Abstract
Aeolidida is one of the largest clades of nudibranchs with at least 560 known species. However, its systematics has not been studied in a comprehensive manner. Phylogenetic analyses of larger clades such as Nudibranchia or Cladobranchia have usually included a poor sample of aeolids. Furthermore, phylogenetic studies at the family or generic level in Aeolidida are a few and far between. The first molecular phylogeny of the aeolid family Aeolidiidae is presented here. This study, the most comprehensive for Aeolidida to date, uses new sequences of two mitochondrial (COI and 16S) genes and one nuclear gene (H3). 251 specimens from members of seven families of Aeolidida, including 39 species of Aeolidiidae were studied. Excluding Pleurolidia juliae, Aeolidiidae is monophyletic. Our results resolve the systematic relationships within the Aeolidiidae at a generic level, requiring changes in the systematics of this family. Spurilla, Anteaeolidiella, Limenandra and Aeolidia are well-supported and monophyletic clades. Aeolidiella stephanieae is transferred to Berghia and Aeolidiopsis ransoni and Spurilla salaamica to Baeolidia, to maintain the monophyletic lineages reflected in this study. The systematics of Cerberilla remains unclear. Some species earlier attributed to Aeolidiella are now grouped in a previously unnamed clade that we designate as Bulbaeolidia gen. nov.
Introduction
Aeolidida contains about a dozen families constituted by approximately 560 described species [1]. Members of this taxon are easily distinguished by their elongated and tapering bodies, lacking distinct gills. All members have specialized dorsal appendages, called cerata, that are used in respiration and defence. The cerata contain branches of the digestive gland that transports nematocysts (acquired from the prey) to their tips where they are stored in the so-called cnidosacs. The vast majority of the aeolids possess a uniseriate radula, although some genera also have lateral teeth (e.g. Flabellina spp, Eubranchus spp, Notaeolidia spp). Although it constitutes the second largest group of Nudibranchia [2], few phylogenetic studies have been undertaken so far. Several contributions are focused on some aeolid genera or families [3]–[13]. Some of them also include phylogenetic analyses, but only Moore & Gosliner [12] and Carmona et al. [13] employed a molecular approach. Most of the phylogenetic studies focused on higher taxa, such as Heterobranchia, Euthyneura, Opisthobranchia, Nudibranchia or Cladobranchia include some aeolid species [14]–[26]. These studies support the monophyly of the aeolids, identifying the presence of cnidosacs and the transformation of the oral veil into oral tentacles as synapomorphies [26]. Only Martin et al. [27] rejected the monophyletic status of Aeolidida after including Hancockia and Embletonia.
Within Aeolidida, Aeolidiidae is one of the largest families, whose members differ from the rest of aeolids by their pectinate radular teeth [4], [8], [28]–[30] and their diet –most of them feed on anemones. Indeed, many species sequester zooxanthellae from their prey and maintain the dinoflagellates alive in their tissues in order to use the photosynthetic products for their own nutrition (e.g. Spurilla neapolitana, Berghia verrucicornis, B. coerulescens, “S. salaamica”,“B. major”, Aeolidiella alderi, “A. stephanieae”; see Kempf [31] and Marín & Ros [32]). This family is composed of the following genera: Aeolidia Cuvier, 1798; Spurilla Bergh, 1864; Aeolidiella Bergh, 1867; Cerberilla Bergh, 1873; Berghia Trinchese, 1877; Baeolidia Bergh, 1888; Protaeolidiella Baba, 1955; Aeolidiopsis Pruvot-Fol, 1956; Limenandra Haefelfinger & Stamm, 1958; Anteaeolidiella Miller, 2001; Burnaia Miller, 2001 and Milleraeolidia Ortea, Caballer & Espinosa, 2004, although the validity of some of these taxa has been questioned [4], [8], [33]. Furthermore, the proposed wide geographical distribution for several species (Aeolidia papillosa, Anteaeolidiella indica, Berghia coerulescens, Limenandra nodosa, Spurilla neapolitana) is controversial and raises doubts about the conspecificity of the different populations that are geographically isolated.
Recent morphological and molecular approaches provided a preliminary idea about the phylogenetic relationships between Aeolidiidae and other members of Aeolidida [10], [13]. The systematic relationships between genera and/or species of Aeolidiidae itself have been subject of controversy for the last seventy years [3]–[4], [8], [30], [34]–[43]. In 1939, based on differences in the position of the nephroproct, Odhner [27] removed Spurilla and Berghia from Aeolidiidae and placed them in a new family called Spurillidae. Haefelfinger & Stamm [35] erected the genus Limenandra with L. nodosa as the type species and transferred Baeolidia fusiformis Baba, 1949 to this new genus. Ten years later, Odhner (in [37]) included Baeolidia in Spurillidae but overlooked Limenandra. Gosliner [41] regarded Limenandra as a junior synonym of Baeolidia and Rudman [30] considered Berghia as a junior synonym of Spurilla. In the same year, Schmekel & Portmann [42], considering only the Mediterranean species, split Aeolidiidae in two subfamilies: Aeolidiinae (Aeolidiella) and Spurillinae (Spurilla, Berghia and Limenandra). In 1985, Gosliner [4] used his previous classification with the aeolidiids from South Africa. Five years later, Rudman [44] regarded Pleurolidia as a junior synonym of Protaeolidiella and considered Protaeolidiella atra Baba, 1955 and Pleurolidia juliae Burn, 1966 to be conspecific. Finally, in the 21st century Anteaeolidiella, Burnaia and Milleraeolidia were included within Aeolidiidae [8], [45], although Milleraeolidia ritmica Ortea, Caballer & Espinosa 2004, the sole species of this genus, has been considered as a synonym of Berghia creutzbergi (Marcus & Marcus, 1970) by Valdés et al. [46] and García-García et al. [33].
The primary objective of this study is to elucidate the phylogenetic relationships of the family Aeolidiidae. Molecular phylogenetic analyses were undertaken based on two mitochondrial and one nuclear gene (mitochondrial COI and 16S rRNA, and nuclear H3). Representatives of seven currently recognized families of Aeolidida have been included. The specific aims are: (i) to test if “Spurillidae”/“Spurillinae” and “Aeolidiinae” are monophyletic; (ii) to test the monophyly of the different genera of Aeolidiidae, reviewing their phylogenetic relationships.
Materials and Methods
Sampling
Samples were obtained using standard SCUBA diving sampling techniques for opisthobranchs and through the study of museum collections. Two hundred and fifty-one specimens including 39 nominal species of Aeolidiidae, 3 species of Babakinidae, 33 species of Facelinidae, 7 species of Flabellinidae, 1 species of Fionidae, 3 species of Piseinotecidae and 4 species of Tergipedidae were studied. The classification of all the species used in this study is listed in Table S1 and arranged based on Gosliner et al. [47] classification. Numbers following “sp.” in the names of undescribed species refer to the identification system used by Gosliner et al. [47]. Undescribed species labelled as “sp.” followed by a capital letter refer to undescribed species not included in Gosliner et al. [47]. Voucher specimens are held in the collections of the California Academy of Sciences, CASIZ (San Francisco, USA), Museo Nacional de Ciencias Naturales, MNCN (Madrid, Spain), Museu de Zoologia da Universidade São Paulo, MZSP (São Paulo, Brazil), Museu Nacional/Universidade Federal do Rio de Janeiro, MNRJ (Rio de Janeiro, Brazil), University Museum of Bergen, ZMBN (Bergen, Norway) or Zoologische Staatssammlung München, ZSM (Munich, Germany).
Ethic Statements
The majority of the specimens used in this study belongs to the California Academy of Sciences Invertebrate Zoology (CASIZ) collection and the DNA collection of the Museo Nacional de Ciencias Naturales (MNCN) of Madrid. We had the permission to take tissue samples from all the specimens for DNA analyses, regardless where the specimen was deposited. As stated in the CASIZ collections policy: ‘No specimens will be accessioned without adequate labelling, collection notes, field notes, or other locality information, nor without appropriate legal documentation (collecting permits, export permits from country of origin, etc.) when applicable.’ Those DNA samples deposited in the MNCN were obtained from pieces of foot from specimens collected during field trips. These specimens will provisionally remain at the University of Cádiz until their anatomical study is completed. Thereafter, each specimen will be deposited in the MNCN. All the museums listed in point 2.1, from which material has been examined in the present study, have strict collection policies in order to avoid illegal practices. Donors are required to provide all the available information about their field studies, including collecting and export permits from country of origin, as well as the number of species and specimens exported. Therefore, the legality of all this material is assumed. On the other hand, all necessary permits were obtained for the field studies carried out in Cuba, Bermuda, Azores, Madeira, Chafarinas Is. and Balearic Is. by the authors. These permits were facilitated by Cuban environmental authorities through the Centro de Investigaciones Marinas (C. I. M.) of the University of La Habana, by the Department of Environmental Protection of the Government of Bermuda, by Secretaria Regional do Ambiente e do Mar of the Regional Government of Azores, by the Secretaria Regional do Ambiente e dos recursos Naturais of the Regional Government of Madeira, by the Organismo Autónomo de Parques Nacionales (Government of Spain) for the Chafarinas Islands and the Dirección General de Pesca of the Regional Government of Balearic Islands. For the remaining localities (Japan, Sweden, France, Morocco, Canary Is., Senegal, Cape Verde, Greece and Italy), the specimens were collected and sent by several gracious collaborators. These locations were not privately-owned or legally protected. Finally, none of the studied species are protected, listed as endangered, or included in the CITES list.
DNA Extraction, Amplification and Sequencing
A total of 194 specimens were successfully sequenced for the cytochrome c oxidase subunit I (COI), 228 for the 16S rRNA (16S) and 229 for the Histone-3 (H3) genes. Thirty-two additional sequences were obtained from GenBank (see Table S1 for full list of samples, localities, and voucher references).
Tritonia antarctica was chosen as the outgroup, due to its basal placement within Cladobranchia [23].
Excluding Anteaeolidiella (see Anteaeolidiella discussion), Baeolidia and Cerberilla, the identities of the different clades within Aeolidiidae were conducted based on the type species of each genus. For those inconclusive clades, current names were retained. In order to minimize disruption to nomenclature, synonyms were resurrected whenever possible.
DNA was extracted from foot tissue of specimens preserved with 70–100% ethanol, except in the cases of small individuals that were used whole. The DNeasy Blood and Tissue Kit of Qiagen (Qiagen, Valencia, CA, USA; 09/2001) was used for DNA extraction.
Partial sequences of COI, 16S and H3 were amplified by polymerase chain reaction (PCR) using the primers: LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAATCA-3′) [48] for COI; 16S ar-L (5′-CGCCTGTTTATCAAAAACAT-3′) and 16S br-H (5′-CCGGTCTGAACTCAGATCACGT-3′) [49] for 16S rRNA; and H3AD5′3′ (5′- ATGGCTCGTACCAAGCAGACVGC-3′) and H3BD5′3′ (5′-ATATCCTTR GGCATRATRGTGAC-3′) [50] for H3. These three gene regions are commonly used in systematic studies of gastropods (e.g. [23], [51]–[55]). However, several internal primers for COI and H3 were designed for those specimens that could not be amplified using the universal ones (Table 1).
Table 1. Forward (F) and reverse (R) PCR specific primers.
| Name | Sequence 5′-3′ |
| COI | |
| Apapi_COI_IntF | TGTGGTGTGGATTAGCAGGA |
| Apapi_COI_IntR | CAGCCAAAACCGGTAAAGAT |
| Bergh_COI_IntF | ATTRGGAATGTGATGTGGGT |
| Bergh_COI_IntR | CCAGCAGGRTCRAAAAACCT |
| Lime_COI_IntrF | TGTTTTAYTAGGRATGTGATGTGG |
| Lime_COI_IntrR | TTGTAGTAATAAAATTAATTGCCCCA |
| Snea_COI_IntF | TTCGTTTTGAACTYGGAACRG |
| Snea_COI_IntR | CACCAGCTAAAACAGGTAGMG |
| H3 | |
| Apapi_H3_IntF | TAAATCCACCGGAGGAAAGG |
| Apapi_H3_IntR | CCTCAANCAGACCGACCAAG |
PCRs were conducted in a 50 µl volume reactions containing 2 µl of both forward and reverse primers (10 µM), 5 µl of dNTP (2 mM), a gene-dependent amount of magnesium chloride (25 mM), 0.5 µl of Qiagen DNA polymerase (250 units), 10 µl of “Q-solution” (5×), and 5 µl of Qiagen buffer (10×) (Qiagen Taq PCR Core Kit cat. no. 201225). Magnesium chloride amounts were 7 µl for COI and 16S, and 4 µl for H3. Amplification of COI was performed with an initial denaturation for 5 min at 94°C, followed by 35 cycles of 1 min at 94°C, 30 s at 44°C (annealing temperature) and 1 min at 72°C with a final extension of 7 min at 72°C. The 16S amplification began with an initial denaturation for 5 min at 95°C followed by 35 cycles of 30 s at 94°C, 30 s at 44°C (annealing temperature), 1 min at 72°C with a final extension of 7 min at 72°C. H3 amplification was performed with an initial denaturation for 3 min at 95°C, followed by 40 cycles of 45 s at 94°C, 45 s at 50°C (annealing temperature), 2 min at 72°C, with a final extension of 10 min at 72°C.
Successful PCR product was purified mixing 5 µl of PCR product with 2 µl of ExoSAP-IT (usb.affymetrix.com). Samples were incubated at 37°C for 15 min followed by an inactivation step at 80°C for 15 min Sequence reactions were run on a 3730XL DNA sequencer, Applied Biosystems. All new sequences have been deposited in GenBank.
Sequence Alignment and Phylogenetic Analyses
DNA sequences were assembled and edited using Geneious Pro 4.7.6 [56]. All the sequences were checked for contamination with BLAST [57] implemented in the Genbank database. Geneious and MAFFT [58] were employed to align the sequences, using the default settings in both programs. The alignments were checked by eye using MacClade (version 4.06) [59]. Protein-coding sequences were translated into amino acids for alignment confirmation. Saturation was visually inspected in MEGA 5.0 [60] by plotting for all specimens including outgroup the total number of pairwise differences (transitions and transversions) against uncorrected p-distances. For the COI and H3 genes, saturation was further examined separately for the first, second and third codon positions.
The most variable regions from the 16S rRNA alignment were removed using both the default settings and the standard options for stringent and less stringent selection in Gblocks [61]. Excluding “indel-rich” regions, the tree was in general poorly resolved with lower node support. Therefore, final analyses were performed including all bases. Sequences of COI, 16S, and H3 were trimmed to 669, 484, and 328 base pairs, respectively.
Individual gene analyses and a concatenated analysis were performed. To test for conflicting phylogenetic signal between genes, the incongruence length difference test (ILD) [62] was conducted as the partition homogeneity test in PAUP* 4.0b10 [63]. Test settings consisted of 10 random stepwise additions (100 replicates) with TBR branch swapping.
The best-fit models of evolution for each gene were determined using the Akaike information criterion [64] implemented in MrModeltest 2.3 [65]. The GTR+I+G was selected for the three genes.
Maximum likelihood (ML) analyses were performed using the software RAxML v7.0.4 [66] and node support was assessed with non-parametric bootstrapping (BS) with 60000 replicates, random starting trees, and parameters estimated from each dataset under the model selected for the original dataset. Bayesian inference analyses (BI) were conducted using MrBayes version 3.1.2b [67] for thirty nine million generations and four chains. Markov chains were sampled every 1000 generations.
The models implemented were those estimated with MrModeltest 2.3. The combined dataset was partitioned among genes and the “unlink” command was used to allow all parameters to vary independently within each partition.
Convergence was diagnosed graphically by plotting for each run the likelihood against the number of generations using the software Tracer version 1.4.1 [68]. For each analysis the first 15000 trees were discarded (‘burn-in’ period) and node support was assessed with posterior probabilities (PP). Only nodes supported by BS ≥75 and PP≥0.90 are discussed. All the alignments and trees published in this study are deposited in TreeBase under project number S14014 (http://purl.org/phylo/treebase/phylows/study/TB2:S14014).
Genetic Distances
In order to compare the genetic distances amongst specimens of Aeolidiidae, we calculated the pairwise uncorrected p-distances for COI using PAUP* 4.0 b 10.0. (Table 2). All codon positions were considered for the analysis.
Table 2. Minimum COI gene pairwise uncorrected p-distances amongst key species.
| Species | COI genetic distances (%) | ||
| A. lurana Naples | vs. | A. lurana Brazil | 0 |
| A. papillosa Sweden | vs. | A. papillosa Maine | 0.3 |
| L. nodosa France | vs. | L. nodosa Bahamas | 1 |
| S. braziliana Costa Rica (PAC) | vs. | S. braziliana Japan | 1.9 |
| B. rissodominguezi Cuba | vs. | B. stephanieae Florida | 5.7 |
| S. neapolitana Naples | vs. | S. sargassicola Bahamas | 6.4 |
| Limenandra sp. A Mexico (PAC) | vs. | L. nodosa Bahamas | 7.3 |
| A. papillosa WA | vs. | Aeolidia sp. B California | 8.4 |
| A. saldanhensis South Africa | vs. | A. lurana Brazil | 8.4 |
| S. braziliana Costa Rica (PAC) | vs. | Spurilla sp. A Morocco (ATL) | 10.8 |
| A. papillosa Maine | vs. | Aeolidia sp. A France (ATL) | 11 |
| A. saldanhensis South Africa | vs. | A. cacaotica Australia | 11.7 |
| A. lurana Naples | vs. | A. cacaotica Australia | 11.8 |
| A. takanosimensis Japan | vs. | A. lurana Brazil | 12.4 |
| B. japonica Marshall Is. | vs. | Baeolidia sp. C Marshall Is. | 12.6 |
| A. saldanhensis South Africa | vs. | A. takanosimensis Japan | 13 |
| A. takanosimensis Japan | vs. | A. cacaotica Australia | 13 |
| Aeolidia sp. A France (ATL) | vs. | Aeolidia sp. B California | 13.3 |
| B. alba Malaysia | vs. | Bulbaeolidia sp. A Brazil | 13.5 |
| S. braziliana Japan | vs. | S. sargassicola Bahamas | 13.6 |
| S. neapolitana Portugal | vs. | S. braziliana Brazil | 14.1 |
| A. takanosimensis Japan | vs. | Anteaeolidiella sp. A Clipperton Is. | 14.2 |
| S. sargassicola Bahamas | vs. | Spurilla sp. A Morocco (ATL) | 14.8 |
| S. neapolitana Naples | vs. | Spurilla sp. A Morocco (ATL) | 14.8 |
Names are final identifications after the analyses.
ATL = Atlantic Ocean;
PAC = Pacific Ocean;
WA = western Atlantic Ocean.
Species Concept and Genetic Divergence Thresholds
To define species, we used the criteria of divergence and reciprocal monophyly supported by independent genetic markers [55], [69]–[72]. Since different groups of organisms may present distinct rates of evolution, the use of genetic threshold is difficult to apply [73]–[74]. However, based on our molecular as well as our morphology results, we established a cut-off range of 5.5–16% between sister species of Aeolidiidae (uncorrected p-distance for COI gene). Table S2 shows the minimum uncorrected p-distance for COI between sister species of each genus.
Nomenclatural Acts
The electronic edition of this article conforms to the requirements of the amended International Code of Zoological Nomenclature, and hence the new names contained herein are available under that Code from the electronic edition of this article. This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN. The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information viewed through any standard web browser by appending the LSID to the prefix "http://zoobank.org/". The LSID for this publication is: LSID urn:lsid:zoobank.org:pub:33A960E8-2106-41DF-A5C8-4FA68B0974CB. The electronic edition of this work was published in a journal with an ISSN, and has been archived and is available from the following digital repositories: It will be available on the Research website of the California Academy of Sciences http://research.calacademy.org/.
Results
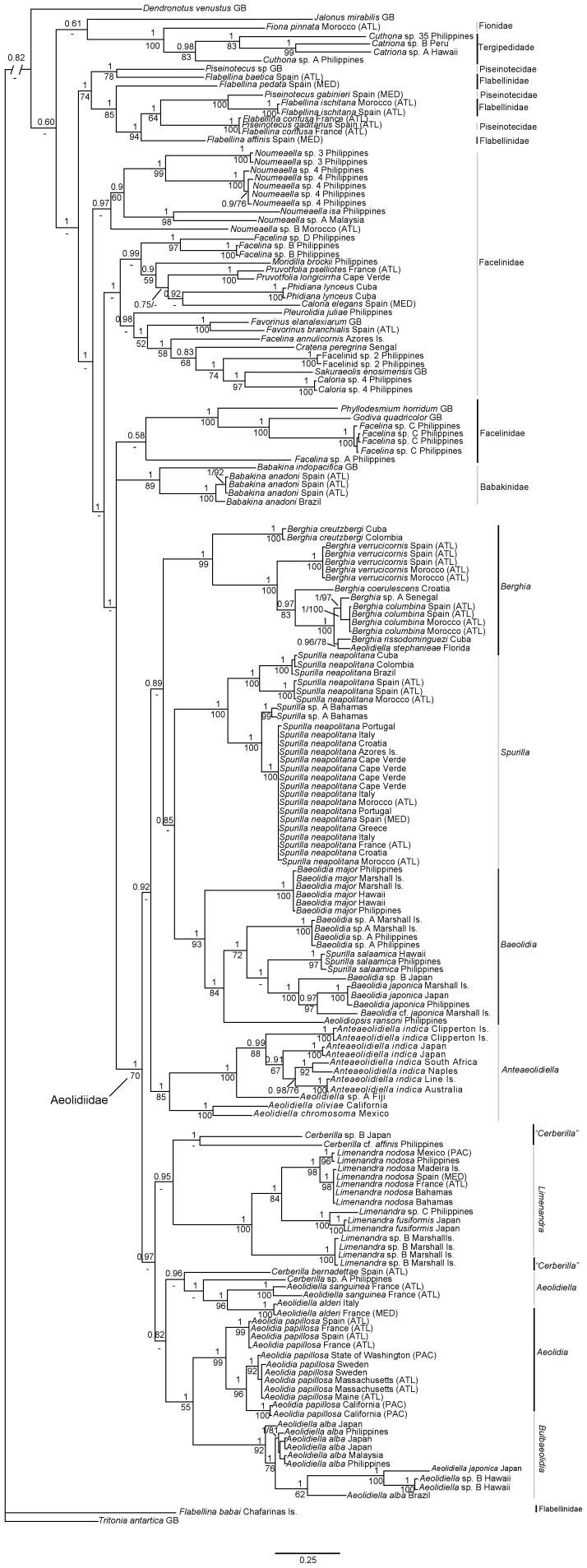
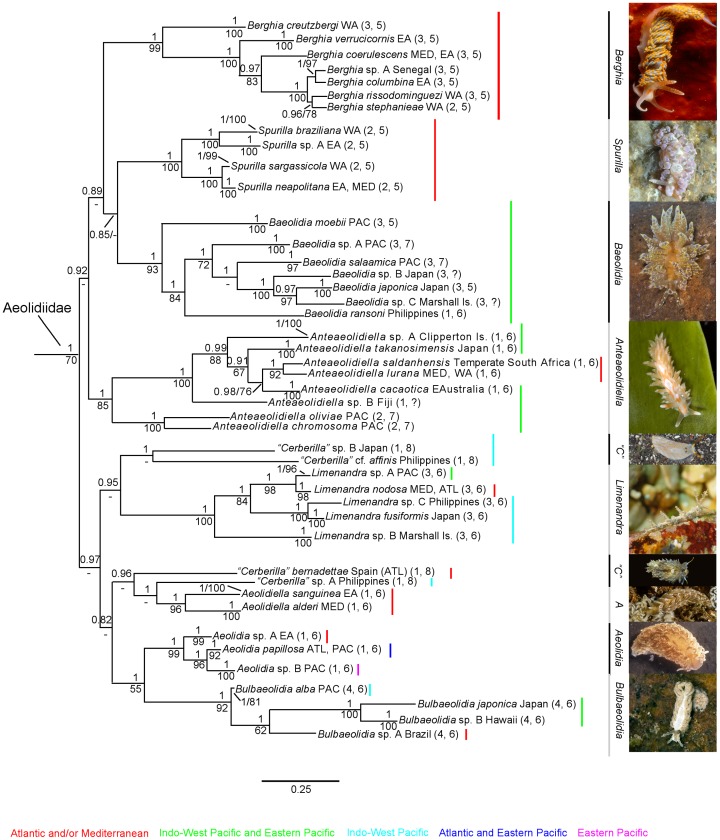
The combined dataset yielded a sequence alignment of 1,481positions. The ILD test showed no significant conflicting signal between the three genes (P = 0.15). No saturation was observed across genes and codon positions (not shown). The combined tree provided better resolution than H3, COI, or 16S separately (only the COI tree is provided, see Figure S1). Although bootstrap values were lower than posterior probabilities in larger clades, the topologies of the ML trees were congruent with the results yielded by Bayesian analyses. The ML tree is provided as Supplementary material (Figure S2). Figures 1 and 2 show the resulting phylogenetic hypothesis based on the combined dataset represented by BI. Figure 1 depicts species names with the preliminary identifications, whereas Figure 2 excludes the whole outgroup and shows only one representative of each species with the final revised names.
Figure 1. Phylogenetic hypothesis based on the combined dataset (H3+COI+16S) inferred by Bayesian analysis (BI).
Numbers above branches represent posterior probabilities from BI. Numbers below branches indicate bootstrap values for ML. Abbreviations: ATL, Atlantic Ocean; EA, eastern Atlantic Ocean; GB, GenBank; MED, Mediterranean; PAC, Pacific; WA, western Atlantic Ocean. Genera names on right side of vertical bars refer to revised classification.
Figure 2. Molecular phylogeny based on the combined dataset (H3+COI+16S) inferred by Bayesian analysis (BI).
Only one specimen per species of Aeolidiidae is shown. Numbers above branches represent posterior probabilities from BI. Numbers below branches indicate bootstrap values for ML. Numbers in brackets are code numbers for different states of the morphological characters listed in Table 3. Excluding Anteaeolidiella and Cerberilla, photographs depict the type species of each genus. Photo credits and details: Berghia coerulescens, Guido Villani, Italy; Spurilla neapolitana, Marina Poddubetskaia, France, Cap Ferret; Baeolidia moebii, Terrence M. Gosliner, Philippines; Anteaeolidiella lurana, Terrence M. Gosliner, Bermuda; Cerberilla sp. B, A. Kawahara, Japan; Limenandra nodosa, Marina Poddubetskaia, France, Cap Ferret; Cerberilla sp. A, Terrence M. Gosliner, Philippines; Aeolidiella alderi, Guido Villani, Italy; Aeolidia papillosa, Marta Pola, Sweden; Bulbaeolidia alba, Terrence M. Gosliner, Philippines. Abbreviations: A, Aeolidiella; ATL, Atlantic Ocean; “C”, “Cerberilla”; EA, eastern Atlantic Ocean; MED, Mediterranean; PAC, Pacific; WA, western Atlantic Ocean. Genera names on right side of vertical bars refer to revised classification; ?, missing data.
The relationship between the monophyletic Babakinidae (PP = 1, BS = 89) and some facelinid species (Phyllodesmium horridum, Godiva quadricolor, Facelina sp. C, and Facelina sp. A) with Aeolidiidae was not resolved (PP = 1, not recovered in ML). Excluding Pleurolidia, which clustered with some Facelinidae, Aeolidiidae was monophyletic and strongly supported in Bayesian analyses (PP = 1, BS = 70). Furthermore, Aeolidiidae was divided in two major subclades: one including Berghia, Spurilla, Baeolidia and Anteaeolidiella (PP = 0.92, not recovered in ML) and the other with Ceberilla, Limenandra, Aeolidiella, Aeolidia and an unnamed subclade (PP = 0.97, not recovered in ML). Within Aeolidiidae, and excluding Aeolidiella, Cerberilla, Berghia, Baeolidia and Aeolidiopsis, all the traditional genera included in this analysis were monophyletic and well supported by the Bayesian and Maximum likelihood analyses.
Aeolidiella stephanieae was the sister species of Berghia rissodominguezi (PP = 0.96, BS = 78; minimum uncorrected p-distance = 5.7% for COI), resulting in a necessary taxonomic change of the former species to preserve the monophyly of Berghia. In addition to the two species mentioned above, the Berghia clade included B. creutzbergi, B. verrucicornis, B. coerulescens, an undescribed species from Senegal and B. columbina (PP = 1, BS = 99).
Spurilla specimens all clustered together in a clade with the maximum support (PP = 1, BS = 100). This clade was divided into two branches. The first one was composed by western Atlantic specimens of Spurilla neapolitana plus a small population from western Andalusia (Spain) and Morocco (Atlantic coasts) (PP = 1, BS = 100; minimum uncorrected p-distance = 10.8% for COI between eastern and western Atlantic specimens). The second branch included Spurilla sp. A from Bahamas and the Mediterranean Sea as well as eastern Atlantic specimens of Spurilla neapolitana (PP = 1, BS = 100; minimum uncorrected p-distance = 6.4% for COI).
The Bayesian inference tree shows Baeolidia (PP = 1, BS = 93) as the sister group of Spurilla, whereas in the Maximum likelihood analyses Baeolidia was the sister species of Berghia (Figure S2). However, none of these relationships were supported (PP = 85, not recovered in ML). Aeolidiopsis ransoni and Spurilla salaamica clustered among Baeolidia species. Baeolidia major specimens clustered in a single subclade (PP = 1, BS = 84). The other subclade was composed by an undescribed species from the Philippines and the Marshall Is., Spurilla salaamica, an undescribed species from Japan, Baeolidia japonica from Marshall Is., Japan and the Philippines and one specimen of Baeolidia cf. japonica from Marshall Is. (minimum uncorrected p-distance = 12.6% for COI between this specimen and B. japonica). Aeolidiopsis ransoni was basal to the latter subclade (PP = 1, BS = 84).
Anteaeolidiella was the sister group of Berghia, Spurilla and Baeolidia (PP = 0.92, not recovered in ML), appearing as a well-supported lineage (PP = 1, BS = 85). Anteaeolidiella was split into two groups: Aeolidiella chromosoma and Aeolidiella oliviae formed the first and basal clade (PP = 1, BS = 100), whereas an undescribed species from Fiji together with Anteaeolidiella indica specimens constituted the second clade (PP = 1, BS = 100). These specimens previously identified as A. indica were divided into four different subclades (PP = 1, BS = 88; minimum uncorrected p-distance = 8.4% for COI among clades) and likely include five distinct species.
Cerberilla was not recovered as monophyletic. Cerberilla affinis and Cerberilla sp. B from Japan formed the sister group of Limenandra (PP = 0.95, not recovered in ML). However, Cerberilla bernadettae and Cerberilla sp. A from the Philippines clustered in a separate clade basal to some Aeolidiella species (PP = 0.96, not recovered in ML).
Limenandra specimens clustered in a clade with maximum support (PP = 1, BS = 100). This clade was divided into two subgroups. All Limenandra nodosa specimens formed a monophyletic group with maximum support (PP = 1, BS = 98), subdivided in Pacific and Mediterranean-Atlantic clusters (minimum uncorrected p-distance = 7.3% for COI between Pacific and Mediterranean-Atlantic specimens). The maximum genetic distance between eastern and western Atlantic population was 1%. Limenandra fusiformis and an undescribed species from the Philippines (PP = 1, BS = 100) appeared as the sister group of “L. nodosa” complex (PP = 1, BS = 84). Another undescribed species from the Marshall Islands completed this clade, forming the second subgroup that is sister to the remainder of Limenandra.
Specimens previously ascribed to Aeolidiella clustered in two distinct and separated clades with maximum support, rendering this taxon polyphyletic. Aeolidiella alderi (type species) clustered with Aeolidiella sanguinea (PP = 1, BS = 96), whereas Aeolidiella alba, Aeolidiella japonica, an undescribed species from Hawaii and one specimen of Aeolidiella alba from Brazil formed the second clade (PP = 1, BS = 92). These latter taxa formed the sister group to Aeolidia (PP = 1, not recovered in ML). The basal position of one of the specimens of A. alba from Japan can be explained because its 16S sequence presented some insertions. Genetic distance between Aeolidiella alba from the Indo-Pacific and from the western Atlantic was 13.5% (minimum uncorrected p-distances for COI).
Finally, specimens of Aeolidia papillosa were placed in two different clades both with maximum support. The first clade included specimens from the Atlantic coast of France and from Galicia (Spain). The second clade was further divided into two subclades: specimens from Sweden, one specimen from Maine (USA), two from Massachusetts (USA) and one specimen from the state of Washington (USA) clustered together; the second subclade included two specimens from the California coast. The minimum p-distance among all Aeolidia clades was 8.4% (uncorrected p-distances for COI).
Flabellinidae, Facelinidae and Piseinotecidae all appeared to be paraphyletic or polyphyletic. Fionidae appeared as the sister group of Tergipedidae (PP = 1, BS = 100).
Discussion
Aeolidiidae
Our results support Aeolidiidae as a monophyletic family, as long as Pleurolidia juliae is excluded. This contrasts with the results obtained by Carmona et al. [13], who stated that Aeolidiidae was not monophyletic due to Facelina punctata (Alder & Hancock, 1845) -a junior synonym of F. annulicornis (Chamisso & Eysenhardt, 1821)- clustered among the aeolidiid species included in their study. The Facelina punctata COI sequence used in the above paper was taken from GenBank [16]. In order to avoid the possibility of a misidentification of such facelinid, in the present study, we sequenced mitochondrial (COI and 16S) and nuclear (H3) genes from a Facelina punctata specimen collected from the Azores Is. and identified by the authors. The inclusion of these sequences in our analyses did not affect the monophyly of Aeolidiidae. This suggests caution when using the specimen of Facelina punctata from GenBank in analysing the phylogenetic relationships of this group.
The close relationship among Aeolidiidae, Babakinidae and a clade composed by several Facelinidae, suggested in previous studies [10], [13], was confirmed using a much broader taxon sampling.
The monophyly of Aeolidia, Anteaeolidiella, Limenandra and Spurilla was highly supported. Intergeneric relationships were also well supported in the Bayesian analyses but not in the Maximum likelihood analyses. Neither Aeolidiidae and Spurillidae sensu Odhner [29] and Odhner (in [37]), nor Spurillinae or Aeolidiinae sensu Schmekel & Portmann [42] were recovered as monophyletic taxa Therefore, in order to minimize disruption of the nomenclature and to choose the most parsimonious option, the validity of these taxa is rejected.
Pleurolidia
In our study, Pleurolidia clusters with some facelinids, but not within the traditional Aeolidiidae. Protaeolidiella and Pleurolidia are two monospecific genera that feed on hydroids instead of anemones. They have been considered as primitive aeolidiids [4], [75]–[76]. Burn [76] described Pleurolidia juliae based on two specimens, although most of the information was referred to the holotype, which possesses a triseriate radula. He pointed out that no details of the buccal bulb of the second specimen were known due to the fact that it was lost. Burn [76] also mentioned that the holotype and the second specimen of his species might not be conspecific. P. juliae would be the only aeolidiid with a triseriate radula because Protaeolidiella atra has an uniseriate radula [77]. But, since no traces of the lateral teeth of Pleurolidia juliae radula were found, Rudman [44] concluded that both were conspecific and therefore rendered P. juliae as junior synonym of Protaeolidiella atra. Two years later, Baba [78] stated that both species were valid although he did not examine the radula of Pleurolidia juliae. Here, we re-examined material of P. juliae and found no trace of lateral teeth (not shown). Hence, there are three possible hypotheses for the triseriate radula of P. juliae: intraspecific variation; an aberrant radula, which has been observed in other Aeolidida species [9], [79] or; a misidentification of this specimen with another aeolid that has a triseriate radula. P. juliae’s diet matches that of most facelinds’ (hydroids rather than sea anemones). This fact further supports the molecular results we obtained where P. juliae is included in Facelinidae. Inclusion of Protaeolidiella atra specimens in future analyses would be worth in order to test this apparent relationship and distinctness in relation to Pleurolidia juliae.
Berghia
The monophyly of the genus Berghia was highly supported when Aeolidiella stephanieae was included. Berghia is entirely restricted to the Atlantic and interestingly, none of the species studied here have an amphiatlantic distribution. This genus has been considered a junior synonym of Spurilla by some authors [30], [80]. Nevertheless, our results show that Berghia is not as closely related to Spurilla, as previously thought. In our analysis, this genus includes B. creutzbergi, B. verrucicornis, B. coerulescens, an undescribed species from Senegal, B. columbina, B. rissodominguezi and Aeolidiella stephanieae. Concerning the latter species, Valdés [81] placed it within Aeolidiella following the diagnosis of Gosliner [4]. This species, which is commercially important as a biological control of Aiptasia infestations in aquaria [46], [81]–[83], has been misidentified as the Mediterranean species Berghia verrucicornis (see [31], [82]–[84]) and Berghia coerulescens [46] for many years. However, A. stephanieae clusters together with Berghia rissodomiguezi, which also has been suggested by Valdés et al. [46] as a possible junior synonym of Berghia coerulescens. Both species, B. rissodominguezi and A. stephanieae, have differences in coloration, ceratal arrangement, rhinophorial ornamentation, genital and anal apertures as well as the masticatory border and the central cusp denticle [81], [85]. Therefore, although the uncorrected p-distance between both specimens is at the lower limit (5.7%) of our range for sister species, we concluded that those morphological differences are significant enough to consider both species as distinct. Hence, Aeolidiella stephanieae should be renamed as Berghia stephanieae.
Spurilla
Spurilla is monophyletic with maximum support. Its type species, Spurilla neapolitana, has been intensively studied by several authors [41]–[42], [86]–[95]. According to the literature, the intraspecific variation of the denticulation of the jaws, the shape of the central cusp and its coloration are widely accepted. Furthermore, Spurilla neapolitana has been considered a circumtropical species (e.g. [41]). Nevertheless, our results clearly demonstrate the existence of a complex of at least three cryptic species under the name S. neapolitana and reject its amphiatlantic or circumtropical distribution. The genetic distance between eastern and western Atlantic specimens attributed to this species (minimum uncorrected p-distance = 14.1%) is at the upper limit of the range here considered for different species. Hence, the true Spurilla neapolitana is distributed along the Mediterranean Sea as well as the eastern Atlantic Ocean (from Portugal to Cape Verde, including the Azores Is.). Regarding the western Atlantic population, MacFarland [96] erected the name of Spurilla braziliana for the Brazilian specimens of this genus. This species has been considered as a junior synonym of S. neapolitana [33], [42]–[43], [46], [95], [97]–[99]. The present study clearly demonstrates that both names correspond to valid species and highlights the difficulties of identifying these species without a molecular phylogenetic framework. Two specimens from Japan and two specimens from the Pacific coast of Costa Rica clustered together with the western Atlantic specimens (Figure S1), showing a 1.9% difference in their COI gene among them. These specimens do not appear in Figure 1 because sequencing of 16S and H3 failed (see Table S1). This outcome suggests that those “S. neapolitana” reported from the Pacific Ocean [41], [100]–[103] may be S. braziliana.
A second sibling species of S. neapolitana was detected from specimens from Huelva (SW Spain, Atlantic Ocean) and Agadir (Morocco, Atlantic Ocean). Considering its geographical distribution, this second species could be ascribed to Spurilla mograbina or Spurilla dakariensis. Pruvot-Fol [104] described Spurilla mograbina and Spurilla dakariensis from material from Temara (Morocco) and Dakar (Senegal) respectively. However, it is not possible to positively identify the specimens from Agadir and Huelva as S. mograbina. Pruvot-Fol’s figure (pl. 1, fig. 12) [104] depicts a single Spurilla specimen with opaque cream spots scattered along the dorsal side of the cerata and the back, especially behind the pericardium, but there are no traces of these cream spots in our specimens. Due to the fact that Pruvot-Fol classified S. mograbina as a variety of S. neapolitana and some of our specimens from Agadir and Temara cluster within the clade of the true S. neapolitana, S. mograbina is probably a junior synonym of S. neapolitana.
The original description of Spurilla dakariensis [104] is very brief and vague. With the scarce information provided by Pruvot-Fol [104] some colour types of Spurilla neapolitana, Spurilla sp. A and Spurilla braziliana could be attributed as Spurilla dakariensis. Thus, since this name is unidentifiable, we conclude that S. dakariensis should be treated as nomen dubium.
Finally, our results show that the sister species of S. neapolitana is S. sargassicola (Kröyer in Bergh, 1861). This latter species has been considered a junior synonym of S. neapolitana by most authors [33], [93], [95], [99]. Recently, Valdés et al. [46] defended its validity by comparing a specimen of each species in the same photograph (see p. 270). Based on differences in coloration, ceratal arrangement and our molecular data, we confirm that both names correspond to closely related, but separate species.
Baeolidia
Baeolidia moebii Bergh, 1888, the type species of this genus, supposedly has not been found since it was described from the Mauritius Is. When Eliot [105] described Baeolidia major from Zanzibar, he stated that the main difference between both species was their size (8 mm long for B. moebii and 40 mm for B. major). Even though he pointed out that B. major could be merely a full-grown individual of B. moebii, Eliot [105] finally considered B. major as a valid species. Besides habitat similarities [105], both species also share some morphological features such us colouration, the leaf-like cerata and the radular morphology. Hence, we suggest that both names are conspecific and consider Baeolidia major to be a junior synonym of Baeolidia moebii.
The monophyly of Baeolidia is well supported only when Aeolidiopsis ransoni and Spurilla salaamica are included in Baeolidia. Since Aeolidiopsis has never been considered as a junior synonym of Baeolidia, the placement of Aeolidiopsis ransoni within Baeolidia represents an intriguing outcome. Aeolidiopsis ransoni has been retained as the sole species of Aeolidiopsis due to its acleioproctic anus dorsal to the notal brim and its smooth rhinophores [4], [8], [106]. Our results suggest that the position of the anus and the ornamentation of the rhinophores cannot be diagnostic for Aeolidiidae genera (see Cerberilla discussion). Therefore, we suggest transferring Aeolidiopsis ransoni to Baeolidia that includes the other two species that feed on the zooanthid Palythoa, Baeolidia harrietae (Rudman, 1982) and Baeolidia palythoae Gosliner, 1985 [4]. Further studies including molecular analyses with B. harrietae and B. palythoae are needed to clarify the relationships among these species.
Since Rudman [30] considered Berghia and Baeolidia as junior synonyms of Spurilla and despite its papillate rhinophores, he ascribed Spurilla salaamica to Spurilla. Gosliner [4] transferred this species to Berghia based on its papillate rhinophores and cerata in arches. Despite the traditional use of ceratal arrangement and rhinophorial ornamentation to separate genera, the present study highlights that these characters cannot be diagnostic for most aeolidiid genera (see the last section of the discussion). Hence, the most parsimonious alternative is to rename Spurilla salaamica as Baeolidia salaamica although further morphological studies are needed in order to find synapomorphies. Members of this lineage are restricted to the Indo-Pacific tropics and the eastern Pacific.
Anteaeolidiella
Our results confirmed the validity of the recent genus Anteaeolidiella [8] from a molecular point of view. The type species, Anteaeolidiella indica (Bergh, 1888), has been considered to have a worldwide distribution [2]–[3], [10], [46]. Several synonyms have been attributed to this species: Aeolis foulisi Angas, 1864, Aeolidiella orientalis Bergh, 1888, A. saldanhensis Barnard, 1927, A. hulli Risbec, 1928, A. takanosimensis Baba, 1930, A. multicolor Macnae, 1954 and A. lurana Marcus & Marcus, 1967 [3], [107]. However, our study demonstrates that at least five distinct allopatric species constitute the Anteaeolidiella indica complex, which presents to a consistent biogeographic pattern. In 1855, Stimpson [108] described Eolis cacaotica from Port Jackson (Sydney Harbour, eastern Australia), while Burn [109] recently suggested that this could be the older name of Aeolis foulisi, which was also described from Port Jackson. Upon examining the original descriptions of these species, we concluded that both descriptions refer to the same taxon and concur with our specimens from Australia. Therefore, we consider Aeolis foulisi as a junior synonym of Eolis cacaotica and transfer the latter to Anteaeolidiella. A Japanese specimen that shares the colouration pattern of A. cacaotica (not shown) clusters among the Australian specimens of A. cacaotica. This outcome suggests that this species is also present in Japan. The Japanese specimen does not appear in Figure 1 because sequencing of COI and H3 failed (see Table S1).
Regarding our specimen from South Africa, Bergh [110] described Aeolidiella indica from the Mauritius Is. as “whitish-yellow or greyish-yellow body; grey rhinophores with white tips; white tentacles with yellow tips and grey or yellow cerata”. Later, Barnard [111] erected the South African species Aeolidiella saldanhensis from a preserved specimen and only provided information about the radula and foot shape. Macnae [112] described Aeolidiella multicolor with material from South Africa based on differences in coloration, size and radular teeth with A. indica and differences in foot corners shape with A. saldanhensis. Nevertheless, Macnae pointed out that the coloration of this species was very variable. Indeed, the most remarkable characteristic of this Aeolidiella, the bluish background of the cerata, presents high intraspecific variation. Just considering coloration, Bergh’s species matches the tropical South African “Aeolidiella indica” specimens, whereas our South African specimen from temperate waters is concurrent with A. multicolor (not shown). Hence, we think that both names are non-conspecific but we treat A. multicolor as a junior synonym of A. saldanhensis and conclude that our specimen from temperate South Africa belongs to this last species. A redefinition of A. indica is needed in order to clarify the morphological characteristics of this species and its intraspecific variability.
The coloration of our Japanese specimens matches with the original drawings of Anteaeolidiella takanosimensis [113]. Although A. takanosimensis is reported as a cosmopolitan species [3], our data suggest that this species is restricted to Japan.
Genetic distance, as well as external differences (not shown), suggest that “Anteaeolidiella indica” from Naples (Mediterranean Sea) is Anteaeolidiella lurana. This species was described based on material from Brazil by Marcus & Marcus in 1967 [93]. However, in our analyses one specimen from Brazil and another from Bermuda clustered together with the specimen from Naples (maximum uncorrected p-distance = 0%). The Brazilian and Bermudan specimens do not appear in Figure 1 because sequencing of some of the genes failed (see Table S1 and Figure S1). Therefore, Anteaeolidiella lurana seems to be an amphiatlantic species closely related to A. saldanhensis, differing from this species molecularly and in their rhinophores and oral tentacles’ colour pattern.
We could not determine the identity of those specimens from the Clipperton Island. They present differences in body shape and coloration with respect to the rest of Anteaeolidiella species, including A. chromosoma and A. oliviae. The eastern Pacific taxon most likely represents an undescribed species, based on its genetic distance from other members of this clade (minimum uncorrected p-distance = 14.2%).
Cerberilla
Cerberilla is the only aeolidiid genus whose systematics remains unresolved. Because of their burrowing behaviour, not many specimens of Cerberilla [79]–[80], [114] are encountered and very few are properly preserved for molecular work. For this reason only seven species have been included in our analyses (Table S1).
The marginal denticle, the occurrence of small denticles between larger denticles of the radula and its pleuroproctic anus have traditionally distinguished this genus from the rest of aeolidiids [4], [8], [76], [115]. Due to its pleuroproctic anus, species of Cerberilla have been considered as primitive aeolidiids together with Protaeolidiella and Pleurolidia [4], [75]–[76]. Nevertheless, our analyses clearly showed that the position of the anus does not have any phylogenetic significance within Aeolidiidae (e.g. Aeolidiopsis ransoni, here transferred to Baeolidia). Tardy [75] pointed out that a classification based on the position of the anus “leads to the formation of tribes that may often appear to be artificial in the light of new investigations”. Furthermore, within each “Cerberilla” clade, a huge intrageneric variability of the radula can be observed. A “classical” Cerberilla radula (e.g. Cerberilla affinis) clusters together with a “classical” Aeolidiella radula (e.g. Ceberilla sp. B from Japan).
A more comprehensive study including the type species of the genus (Cerberilla longicirrha Bergh, 1873), more Cerberilla species and a detailed examination of their morphology is certainly required in other to clarify its phylogenetic status.
Limenandra
Many authors have considered Limenandra as a junior synonym of Baeolidia [4], [41], [46]–[47]. Nevertheless, our study confirmed the validity of this genus and its monophyly. Baeolidia and Limenandra are not even sister groups. Rather, Limenandra is more closely related to some “Cerberilla” species. Limenandra nodosa, the type species of the genus, is amphiatlantic but is not so widely distributed as it is referred to in the bibliography [4], [41], [46]–[47]. Specimens from the Pacific (the Philippines and Mexico) attributed to Limenandra nodosa appear to be a distinct species that is sister to L. nodosa (the Atlantic and the Mediterranean) (minimum uncorrected p-distance = 7.3% between both species).
On the other hand, the sister group of the above clade is composed of L. fusiformis and an undescribed species, Limenandra sp. C (Baeolidia sp. 3 in [47]), from the Philippines. All these taxa form a clade that is the sister taxon to Limenandra sp. B (Baeolidia sp. 2 in [47]), from the Marshall Islands and the Philippines. Therefore, Limenandra is clearly far more diverse than previous studies have indicated.
Aeolidiella
Aeolidiella is the aeolidiids genus to which most nominal species have been ascribed (see Table S3). Nevertheless, our data depict a completely different scenario. Aeolidiella alderi (type species) and A. sanguinea constitute a small and monophyletic group included in a larger clade that includes some species of “Cerberilla”. The remaining Aeolidiella species included in this study (Aeolidiella alba, Aeolidiella japonica, Aeolidiella sp. B and the Brazilian specimens attributed to Aeolidiella alba) cluster together as the sister group of Aeolidia. Therefore, the traditional Aeolidiella is rendered as polyphyletic. In order to recover its monophyly, a new name for the clade constituted by A. alba, A.japonica, Aeolidiella sp. B and the specimens attributed to Aeolidiella alba from Brazil is erected. The relationship of Aeolidiella with “Ceberilla” bernadettae and “Ceberilla” sp. A still needs further study. So far, the genus Aeolidiella is restricted to the Atlantic-Mediterranean.
Aeolidia
Aeolidia is the type genus of the family and the sister group of a clade named below and composed of several species previously attributed to Aeolidiella. The monophyly of Aeolidia is highly supported. Our results show the existence of a complex of three sibling species under the name of A. papillosa besides confirming the amphiatlantic status of this species. Thus, the true A. papillosa is distributed from Sweden (close to the type locality) to the Northeastern Pacific coasts (state of Washington), including the western Atlantic (maximum uncorrected p-distance = 1.6%). Its sister taxon is a sibling species from California (minimum uncorrected p-distance = 8.4% between both clades). A third cryptic species includes specimens early attributed to “Aeolidia papillosa” from Cap Ferret (France, Atlantic) and Galicia (NW Spain, Atlantic) (minimum uncorrected p-distance = 11% between this clade and the true A. papillosa clade). Other Aeolidia species, such as A. herculea Bergh, 1894 and A. collaris Odhner, 1921 were not included in this study as material properly preserved for molecular purposes was not available.
Bulbaeolidia gen. nov
LSID urn:lsid:zoobank.org:act:EA7635CA-F380-4BED-9663-298641A2C1B8.
Aeolidiella alba and Aeolidiella japonica were originally ascribed to the genus Aeolidiella by Risbec [116] and Eliot [117] respectively. However, in our study these species constitute a monophyletic and high supported group (PP = 1, BS = 92) together with an undescribed species from Hawaii and one specimen attributed to A. alba from Brazil. This outcome renders the traditional Aeolidiella polyphyletic. On the other hand, this clade should not be joined with Aeolidia, its sister clade, (Figures 1 and 2) due to the anatomical differences as well as these “Aeolidiella” species have never been ascribed to Aeolidia. Hence, in order to choose the most parsimonious option “A”. alba, “A”. japonica, “Aeolidiella” sp. B and the Brazilian “Aeolidiella alba” were grouped in the new genus Bulbaeolidia gen. nov. This genus is characterized by a low, wide body, with an inconspicuous pericardial swelling. Head large with short oral tentacles that are constricted at the midpoint. Rhinophores with a pair of prominent bulbous swellings. Cerata club-shaped, in single rows, with the number decreasing posteriorly and slightly curved inwards towards dorsal mid line. Cleioproctic, reproductive aperture immediately below the second ceratal row. Oral glands, composite, moderately large, tubular and tapering. Radular tooth bilobed and deeply indented, with a prominent central cusp. Jaw masticatory process smooth. The new name combines the bulbous swellings in the rhinophores of the species in the clade with the name of its sister group, Aeolidia.
Concerning the cryptic species from Brazil previously attributed to Aeolidiella alba, more Atlantic specimens from Florida, as well as deep examination of their morphology, are needed in order to clarify its geographical distribution.
Are Traditional Morphological Characters Useful for Phylogenetic Inferences in Aeolidiidae?
Traditionally, the position of the anus, the shape of the radular tooth, the ceratal arrangement and the ornamentation of the rhinophores have been the most important characters within Aeolidiidae [4], [8], [30], [118]. However, there is little consensus regarding which of these characters are most systematically and phylogenetically informative. Indeed, depending on the author the emphasis on each morphological character varies. These disagreements have questioned the validity of some aeolidiid genera and the addition of new species has blurred the differences between these genera, especially with regard to species placed in Spurilla, Berghia, Limenandra and Baeolidia.
Our results clearly show that none of these characters have any unique phylogenetic signal within Aeolidiidae (see Berghia, Baeolidia and Cerberilla discussion). In order to illustrate these results, Table 3 shows the different states of ceratal arrangement and the rhinophoral ornamentation in Aeolidiidae while Figure 2 maps their evolution within this family. Thus, we can conclude that ornamented rhinophores and cerata in rows or/and arches have evolved independently within different lineages. For instance, considering smooth rhinophores as the plesiomorphic state [6], papillate rhinophores have evolved separately three times in Aeolidiidae. Recent molecular analyses highlight the need to re-examining the morphological characters according to relationships revealed a posteriori [119]–[124]. The finding of new morphological synapomorphies requires additional studies and is beyond the scope of this study.
Table 3. Code numbers for different states of the morphological characters traditionally used in Aeolidiidae.
| Code | Morphological character |
| 1 | Smooth rhinophores |
| 2 | Perfoliate rhinophores |
| 3 | Papillate rhinophores |
| 4 | Rhinophores with a pair of prominent bulbous swellings |
| 5 | Cerata in arches |
| 6 | Cerata in rows |
| 7 | Cerata in arches and rows |
| 8 | Cerata on elevated cushions |
Conclusions
This study includes only a small representation of the aeolid heterobranchs diversity, but triplicates the number of species and quintuplicates the number of specimens included by Carmona et al. [13], the largest aeolid dataset analysed to date. Together with Johnson & Gosliner [125], this study constitutes the most extensive molecular data set for a group of heterobranchs and includes three molecular markers (COI, 16S rRNA and histone H3). Table S3 lists the ninety-seven described Aeolidiidae species. Our study includes the fifty per cent of the known species. Although our study covers only half of them, lack of material properly preserved for genetic research makes it presently impossible to undertake a more comprehensive molecular phylogenetic study of this clade. Moreover, many species such as Milleraeolidia ritmica, Burnaia helicochorda, Baeolidia cryoporos and Berghia chaka have not been reported since they were originally described. Likewise, many old taxa were poorly described and it will be a challenge to recognize such taxa even if they were encountered again. In conclusion, this study confirms the monophyly of Aeolidiidae, excluding Pleurolidia. Neither Spurillidae/Spurillinae nor Aeolidiinae were recovered as monophyletic taxa. After some changes in the systematics of the family here proposed, Aeolidiidae includes eight monophyletic genera: Berghia, Spurilla, Baeolidia, Anteaeolidiella, Limenandra, Aeolidiella, Aeolidia and Bulbaeolidia gen. nov. The taxonomic status of Cerberilla, however, remains unresolved. Our results also suggest the existence of four sibling-species complexes within Aeolidiidae, which may increase to a total of 115 species, including 18 undescribed species and the resurrection of six species previously in synonymy. Finally, this contribution highlights the necessity of additional research to identify and characterize new morphological synapomorphies to better explain the phylogenetic relationships within this family.
Supporting Information
Molecular phylogeny inferred from partial sequences of the individual mitochondrial COI gene by Bayesian analysis. Numbers above branches represent posterior probabilities from BI. Numbers below branches indicate bootstrap values for ML. Abbreviations: ATL, Atlantic Ocean; EA, eastern Atlantic Ocean; GB, GenBank; MED, Mediterranean; PAC, Pacific; WA, western Atlantic Ocean.
(TIF)
Phylogenetic hypothesis based on the combined dataset (H3+COI+16S) inferred by Maximum likelihood analysis (ML). Numbers above branches indicate bootstrap values for ML. Abbreviations: ATL, Atlantic Ocean; EA, eastern Atlantic Ocean; GB, GenBank; MED, Mediterranean; PAC, Pacific; WA, western Atlantic Ocean. Genera names on right side of vertical bars refer to revised classification.
(TIF)
List of specimens used for phylogenetic analyses. We include both the species names resulting from our morpho-chromatic identification (provisional ids) and the names after analyses (final ids; this only when changes have occurred). EA = eastern Atlantic Ocean; EP = eastern Pacific; GB = GenBank; MED = Mediterranean; WA = western Atlantic Ocean.
(DOCX)
Minimum COI gene pairwise uncorrected p -distances between sister species of each genus.
(DOCX)
List of nominal species of Aeolidiidae. Final identifications are determined after a review of the pertinent literature. ✓ = species analysed here; X = species not found properly preserved for molecular analyses; − = validity and/or identity of this species not studied here.
(DOCX)
Acknowledgments
We are deeply grateful to all individuals that helped to collect and provided specimens and images to this study, such as S. García-Gómez, C. Pittman, C. Redfern, R. Nakano, G. Calado, G. Villani, M. Poddubetskaia, N. Ardila, N. Tamsouri, D. Behrens, S. Johnson, C. Magenta and V. Padula. Many thanks are due to W. Sterrer (Bermuda Aquarium and Museum of Zoology, BAMZ), B. Williams (Bermuda Institute of Oceanographic Sciences) and T. M. Trott (Acting Senior Marine Resources Officer, Bermuda Government), M. Biscoito and T. M. Freitas (Marine Biological Station of Funchal), M. E. Ibarra and M. Ortiz-Touzet (Centro de Investigaciones Marinas, Universidad de la Habana) and J. Gonçalves, G. Carreira and R. S. Santos (Department of Oceanography and Fisheries, University of Azores) for welcoming, housing, and facilitating permits during the expeditions in Bermuda, Madeira, Cuba and the Azores. A. Sellas helped us with some molecular lab work. Thanks to J. Vierna, A. Valdés, J. Reis and M. Benavides, for their invaluable assistance. J.L. Cervera and M. Pola are indebted to P. Sundberg and the Swedish Taxonomy Initiative for the invitation to the Lovén Centre for Marine Science (Tjärno, Sweden). This is Contribution #208, Bermuda Biodiversity Project (BBP), Bermuda Aquarium, Natural History Museum and Zoo, Department of Conservation Services. This is CEI·MAR journal publication 22.
Funding Statement
This work is supported by the University of Cádiz, the California Academy of Sciences, the National Science Foundation DEB 0329054 to TMG, and grants from the Spanish Ministry of Economy and Competitiveness (includes the early Ministry of Sciences and Innovation) CGL2006-05182/BOS, CTM2008-05228-E/MAR and CGL2010-17187; Institut Menorquí d’Estudis, Balearic Is; Bermuda Zoological Society; AECID-PCI A/8688/07 (Spanish Agency of International Cooperation and Development) to JLC. The funders had no role in the study, design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wägele H (2004) Potential key characters in Opisthobranchia (Gastropoda, Mollusca) enhancing adaptive radiation. Org Divers Evol 4: 175–188. [Google Scholar]
- 2.Debelius H, Kuiter RH (2007) Nudibranchs of the world. IKAN-Unterwasserarchiv. Frankfurt, Germany. 361 p.
- 3. Gosliner TM, Griffiths RJ (1981) Description and revision of some South African aeolidacean Nudibranchia (Mollusca, Gastropoda). Ann S Afr Mus 82: 102–151. [Google Scholar]
- 4. Gosliner TM (1985) The aeolid nudibranchs family Aeolidiidae (Gastropoda, Opisthobranchia) from tropical southern Africa. Ann S Afr Mus 95: 233–267. [Google Scholar]
- 5. Wägele H (1990) Revision of the Antarctic genus Notaeolidia (Gastropoda, Nudibranchia), with a description of a new species. Zool Scr 19: 309–330. [Google Scholar]
- 6. Gosliner TM, Kuzirian AM (1990) Two new species of Flabellinidae (Opisthobranchia: Aeolidacea) from Baja California. Proc Calif Acad Sci 47: 1–15. [Google Scholar]
- 7. Gosliner TM, Willan RC (1991) Review of the Flabellinidae (Nudibranchia: Aeolidacea) from the tropical Indo-Pacific with descriptions of five new species. Veliger 34: 97–133. [Google Scholar]
- 8. Miller MC (2001) Aeolid nudibranchs (Gastropoda: Opisthobranchia) of the family Aeolidiidae from New Zealand waters. J Nat Hist 35: 629–662. [Google Scholar]
- 9. Valdés Á, Angulo-Campillo O (2004) Systematics of pelagic aeolid nudibranchs of the family Glaucidae (Mollusca, Gastropoda). B Mar Sci 75: 381–389. [Google Scholar]
- 10. Gosliner TM, González-Duarte MM, Cervera JL (2007) Revision of the systematics of Babakina Roller, 1973 (Mollusca: Opisthobranchia) with the description of a new species and a phylogenetic analysis. Zool J Linn Soc 151: 671–689. [Google Scholar]
- 11. Moore EJ, Gosliner TM (2009) Three new species of Phyllodesmium Ehrenberg (Gastropoda: Nudibranchia: Aeolidoidea), and a revised phylogenetic analysis. Zootaxa 2201: 30–48. [Google Scholar]
- 12. Moore EJ, Gosliner TM (2011) Molecular phylogeny and evolution of symbiosis in a clade of Indopacific nudibranch. Mol Phylogenet Evol 58: 116–123. [DOI] [PubMed] [Google Scholar]
- 13. Carmona L, Gosliner TM, Pola M, Cervera JL (2011) A molecular approach to the phylogenetic status of the aeolid genus Babakina Roller, 1973 (Nudibranchia). J Moll Stud 77: 417–422. [Google Scholar]
- 14. Thollesson M (1999) Phylogenetic analysis of Euthyneura (Gastropoda) by means of the 16S rRNA gene: use of a ‘fast’ gene for ‘higher-level’ phylogenies. Proc R Soc Lond 266: 75–83. [Google Scholar]
- 15. Wollscheid-Lengeling E, Wägele H (1999) Initial Results on the Molecular Phylogeny of the Nudibranchia (Gastropoda, Opisthobranchia) Based on 18S rDNA data. Mol Phylogenet Evol 13: 215–226. [DOI] [PubMed] [Google Scholar]
- 16. Wollscheid-Lengeling E, Boore J, Brown W, Wägele H (2001) The phylogeny of Nudibranchia (Opisthobranchia,Gastropoda, Mollusca) reconstructed by three molecular markers. Org Divers Evol 1: 241–256. [Google Scholar]
- 17.Wägele H, Vonnemann V, Wägele JW (2003) Toward a phylogeny of the Opisthobranchia. In: Lydeard C, Lindberg D, editors. Molecular systematics and phylogeography of mollusks Smithsonian Institution Press, Washington. 185–228.
- 18.Wägele H, Klussmann-Kolb A, Vonnemann V, Medina M (2008) Heterobranchia I: The Opisthobranchia. In: Ponder WF, Lindberg D, editors. Phylogeny and Evolution of the Mollusca. University of California Press, Berkeley. 385–408.
- 19. Grande C, Templado J, Cervera JL, Zardoya R (2004a) Molecular phylogeny of Euthyneura (Mollusca: Gastropoda). Mol Biol Evol 21: 303–313. [DOI] [PubMed] [Google Scholar]
- 20. Grande C, Templado J, Cervera JL, Zardoya R (2004b) Phylogenetic relationships among Opisthobranchia (Mollusca: Gastropoda) based on mitochondrial cox 1, trnV, and rrnL genes. Mol Phylogenet Evol 33: 378–388. [DOI] [PubMed] [Google Scholar]
- 21. Wägele H, Klussmann-Kolb A (2005) Opisthobranchia (Mollusca, Gastropoda) – more than just slimy slugs. Shell reduction and its implications on defence and foraging. Front Zool 2: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vonnemann VMS, Schrödl M, Klussmann-Kolb A, Wägele H (2005) Reconstruction of the phylogeny of the Opisthobranchia (Mollusca: Gastropoda) by means of 18S and 28S rRNA gene sequences. J Moll Stud 71: 113–125. [Google Scholar]
- 23. Pola M, Gosliner TM (2010) The first molecular phylogeny of cladobranchian opisthobranchs (Mollusca, Gastropoda, Nudibranchia). Mol Phylogenet Evol 56: 931–941. [DOI] [PubMed] [Google Scholar]
- 24. Göbbeler K, Klussmann-Kolb A (2011) Molecular phylogeny of the Euthyneura (Mollusca, Gastropoda) with special focus on Opisthobranchia as a framework for reconstruction of evolution of diet. Thalassas 27: 121–154. [Google Scholar]
- 25. Wägele H, Klussmann-Kolb A (2005) Opisthobranchia (Mollusca, Gastropoda) – more than just slimy slugs. Shell reduction and its implications on defence and foraging. Front in Zool 2: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wägele H, Willan RC (2000) Phylogeny of the Nudibranchia. Zool J Linn Soc 130: 83–181. [Google Scholar]
- 27. Martin R, Tomaschko KH, Heb M, Schrödl M (2010) Cnidosac-Related Structures in Embletonia (Mollusca, Nudibranchia) Compared with Dendronotacean and Aeolidacean Species. Open Mar Biol J 4: 96–100. [Google Scholar]
- 28.Vayssière Á (1913) Mollusques de la France and des Régions Voisines. 1. Encyclopédie Scientifique, G. Doin and Cia., Paris. 418 p.
- 29. Odhner NH (1939) Opisthobranchiate Mollusca from the western and northern coasts of Norway. Det Kgl Norske Vidensk Selsk Skrift 1: 1–93. [Google Scholar]
- 30. Rudman WR (1982) The taxonomy and biology of further aeolidacean and arminacean nudibranch molluscs with symbiotic zooxanthellae. Zool J Linn Soc 74: 147–196. [Google Scholar]
- 31. Kempf SC (1991) A “primitive” symbiosis between the aeolid nudibranch Berghia verrucicornis (A. Costa, 1867) and a Zooxanthella. J Moll Stud 57: 75–85. [Google Scholar]
- 32. Marín A, Ros J (1991) Presence of intracellular zooxanthellae in Mediterranean nudibranches. J Moll Stud 57: 88–101. [Google Scholar]
- 33.García-García FJ, Domínguez M, Troncoso JS (2008) Opistobranquios de Brasil. Feito S. L., Vigo. 215 p.
- 34. Marcus Er (1958) On western Atlantic opisthobranchiate gastropods. Am Mus Novit 1906: 1–82. [Google Scholar]
- 35.Haefelfinger HR, Stamm RA (1958) Limenandra nodosa gen. et spec. nov. (Nudibranchia, Aeolidiidae prop.), un opisthobranche nouveau de la Méditerranée. Vie Milieu, 418–423.
- 36. Tardy JP (1962) A propos des espèces de Berghia (Gastéropodes Nudibranches) des côtes de France et de leur biologie. Bull Inst Océanogr 59: 1–20. [Google Scholar]
- 37.Franc A (1968) Sous-classe des opisthobranches, In: Grassé PP editors. Traite de Zoologie. Masson, Paris. 608–893.
- 38. Burn R (1969) A memorial report on the Tom Crawford collection of Victorian Opisthobranchia. J Malac Soc Aust 12: 64–106. [Google Scholar]
- 39. Edmunds M (1969) Opisthobranchiate Mollusca from Tanzania. I. Eolidacea (Eubranchidae and Aeolidiidae). Proc Malac Soc Lond 38: 451–469. [Google Scholar]
- 40. Marcus Er, Marcus Ev (1970) Opisthobranch from Curaçao and faunistically related regions. Stud. Fauna. Curaçao Caribbean Isl 33: 1–129. [Google Scholar]
- 41. Gosliner TM (1980) The systematics of the Aeolidacea (Nudibranchia: Mollusca) of the Hawaiian Islands, with description of two new species. Pac Sci 33: 37–77. [Google Scholar]
- 42.Schmekel L, Portmann A (1982) Opisthobranchia des Mittelmeeres. Springer Verlag Berlin. 410 p.
- 43. Domínguez M, Troncoso JS, García FJ (2008) The family Aeolidiidae Gray, 1827 (Gastropoda Opisthobranchia) from Brazil, with a description of a new species belonging to the genus Berghia Trinchese, 1877. Zool J Linn Soc 153: 349–368. [Google Scholar]
- 44. Rudman WR (1990) Protaeolidiella atra Baba, 1955 and Pleurolida juliae Burn, 1966; one species, two families (Nudibranchia). J Moll Stud 56: 505–514. [Google Scholar]
- 45. Ortea J, Caballer M, Espinosa J (2004) Nuevos aeolidaceos de Costa Rica. Rev Acad Canar Cienc 15: 3–4. [Google Scholar]
- 46.Valdés Á, Hamann J, Behrens DW, DuPont A (2006) Caribbean Sea Slugs. A field guide to the opisthobranch mollusks from the tropical northwestern Atlantic. Sea Challengers Natural History Books, Gig Harbor, Washington. 289 p.
- 47.Gosliner TM, Behrens DW, Valdés Á (2008) Indo-Pacific nudibranchs and sea slugs. Sea Challengers Natural History Books, Gig Harbor, WA. 426 p.
- 48. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech 3: 294–299. [PubMed] [Google Scholar]
- 49.Palumbi SR, Martin A, Romano S, Owen MacMillan W, Stice L, et al.. (1991) The Simple Fool’s Guide to PCR. Department of Zoology, University of Hawaii, Honolulu. 45 p.
- 50. Colgan D, McLauchlan A, Wilson GDF, Livingston SP, Edgecombe GD, et al. (1998) Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Aust J Zool 46: 419–437. [Google Scholar]
- 51. Meyer CP (2003) Molecular systematics of cowries (Gastropoda: Cypraeidae) and diversification patterns in the tropics. Biol J Linn Soc 79: 401–459. [Google Scholar]
- 52. Williams ST, Reid DG (2004) Speciation and diversity on tropical rocky shores: a global phylogeny of snails of the genus Echinolittorina. . Evolution 58: 2227–2251. [DOI] [PubMed] [Google Scholar]
- 53. Dinapoli A, Tamer C, Frassen S, Naduvilezhath L, Klussmann-Kolb A (2006) Utility of H3-Genesequences for phylogenetic reconstruction – a case study of heterobranch Gastropoda –. Bonn Zool Beitr 55: 191–202. [Google Scholar]
- 54. Frey MA, Vermeij GJ (2008) Molecular phylogenies and historical biogeography of a circumtropical group of gastropods (Genus: Nerita): implications for regional diversity patterns on the marine tropics. Mol Phylogenet Evol 48: 1067–1086. [DOI] [PubMed] [Google Scholar]
- 55. Malaquías MAE, Reid DG (2009) Tethyan vicariance, relictualism and speciation: evidence from a global molecular phylogeny of the opisthobranch genus Bulla . J Biogeogr 36: 1760–1777. [Google Scholar]
- 56.Drummond AJ, Ashton B, Cheung M, Heled J, Kearse M, et al. (2009) Geneious v4.6. Available: http://www.geneious.com/. September 2012.
- 57. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 58. Katoh K, Asimenos G, Toh H (2009) Multiple Alignment of DNA Sequences with MAFFT. In Bioinformatics for DNA Sequence Analysis edited by D. Posada. Methods Mol Biol 537: 39–64. [DOI] [PubMed] [Google Scholar]
- 59.Maddison DR, Maddison WP (2005) MacClade 4., v. 4.08 for OSX. Sinauer Associates, Sunderland, MA.
- 60. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Phylogenet Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56: 564–577. [DOI] [PubMed] [Google Scholar]
- 62. Farris JS, Källersjö M, Kluge AG, Bult C (1994) Testing significance of incongruence. Cladistics 10: 315–319. [Google Scholar]
- 63.Swofford DL (2002) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, MA: Sinauer. 142 p.
- 64. Akaike H (1974) A new look at the statistical model identification. IEEE T Automat Contr 19: 716–723. [Google Scholar]
- 65.Nylander JAA (2004) MrModeltest v2.3. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- 66. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 67. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 68. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Knowlton N (2000) Molecular genetic analyses of species boundaries in the sea. Hydrobiologia 420: 73–90. [Google Scholar]
- 70.Wheeler QD, Meier R. (eds), (2000) Species concepts and phylogenetic theory: a debate. Columbia University Press, New York. 230 p.
- 71.Avise JC (2004) Molecular markers, natural history, and evolution. Edn 2. Sinauer Associates, Sunderland, MA. 532 p.
- 72. Reid DG, Lal K, Mackenzie-Dodds J, Kaligis F, Littlewood DTJ, et al. (2006) Comparative phylogeography and species boundaries in Echinolittorina snails in the central Indo-West Pacific. J Biogeogr 33: 990–1006. [Google Scholar]
- 73. Hebert PDN, Ratnasingham S, Waard JRD (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc 270: S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Williams ST, Reid DG, Littlewood DTJ (2003) A molecular phylogeny of the Littorininae (Gastropoda: Littorinidae): unequal evolutionary rates, morphological parallelism and biogeography of the Southern Ocean. Mol Phylogenet Evol 28: 60–86. [DOI] [PubMed] [Google Scholar]
- 75.Tardy JP (1965) Description et biologie de Cerberilla bernadetti, espèce nouvelle de Gasteropode Nudibranche de la côte atlantique française. Discussion sur la position systématique du genre. Bull Inst Océanogr 65: 1–22, pls. 1–6.
- 76. Burn R (1966) Descriptions of Australian Eolidacea (Mollusca: Opisthobranchia) 4. The genera Pleurolidia, Fiona, Learchis and Cerberilla from Lord Howe Island. J Malac Soc of Aust 10: 21–34. [Google Scholar]
- 77.Baba K (1955) Opisthobranchia of Sagami Bay supplement. Iwanami Shoten, Tokyo. 59 p.
- 78. Baba K (1992) Comment on the taxonomy of Protaeolidiella atra Baba, 1955 and an allied species (Mollusca: Nudibranchia: Aeoidiidae) from Japan. Rep Sado Mar Biol Stat Niigata Univ 22: 29–35. [Google Scholar]
- 79. Odhner NH (1937) Coryphella islandica n. sp., a new nudibranchiate mollusc from Iceland. Vidensk Medd Dan Nat hist Foren 101: 253–257. [Google Scholar]
- 80. García-Gómez JC, Thompson TE (1990) North Atlantic spurillid nudibranch, with a description of a new species, Spurilla columbina, from the Andalusian coast of Spain. J Moll Stud 56: 323–331. [Google Scholar]
- 81. Valdés Á (2005) A new species of Aeolidiella Bergh, 1867 (Mollusca: Nudibranchia: Aeolidiidae) from Florida Keys, USA. Veliger 43: 218–223. [Google Scholar]
- 82.Kempf SC, Brittsan M (1996) Berghia verrucicornis, a nudibranch predator of the aquarium “weed” anemone Aiptasia. Reg conf proc - Am Zoo Aquar Assoc, Audubon Park and Zoological Garden, New Orleans, Lousiana.
- 83.Kempf SC (2002) Photo of “Berghia verrucicornis” from Florida. Sea Slug Forum. Available: http://www.seaslugforum.net/find/6472. September 2012.
- 84. Carroll DJ, Kempf SC (1990) Laboratory culture of the aeolid nudibranch Berghia verrucicornis (Mollusca, Opisthobranchia): Some aspects of its development and life history. Bio Bull 179: 243–253. [DOI] [PubMed] [Google Scholar]
- 85. Muniain C, Ortea J (1999) First records of the genus Berghia Trinchese, 1977 (Opisthobranchia: Aeolidiidae) from Argentina, with description of a new species. Avicennia10/11: 143–150. [Google Scholar]
- 86. Bergh R (1877) Beiträge zur Kenntniss der Aeolidiaden. 4. Verhandlungen der k.k. Zoologish-botanischen Gesellschaft in Wien (Abhandlungen) 26: 737–764. [Google Scholar]
- 87. Trinchese S (1878) Anatomia e fisiologia della Spurilla neapolitana . Mem Accad Sci Bologna (3)9(3): 405–450. [Google Scholar]
- 88. Bergh R (1882) Beiträge zur Kenntniss der Aeolidiaden. 7. Verhandlungen der k.k. Zoologish-botanischen Gesellschaft in Wien (Abhandlungen) 31: 7–74. [Google Scholar]
- 89. Vayssière Á (1888) Recherches zoologiques et anatomiques suer les mollusques opisthobranches du Golfe de Marseille. 2. Nudibranches (Cirrobranches) et ascogloses. Ann Mus Hist Nat Marseille 3: 1–160. [Google Scholar]
- 90. Engel H (1925) Westindische Opisthobranchiate Mollusken I. Bijdr Dierk. 24: 33–80. [Google Scholar]
- 91.Pruvot-Fol A (1954) Mollusques opisthobranches. Faune de France. 58. Paris, Paul Lechevalier. 460 p.
- 92. Swennen C (1961) On a collection of Opisthobranchia from Turkey. Zool Meded 38: 42–75. [Google Scholar]
- 93. Marcus Ev, Marcus Er (1967) American opisthobranch mollusks. Stud Trop Oceanogr 6: 1–256. [Google Scholar]
- 94. Bebbington A, Thompson TE (1968) Note sur les Opistobranches du bassin d’Arcachon. Actes Soc Linn Bordeaux 105: 1–35. [Google Scholar]
- 95. García-Gómez JC, Cervera JL (1985) Revisión de Spurilla neapolitana Delle Chiaje 1823 (Mollusca: Nudibranchiata). J Moll Stud 51: 138–156. [Google Scholar]
- 96. MacFarland FM (1909) The opisthobranchiate Mollusca of the Branner-Agassiz expedition to Brazil. Leland Stanford Jr Univ Publ 2: 1–104. [Google Scholar]
- 97. Marcus Er (1957) Opisthobranchia from Brazil (2). J Linn Soc London Zoology 43: 390–487. [Google Scholar]
- 98.Marcus Ev (1977) An annotated checklist of the western Atlantic warm water opisthobranch molluscs. J Moll Stud Suppl 4: 1–22.
- 99. Edmunds M, Just H (1983) Eolid nudibranchiate mollusca from Barbados. J Moll Stud 49: 185–203. [Google Scholar]
- 100.Kerstitch A (1989) Sea of Cortez marine invertebrates: A Guide of the Pacific Coast, Mexico to Ecuador. Sea Challengers. Monterey, California. 114 p.
- 101. Hamatani I (2000) A new recorded species of the genus Spurilla Bergh, 1864 from Osaka Bay, Middle Japan (Opisthobrachia, Aeolidacea). Venus 59: 263–265. [Google Scholar]
- 102. Willan R (2006) Globe-trotting nudibranch arrives in Australia. Aust shell news 130: 1–3. [Google Scholar]
- 103. Uribe RA, Pacheco AS (2012) First record of Spurilla neapolitana (Mollusca: Nudibranchia: Aeolidiidae) on the central coast of Peru (Humboldt Current Upwelling Ecosystem). MBR 5: 1–5. [Google Scholar]
- 104. Pruvot-Fol A (1953) Étude de quelques opisthobranches de la côte atlantique du Maroc et du Sénégal. Trav Inst Sci Chérifien Zool 5: 1–105. [Google Scholar]
- 105. Eliot CNE (1903) On some nudibranchs from east Africa and Zanzibar, part II. Proc Zool Soc Lond 1: 250–257. [Google Scholar]
- 106. Pruvot-Fol A (1956) Un Aeolidien nouveau des mers tropicales: Aeolidiopsis ransoni n. g., n. sp. Bull Mus Nat Hist Nat 28: 228–231. [Google Scholar]
- 107. Burn R (2006) A checklist and bibliography of the Opisthobranchia (Mollusca: Gastropoda) of Victoria and the Bass Strait area, south-eastern Australia. Mus Vic Sci Rep 10: 1–42. [Google Scholar]
- 108. Stimpson W (1855) Descriptions of some new marine Invertebrata. Proc Acad Nat Sci Phila 7: 385–395. [Google Scholar]
- 109. Burn R (1964) A centennial commentary and zoogeographical remarks on Angas’ Sydney nudibranchs (Molluscs, Gastropoda). Journ Conchyl CIV: 85–93. [Google Scholar]
- 110. Bergh R (1888) Malacologische Untersuchungen. In: Reisen im Archipel der Philippinen von Dr. Carl Gottfried Semper. Zweiter Theil. Wissenschaftliche Resultate 3: 755–814. [Google Scholar]
- 111. Barnard KH (1927) South African nudibranch Mollusca, with descriptions of new species and a note on some specimens from Tristan d’Acunha. Ann S Afr Mus 25: 171–215. [Google Scholar]
- 112. Macnae W (1954) On some eolidacean nudibranchiate molluscs from South Africa. Ann Natal Mus 13: 1–50. [Google Scholar]
- 113. Baba K (1930) Studies on Japanese nudibranchs 3. Venus 2: 117–125. [Google Scholar]
- 114. Padula V, Delgado M (2010) A new species of Cerberilla (Gastropoda: Nudibranchia: Aeolidiidae) from northeastern Brazil. Nautilus 124: 175–180. [Google Scholar]
- 115. McDonald GR, Nybakken J (1975) Cerberilla mosslandica, a new eolid nudibranch from Monterey Bay, California. Veliger 17: 378–382. [Google Scholar]
- 116.Risbec J (1928) Contribution a l’etude des nudibranches Néo-Calédoniens. Thèse présentée à la faculté des sciences de l’Université de Paris pour obtenir le grade de docteur ès-sciences naturelles. Faune des Colon Franç 2: 1–328, pls. 1–12.
- 117.Eliot CNE (1913) Japanese nudibranchs. Journ Coll Sci Tokyo 35: 1–47, pls. 1–2.
- 118. Marcus Er (1961) Opisthobranch molluscs from California. Veliger 3: 1–84. [Google Scholar]
- 119. Pola M, Cervera JL, Gosliner TM (2007) Phylogenetic relationships of Nembrothinae (Mollusca: Doridacea: Polyceridae) inferred from morphology and mitochondrial DNA. Mol Phylogenet Evol 43: 726–742. [DOI] [PubMed] [Google Scholar]
- 120. Pola M, Cervera JL, Gosliner TM (2008) Description of the first Roboastra species (Nudibranchia, Polyceridae, Nembrothinae) from the western Atlantic. Bull Mar Sci 83: 391–399. [Google Scholar]
- 121. Ornelas-Gatdula E, Dupont A, Valdés A (2011) The tail tells the tale: taxonomy and biogeography of some Atlantic Chelidonura (Gastropoda: Cephalaspidea: Aglajidae) inferred from nuclear and mitochondrial gene data. Zool J Linn Soc 163: 1077–1095. [Google Scholar]
- 122. Schrödl M, Jörger KM, Klussmann-Kolb A, Wilson NG (2011) Bye bye “Opisthobranchia”! A review on the contribution of mesopsammic sea slugs to euthyneuran systematics. Thalassas 27: 101–112. [Google Scholar]
- 123. Johnson RF, Gosliner TM (2012) Traditional taxonomic groupings mask evolutionary history: A molecular phylogeny and new classification of the chromodorid nudibranchs. PloS One 7: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Jöger KM, Stöger I, Kano Y, Fukuda H, Knebelsberger T, et al. (2010) On the origin of Acochlidia and other enigmatic euthyneuran gastropods, with implications for the systematics of Heterobranchia. BCM Evol Biol 10: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Johnson RF, Gosliner TM (2012) raditional Taxonomic Groupings Mask Evolutionary History: A Molecular Phylogeny and New Classification of the Chromodorid Nudibranchs. PloS One 7: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Molecular phylogeny inferred from partial sequences of the individual mitochondrial COI gene by Bayesian analysis. Numbers above branches represent posterior probabilities from BI. Numbers below branches indicate bootstrap values for ML. Abbreviations: ATL, Atlantic Ocean; EA, eastern Atlantic Ocean; GB, GenBank; MED, Mediterranean; PAC, Pacific; WA, western Atlantic Ocean.
(TIF)
Phylogenetic hypothesis based on the combined dataset (H3+COI+16S) inferred by Maximum likelihood analysis (ML). Numbers above branches indicate bootstrap values for ML. Abbreviations: ATL, Atlantic Ocean; EA, eastern Atlantic Ocean; GB, GenBank; MED, Mediterranean; PAC, Pacific; WA, western Atlantic Ocean. Genera names on right side of vertical bars refer to revised classification.
(TIF)
List of specimens used for phylogenetic analyses. We include both the species names resulting from our morpho-chromatic identification (provisional ids) and the names after analyses (final ids; this only when changes have occurred). EA = eastern Atlantic Ocean; EP = eastern Pacific; GB = GenBank; MED = Mediterranean; WA = western Atlantic Ocean.
(DOCX)
Minimum COI gene pairwise uncorrected p -distances between sister species of each genus.
(DOCX)
List of nominal species of Aeolidiidae. Final identifications are determined after a review of the pertinent literature. ✓ = species analysed here; X = species not found properly preserved for molecular analyses; − = validity and/or identity of this species not studied here.
(DOCX)