderophore synthetase for ferricrocin (L. Johnson et al., unpublished results). NRPS2, renamed as sidN, was confirmed to be a siderophore-synthesizing NRPS from three-dimensional structural elucidation and biochemical studies of the third adenylation domain of SidN [52]. These studies showed that this domain activates anhydromevalonyl-N6-hydroxy-L-ornithine (AMHO), a large hydroxamate amino acid known to be a component of fungal siderophores [52]. More recently, the structure of a novel ferrichrome-type siderophore containing AMHO moieties which we have designated as epichloenin A, has been elucidated by high resolution tandem mass spectrometry (HRMSMS) and NMR from iron-depleted cultures of E. festucae [53]. An additional minor variant, epichloënin B, has also been detected in cultures of WT and C-sidN strains [53]. Here, we report a study establishing that sidN (formerly NRPS2) encodes a siderophore synthetase required for the biosynthesis of the extracellular siderophore, epichloënin A. Our research shows that sidN is required for maintaining the mutualistic interaction of the endophyte E. festucae with perennial ryegrass.
Results
sidN Encodes a Siderophore Synthetase Involved in the Biosynthesis of the Extracellular Siderophore, Epichloënin A
To investigate whether sidN (formally NRPS2) is involved in the biosynthesis of epichloënin A and its role in endophyte-grass symbioses, a gene disruption construct was designed and the gene disruption performed by homologous recombination in E. festucae wild-type (WT) strain Fl1 (see Methods for details). Four independent ΔsidN mutants (ΔsidN 85, 82, 54, and 26) were identified at a homologous recombination frequency of 10%. These four mutants displayed similar phenotypes as reported throughout this study.
The axenic vegetative growth characteristics of the ΔsidN E. festucae mutants were examined on defined medium (DM) and on DM supplemented with the ferrous iron chelator bathophenanthrolinedisulfonic acid (BPS), which is impermeable to cell membranes [54], [55] (Figure 1A). BPS supplementation sequesters trace ferrous iron from the extracellular medium, subsequently inhibiting RIA iron uptake by removing the substrate for this process. For the WT, addition of BPS to DM moderately reduced the radial growth rate (by approximately 20%), whereas radial growth of the ΔsidN mutants was almost completely inhibited (shown for two independent ΔsidN mutants in Figure 1A). The lack of growth by the mutants on this medium is explainable by the loss of both RIA-mediated (via ferrous iron BPS chelation) and siderophore mediated iron uptake from the extracellular medium (i.e. loss of biosynthesis of the extracellular siderophore), and demonstrates the presence of RIA in E. festucae. The inhibition of radial growth on DM+BPS medium could be moderately complemented by the addition of enriched culture supernatants (Sephadex-column purified culture filtrate - CF) from WT (Figure 1A), but not from ΔsidN 85 (data not shown), indicating the presence of the native extracellular siderophore in WT CF only. Growth of all ΔsidN mutant strains could also be fully restored by addition of FeSO4, or FeCl3 to DM+BPS medium (Figure 1A).
Figure 1. Iron depletion renders ΔsidN mutants incapable of axenic vegetative growth.
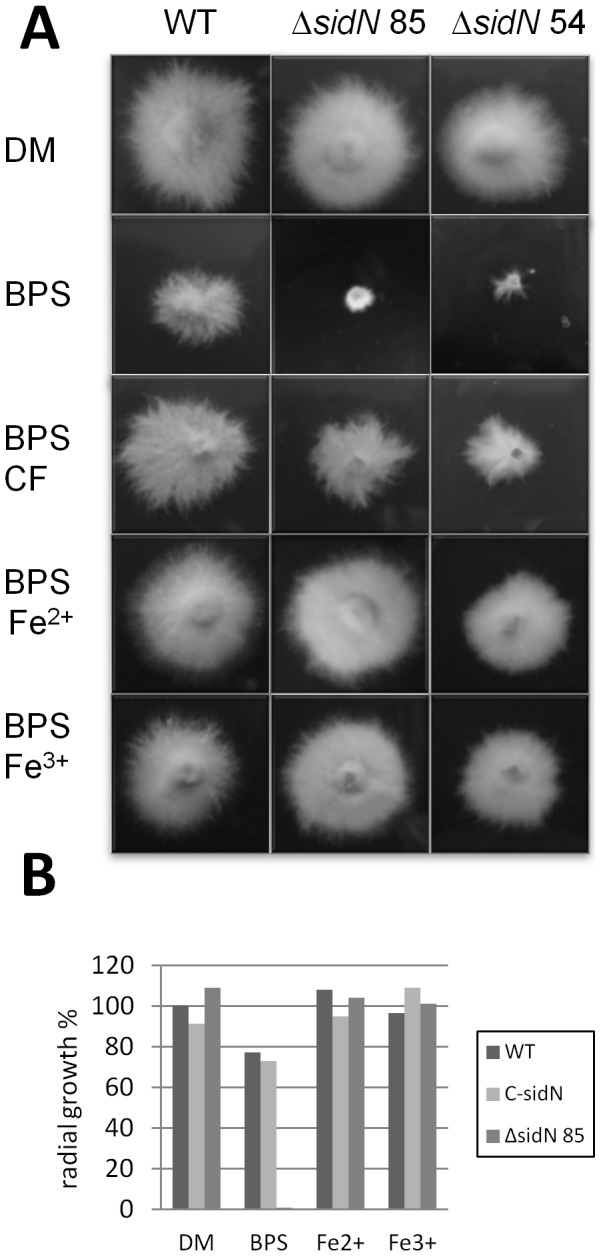
A. 16-day-old cultures of wild-type E. festucae Fl1 (WT) and ΔsidN mutant strains (ΔsidN 54 and ΔsidN 85) were grown on iron depleted defined media (DM), and DM media supplemented with 100 µM BPS, or 100 µM BPS and culture filtrate from WT (BPS CF), or 100 µM BPS and 20 µM FeS04 (BPS Fe2+) or 100 µM BPS and 20 µM FeCl3 (BPS Fe3+) respectively. B. Radial growth measurements of WT, complement (C-sidN) and ΔsidN 85 were determined by inoculating mycelial plugs in triplicate onto DM media, or DM supplemented with 100 µM BPS, DM with 100 µM BPS and 20 µM FeS04 (Fe2+) and DM with 100 µM BPS and 20 µM FeCl3 (Fe3+) respectively. Colonies were measured at 10 days. The results represent the mean of three independent experiments. The radial growth is normalized to that of WT grown on DM media.
To validate that the phenotypic effects described in the ΔsidN strains arose from gene inactivation of sidN, a complementation was performed by co-transforming protoplasts from ΔsidN 85 with fosmid clone G8-5 (containing the full-length genomic copy of E. festucae sidN) and a plasmid containing the geneticin resistance gene (pII99). Screening for complementation was carried out using an in vitro plate assay that exploited the phenotypic trait of the ΔsidN mutants to have extremely poor mycelial growth on BPS-containing, iron-depleted DM (see Figure 1A). Four independent transformants formed large colonies on BPS medium. We designated the complemented strain used for further studies C-sidN; measurements of radial growth of ΔsidN 85, WT and C-sidN show that C-sidN is able to grow at 94% of WT on BPS supplemented DM (Figure 1B). This indicates that transformation with the full-length genomic copy of sidN restored growth of ΔsidN mutants on BPS supplemented medium and congruent with sidN encoding a biosynthetic enzyme for high-affinity siderophore uptake of iron from extremely iron-depleted medium. To determine if the siderophore assembled by the biosynthetic pathway involving SidN was epichloënin A, a chemical analysis was performed on cultures of the WT, ΔsidN mutant and complemented C-sidN strains grown under iron depleted conditions (Figure 2). Culture supernatant and mycelial extracts from these strains were analyzed by liquid chromatography – mass spectrometry (LCMS). Epichloënin A (MS1 m/z 542 ([MH2]2+) was the predominant form in the supernatant of WT cultures (Figure 2), while ferriepichloënin A (MS1 m/z 569 ([MH2]2+) predominated in the mycelial extracts. Since the ratio of the iron-free to iron-bound form is much higher in the extracellular supernatant than in the mycelium, we conclude that epichloënin A is an extracellular siderophore. A similar pattern of occurrence was observed for the C-sidN supernatants, albeit at reduced concentrations compared to WT. However, both forms were absent from supernatants and mycelia of the ΔsidN mutant strains, clearly demonstrating that SidN is responsible for the assembly of epichloënin A. Figure 3A presents the postulated siderophore biosynthetic pathway according to Plattner and Diekmann [56] adapted for epichloënin A biosynthesis in E. festucae.
Figure 2. Synthesis of epichloënin A is dependent on sidN.
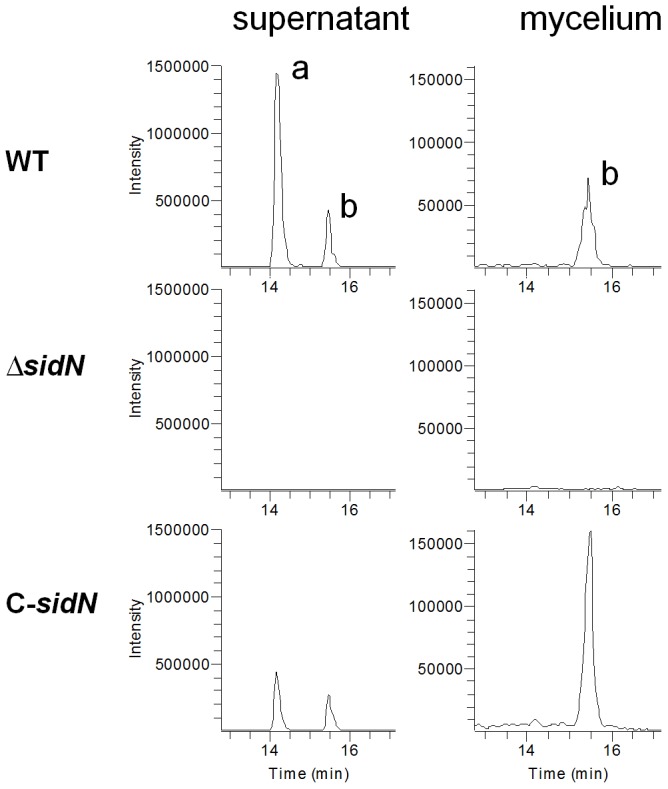
LC-MS analysis showing MS1extracted ion chromatograms for both epichloënin A (a, m/z 542) and ferriepichloënin A (b, m/z 569) in supernatant and mycelium from two week old iron-depleted cultures of wild-type E. festucae Fl1 (WT), ΔsidN mutant 85 (ΔsidN), and a complemented ΔsidN strain (C-sidN). Note scale for supernatant is 10× of that for mycelium.
Figure 3. SidN modular structure and postulated siderophore biosynthetic pathway for epichloënin A.
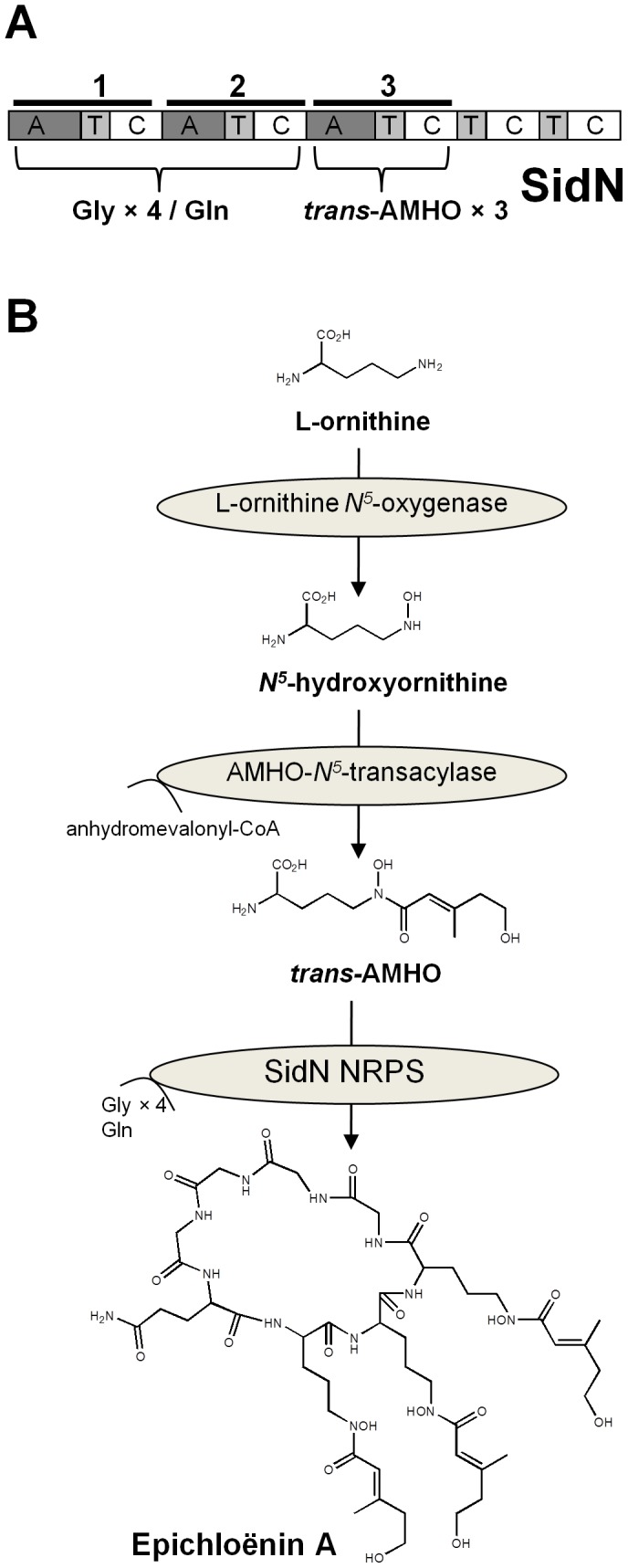
A. NRPS enzyme, SidN assembles Epichloënin A by activating and incorporating three trans-anhydromevalonylhydroxyornithine (trans-AMHO), 1 glutamine and 4 glycine moieties. This pathway is according to Plattner and Diekmann [56]. B. The three modules of SidN are composed of an A (adenylation) domain, a T (peptidyl carrier) domain and a C (condensation) domain. Proposed substrates for modules 1 and 2 are glycine (Gly) and glutamine (Gln) residues, while for module 3 are trans-AMHO residues.
The Modular Structure of SidN and Genome Analyses
To enable further investigation of sidN (formally NRPS2), including the determination of the modular architecture of this siderophore-synthesizing NRPS, we obtained the full-length gene sequence by constructing an E. festucae (strain Fl1) fosmid library. Screening of this library by PCR (using NRPS2 specific primers) gave three positive clones, of which one (clone G8-5) contained the entire genomic copy of the full-length gene. Sequencing of the E. festucae Fl1 sidN fosmid clone revealed an open reading frame (ORF) of 14,073 bp with no introns. A BLASTX analysis of the deduced amino acid sequence of sidN revealed similarities to fungal NRPS genes involved in siderophore biosynthesis. Top hits to SidN (with an expect value of 0.0) included functionally characterized siderophore NRPSs involved in the biosynthesis of ferricrocin or ferrichrome. The top hit was to A. fumigatus SidC [36] at 34% amino acid identity (52% positives). Out of the six modular architectures described for ferrichrome synthetase NRPSs by Bushley et al. [57], the most significant hits using sidN as the query sequence were to 12 of the 13 Type II ferrichrome NRPSs, followed by less similarity to other ferrichromes. These results support our previous findings that SidN encodes a siderophore biosynthetic enzyme of the ferrichrome family [52], and upholds our current discovery that SidN assembles epichloënin A, a novel ferrichrome family member. It is however interesting that the amino acid sequence analyses indicates SidN has the highest similarity to SidC which assembles ferricrocin, but the acyl groups of the hydroxamates of both epichloënin A and ferricrocin are different: for SidN it is trans- anhydromevalonyl [52], [53], whereas for ferricrocin, a simple acetyl group is incorporated into its hydroxamate group [11].
Analysis of the SidN amino acid sequence revealed three complete A-T-C modules and a terminal T-C repeat identical to Type II ferrichrome NRPSs (where A = adenylation domain, T = peptidyl carrier domain and C = condensation domain) (Figure 3B). From the work by Lee et al. [52] the third domain (A3) of SidN incorporates the hydroxamate groups of the siderophore which forms an octahedral iron complex. The other component amino acids of epichloënin A are one glutamine and four glycines [53], which we postulate are assembled by SidN A domains one (A1) and two (A2). Analysis of the putative binding pocket residues of the SidN A1 and A2 domains showed that they were dissimilar to those of other characterized fungal siderophore synthetases [28], [57] and did not allow a prediction of which A domain activated glutamine or glycine or the number of iterative cycles of their addition into the peptide. Figure 3B shows that A1 and A2 domains of SidN activate either the addition of one glutamine or four cycles of glycine addition, while the A3 domain is predicted to activate three cycles of addition of trans-AMHO residues.
Recently, the genomes of two E. festucae strains (Fl1 and E2368) have been sequenced (http://www.endophyte.uky.edu/). A BLASTN analysis of the E2368 and Fl1 sidN sequences revealed a 99% nucleotide identity with an expect value of 0.0. We also previously identified a second siderophore-like NRPS from N. lolii (NRPS9; [51]) which was located on a different supercontig to sidN in both E. festucae genomes and putatively encodes an intracellular siderophore synthetase, ferricrocin (L. Johnson et al., unpublished results). We have not identified any other siderophore-like NRPS genes in the E. festucae Fl1 and E2368 genomes.
To determine what other genes are clustered with sidN we analyzed the predicted ORFs surrounding the sidN homologue from the E. festucae Fl1 and E2368 genome databases. These analyses revealed that just one ORF (abcI), located upstream of sidN appears to be grouped with sidN forming a partial siderophore biosynthetic gene cluster, possibly sharing a divergent promoter with sidN. This ORF encodes a putative member belonging to the P-glycoprotein-like multidrug resistance (MDR) subfamily of ATP-binding cassette (ABC) transporters. A similarity search revealed a highly significant hit to ABC1 from H. capsulatum, located in the H. capsulatum SID1 gene cluster in an analogous position and orientation with respect to its siderophore synthetase, NPS1 [29]. There were no other shared regions of synteny. Of note, ABC1 is iron-regulated and contains a putative Sre1 GATA motif consensus site, for regulation by the GATA type iron regulator [29].
In vitro trans-AMHO Accumulation
Schrettl et al. [36] reported the accumulation of the siderophore precursor cis-AMHO, in the supernatant of an iron-depleted culture of a sidD deletion mutant; sidD encodes the NRPS responsible for fusarinine C biosynthesis in A. fumigatus. We therefore examined supernatants of cultures of ΔsidN mutants grown under iron-depleted conditions by LCMSMS for the presence of likely precursors of epichloënin A, and found a product of m/z 261 which we identified putatively as trans-AMHO by comparison with the retention and MS2 spectrum of cis-AMHO prepared from fusarinine C [52] (Figure S1A). This is consistent with the finding that epichloënin A incorporates trans-AMHO moieties [53]. Targeted LCMSMS analyses of mycelial extracts of the WT, ΔsidN 85 mutant and the complemented strain showed that trans-AMHO accumulation was very low in the WT, slightly higher in the complemented strain, but nearly 40-fold higher in ΔsidN 85 compared to WT, indicating that trans-AMHO is a precursor that is significantly accumulating in the mutant (Figure S1B).
In vitro Growth Characteristics of ΔsidN Mutants
Light microscopy was employed to observe the vegetative hyphal growth characteristics of the ΔsidN mutants on water agar as it is a useful medium for studying the morphology of individual hyphal strands within a colony. It is also a nutrient deficient medium. The mutants displayed various abnormalities compared to WT such as increased lateral branching, coils (not shown), atypical hyphal convolutions with a high frequency of hyphal tip and compartmental swellings (Figure 4); complementation in the C-sidN strain restored normal hyphal growth (data not shown).
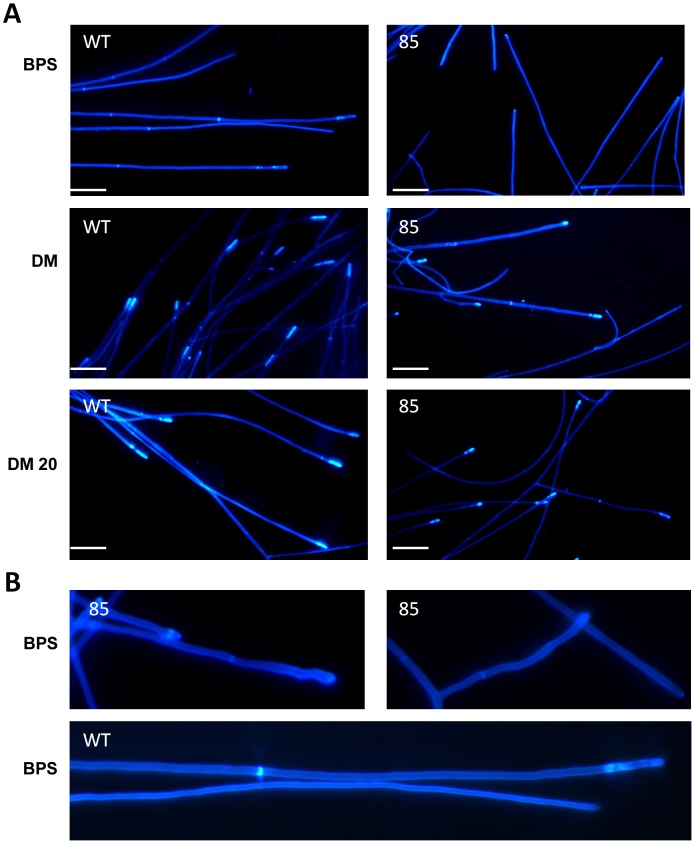
Figure 4. ΔsidN mutants display abnormal hyphal morphologies on water-agar.
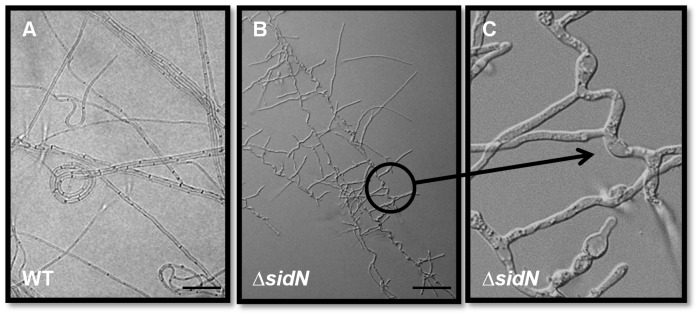
A. Mycelia of wild-type E. festucae Fl1 (WT) on water agar. B. Mycelia of ΔsidN 85 mutant proliferating on water agar by lateral branches. C. Close up of ΔsidN 85 mutant mycelia showing hyphal convolutions and swellings. Bar is 50 µM.
To evaluate the cell wall integrity of the ΔsidN mutants, staining with Calcofluor White was used to analyze the distribution of chitin (and other β-linked fungal cell wall polysaccharides) [58] in hyphae grown in liquid DM, and DM amended with BPS or 20 µM iron (Figure 5). For the WT, there were no significant differences in tip, septa, or hyphal wall staining in DM or DM supplemented with iron, however, the addition of the ferrous iron chelator BPS to the medium resulted in the majority of tips displaying no or weak speckled staining near the tip region, with septa staining not affected. Also, the hyphal contents appeared to have a much higher background stain compared to staining of hyphae in DM or DM with iron supplementation. The complement was comparable to the WT (results not shown). It is remarkable that addition of BPS, an agent that chelates ferrous iron and as a consequence inhibits RIA uptake, can dramatically alter chitin localization. A similar response to BPS supplementation was observed with stained mycelium from ΔsidN 85, yet some specific differences with respect to WT were observed under all 3 conditions tested. In DM, mutant hyphae displayed slightly less tip staining compared to the WT, and often the length of staining at the tip was reduced (Figure 5). Addition of iron appeared to improve overall tip staining length and brightness to that observed for WT hyphae. Supplementation with BPS, also radically affected the chitin distribution of mutant hyphae (similar to WT), however the pattern of hyphal growth on this medium was highly branched (as observed for ΔsidN mutants on water agar), and notably some of these branches were extending in a meandering manner (see Figure 5B). Collectively, these observations suggest that the ΔsidN mutants are defective in maintaining polar growth when iron availability is low; to our knowledge, this is a feature not previously linked to fungal siderophore mutants.
Figure 5. Chitin accumulation is abnormally distributed in ΔsidN mutants.
A. Chitin distribution pattern in hyphal tips of wild-type E. festucae Fl1 (WT) and ΔsidN mutant 85 (85) grown in liquid of iron-depleted DM (defined medium) supplemented with BPS (BPS), DM (DM) and DM supplemented with 20 µM Fe2+ (DM 20). Bars = 50 µM. B. Enlargement of meandering hyphal tips from A above of wild-type E. festucae Fl1 (WT) and ΔsidN mutant 85 (85) grown in liquid DM supplemented with BPS (BPS).
To test for sensitivity of the ΔsidN mutants to oxidative stress, hydrogen peroxide was applied to the growth medium. On complete medium (PD), only ΔsidN mutant 85 showed a slight sensitivity to H2O2, whereas on iron-depleted DM both ΔsidN mutants 54 and 85 were significantly more sensitive to H2O2 than WT (Table 1).
Table 1. Colonies of ΔsidN mutants are sensitive to hydrogen peroxide on DM.
| Growth medium | Fungal strain | Statistics | ||||
| WT | ΔsidN85 | ΔsidN54 | LSD | SED | p-value | |
| DM+H2O2 | 0.707 | 0.495 | 0.52 | 0.1115 | 0.0554 | 0.002 |
| PD+H2O2 | 0.878 | 0.724 | 0.84 | 0.1037 | 0.0408 | 0.016 |
Values given are ratios of radial growth measurements of colonies grown for 7 days at 22°C on DM (defined medium) or PD (potato dextrose) medium supplemented with 0.7 mM H2O2 versus DM or PD. Statistics were generated from an analysis of variance. LSD is the Least Significant Difference between any two means and the SED represents the Standard Error of the Difference.
A reduction in asexual sporulation (in iron-depleted medium) had been previously documented for deletion strains of C. heterostrophus NPS6 [18]. To study sporulation in the ΔsidN mutants, it was necessary to find suitable asexual sporulation conditions to generate sufficient numbers of conidia from the WT strain for comparison. Only growth on water-agar (with a cold-inducing incubation step) produced satisfactory levels of sporulation in the WT on coil structures and hyphal strands, while in comparison, colonies of the ΔsidN mutants produced very low numbers of conidia under these conditions (see Figure S2A and S2B). Asexual spore morphology did not appear to be affected by the sidN mutation (Figure S2B). It is not currently possible to determine if ascospore formation is affected in these mutants since the pre-sexual choke state required for the commencement of the sexual cycle has never been observed with artificial infection of perennial ryegrass (L. perenne) with E. festucae [59].
Iron Represses sidN Expression
Fungal siderophore biosynthetic genes are typically negatively regulated by iron, and we therefore compared the expression of sidN from liquid cultures grown under iron depleted conditions versus iron supplemented conditions. Expression of sidN in the WT was strongly repressed under iron supplemented conditions and only detectable by reverse transcriptase polymerase chain reaction (RT-PCR) when grown in iron-depleted medium (Figure 6). As expected, expression of sidN was not detected in the ΔsidN mutants under any of the conditions tested.
Figure 6. Expression of sidN is repressed under iron replete conditions.
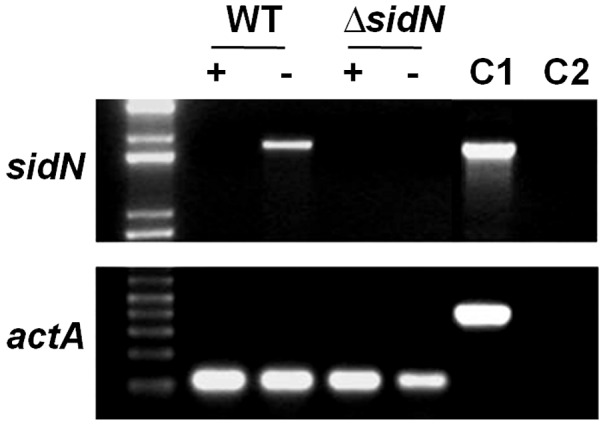
RT-PCR expression analysis of ΔsidN mutant 85 (ΔsidN) and wild-type E. festucae Fl1 (WT) grown in liquid cultures under iron replete (+) conditions (DM supplemented with 20 µm FeS04) or iron-depleted (−) conditions (DM - no iron supplementation). gDNA is used as a positive control (C1) and water as a negative control (C2) for sidN and actA (actin) gene expression.
Loss of sidN Adversely Affects the Mutualistic Relationship Between E. festucae and Perennial Ryegrass
To explore the effect of loss of the sidN product on the symbiotic relationship of E. festucae with perennial ryegrass, ΔsidN mutants, WT and C-sidN complemented strains were inoculated into perennial ryegrass seedling lines (Figure 7A). Onset of symptom development with the ΔsidN mutants was apparent within two weeks of planting inoculated seedlings into soil. Infection with the ΔsidN mutants resulted in a range of stunted, erect (versus prostrate) plant phenotypes, all with a large increase in tiller numbers (Figure 7A).
Figure 7. The Endophyte-Grass symbiotic interaction phenotype is disrupted in ΔsidN infected plants.
A. Phenotypes of perennial ryegrass plants infected with wild-type E. festucae Fl1 (WT), complemented ΔsidN strain (C-sidN) and ΔsidN mutant 85 (ΔsidN). B. Root systems of sidN infected plants are reduced compared to WT infections. A perennial ryegrass plant infected with E. festucae (WT) is compared against three ΔsidN 85 infected plants displaying increased levels of plant stunting from left to right. Plants are 14 weeks old. C, D and E. Light micrograph DIC images of aniline-blue stained hyphae of wild-type E. festucae Fl1 (WT). C. ΔsidN mutant 85. D. ΔsidN mutant 54. E. in mature leaf sheaths.
All plants infected with ΔsidN mutants showed abnormal underdeveloped root systems, and with the more severely stunted plants having markedly reduced root systems (Figure 7B). Severely stunted ΔsidN infected plants frequently died when transplanted from root trainers to larger pots, in contrast to the more moderately stunted plants, which could be maintained for long periods in the glasshouse.
To examine hyphal growth in planta, light microscopy of aniline-blue stained leaf sheaths of grass pseudostems was performed (Figure 7C, D, E). Hyphae of WT infected plants grow mainly by intercalary growth typified by infrequently branched hyphae that are generally aligned parallel to the longitudinal leaf axis of the elongating grass leaf [43]. Microscopic examination of ΔsidN mutants in perennial ryegrass revealed extensive colonization of leaf sheath tissues in comparison to WT (Figure 7C, D, E). There were large regional areas of synchronized hyphal growth (as described by Christensen et al. [43]) alongside unrestricted convoluted hyphae (Figure 7D, E). Less frequent patches of excessively convoluted hyphae extending out in all directions were also observed (see Figure 7E for an example).
Toluidine blue stained 1 µM cross sections of pseuodostems from WT, C-sidN and ΔsidN mutants 54 and 85 were examined to analyze the distribution of hyphae within different ages of leaf sheaths and blades (Figure 8A, B). We observed that hyphae from the WT or the complemented strains showed an even distribution of hyphae across all tissue types and tissue ages, but that hyphae from stunted plants infected with ΔsidN 54 or 85 were abnormally distributed. Specifically, the number of ΔsidN mutant hyphae were found to be the lowest in the inner developing leaf blade, and increased with age so that more hyphae were observed in the older outer leaf blade and sheaths (results not shown). We could also gain information about the cytoplasmic density of hyphae in specific tissues based on penetration of toluidine blue. Hyphae from the WT or complemented strain were generally dense in all tissues examined, but not so for the mutant strains. Dense hyphae were only observed in ΔsidN 54 or 85 infections from within the youngest tissue, the inner leaf blade, as well as hyphae located in the phloem of the vascular bundles. However, the majority of mutant hyphae located in the older leaf blade and all three sheaths were empty (Figure 8A shows hyphae located in the inner leaf sheath in both mesophyll and vascular tissue). Unlike the WT in which no hyphae were observed in the vasculature, mutant hyphae were found in large numbers in some of the vascular bundles (mostly in the phloem) of all of the tissue types examined (Figure 8A). Additionally, we noted many epiphyllous hyphae present with the mutant infection compared to few for the WT or complement, with most being large in diameter and apparently empty as hyphal contents were not stained at all with toluidine blue (Figure 8A). These light microscopy pictures indicate that the ΔsidN mutant is proliferating mostly in only the older parts of the plant, with typical WT-like growth confined to the inner leaf blade, where hyphae are few and stain densely. Generally, the densely-stained mutant hyphae were only seen in nutrient rich vasculature or in the inner leaf blade.
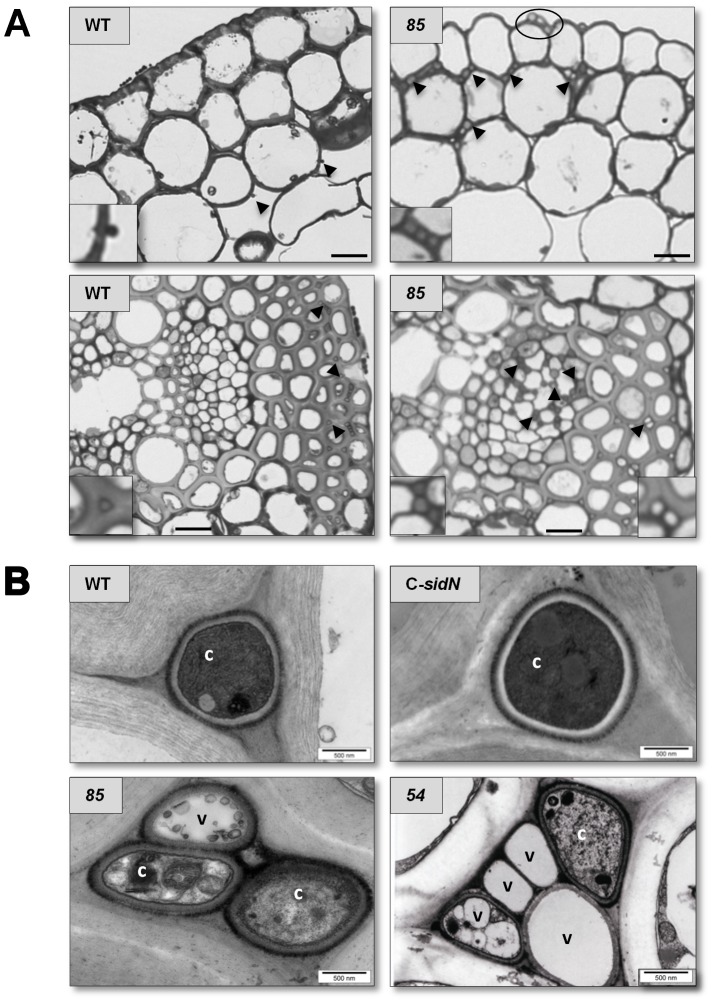
Figure 8. Abnormalities in the hyphal distribution and ultrastructure of ΔsidN mutants in perennial ryegrass plants.
A. Light micrographs of 1 µM cross sections of the inner leaf sheath of perennial ryegrass infected with wild-type E. festucae Fl1 (WT) and ΔsidN mutant 85 (85) are shown. The top panel is a cross section of mesophyll cells, whereas the lower panel is a close up of vascular tissue. Representative hyphae are indicated by arrows and the circle on the 85 panel indicates epiphyllous hyphae. Inserts show higher magnification of the endophyte hyphae indicated by the arrowheads in the main panels. Bars = 20 µM. B. Transmission electron micrographs of cross sections of endophyte hyphae in the intercellular spaces of perennial ryegrass. Wild-type E. festucae Fl1 (WT), complemented ΔsidN strain (C-sidN) ΔsidN mutant 54 (54), ΔsidN mutant 85 (85) are shown. Samples shown were photographed from leaf sheath sections, with WT, C-sidN, and ΔsidN 85 hyphae located in mesophyll tissue, whereas ΔsidN 54 is in sclerenchma tissue. c, cytoplasm, v, vacuole. Bars = 500 nm.
Transmission electron microscopy (TEM) of perennial ryegrass plants infected with WT, complement and ΔsidN mutants 54 and 85 confirmed the light microscopy findings (described above) and provided detailed images of various abnormalities apparent in the hyphal ultrastructure of ΔsidN mutants (Figure 8B). Frequently observed irregularities were atypical vacuolation as well as poorly stained hyphae versus rarely vacuolated and frequently densely-stained WT/complemented hyphae indicating that a characteristic feature of the hyphae of ΔsidN mutants is diffuse cytoplasm (Figure 8B, Figure 9). As already noted by light microscopy, hyphae located in the vasculature contained dense cytoplasm, and the contents as determined by TEM appeared very normal (WT-like) and hyphae were also mostly regularly shaped like WT (Figure 9). Additionally, we often observed asymmetrically shaped mutant hyphae of variable diameter in the mesophyll of various plant tissue types and occasionally, but only in mutant infected plants, the presence of clusters of hyphae orientated both longitudinally, obliquely and transversely surrounding host cells, but did not enter host cells (Figure 9). Epiphyllous hyphae were also able to be visualized by TEM in ΔsidN infections as they were so abundant, and were observed to always be highly vacuolated and encapsulated in plant host derived material (Figure 9).
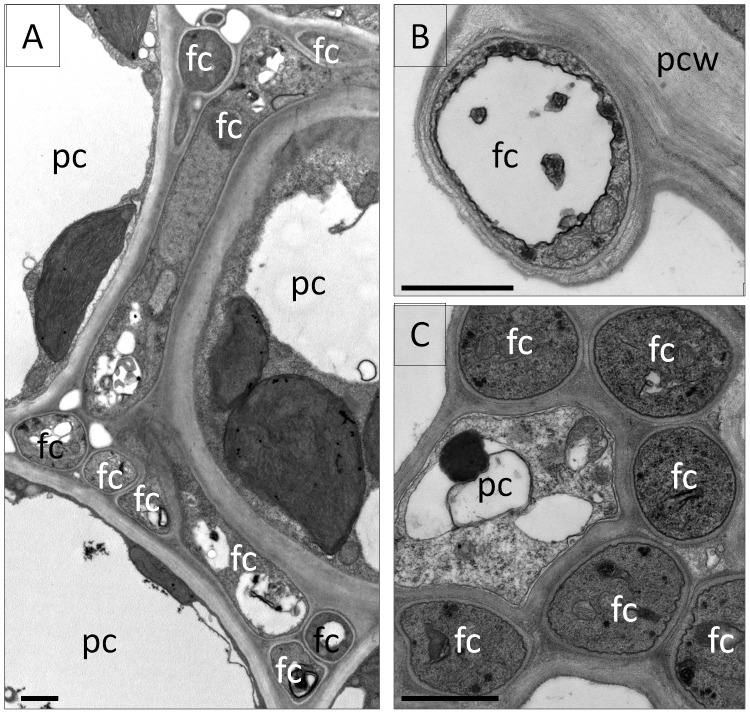
Figure 9. Ultrastructural features of the ΔsidN mutant - plant infection phenotype.
A. Aberrant ΔsidN 85 mutant hyphae in the mesophyll of the inner leaf sheath surrounding host cells. B. An epiphyllous ΔsidN 85 mutant hypha on the outside of the outer leaf blade. C. ΔsidN 85 mutant hyphae in the main vascular bundle of the inner leaf sheath. Bar indicates 1000 nm. fc indicates fungal cell, pc for plant cell, pcw for plant cell wall.
Light microscopy and TEM observations showed that the ΔsidN mutants colonized the apoplastic space with multiple hyphae compared to single or few hyphae in the WT (Figures 8A, 8B, 9). This apparent increase in fungal biomass was quantified by real-time quantitative PCR (qPCR) of the single copy fungal gene, NRPS1, in total gDNA (plant and fungal; [60]). Relative to WT infected plant pseudostems (that comprise a mixture of enclosed immature emerging leaf blades and surrounding mature sheaths), the fungal biomass in plants infected with ΔsidN 54 was 1.4 fold higher, while in plants infected with ΔsidN 85 fungal biomass was 1.8 fold higher.
Effects on the Chemical Phenotype of the Symbiotum
To determine whether the fungal extracellular siderophore migrated into the plant, we analyzed guttation fluid from perennial ryegrass infected with WT, ΔsidN 85 mutant and complemented C-sidN strains by targeted LCMSn, as we have found that this fluid provides a clean matrix for the detection of endophyte metabolites in planta [61]. While epichloënin A could not be detected in planta, ferriepichloënin A was detectable at trace levels in several samples from plants infected with the WT or C-sidN strains but could not be detected in plants infected with the ΔsidN 85 mutant (results not shown).
Indirect effects of the ΔsidN 85 mutant on the chemical phenotype of the symbiotum were more marked. E. festucae strain Fl1, in association with perennial ryegrass, synthesizes three major classes of alkaloids: indolediterpenoids, of which lolitrem B is the major product; ergot alkaloids, of which ergovaline is the major product, and the pyrrolopyrazine peramine. Production of all three is minimal or undetectable in axenic cultures, but all are produced in significant amounts (of the order of µg/g) in planta [62]–[64]. Many biotic and abiotic factors influence the production of fungal alkaloids in planta, including mineral stress, for example nitrogen and phosphorous availability [60], [65], [66]. To test if the altered phenotype of ΔsidN 85 infected plants may have an effect on alkaloid production, we determined alkaloid concentrations in infected plants on at least 3 occasions. While considerable variation was observed in concentrations of lolitrem B, and to a lesser extent of peramine, ergovaline consistently accumulated to much higher levels in the ΔsidN 85 mutant plants compared to WT (10-fold for ΔsidN 85, Figure 10), a factor much larger than the 1.8-fold increase in fungal biomass in the identical plant tissue as described above. Other ΔsidN mutant strains showed a similar elevation in ergovaline levels (data not shown). In agreement with this, a transcriptomic experiment that compared plants either infected with WT or ΔsidN 54 strains by a custom designed Affymetrix GeneChip revealed that the expression of genes involved in the biosynthesis of ergovaline were highly elevated in the plants infected with the ΔsidN mutant (L. Johnson et al., unpublished results).
Figure 10. Elevated ergovaline levels detected in ΔsidN infected plants.
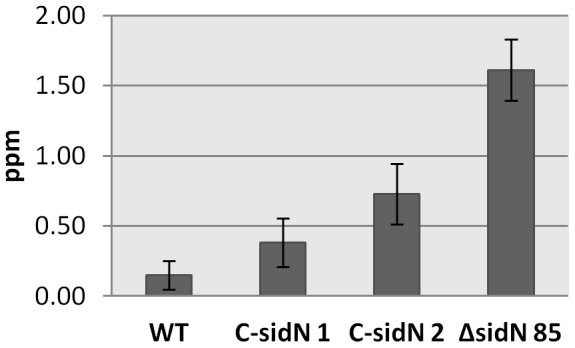
HPLC analysis of ergovaline production was carried out on perennial ryegrass pseudostems infected with wild-type E. festucae Fl1 (WT), ΔsidN mutant 85 (ΔsidN), and complemented ΔsidN strains (C-sidN 1 and C-sidN 2). The numbers of plant reps used for analysis were 3–5 for each sample. Error bars indicate standard deviation.
Quantitative RT-PCR (RT-qPCR) Analysis of Iron-Regulated Genes
Siderophore mediated iron-uptake does not appear to be the only high affinity iron uptake system operational in Epichloë as a search of the E. festucae (E2368, Fl1) genome sequences revealed the presence of a putative ferroxidase, fetC and a putative high affinity iron permease, ftrA that form the bipartite Fet3/Ftr1 complex responsible for reductive iron assimilation (ferroxidation/permeation) [23] in these endophyte strains. Only one putative ortholog of fetC and ftrA, respectively, was identified. These two genes are co-localized in the genome and share a bidirectional promoter as is commonly found for other fungi [15], [16], [67]. Two other genes regulating iron homeostasis were also found in the genome; sreA, the GATA-type transcriptional repressor of iron uptake during iron-replete conditions [68], [69], and hapX involved in the repression of iron-utilizing proteins under iron deficiency mediated via the CCAAT-binding complex [70]–[73]. See Table S2 for detailed descriptions of identified genes used for RT-qPCR below.
To test our hypothesis that the ΔsidN mutants in perennial ryegrass, due to loss of their extracellular siderophore, are sensing changes in their cellular iron status, we quantified mRNA abundances of the putative E. festucae iron-repressed genes fet3, ftr1, and hapX. Significant up-regulation of all three iron-repressed genes in pseudostem tissue of ΔsidN 54 and 85 compared to WT infected plants was found, suggesting reduced intracellular iron levels in the ΔsidN mutants as a result of the loss of epichloënin A production (Figure 11).
Figure 11. RT-qPCR of E. festucae iron-regulated genes and Nox genes in ΔsidN infected perennial ryegrass.
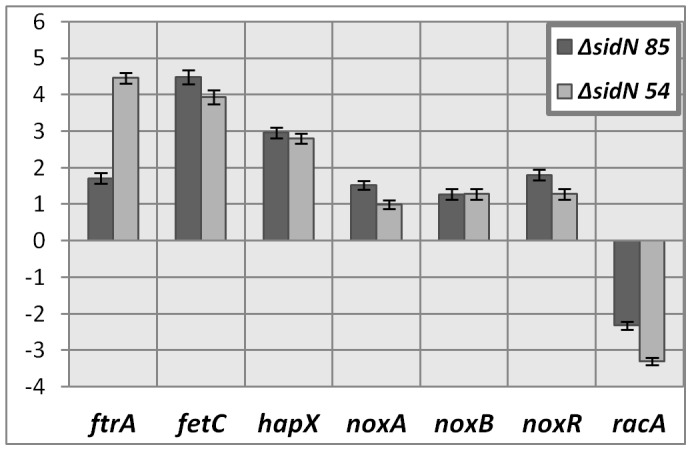
Relative mean abundance relative to wild-type (fold difference displayed) of iron regulated gene expression (ftrA, fetC, hapX) and Nox gene expression (noxA, noxB, noxR, racA) in perennial ryegrass infected with ΔsidN mutants 54 and 85 are shown. For ftrA, fetC, noxA and racA results have a p-value of <0.001 and for hapX, noxB and noxR, the p-values are 0.002, 0.005 and 0.017 respectively. Error bars indicate SED.
RT-qPCR Analysis of NADPH Oxidase (Nox) Genes
The roles of genes of the superoxide-generating NADPH oxidase (Nox) family have been well characterised in E. festucae and shown to be critical for symbiosis maintenance [74]–[77]. Perturbations in levels of reactive oxygen species (ROS) via mutation of components of the Nox complex in E. festucae affect regulation of hyphal growth and branching in the host and cause plant stunting. A number of shared phenotypic features of the in planta phenotypes of the ΔsidN mutants with the E. festucae Nox mutants, such as unrestricted hyphal growth and swollen hyphal tips, suggested that alterations in ROS could play a role in the ΔsidN mutant phenotype.
To investigate whether there were any alterations in the transcriptional regulation of the Nox complex, we quantified mRNA abundances by RT-qPCR of the Nox genes, noxA (required for symbiosis, [75]) and noxB (no known function, symbiosis not affected, [75]), as well as the regulators of this complex noxR [74] and the small GTPase racA (required for NoxA activation and regulated ROS production, [76]) in pseudostem tissue of plants either infected with WT or ΔsidN mutant strains. For perennial ryegrass plants infected with ΔsidN mutants, noxA, noxB and noxR genes were not differentially expressed relative to WT infected plants, but racA was down-regulated 2.2 to 3.3 fold in ΔsidN 85 and ΔsidN 54 infected plants respectively (Figure 11). To determine if altered racA expression resulted in a measureable difference in ROS production in the ΔsidN mutants, we examined ROS production in axenic cultures by both nitroblue tetrazolium (NTB) and DAB (3,3′-diaminobenzidine) staining for detection of superoxide and H2O2 respectively. However, the production of both superoxide and H2O2 of ΔsidN mutant colonies grown on iron-depleted (DM or DM with BPS) or iron-replete (DM supplemented with iron or PD) media showed no consistent significant difference compared to WT (results not shown). We did on the other hand observe an obvious increase in H2O2 (by DAB staining) across all mutant and control colony strains when grown on iron supplemented DM compared to iron-depleted DM alone (results not shown) indicating the general influence of free iron supplementation to increase ROS levels.
Discussion
E. festucae sidN Encodes the Novel Extracellular Siderophore Epichloënin A
This study presents the characterization of extracellular siderophore biosynthesis in the foliar grass endophytic fungus E. festucae of the Clavicipitaceae. Through targeted gene disruption of the NRPS gene sidN (formerly NRPS2) and complementation of a mutated ΔsidN strain, we confirmed that sidN encodes a siderophore synthetase required for the production of the novel extracellular siderophore epichloënin A, a variant of ferrirubin of the ferrichrome family. In silico analysis of the SidN domain architecture indicates that the protein consists of three complete A-T-C modules, two terminal T-C repeats and is identical to Type II ferrichrome NRPSs [57]. We can therefore infer that SidN activates three component amino acids required for the biosynthesis of a ferrichrome. This interpretation is consistent with the recent structural characterization of epichloënin A that showed it to be a cyclic octapeptide ferrichrome comprised of three contiguous units of trans-AMHO (the iron-chelating residues of hydroxamate siderophores [28], [57]), a glutamine and four glycine residues [53]. The third A domain (A3) of SidN has been experimentally confirmed to activate AMHO residues [52]. The other two A domains (A1 and A2) are therefore predicted to activate the two other remaining component amino acids [glutamine (novel component) and glycine (commonly incorporated)] of epichloënin A.
Investigations into the cellular location of epichloënin A and its ferri-form in the WT fungus grown on iron depleted defined medium (DM+BPS) showed that while epichloënin A was predominant in the culture supernatant, ferriepichloënin A was relatively more abundant in the mycelium. These results suggest epichloënin A produced by the fungus is secreted into the extracellular fluids where it binds available iron as ferriepichloënin A which is subsequently taken up by the fungus where the iron is retrieved (referred to as the “shuttle” mechanism; see Howard [78] for details on siderophore transport mechanisms). We are investigating whether ferriepichloënin A could also have an intracellular role, in addition to that of ferricrocin (putatively encoded by NRPS9), a well-known cellular siderophore which we have found only in mycelial extracts (L. Johnson et al., unpublished results).
Loss of Epichloënin A Biosynthesis Adversely Affects the Mutualistic Perennial Ryegrass – Endophyte Association
Colonization of grass leaves by epichloae endophytes occurs by a unique process described in an intercalary growth model that has been experimentally verified by Christensen et al. [43]. The model illustrates how hyphae in the leaf extension zone, extend by intercalary growth (non-tip hyphal extension) at a rate that is synchronized with the expansion and migration of leaf cells. Once the leaf ceases to expand, hyphal growth also ceases, but hyphae remain metabolically active, continuing to be of benefit to their host [79]. This mode of endophytic growth in planta ensures that hyphal growth is synchronized with plant growth. The maintenance and regulation of this lifestyle is pivotal for the mutualistic nature of the associations studied here.
We have investigated the effects of loss of production of epichloënin A on the mutualistic association formed between E. festucae with its host grass perennial ryegrass and showed that loss of the extracellular siderophore is deleterious for the maintenance of mutualism. Plants infected with ΔsidN mutants were variably stunted, with altered tiller and root morphology compared to WT. The endophytic hyphae in ΔsidN plants are evidently no longer colonizing only by intercalary hyphal extension but also growing from hyphal tips in an unrestricted and sometimes disorientated manner with respect to the leaf axis, typified by hyper-branching and variable diameter. All of these effects are indicative of a change in the nature of the symbiotic relationship between the fungal endophyte and its host from being mainly mutualistic to detrimental. This study is the first account of the consequences of the loss of an extracellular siderophore in a mutualistic system and indicates that balancing of symbiotic iron homeostasis is an important factor in the maintenance of the mutualistic nature of grass-endophyte associations.
Despite the fact that loss of extracellular siderophores sometimes causes microbial pathogens to become less pathogenic, or a mutualistic symbiont to turn pathogenic-like, the commonality in this apparent paradox is that iron uptake appears obligatory for survival of all microbes, whether friend or foe. The symbiotic endophyte studied here is confined to the apoplastic spaces of above ground plant parts; it therefore depends completely on host iron for survival and our results suggest it must scavenge for iron via extracellular siderophore-mediated uptake. The inability of ΔsidN mutants to grow on iron depleted (DM+BPS) media indicates epichloënin A biosynthesis contributes significantly to iron assimilation in E. festucae. This is substantiated by the expression analysis of putative components of reductive iron assimilation (RIA) that are up-regulated in the ΔsidN infected plants, but appear unable to recompense for the lack of epichloënin A. RIA is therefore a functioning high affinity iron uptake system in E. festucae during in planta vegetative growth, but it does not appear to be the major system required for regulated iron uptake. Accordingly, up-regulation of hapX, the transcription factor that represses iron utilizing proteins in ΔsidN infected plants implies that the ΔsidN mutants require more iron than they are receiving via RIA alone. Another possible explanation is that RIA is bringing in sufficient iron but cellular iron handling processes have become deregulated due to the loss of epichloënin A, which could be acting as a regulatory iron sensor and/or cellular iron store (since epichloënin A is both secreted outside of the cell (iron-free) and also found intracellularly bound to iron), and consequently the incoming iron cannot be handled properly and thus utilized effectively. The observed proliferation of ΔsidN mutant hyphae in planta, in which the majority of hyphae (located in the older parts of the plant) are abnormal (frequently devoid of cytoplasm) and some appear dead (empty of cytoplasm), apart from those extensively colonizing the vasculature which have dense healthy looking cytoplasm suggests that hyphae are growing at a rate greater than can be sustained. The effect of the iron chelator BPS on the distribution of chitin in hyphae of the WT and mutant grown in vitro was also revealing in that it suggests that loss of RIA (via BPS amendment to the media) drastically changes chitin distribution and therefore cell polarity. We therefore conclude that changes in iron sensing and regulation are presumably the underlying agents responsible for the abnormal appearance of many mutant fungal hyphae in planta.
We observed an interesting increase on the endophyte in planta induced alkaloid ergovaline, in plants infected with ΔsidN mutant strains. This could be as a consequence of iron starvation or may be associated with the extensive hyphal tip growth and branching of fungal hyphae observed in the leaf sheaths of these plants. This pattern of growth is normally restricted to basal meristematic tissues, and a detailed dissection study of N. lolii-infected plants [64] found concentrations of ergovaline to be highly elevated in these tissues.
Oide et al. [19] present consequences of extracellular fungal siderophore loss in several fungal phytopathogens through NPS6 gene deletion that resulted in reduced virulence and hypersensitivity to H2O2 [80], the latter being consistent with our findings of increased H2O2 sensitivity in ΔsidN mutants. The host penetration ability of these Δnps6 strains was not affected, however plant colonization was defective [19]. Application of iron restored this defect indicating that iron deficiency caused the lack of virulence in these fungi. Our work demonstrates that extracellular siderophores play a critical role in fungal-host relationships, but the consequences of loss of siderophore differ depending on the nature of that relationship (pathogenic vs. mutualistic).
Symbiotic Iron Homeostasis, Oxidative Stress and ROS
The endophyte is housed in the apoplastic space where free iron is presumed to be a limiting factor. Our findings indicate that the endophyte requires its extracellular siderophore for mutualistic growth and supports the idea that iron is not readily accessible within the apoplast. The symbioses formed between epichloae endophytes and the Poaceae have co-evolved over evolutionary time, and maintaining symbiotic iron homeostasis is likely to be an intricately balanced process, and a key factor in keeping hyphal growth controlled. Iron homeostasis of the whole endophyte-grass symbiotum must be balanced for two primary reasons. Enough iron must be supplied for metabolism of both plant and fungal partners and excess must be avoided since free iron is toxic due to the formation of reactive oxygen species produced by the Fenton reaction giving rise to oxidative stress [4]. In vitro studies indicate the ΔsidN mutants are significantly more sensitive to H2O2 on iron-depleted medium than on iron-replete medium. This could simply be explained by a reduction in iron-dependent antioxidative enzymes, such as catalases and peroxidases, which require heme. A similar phenomenon was reported for Δnps6 strains where iron application enhanced tolerance to H2O2 and KO2 [19]. Intriguingly, the Fenton reaction does not appear to be the source of oxidative stress sensitivity since iron application would be expected to promote the Fenton reaction. Furthermore, Oide et al. [19] were able to show that both iron-saturated and nonsaturated siderophore (desferrioxamine) alleviated reactive oxygen species hypersensitivity indicating a possible protective role for siderophores against reactive oxygen species (ROS) in vitro; however their studies to demonstrate this role in planta were inconclusive. Further insights into the connections between ROS and iron regulation have been demonstrated from research into the monothiol glutaredoxins Grx3 and Grx4 of Sacchromyces cerevisiae, which show they function in the defense against oxidative stress through the regulation of iron homeostasis [81].
Research on the NADPH oxidase (Nox) complex in E. festucae associations with perennial ryegrass (through fungal mutants NoxA, NoxB, NoxR, RacA and BemA) have shown that Nox produced ROS plays a key role in regulating hyphal growth and branching in the host [74]–[77]. Disruption of the spatial and temporal production of fungal ROS leads to the loss of the characteristic features of endophyte-grass mutualistic associations. As the ΔsidN mutant infected plants shared some phenotypic features with the E. festucae Nox mutants, such as hyperbranching of hyphae within host leaves and plant stunting along with increases in tiller number, we explored whether the transcriptional regulation of the E. festucae Nox complex genes was affected in the ΔsidN infected plants. The Nox regulator racA was significantly down-regulated in both ΔsidN mutants studied, and although noxR gene expression was not differentially expressed in the ΔsidN mutants, NoxR requires a functional RacA to spatially regulate ROS production and control hyphal branching [74], [76]. However, this did not result in changes in ROS production levels in colonies of ΔsidN mutants grown on iron-depleted (or replete) medium. Based on these in vitro results, it seems less likely to find a significant difference in ROS production in the ΔsidN mutant infected host plants and was therefore not pursued.
Ultimately, to ensure continuance of mutualism, we postulate that maintenance of restricted hyphal growth of E. festucae in planta does not only require a functional Nox complex, but also the maintenance of iron homeostasis which is mediated via epichloënin A. To recapitulate, our results demonstrate that iron acquisition through siderophore-mediated iron uptake is necessary to maintain endophyte mutualism with perennial ryegrass.
Methods
Biological Materials and Growth Conditions
Epichloë festucae strain Fl1 (ex cultivar SR3000) and derivatives (this study) were grown on 2.4% potato dextrose agar (PDA, Difco Laboratories) and maintained as previously described [82], [83]. Defined medium (DM) for iron growth studies and chemical analyses were modified from Mantle and Nisbet [84], with yeast extract replaced with 0.6 µM thymine and iron was omitted. DM medium was also supplemented with the following as indicated: 100 µM BPS (bathophenanthrolinedisulfonic acid; Sigma), 20 µM FeSO4, 20 µM FeCl3.
Plant Inoculations and Growth Conditions
Inoculation of seedlings of perennial ryegrass ‘Nui’ was performed using the method of Latch and Christensen [85]. Plant growth from seed to 6 weeks-old is as described by Tanaka et al. [86], except the potting mix was of the following composition: 60% peat, 40% coarse sand with slow release fertiliser (nutricote with F.T.E.) and dolomite lime at 2.4 kg/m. Determination of endophyte infection was performed by immunoblotting [87]–[89] and light microscopy of epidermal leaf sheaths by aniline blue staining [89].
Examination of Asexual Sporulation In Vitro
Plates containing 2% water agar were inoculated with small agar plugs of mycelium and grown for 2 weeks at 22°C, followed by 2 weeks at 4°C. Slides were then mounted with agar blocks using a stereomicroscope to locate appropriate regions for counting of conidia by light microscopy using bright field optics. To determine conidia number from each colony mounted, 5 regions were counted starting from the colony edge and moving towards the colony centre. Conidia numbers were recorded and the data graphed is the mean spore count obtained from three independent colonies and three technical replicates.
Oxidative Stress Test
Sensitivity to oxidative stress was examined using a final concentration of 0.7 mM H2O2 in the medium (DM or PD) and tests were repeated at least 3 times. The colony diameters obtained from growth on DM or PD with or without H2O2 supplementation were measured and recorded at 7 days (at 22°C). The ratios of DM/DM+H2O2 or PD/PD+H2O2 were recorded for each fungal strain. Data were analysed by an analysis of variance (ANOVA) and the least significant difference used to compare stress sensitivity of strains.
DNA and Fosmid Library Preparations and Standard Molecular Techniques
Plasmid or fosmid DNA was isolated using the QIAprep Spin Miniprep Kit (Qiagen). Fosmid DNA prior to extraction was induced to high copy number using the manufacture's protocol (Epicentre Biotechnologies). Escherichia coli TOPO strain (Invitrogen) was used to propagate plasmids using standard techniques [90].
Fungal genomic DNA (gDNA) for Southern Blot analysis was isolated using freeze-dried or fresh mycelium by the method of Yoder [91]. For identifying a positive disruption event by PCR, a small scale DNA protocol was used. Transformants were inoculated from one small mycelial plug (taken from the outer colony margin) into 100 µl of PD broth in a 1.5 ml tube and grown at 26°C for 3 days. Supernatant was removed and freeze-dried mycelia were ground and extracted in 150 µl of lysis buffer (100 mM Tris-Cl, pH 8.0, 100 mM EDTA and 1% SDS). Following incubation at 70°C for 30 min, the lysates were mixed with 150 µl of 5 M potassium acetate solution and incubated on ice for 10 min. After centrifugation for 10 min, the supernatant containing the fungal DNA was precipitated with 0.7 volumes of isopropanol. The DNA pellet was washed once with 70% ethanol and finally dissolved in 20 µl of water. 1 µl of DNA was used for PCR analysis. Isolation of high molecular weight DNA for fosmid library preparation from protoplasts was performed based on the method by Denning et al. [92] and modified as follows: 500 µl of protoplasts in STC buffer were prepared as described by Young et al. [93], [94], lysed, and then proteinase K treated and phenol-chloroform purified as stated by Denning et al. [92]. The resulting aqueous phase was precipitated with isopropanol, followed by RNase A treatment (0.06 mg of RNase A (Invitrogen); incubation at 37°C for 30 min), then precipitated with 100% ethanol, and the resulting pellet washed with 70% ethanol, air dried and finally resuspended in water.
For Southern blot analysis, restriction enzyme digested gDNA was transferred to Hybond N+ (Amersham Pharmacia Biotech) overnight with 0.4 M NaOH. Filters were hybridized at 42°C with digoxigenin (DIG)-labelled DNA probes which were labelled by PCR incorporation of DIG-11-dUTP according to the manufacturer's instructions (Roche).
Standard PCR conditions for amplification from DNA templates were performed in a 15 µl reaction volume containing 20 mM Tris-HCl (pH 8.0), 50 mM KCl, 1 mM MgCl2, 200 µM dNTPS, 0.3 µM of each primer, 0.9 U of Taq DNA polymerase (Invitrogen). Cycling program (for products less than 2.0 kb) was as follows: one cycle at 95°C for 2 mins; 30 cycles at 95°C for 30 s, 58°C (dependant on primer TM) for 30 s and 72°C for 30 s; 72°C for 10 min.
An E. festucae fosmid library using gDNA isolated from protoplasts of E. festucae strain FL1 was constructed using the CopyControl Fosmid Production Kit (Epicentre Biotechnologies) according to the manufacturer's instructions. Broth cultures from 3840 independent colonies were obtained and a single pooled 384-well plate (containing 3840 colonies) was screened by PCR using primers Sid1F & Sid1R. See Table S1 for details of primers used in this study. Standard PCR conditions were used as cited above.
Generation of Disruption and Complementation Vectors, E. festucae Transformation and Characterization of Transformants
We had previously probed a N. lolii Lp19 lambda library with a NRPS2 derived PCR product, and identified one lambda clone of approximately 5.8 kb with the following partial domain structure: A (truncated)-T-C-A-T-C-T-C-T-C. This information was used for performing a gene disruption by homologous recombination in the sexual relative, E. festucae wild-type (WT) strain Fl1. The gene disruption vector was constructed using Multisite Gateway Three-fragment Vector Construction Kit (Invitrogen) so that a portion of the genomic region encoding the third A domain of sidN was replaced with the hygromycin B resistance gene (see Figure S3A). The 5′ and 3′ entry clones were respectively PCR amplified from E. festucae (primer pairs Sid5F and Sid5R/Sid6F and Sid6R), then combined to produce a destination vector containing inserts of approximately 3.0 kb of 5′ NRPS coding sequence, followed by a 4.0 kb hygromycin B resistant gene (originally amplified from pAN7-1) and approximately 3.0 kb of consecutive 3′ NRPS sequence after a deleted region of 35 bp.
Protoplasts were prepared as described by Young et al. [93], [94]. PEG-mediated transformation of protoplasts was carried out with a linear PCR product of ∼10 kb derived from the sidN gene disruption construct and obtained by PCR using the ΔsidN disruption vector as a template with primers sid2F and sid2R. PCR amplification in a 50 µl reaction volume contained 1× Tuning buffer with 5 mM Mg2+ (Eppendorf), 500 µM dNTPs, 400 nM of each primer, 1 ng of plasmid DNA and 2 units of TripleMaster Polymerase Mix (Eppendorf). Cycling program was as follows: one cycle at 93°C for 3 min; 20 cycles at 95°C for 15 s, 56–58°C for 30 s, 68°C for 8 min; 68°C for 10 min.
Transformants generated from the gene disruption experiment were initially screened by PCR with primers specific to sidN (Sid4F and Sid4R) which flank the hygromycin B resistance gene (wild-type 0.24 kb, disruption 4.24 kb; data not shown). Southern blot analysis using a probe (amplified with primers Sid2R and Sid3F) specific to sidN which also spans the hygromycin B resistance gene in the disruption construct confirmed the disruption event (mutant strains showed loss of WT 5.45 kb band and a new band of 9.45 kb) (see Figure S3B). Putative ΔsidN mutants were subsequently confirmed by southern blotting to have the disruption event (see Figure S3B).
Complementation of ΔsidN 85 strain was carried out by co-transforming 5 µg of fosmid DNA [containing the entire promoter and open reading frame of sidN (as well as 2 additional ORFs located 3′ of sidN)] along with 1 µg of circular PII99 [95] carrying the selectable antibiotic resistant marker geneticin.
The transformation methodology of Vollmer and Yanofsky [96] with modifications by Itoh et al. [97] was used for gene disruption and complementation experiments. Disruption transformants were selected on regeneration (RG) medium containing hygromycin (150 µg ml−1), whereas geneticin resistant transformants derived from the complementation experiment were selected on RG medium containing 200 µg ml−1 of geneticin. Mycelium was subcultured three times for nuclear purification of transformants.
Complementation transformants were identified by their ability to grow on iron-depleted medium containing the iron chelator BPS. Confirmation was obtained by assaying culture filtrates from a subset of positive transformants for siderophore production by LCMSMS.
Determination of Fungal Concentration in planta by qPCR
qPCR on gDNA isolated from endophyte-infected plants (pseudostem) was performed on a MyiQ cycler (Bio-Rad) using primers designed to a nonribosomal peptide synthetase (NRPS-1) gene as described in Rasmussen et al. [60]. Concentration of endophyte in infected tissues is expressed as the number of copies of the single copy NRPS1 gene per total gDNA (from plant and fungus).
RNA Preparation and Expression Analysis
Total RNA was extracted from frozen fungal mycelium using TRIzol reagent (Invitrogen). For extraction from perennial ryegrass tissues, RNA was at first isolated using TRIzol and further purified through the Plant RNeasy Kit (Qiagen) as follows: ∼30 mg of frozen plant tissue was ground to a fine powder, mixed well with 750 µl of TRIzol and incubated at room temperature for 5 min. Following centrifugation at 12K rpm for 10 min, the supernatant was transferred to a new tube and 150 µl of chloroform added. After mixing well for 15 s and incubating at room temperature for 3 min, samples were centrifuged at 13.2 K rpm for 5 min. The aqueous phase was transferred to a fresh tube and an equal volume of 70% ethanol added before loading onto a Qiagen RNeasy mini column (pink). From this point onwards, total RNA was column purified using the manufacturer's instructions (Qiagen). The optional DNase digestion on the column step was also carried out for plant samples.
Any DNA still remaining was removed from total RNA by treating 10 µg of RNA with 20 U of DNase I, RNase-free (Roche) and 5 mM MgSO4 at 37°C for 30 min, followed by 5 min at 75°C. First strand cDNA primed with oligo(dT) was synthesized using the ThermoScript RT-PCR system (Invitrogen) from 1 or 2 µg of denatured total RNA by incubation at 50°C for 60 min, followed by 85°C for 5 min.
For RT-PCR expression analysis of sidN from axenic liquid DM cultures, the primer pair Sid4F and Sid4R was used to amplify cDNA using standard PCR conditions. Primers to the Neoptyphodium lolii actin gene (Acting and ActinR) were used to check for cDNA quality.
Two-step RT-qPCR analysis of perennial ryegrass tissue was performed on an iCycler (MyiQ Single Color Real-Time PCR Detection System, Bio-Rad) using Power SYBR Green PCR Master Mix (AB applied biosystems). The following cycle parameters were applied: 95°C for 5 min and then 40 cycles of 95°C for 20 s, 54–56°C for 20 s, and 72°C for 30 s followed by a melt curve analysis. Primer efficiency ranged between 94% and 110%. Primers for RT-qPCR (see Table S1) of putative iron-regulated genes, Ftr1, Fet3 and HapX were designed to sequences from the E. festucae genome strain E2368. These genes were identified by performing tblastx queries of the E. festucae (E2368) genome with characterized iron-regulated genes from other fungal species (http://csbio-l.csr.uky.edu/ef2011/blast/blast.html). The deduced amino acid sequences of the identified genes from E. festucae (strain E2368) were then compared to the NCBI protein database by blastx (see Table S2 for descriptions of identified genes used for RT-qPCR).
Primers for the RT-qPCR analysis of NADPH Oxidase (Nox) genes (see Table S1) were designed to E. festucae sequences sourced from accession numbers: AB236860 (noxA), AB236861 (noxB), AB260938 (noxR) and AB260937 (racA). Purified PCR products were used to create a calibration curve for each gene-specific primer pair. Two endophyte housekeeping genes (a 60S ribosomal protein L35 and gamma actin, see Table S1 for details) were used to normalize the expression levels of the different genes. An analysis of variance (ANOVA) was performed from the geometric mean obtained from three biological replicates and two or three technical replicates for each primer pair. The least significant difference was used to compare the samples. P-values obtained for each transcript were: ftrA P<0.001; fetC, P<0.001; hapX P<0.002, noxA P<0.001, noxB P = 0.005, noxR P = 0.017; racA P<0.001.
DNA Sequencing and Bioinformatics
Sequencing was performed with the Big-Dye (Version 3) chemistry (PE biosystems) and the products separated on an ABI Prism 3100 automated sequencer (Applied Biosystems).
Sequence comparisons were performed against local mirrors of a number of public databases, including GenBank, RefSeq, Cogeme and a selection of fungal genomes from the Broad Institute. Algorithms tblastx, blastx and blastn were employed to generate alignments [98]. Contigs were assembled using Vector NTI Advance 9.1.0, ContigExpress (Invitrogen) and SEQUENCER 4.6 (Gene Codes Corporation). NRPS domain structure was determined using a local mirror of InterProScan combined with a manual annotation based on the identification of motifs within domains as described by Schwarzer et al. [99].
Analysis of Siderophores in Culture by LCMS
Samples of supernatant from liquid cultures grown for 2 weeks were separated for analysis by centrifugation. The residual mycelium was freeze-dried and finely ground, and extracted with water, and siderophores separated by solid phase extraction [100]. All samples were stored at −20°C prior to analysis. Milli-Q water and HPLC grade solvents were used for LCMS. The samples were thawed prior to analysis and transferred to a HPLC vial with 200 µl insert. Samples were kept at 5°C in the autosampler, and 10 µl subsamples were injected. Analytes were eluted through a C18 Luna column (Phenomenex Torrence, CA, USA) (150×2 mm, 5 µm) at a flow rate of 200 µl min−1 using a Thermo Finnigan Surveyor HPLC system with a solvent gradient (solvent A: H2O 0.1% formic acid; B: MeCN 0.1% formic acid), starting with 5% B, 95% A for 5 min and then increasing to 33% B after 15 min, then to 95% B by 20 min where the composition was held for 5 min to wash the column before being returned to 5% B to re-equilibrate the column. Mass spectra were detected with a linear ion trap mass spectrometer (Thermo LTQ) using ESI in positive ion mode. The spray voltage was 4.5 kV and the capillary temperature 275°C. The flow rates of nitrogen sheath gas, auxiliary gas, and sweep gas were set to 20, 5, and 10 (arbitrary units), respectively. Ferriepichloënin A was detected as an MS1 ion of m/z 569 [MH2]2+; epichloënin A was detected as an MS1 ion of m/z 542 [MH2]2+. Cis-AMHO in a standard solution (kindly provided by T. Verne Lee, University of Auckland, New Zealand) and trans-AMHO in culture supernatants were detected by monitoring the MS1 ion of m/z 261, and MS2 spectra were recorded by selecting and fragmenting this ion.
Analysis of Siderophores in Guttation Fluid by LCMSMS
Guttation fluid was collected from the association of L. perenne G1057 with E. festucae Fl1, as previously described by Koulman et al. [61]. In brief, plants were placed overnight in a closed container and in the early morning the fluid accumulated at the leaf ends of a plant was collected with a glass pipette, transferred to a plastic container, and stored at −20°C until analysis.
Samples were analysed by direct injection of a 10 µl subsample of guttation fluid into a Thermo Finnigan Surveyor HPLC system attached to a linear ion trap mass spectrometer (Thermo LTQ) operated as described above. Ferriepichloënin A was detected by selecting and fragmenting the parent [MH2]2+ ion m/z 569±2 (35% relative collision energy), and selecting and fragmenting the product ion m/z 1024±2 (35% relative collision energy) and monitoring the total ion current.
Alkaloid Analysis
HPLC was used to measure in planta levels of lolitrem B, ergovaline and peramine alkaloids as previously described [101], [102].
Light and Transmission Electron Micrographs
Hyphae within leaf sheaths were examined by light microscopy of epidermal leaf sheaths by aniline blue staining [89].
For calcofluor (1 µg/ml) staining, mycelia were grown on microscopy slides covered with a thin layer of DM medium. Samples were directly incubated with the dye for up to 20 minutes at room temperature and washed with water to remove background fluorescence.
Superoxide production was detected by NTB staining using the method described by Takemoto et al. [74] except slides were overlaid with DM or PD media.
H2O2 production was examined by DAB staining (DAB forms a brick-red precipitate upon reaction with H2O2 [103] of cultures grown on DM and PD media as described by [104].
Examination of sections from pseudostem samples (approximately 1 mm long) required fixation in 3% glutaraldehyde and 2% formaldehyde in 0.1 M phosphate buffer pH 7.2 for 2 h at room temperature, followed by treatment in 1% osmium tetroxide in 0.1 M phosphate buffer pH 7.2 for 0.5 h at room temperature. The tissues were then washed 3 times in 0.1 M phosphate buffer pH 7.2, dehydrated in an acetone/water series and two times in 100% acetone. Samples were infiltrated with an acetone/polarbed 812 resin mixture (50/50, v/v), and then embedded in fresh resin mixture in silicone rubber moulds and cured for 48 h at 60°C. To examine the distribution of hyphae within different ages of leaf sheaths, cross sections (approximately 1 µm thick) were prepared from the treated pseudostem samples, and stained with toluidine blue for examination by bright field light microscopy. For examination with a Philips 201C transmission electron microscope, ultra-thin sections were cut and stained with saturated uranyl acetate in 50% ethanol for 4 min followed by lead citrate for 4 min.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: N. lolii NRPS2 (EF19536), N. lolii (strain Lp19) sidN (JN132404), E. festucae (strain Fl1) sidN (JN132407), E. festucae (strain E2368) sidN (JN132403), E. festucae (strain E2368) ftrA (JN132405), E. festucae (strain E2368) fetC (JN132406), E. festucae (strain E2368) hapX (JN132401), E. festucae (strain E2368) actG (FJ826616.1), E. festucae (strain E2368) actA (FJ379533.1), N. lolii RP(L35) (JN132402).
Supporting Information
Δ sidN Mutants Accumulate the Epichloënin A Precursor Trans -AMHO. A. Positive electrospray LS-MS extracted MS1 ion chromatograms of the parent [M+H]+ion (m/z 261) and CID MS2 spectra of authentic cis-AMHO standard and putative trans-AMHO from ΔsidN 85 mutant. B. Relative concentrations of trans-AMHO in extracts of mycelium from cultures of wild-type E. festucae Fl1 (WT), ΔsidN mutant 85 (ΔsidN), and a complemented ΔsidN strain (C-sidN) grown under Fe-depleted conditions detected by LCMS (MS1 m/z 261) (arbitrary units).
(TIF)
Conidiation in E. festucae is Reduced by Loss of Epichloënin A. A. Spore counts produced from colonies of E. festucae Fl1 (WT), complement (C-sidN) and ΔsidN mutant 85 (ΔsidN) grown on water-agar for 2 weeks at 22°C, followed by 2 weeks at 4°C. Data were generated from three independent colonies and three technical replicates. Error bars = standard error. B. Microscopic examination of WT and ΔsidN water-agar colonies used for spore counts showing spores on coil structures and hyphal strands
(TIF)
Gene Disruption Strategy and Southern Blot Analysis. A. A linear sidN disruption vector is shown annotated with corresponding modular NRPS domain structure [A-domain (adenylation), T-domain (peptidyl carrier) and C-domain (condensation)] and restriction enzyme sites for HindIII (H). The hygromycin (HYG) cassette is indicated to insert into the A-domain of SidN (which was deduced from partial sidN sequence). B. An autoradiograph of a DNA gel blot of HindIII digested genomic DNA of ΔsidN mutants 54, 82 and 85 along with wild-type (WT) and ectopic (E) controls probed with a DIG-labeled sidN PCR product amplified with primers Sid3F and Sid2R.
(TIF)
Detailed Information of the Primers Used in This Study.
(DOCX)
Iron-Responsive Genes in E. festucae.
(DOCX)
Acknowledgments
We thank Brian Tapper and Wade Mace for HPLC analysis, Zaneta Park for statistical analyses, Douglas H. Hopcroft and Jianyu Chen for TEM support, Iain Lamont for critically reading this manuscript and the following AgResearch personnel for technical support; Catherine Tootill, Hong Xue, Craig Anderson, Wayne Simpson and Anouck de Bonth. We are grateful to Verne Lee for kindly supplying cis-AMHO. The E. festucae E2368 genome sequence was made available by Christopher Schardl through grants EF-0523661 from the US National Science Foundation (to Christopher Schardl, Mark Farman and Bruce Roe) and 2005-35319-16141 from the US Department of Agriculture National Research (to Christopher Schardl).
Funding Statement
Our work is supported through funds from the New Zealand Foundation of Research Science and Technology under contracts AGRX0204, C10X0203 and C10X0815. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Guerinot ML (1994) Microbial iron transport. Annual Review of Microbiology 48: 743–772. [DOI] [PubMed] [Google Scholar]
- 2. Beard JL, Dawson H, Pinero DJ (1996) Iron metabolism: a comprehensive review. Nutrition Reviews 54: 295–317. [DOI] [PubMed] [Google Scholar]
- 3. Curie C, Briat JF (2003) Iron transport and signaling in plants. Annual Review of Plant Biology 54: 183–206. [DOI] [PubMed] [Google Scholar]
- 4. Halliwell B, Gutteridge JMC (1992) Biologically Relevant Metal Ion-Dependent Hydroxyl Radical Generation an Update. FEBS Letters 307: 108–112. [DOI] [PubMed] [Google Scholar]
- 5.Expert D (1999) Withholding and exchanging iron: Interactions between Erwinia spp. and their plant hosts. In: Webster RK, editor. Annual Review of Phytopathology. Palo Alto, California: Annual Reviews Inc., pp. 307–334. [DOI] [PubMed]
- 6.Haas H (2004) Molecular genetics of iron uptake and homeostasis in fungi. In: Brambl R, Marzluf GA, editors. The Mycota III Biochemistry and Molecular Biology. Berlin, Germany: Springer-Verlag.
- 7. Schaible UE, Kaufmann SHE (2004) Iron and microbial infection. Nature Reviews: Microbiology 2: 946–953. [DOI] [PubMed] [Google Scholar]
- 8. Jeong J, Guerinot ML (2009) Homing in on iron homeostasis in plants. Trends in Plant Science 14: 280–285. [DOI] [PubMed] [Google Scholar]
- 9. Clifton MC, Corrent C, Strong RK (2009) Siderocalins: Siderophore-binding proteins of the innate immune system. Bio Metals 22: 557–564. [DOI] [PubMed] [Google Scholar]
- 10. Ganz T (2009) Iron in innate immunity: starve the invaders. Current Opinion in Immunology 21: 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haas H, Eisendle M, Turgeon BG (2008) Siderophores in fungal physiology and virulence. Annual Review of Phytopathology 46: 149–187. [DOI] [PubMed] [Google Scholar]
- 12. Johnson L (2008) Iron and siderophores in fungal-host interactions. Mycological Research 112: 170–183. [DOI] [PubMed] [Google Scholar]
- 13. Hider RC, Kong X (2010) Chemistry and biology of siderophores. Natural Product Reports 27: 637–657. [DOI] [PubMed] [Google Scholar]
- 14. Eisendle M, Oberegger H, Zadra I, Haas H (2003) The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding l-ornithine N 5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC). Molecular Microbiology 49: 359–375. [DOI] [PubMed] [Google Scholar]
- 15. Eichhorn H, Lessing F, Winterberg B, Schirawski J, Kamper J, et al. (2006) A Ferroxidation/Permeation Iron Uptake System Is Required for Virulence in Ustilago maydis . Plant Cell 18: 3332–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, et al. (2004) Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. Journal of Experimental Medicine 200: 1213–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hissen AHT, Wan ANC, Warwas ML, Pinto LJ, Moore MM (2005) The Aspergillus fumigatus Siderophore Biosynthetic Gene sidA, Encoding L-Ornithine N5-Oxygenase, Is Required for Virulence. Infection and Immunity 73: 5493–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oide S, Krasnoff SB, Gibson DM, Turgeon BG (2007) Intracellular siderophores are essential for ascomycete sexual development in heterothallic Cochliobolus heterostrophus and homothallic Gibberella zeae . Eukaryotic Cell 6: 1339–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oide S, Moeder W, Krasnoff S, Gibson D, Haas H, et al. (2006) NPS6, encoding a nonribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell 18: 2836–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hof C, Eisfeld K, Welzel K, Antelo L, Foster AJ, et al. (2007) Ferricrocin synthesis in Magnaporthe grisea and its role in pathogenicity in rice. Molecular Plant Pathology 8: 163–172. [DOI] [PubMed] [Google Scholar]
- 21. Eisendle M, Schrettl M, Kragl C, Muller D, Illmer P, et al. (2006) The intracellular siderophore ferricrocin is involved in iron storage, oxidative-stress resistance, germination, and sexual development in Aspergillus nidulans . Eukaryotic Cell 5: 1596–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaplan J, McVey Ward D, Crisp RJ, Philpott CC (2006) Iron-dependent metabolic remodeling in S. cerevisiae . Biochimica et Biophysica Acta 1763: 646–651. [DOI] [PubMed] [Google Scholar]
- 23. Kosman DJ (2003) Molecular mechanisms of iron uptake in fungi. Molecular Microbiology 47: 1185–1197. [DOI] [PubMed] [Google Scholar]
- 24. Askwith C, Kaplan J (1997) An oxidase-permease-based iron transport system in Schizosaccharomyces pombe and its expression in Saccharomyces cerevisiae . Journal of Biological Chemistry 272: 401–405. [DOI] [PubMed] [Google Scholar]
- 25. Greenshields DL, Liu G, Feng J, Selvaraj G, Wei Y (2007) The siderophore biosynthetic gene SID1, but not the ferroxidase gene FET3, is required for full Fusarium graminearum virulence. Molecular Plant Pathology 8: 411–421. [DOI] [PubMed] [Google Scholar]
- 26. Mei B, Budde AD, Leong SA (1993) sid1, A gene initiating siderophore biosynthesis in Ustilago maydis: molecular characterization, regulation by iron, and role in phytopathogenicity. Proceedings of the National Academy of Sciences, USA 90: 903–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roman DG, Dancis A, Anderson GJ, Klausner RD, Roman DG, et al. (1993) The fission yeast ferric reductase gene frp1+ is required for ferric iron uptake and encodes a protein that is homologous to the gp91-phox subunit of the human NADPH phagocyte oxidoreductase. Molecular and Cellular Biology 13: 4342–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwecke T, Goettling K, Durek P, Duenas I, Kaeufer NF, et al. (2006) Nonribosomal peptide synthesis in Schizosaccharomyces pombe and the Architectures of ferrichrome-type siderophore synthetases in fungi. Chem Bio Chem 7: 612–622. [DOI] [PubMed] [Google Scholar]
- 29. Hwang LH, Mayfield JA, Rine J, Sil A (2008) Histoplasma requires SID1, a member of an iron-regulated siderophore gene cluster, for host colonization. PLoS Pathogens 4: e1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jung WH, Sham A, White R, Kronstad JW (2006) Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans . PLoS Biology 4: 2282–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramanan N, Wang Y (2000) A high-affinity iron permease essential for Candida albicans virulence. Science 288: 1062–1064. [DOI] [PubMed] [Google Scholar]
- 32.Van Der Helm D, Winkelmann G (1994) Hydroxamates and polycarboxylates as iron transport agents (siderophores) in fungi. In: Winkelmann G, Winge DR, editors. Mycology Series; Metal ions in fungi. New York, New York: Marcel Dekker, Inc. pp. 39–98.
- 33. Finking R, Marahiel MA (2004) Biosynthesis of nonribosomal peptides. Annual Review of Microbiology 58: 453–488. [DOI] [PubMed] [Google Scholar]
- 34. Yuan WM, Gentil GD, Budde AD, Leong SA (2001) Characterization of the Ustilago maydis sid2 gene, encoding a multidomain peptide synthetase in the ferrichrome biosynthetic gene cluster. Journal of Bacteriology 183: 4040–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winterberg B, Uhlmann S, Linne U, Lessing F, Marahiel MA, et al. (2010) Elucidation of the complete ferrichrome A biosynthetic pathway in Ustilago maydis . Molecular Microbiology 75: 1260–1271. [DOI] [PubMed] [Google Scholar]
- 36. Schrettl M, Bignell E, Kragl C, Sabiha Y, Loss O, et al. (2007) Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathogens 3: 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hof C, Eisfeld K, Antelo L, Foster AJ, Anke H (2009) Siderophore synthesis in Magnaporthe grisea is essential for vegetative growth, conidiation and resistance to oxidative stress. Fungal Genetics and Biology 46: 321–332. [DOI] [PubMed] [Google Scholar]
- 38. Haselwandter K (1995) Mycorrhizal fungi siderophore production. Critical Reviews in Biotechnology 15: 287–291. [DOI] [PubMed] [Google Scholar]
- 39. Haselwandter K, Winkelmann G (2002) Ferricrocin–an ectomycorrhizal siderophore of Cenococcum geophilum . BioMetals 15: 73–77. [DOI] [PubMed] [Google Scholar]
- 40. Haselwandter K, Passler V, Reiter S, Schmid DG, Nicholson G, et al. (2006) Basidiochrome - a novel siderophore of the orchidaceous mycorrhizal fungi Ceratobasidium and Rhizoctonia spp . Bio Metals 19: 335–343. [DOI] [PubMed] [Google Scholar]
- 41. Hordt W, Romheld V, Winkelmann G (2000) Fusarinines and dimerum acid, mono- and dihydroxamate siderophores from Penicillium chrysogenum, improve iron utilization by strategy I and strategy II plants. Bio Metals 13: 37–46. [DOI] [PubMed] [Google Scholar]
- 42. Schardl CL (2010) The epichloae, symbionts of the grass subfamily Poideae. Annals of the Missouri Botanical Garden 97: 646–665. [Google Scholar]
- 43. Christensen MJ, Bennett RJ, Ansari HA, Koga H, Johnson RD, et al. (2008) Epichloë endophytes grow by intercalary hyphal extension in elongating grass leaves. Fungal Genetics and Biology 45: 84–93. [DOI] [PubMed] [Google Scholar]
- 44. Schardl CL, Leuchtmann A, Spiering MJ (2004) Symbioses of grasses with seedborne fungal endophytes. Annual Review of Plant Biology 55: 315–340. [DOI] [PubMed] [Google Scholar]
- 45. Arachevaleta M, Bacon CW, Hoveland CS, Radcliffe DE (1989) Effect of the tall fescue endophyte on plant response to environmental stress. Agronomy Journal 81: 83–90. [Google Scholar]
- 46. Kimmons CA, Gwinn KD, Bernard EC (1990) Nematode reproduction on endophyte-infected and endophyte-free tall fescue. Plant Disease 74: 757–761. [Google Scholar]
- 47. Gwinn KD, Gavin AM (1992) Relationship between endophyte infestation level of tall fescue seed lots and Rhizoctonia zeae seedling disease. Plant Disease 76: 911–914. [Google Scholar]
- 48. Clay K, Holah J (1999) Fungal endophyte symbiosis and plant diversity in successional fields. Science 285: 1742–1744. [DOI] [PubMed] [Google Scholar]
- 49.Lane GA, Christensen MJ, Miles CO (2000) Chapter 14. Coevolution of fungal endophytes with grasses: the significance of secondary metabolites. In: Bacon CWW, J. F, editor. Microbial Endophytes. New York: Marcel Dekker. pp. 341–388.
- 50. Koulman A, Cao M, Faville M, Lane G, Mace W, et al. (2009) Semi-quantitative and structural metabolic phenotyping by direct infusion ion trap mass spectrometry and its application in genetical metabolomics. Rapid Communications in Mass Spectrometry 23: 2253–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Johnson R, Voisey C, Johnson L, Pratt J, Fleetwood D, et al. (2007) Distribution of NRPS gene families within the Neotyphodium/Epichloe complex. Fungal Genetics and Biology 44: 1180–1190. [DOI] [PubMed] [Google Scholar]
- 52. Lee TV, Johnson LJ, Johnson RD, Koulman A, Lane GA, et al. (2010) Structure of a Eukaryotic Nonribosomal Peptide Synthetase Adenylation Domain That Activates a Large Hydroxamate Amino Acid in Siderophore Biosynthesis. Journal of Biological Chemistry 285: 2415–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Koulman A, Lee TV, Fraser K, Johnson L, Arcus V, et al. (2012) Identification of extracellular siderophores and a related peptide from the endophytic fungus Epichloë festucae in culture and endophyte-infected Lolium perenne . Phytochemistry 75: 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thorstensen K (1988) Hepatocytes and reticulocytes have different mechanisms for the uptake of iron from transferrin. Journal of Biological Chemistry 263: 16837–16841. [PubMed] [Google Scholar]
- 55. Richardson DR, Baker E (1994) Two saturable mechanisms of iron uptake from transferrin in human melanoma cells: the effect of transferrin concentration, chelators, and metabolic probes on transferrin and iron uptake. Journal of Cellular Physiology 161: 160–168. [DOI] [PubMed] [Google Scholar]
- 56.Plattner HJ, Diekmann H (1994) Enzymology of siderophore biosynthesis in fungi. In: Winkelmann G, Winge DR, editors. Metal ions in fungi. New York: Marcel Decker. pp. 99–117.
- 57. Bushley K, Ripoll D, Turgeon BG (2008) Module evolution and substrate specificity of fungal nonribosomal peptide synthetases involved in siderophore biosynthesis. BMC Evolutionary Biology 8: 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Harrington BJ, Hageage GJ Jr (1991) Calcofluor white: Tips for improving its use. Clinical Microbiology Newsletter 13: 3–5. [Google Scholar]
- 59. Schardl CL, Phillips TD (1997) Protective grass endophytes: where are they from and where are they going? Plant Disease 81: 430–437. [DOI] [PubMed] [Google Scholar]
- 60. Rasmussen S, Parsons AJ, Bassett S, Christensen MJ, Hume DE, et al. (2007) High nitrogen supply and carbohydrate content reduce fungal endophyte and alkaloid concentration in Lolium perenne . New Phytologist 173: 787–797. [DOI] [PubMed] [Google Scholar]
- 61. Koulman A, Lane GA, Christensen MJ, Fraser K, Tapper BA (2007) Peramine and other fungal alkaloids are exuded in the guttation fluid of endophyte-infected grasses. Phytochemistry 68: 355–360. [DOI] [PubMed] [Google Scholar]
- 62. Rowan DD (1993) Lolitrems, paxilline, and peramine: mycotoxins of the ryegrass/endophyte interaction. Agriculture, Ecosystems and Environment 44: 103–122. [Google Scholar]
- 63. Craven KD, Blankenship JD, Leuchtmann A, Hignight K, Schardl CL (2001) Hybrid fungal endophytes symbiotic with the grass Lolium pratense . Sydowia 53: 44–73. [Google Scholar]
- 64. Spiering MJ, Lane GA, Christensen MJ, Schmid J (2005) Distribution of the fungal endophyte Neotyphodium lolii is not a major determinant of the distribution of fungal alkaloids in Lolium perenne plants. Phytochemistry 66: 195–202. [DOI] [PubMed] [Google Scholar]
- 65. Malinowski DP, Zuo H, Belesky DP, Alloush GA (2005) Evidence for copper binding by extracellular root exudates of tall fescue but not perennial ryegrass infected with Neotyphodium spp. endophytes. Plant and Soil 267: 1–12. [Google Scholar]
- 66. Liu Q, Parsons AJ, Xue HF, K., Ryan GD, Newman JA, et al. (2011) Competition between foliar Neotyphodium lolii endophytes and mycorrhizal Glomus spp. fungi in Lolium perenne depends on resource supply and host carbohydrate content. Functional Ecology 25: 910–920. [Google Scholar]
- 67. Courty PE, Hoegger PJ, Kilaru S, Kohler A, Buee M, et al. (2009) Phylogenetic analysis, genomic organization, and expression analysis of multi-copper oxidases in the ectomycorrhizal basidiomycete Laccaria bicolor . New Phytologist 182: 736–750. [DOI] [PubMed] [Google Scholar]
- 68. Haas H, Zadra I, Stoeffler G, Angermayr K (1999) The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. Journal of Biological Chemistry 19, 1999. [DOI] [PubMed] [Google Scholar]
- 69. Schrettl M, Kim HS, Eisendle M, Kragl C, Nierman WC, et al. (2008) SreA-mediated iron regulation in Aspergillus fumigatus . Molecular Microbiology 70: 27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mercier A, Pelletier B, Labbe S (2006) A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe . Eukaryotic Cell 5: 1866–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hortschansky P, Eisendle M, Al-Abdallah Q, Schmidt AD, Bergmann S, et al. (2007) Interaction of HapX with the CCAAT-binding complex-a novel mechanism of gene regulation by iron. EMBO Journal 26: 3157–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, et al. (2010) HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus . PLoS Pathogens 6: e1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hsu P-C, Yang C-Y, Lan C-Y (2011) Candida albicans Hap43 Is a Repressor Induced under Low-Iron Conditions and Is Essential for Iron-Responsive Transcriptional Regulation and Virulence. Eukaryotic Cell 10: 207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Takemoto D, Tanaka A, Scott B (2006) A p67Phox-like regulator is recruited to control hyphal branching in a fungal-grass mutualistic symbiosis. Plant Cell 18: 2807–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tanaka A, Christensen MJ, Takemoto D, Park P, Scott B (2006) Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction. Plant Cell 18: 1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tanaka A, Takemoto D, Hyon GS, Park P, Scott B (2008) NoxA activation by the small GTPase RacA is required to maintain a mutualistic symbiotic association between Epichloe festucae and perennial ryegrass. Molecular Microbiology 68: 1165–1178. [DOI] [PubMed] [Google Scholar]
- 77. Takemoto D, Kamakura S, Saikia S, Becker Y, Wrenn R, et al. (2011) Polarity proteins Bem1 and Cdc24 are components of the filamentous fungal NADPH oxidase complex. Proceedings of the National Academy of Sciences, USA 108: 2861–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Howard DH (1999) Acquisition, transport, and storage of iron by pathogenic fungi. Clinical Microbiology Reviews 12: 394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tan YY, Spiering MJ, Scott V, Lane GA, Christensen MJ, et al. (2001) In planta regulation of extension of an endophytic fungus and maintenance of high metabolic rates in its mycelium in the absence of apical extension. Applied and Environmental Microbiology 67: 5377–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lee B, Kroken S, Chou DYT, Robbertse B, Yoder OC, et al. (2005) Functional analysis of all nonribosomal peptide synthetases in Cochliobolus heterostrophus reveals a factor, NPS6, involved in virulence and resistance to oxidative stress. Eukaryotic Cell 4: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pujol-Carrion N, De La Torre-Ruiz MA (2010) Glutaredoxins Grx4 and Grx3 of Saccharomyces cerevisiae play a role in actin dynamics through their trx domains, which contributes to oxidative stress resistance. Applied and Environmental Microbiology 76: 7826–7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Moon CD, Scott B, Schardl CL, Christensen MJ (2000) The evolutionary origins of Epichloë endophytes from annual ryegrasses. Mycologia 92: 1103–1118. [Google Scholar]
- 83. Moon CD, Tapper BA, Scott B (1999) Identification of Epichloë endophytes in planta by a microsatellite- based PCR fingerprinting assay with automated analysis. Applied and Environmental Microbiology 65: 1268–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mantle PG, Nisbet LJ (1976) Differentiation of Claviceps purpurea in axenic culture. Journal of General Microbiology 93: 321–334. [DOI] [PubMed] [Google Scholar]
- 85. Latch GCM, Christensen MJ (1985) Artificial infection of grasses with endophytes. Annals of Applied Biology 107: 17–24. [Google Scholar]
- 86. Tanaka A, Tapper BA, Popay A, Parker EJ, Scott B (2005) A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Molecular Microbiology 57: 1036–1050. [DOI] [PubMed] [Google Scholar]
- 87. Gwinn KD, Collins-Shepard MH, Reddick BB (1991) Tissue print-immunoblot, an accurate method for the detection of Acremonium coenophialum in tall fescue. Phytopathology 81: 747–748. [Google Scholar]
- 88. Christensen MJ, Leuchtmann A, Rowan DD, Tapper BA (1993) Taxonomy of Acremonium endophytes of tall fescue (Festuca arundinacea), meadow fescue (F. pratensis) and perennial rye-grass (Lolium perenne). Mycological Research 97: 1083–1092. [Google Scholar]
- 89. Tanaka A, Tapper BA, Popay A, Parker EJ, Scott B (2005) A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Molecular Microbiology 57: 1036–1050. [DOI] [PubMed] [Google Scholar]
- 90.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- 91. Yoder OC (1988) Cochliobolus heterostrophus, cause of southern leaf blight. Advances in Plant Pathology 6: 93–112. [Google Scholar]
- 92. Denning DW, Clemons KV, Hanson LH, Stevens DA (1990) Restriction endonuclease analysis of total cellular DNA of Aspergillus fumigatus isolates of geographically and epidemiologically diverse origin. Journal of Infectious Diseases 162: 1151–1158. [DOI] [PubMed] [Google Scholar]
- 93. Young C, Itoh Y, Johnson R, Garthwaite I, Miles CO, et al. (1998) Paxilline-negative mutants of Penicillium paxilli generated by heterologous and homologous plasmid integration. Current Genetics 33: 368–377. [DOI] [PubMed] [Google Scholar]
- 94. Young CA, Bryant MK, Christensen MJ, Tapper BA, Bryan GT, et al. (2005) Molecular cloning and genetic analysis of a symbiosis-expressed gene cluster for lolitrem biosynthesis from a mutualistic endophyte of perennial ryegrass. Molecular Genetics and Genomics 274: 13–29. [DOI] [PubMed] [Google Scholar]
- 95. Namiki F, Matsunaga M, Okuda M, Inoue I, Nishi K, et al. (2001) Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis . Molecular Plant-Microbe Interactions 14: 580–584. [DOI] [PubMed] [Google Scholar]
- 96. Vollmer SJ, Yanofsky C (1986) Efficient cloning of genes of Neurospora crassa . Proceedings of the National Academy of Sciences, USA 83: 4869–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Itoh Y, Johnson R, Scott B (1994) Integrative transformation of the mycotoxin-producing fungus, Penicillium paxilli . Current Genetics 25: 508–513. [DOI] [PubMed] [Google Scholar]
- 98. Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Schwarzer D, Finking R, Marahiel MA (2003) Nonribosomal peptides: from genes to products. Natural Product Reports 20: 275–287. [DOI] [PubMed] [Google Scholar]
- 100. McCormack P, Worsfold PJ, Gledhill M (2003) Separation and detection of siderophores produced by marine bacterioplankton using high-performance liquid chromatography with electrospray ionization mass spectrometry. Analytical Chemistry 75: 2647–2652. [DOI] [PubMed] [Google Scholar]
- 101. Gallagher RT, Hawkes AD, Stewart JM (1985) Rapid determination of the neurotoxin lolitrem B in perennial ryegrass by high-performance liquid chromatography with fluorescence detection. Journal of Chromatography A 321: 217–226. [DOI] [PubMed] [Google Scholar]
- 102. Spiering MJ, Davies E, Tapper BA, Schmid J, Lane GA (2002) Simplified extraction of ergovaline and peramine for analysis of tissue distribution in endophyte-infected grass tillers. Journal of Agricultural and Food Chemistry 50: 5856–5862. [DOI] [PubMed] [Google Scholar]
- 103. Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant Journal 11: 1187–1194. [Google Scholar]
- 104. Eaton CJ, Jourdain I, Foster SJ, Hyams JS, Scott B (2008) Functional analysis of a fungal endophyte stress-activated MAP kinase. Current Genetics 53: 163–174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Δ sidN Mutants Accumulate the Epichloënin A Precursor Trans -AMHO. A. Positive electrospray LS-MS extracted MS1 ion chromatograms of the parent [M+H]+ion (m/z 261) and CID MS2 spectra of authentic cis-AMHO standard and putative trans-AMHO from ΔsidN 85 mutant. B. Relative concentrations of trans-AMHO in extracts of mycelium from cultures of wild-type E. festucae Fl1 (WT), ΔsidN mutant 85 (ΔsidN), and a complemented ΔsidN strain (C-sidN) grown under Fe-depleted conditions detected by LCMS (MS1 m/z 261) (arbitrary units).
(TIF)
Conidiation in E. festucae is Reduced by Loss of Epichloënin A. A. Spore counts produced from colonies of E. festucae Fl1 (WT), complement (C-sidN) and ΔsidN mutant 85 (ΔsidN) grown on water-agar for 2 weeks at 22°C, followed by 2 weeks at 4°C. Data were generated from three independent colonies and three technical replicates. Error bars = standard error. B. Microscopic examination of WT and ΔsidN water-agar colonies used for spore counts showing spores on coil structures and hyphal strands
(TIF)
Gene Disruption Strategy and Southern Blot Analysis. A. A linear sidN disruption vector is shown annotated with corresponding modular NRPS domain structure [A-domain (adenylation), T-domain (peptidyl carrier) and C-domain (condensation)] and restriction enzyme sites for HindIII (H). The hygromycin (HYG) cassette is indicated to insert into the A-domain of SidN (which was deduced from partial sidN sequence). B. An autoradiograph of a DNA gel blot of HindIII digested genomic DNA of ΔsidN mutants 54, 82 and 85 along with wild-type (WT) and ectopic (E) controls probed with a DIG-labeled sidN PCR product amplified with primers Sid3F and Sid2R.
(TIF)
Detailed Information of the Primers Used in This Study.
(DOCX)
Iron-Responsive Genes in E. festucae.
(DOCX)