Highlights
► Utrophin actin binding domain crystallises as an open dimer but is monomeric in solution. ► We introduced cysteines in the CH domains to lock the monomer closed. ► Utrophin bound to F-actin with reduced affinity in the closed conformation. ► Differential scanning calorimetry confirmed the open conformation of utrophin on actin.
Abbreviations: ABD, actin binding domain; CH, calponin homology; CD, circular dichroism; DSC, differential scanning calorimetry; EM, electron microscopy; F-actin, filamentous actin; NTCB, 2-nitro-5-thiocyanobenzoic acid; SDS-PAGE, sodium dodecyl sulphate poly-acrylamide electrophoresis; Tm, melting temperature; UTR261, utrophin residues 1-261
Keywords: α-Actinin, Actin binding domain, Calponin homology domain, Differential scanning calorimetry, Dystrophin, Spectrin
Abstract
Structural analyses of actin binding regions comprising tandem calponin homology domains alone and when bound to F-actin have revealed a number of different conformations with calponin homology domains in ‘open’ and ‘closed’ positions. In an attempt to resolve these issues we have examined the properties of the utrophin actin binding domain in open and closed conformations in order to verify the conformation when bound to F-actin. Locking the actin binding domain in a closed conformation using engineered cysteine residues in each calponin homology domain reduced the affinity for F-actin without affecting the stoichiometry furthermore differential scanning calorimetry experiments revealed a reduction in melting temperature on binding to actin. The data suggest the amino-terminal utrophin actin binding domain is in an open conformation in solution and when bound to F-actin.
1. Introduction
Calponin homology (CH) domains are found primarily, but not exclusively, in proteins that interact with the F-actin cytoskeleton. In most cases a functional actin binding domain (ABD) comprises two structurally equivalent but functionally distinct CH domains [1]. Tandem-CH domain ABDs are found in a large number of F-actin binding proteins with roles as structural linkers including the spectrin family of proteins, the spectroplakin family and other F-actin bundling or cross-linking proteins such as filamin and fimbrin. High-resolution atomic structures have been determined for members of each of these groups [2–4]. Furthermore, in an attempt to understand the structural and functional determinants for actin binding, several of these proteins have also been analysed in complex with F-actin by electron microscopy (EM); however, analysis of the utrophin ABD by this route has resulted in a number of conflicting models, reviewed in [5]. Part of the controversy may have arisen because in the original crystal structure of the utrophin ABD [6], despite being a monomer in solution the ABD crystallised as a dimer. Moreover, because of the orientation of the individual chains within the dimer it was suggested that a three-dimensional domain swap may have occurred (Fig. 1). The crystal structures of utrophin and dystrophin ABD reveal dimers in an extended conformation with CH domain 1 (CH1) of one crystallographic dimer interacting with CH domain 2 (CH2) of the other crystallographic dimer (Fig. 1A and B, red and blue structures) [6,7]. The α-actinin ABD on the other hand, which shares considerable sequence and structural homology [4,8], crystallised as a monomer with CH1 and CH2 from the same molecule in close apposition and in an orientation similar to CH1 and CH2 from opposite dimers of the utrophin or dystrophin structure (Fig. 1A and B, green structure).
Fig. 1.
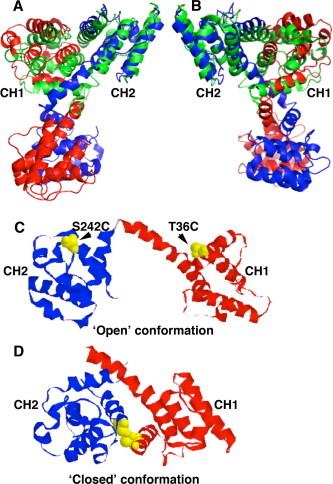
The utrophin ABD structure. (A and B) Ribbon diagrams of two different views of the UTR261 crystal structure 1QAG comprising a dimer with two molecules shown in red and blue. These are overlayed with the structure of α-actinin 1WKU to demonstrate the apparent two-dimensional domain swap. Annotation to CH1 and CH2 refer to the corresponding CH domains of the UTR261 structure and the α-actinin fit. (C and D) Images of utrophin monomers in the open conformation and closed conformation derived from the structure in (A) and (B). The position of threonine 36 and serine 241 are shown in yellow spacefill.
Nonetheless the idea that the utrophin ABD could exist in an open extended and closed compact form was attractive, and to an extent supported by some of the available data [9]. One of the main arguments against the open conformation stemmed from available crystal structures. With the exception of utrophin and its close homologue dystrophin, all other tandem CH domains that had been crystallised, did so in a closed conformation. Because of the apparent domain swap in the utrophin and dystrophin crystals, possibly induced by the crystallisation at low pH, it has been argued that the extended conformation is an artefact of crystallisation and does not reflect a true state in solution [5], whereas more recent studies do suggest an open conformation [10]. We have therefore examined the binding properties of utrophin with F-actin using native utrophin that is allowed to adopt any conformation (open or closed) and in a closed conformation by introducing cysteine residues into each CH domain to form an inter-CH domain disulphide that locks the two CH domains together. The utrophin ABD is easily expressed in bacteria, highly soluble (up to mM concentrations) and is stable. Furthermore, unlike dystrophin and α-actinin the primary sequence contains no cysteine residues making it an ideal model to introduce cysteine residues to address functional changes.
2. Methods
2.1. Purification and characterisation of UTR261 cysteine mutants
The double cysteine mutant construct was generated by QuikChange mutagenesis of UTR261, first the T36C mutation was generated to give UTR261T36C, and then the S242C mutation was introduced into UTR261T36C to generate UTR261T36C/S242C. UTR261T36C and UTR261T36C/S242C are expressed in soluble form in Escherichia coli BL21(DE3) and purified under the same conditions as wildtype UTR261 [8]. Cleavage of UTR261 by 2-nitro-5-thiocyanobenzoic acid (NTCB) was carried out as described previously [11]. Oxidation or reduction of UTR261T36C/S241C was achieved overnight in 20 mM Tris (pH 8.0) in the presence of 4 mM o-phenanthroline and 1 mM CuSO4 or 1 mM Tris(2-carboxyethyl) phosphine hydrochloride, respectively, proteins were then dialysed back into 20 mM Tris for functional studies.
2.2. High speed co-sedimentation actin binding assays
Rabbit skeletal muscle actin was purified as described previously [8]. High speed co-sedimentation of 5 μM F-actin in the presence of increasing concentrations of UTR261, reduced UTR261T36C/S241C and oxidised UTR261T36C/S241C were carried out as previously described [8].
2.3. Fluorescence spectroscopy and differential scanning calorimetry
Tryptophan fluorescence spectroscopy was measured using a Shimadzu RF-5301PC spectrofluorophotometer. Protein samples were excited at 296 nm and fluorescence emission data were recorded between 300 and 450 nm. Differential scanning calorimetry (DSC) experiments were carried out in a N-DSC II differential scanning calorimeter from Calorimetry Sciences Corp. (Provo, UT), at scanning rate of 1 K/min under 3.0 atm of pressure. DSC samples contained 10 μM UTR261 (wildtype or mutants) using buffer conditions identical to those described previously [3]. UTR261T36C and UTR261T36C/S241C samples under reducing conditions were kept with 1.0 mM DTT at all times and diluted 10-fold with DTT-free buffer immediately before loading into calorimeter. Where stated 10 μM F-actin or 20 μM F-actin + 20 μM phalloidin were also added.
3. Results and discussion
Based on the previous studies of de Pereda and colleagues on the plectin ABD [3], and using a notional closed conformation of the utrophin ABD derived from the crystallographic dimer (Fig. 1C and D), we identified threonine 36 in CH1 and serine 242 in CH2 that would be close together in a predicted closed conformation. UTR261 T32 was mutated to cysteine, and then using this UTR261T36C as template, the second site was mutated to give UTR261T36C/S242C. DNA sequencing of the mutated construct confirmed the presence of both cysteine substitutions, which was further demonstrated by chemical cleavage at the cysteines with NTCB. As can be seen from Fig. 2B, compared to UTR261 which contains no cysteines, the UTR261T36C/S242C protein was susceptible to cleavage by NTCB. Furthermore, chemical oxidation of UTR261T36C/S242C revealed a mobility shift on non-reducing SDS-PAGE consistent with the formation of the intra-chain disulphide, with no evidence of inter-chain disulphide formation leading to dimerisation (Fig. 2C). The latter was also confirmed by analytical gel filtration, with the oxidised protein eluting as a monodisperse peak with a calculated mass of 28kDa (data not shown).
Fig. 2.
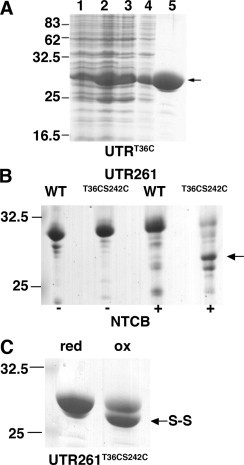
Purification and characterisation of UTR261 cysteine mutants. (A) SDS-PAGE of stages in the purification of UTR261T36C. Lane 1, pre-induction; 2, post-induction; 3, soluble fraction; 4, post ion exchange pool; 5, purified UTR261T36C following size exclusion chromatography. (B) Chemical cleavage at cysteine residues. In the absence of NTCB (−) UTR261 and UTR261T36C/S241C run as single bands, whilst in the presence (+) of NTCB there is no cleavage of UTR261 due too the lack of cysteines, but UTR261T36C/S241C is cleaved into two prominent bands presumed based on relative mass to correspond to the cleavage at C242 (upper band, arrowed) and C36 (lower band). (C) Non-reducing SDS-PAGE of reduced (red) and incompletely oxidised (ox) UTR261T36C/S241C which clearly demonstrates a small size shift on formation of the disulphide, marked by an arrow and S–S. Position of molecular mass standards are indicated in kDa.
Analysis of the F-actin binding properties of wild type and cysteine mutants of UTR261, in either reduced or oxidised form as shown in Fig. 3. UTR261 bound to F-actin with similar stoichiometry (Bmax; 1:1) and dissociation constant as reported previously (19.2 ± 2.2 μM; [12,13]); however, introduction of the two cysteine residues did have an effect on the dissociation constant but without affecting the stoichiometry. Threonine 36 is within the conserved KTFT motif, also termed ‘ABS1’ in earlier mapping studies of actin binding regions within the amino-terminal actin binding domains of dystrophin and utrophin [8]. Whether this region is in direct contact with F-actin or is simply required for structural integrity of CH1 remains equivocal. CD spectra of UTR261 and cysteine mutants showed no significant changes in overall secondary structure (data not shown); however, there was a reduction in tryptophan fluorescence on introduction of T36C and S242C but there was little difference between reduced and oxidised UTR261T36C/S242C (Fig. 4). The reduction in affinity for F-actin could be due to an effect of T36C on ABS1 or this structurally conserved region, and the drop in tryptophan fluorescence may result from cysteine quenching [14] of the nearby W40, and W128 which is close in the structure. Furthermore, as determined by DSC, the Tm for all reduced proteins in solution is within 3 °C (Table 1), suggesting that there are not large scale structural changes. The double cysteine mutants were also slightly red shifted compared to UTR261. The oxidised form of the double cysteine mutant, however bound to F-actin with an even lower affinity than the reduced form (74.8 ± 19 μM reduced, 123 ± 14 μM oxidised) suggesting that either the open form of the ABD bound to actin better, or that a greater degree of flexibility was required for the interaction with F-actin which was inhibited by locking the two CH domains closed.
Fig. 3.
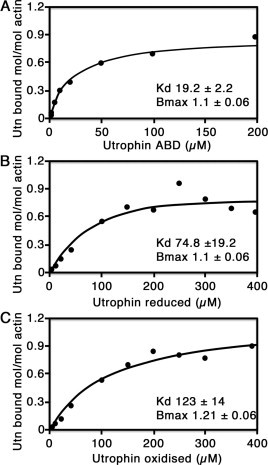
Binding of UTR261 to F-actin. High speed co-sedimentation of 5 μM F-actin in the presence of increasing concentrations of UTR261 (A), reduced UTR261T36C/S241C (B) and oxidised UTR261T36C/S241C (C) were carried out as previously described [8]. Data presented are the mean of three independent experiments (mean ± SEM) with binding parameters shown within each graph.
Fig. 4.
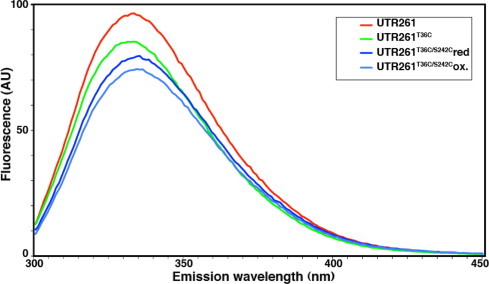
Tryptophan fluorescence of UTR261 and cysteine mutants. Tryptophan fluorescence of 30 μM samples of each of UTR261 (red), UTR261T36C, (green) reduced UTR261T36C/S241C, (dark blue) and oxidised UTR261T36C/S241C (light blue). The introduction of cysteines slightly reduced the fluorescence emission, and furthermore the presence of two cysteines caused a slight red-shift of the spectrum whether the UTR261T36C/S241C was reduced or oxidised.
Table 1.
Denaturation temperatures for DSC scans shown in Fig. 5.
| Proteins | Tm1 (°C) | Tm2 (°C) | Tm3 (°C) |
|---|---|---|---|
| UTR261 WT | 53.3 | – | – |
| UTR261 1C | 52.6 | – | – |
| UTR261 2C reduced | 56.3 | 68.6 | – |
| UTR261 2C oxidised | – | 68.1 | – |
| F-actin | – | – | 69.1 |
| UTR261 WT + F-actin | 55.5 | – | 69.8 |
| UTR261 1C + F-actin | 55.0 | – | 68.7 |
| UTR261 2C reduced + F-actin | – | Shoulder at ∼67 | 69.7 |
| UTR261 2C oxidised + F-actin | – | Shoulder at ∼67 | 69.6 |
| F-actin-phalloidin | – | – | 80.0 |
| UTR261 WT + F-actin-phalloidin | 56.2 | – | 79.8 |
| UTR261 1C + F-actin-phalloidin | 56.5 | Shoulder at ∼69 | 80.6 |
| UTR261 2C reduced + F-actin-phalloidin | – | 68.5 | 80.8 |
| UTR261 2C oxidised + F-actin-phalloidin | – | 68.2 | 79.4 |
The absolute errors in Tm values did not exceed 0.2 °C. WT = wildtype sequence, UTR261 1C = UTR261T36C and UTR261 2C = UTR261T36C/S242C.
In order to test further the conformation of UTR261 when bound to F-actin we carried out differential scanning calorimetry on UTR261 and cysteine mutants, either alone or in the presence of F-actin (Table 1, Fig. 5). UTR261 denatured in DSC experiments as a single peak with Tm = 53.3 °C (Table 1, Fig. 5A). In studies conducted under otherwise identical conditions the Tm of UTR261 was much lower than the Tm of the plectin ABD either in solution or when in complexed with actin: 63.9 °C vs 59.1 °C, [3]. The plectin ABD DSC data were interpreted by Garcia-Alvarez et al., to suggest that uncomplexed plectin ABD was in a closed state and plectin ABD in complex with F-actin was in an open state [3]. The Tm of UTR261 in complex with F-actin increased (rather than decreased as in the case of plectin) but only slightly to 55.5 °C. These observations, along with those of the recent studies performed using spin labelling [10] suggest that UTR261 adopts an open conformation in solution. To verify this we have used UTR261T36C/S242C with two cysteines introduced at T36 and S242 positions, which based on the prediction from Fig. 2, the formation of disulphide bond should lock UTR261 in the closed state. In DSC experiments oxidised UTR261T36C/S242C denatured at much higher temperature than UTR261 (Table 1, Fig. 5D). The Tm = 68.1 °C was as high as that of the analogous plectin ABD T74C/S277C mutant in the oxidised, i.e. closed state [3]. This similarity suggests that we have also succeeded in locking UTR261T36C/S242C in the closed conformation.
Fig. 5.
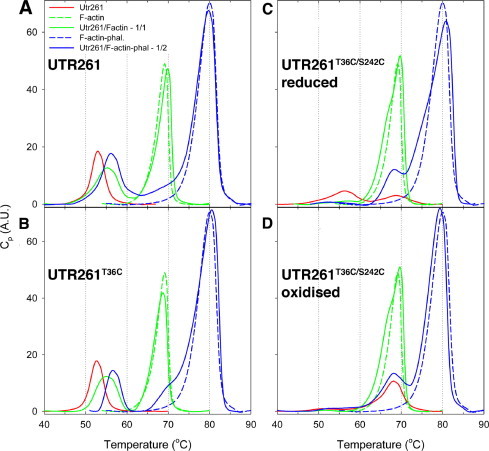
Differential scanning calorimetry. DSC traces of UTR261 and cysteine mutants alone and in the presence of 10 μM F-actin or 20 μM F-actin + 20 μM phalloidin. UTR261 (A), UTR261T36C (B), reduced UTR261T36C/S241C (C), and oxidised UTR261T36C/S241C (D) scans are shown in red in all traces, F-actin alone in green dashed lines, F-actin with the corresponding UTR261 protein in solid green line. F-actin stabilised with phalloidin alone in blue dashed lines, F-actin/phalloidin with the corresponding UTR261 protein in solid blue line.
To verify that the structural effect associated with increased Tm of oxidised UTR261T36C/S242C is due to S–S cross-linking and not cysteine mutation per se, we ran DSC on reduced UTR261T36C/S242C and reduced UTR261T36C (with a single cysteine at T36). The melting profile and Tm of UTR261T36C were very close to those of UTR261 (Table 1, Fig. 5B) suggesting no significant effect of the T36C mutation on the conformation of UTR261 in solution. The melting profile of reduced UTR261T36C/S242C had two peaks (Table 1, Fig. 5C), which indicates the presence of two UTR261 populations with different conformations in this sample. The greater fraction of UTR261T36C/S242C under reduced conditions melted with Tm = 56.3 °C, close to that of UTR261. It is logical to assume that this population is in the open state in solution. The smaller fraction of reduced UTR261T36C/S242C melted with Tm = 68.6 °C, very close to that of oxidised UTR261T36C/S242C. This population likely exists in the closed state. Since no cross-linking was detected on the SDS-PAGE for reduced UTR261T36C/S242C, this result indicate that introduction of two cysteines in the CH1–CH2 interface shifts the equilibrium towards the closed state. Overall, DSC analysis of UTR261, UTR261T36C and UTR261T36C/S242C preparations demonstrated that similar to plectin ABD, utrophin ABD can adopt two conformations, closed and open. However, in contrast to plectin ABD which exists in a predominantly closed state in solution, unmodified utrophin ABD in solution is likely to be in a predominantly open state.
In the presence of F-actin the Tm of UTR261 increased to 55.5 °C (Table 1, Fig 5A). While this increase reflects UTR261 binding to F-actin, the relatively small amplitude of the effect (2.2 °C) indicates that there are no major changes in the conformation of UTR261 and thus, it is likely that it binds F-actin in the same state as in solution ie presumed to be open. Interestingly, unmodified plectin ABD also bound F-actin in the open state [3]. The behaviour of UTR261T36C in the presence of F-actin (Fig. 5D) was very similar to that of UTR261. In the DSC profile of F-actin complexed with oxidised UTR261T36C/S242C (Fig. 5D), the main peak with Tm = 69.7 °C (associated with melting of F-actin) has a shoulder at ∼67 °C. This shoulder likely represents melting of UTR261T36C/S242C. To resolve the peaks of F-actin and oxidised UTR261T36C/S242C we have repeated this experiment in the presence of phalloidin. As reported before [15] and seen in Fig. 5, phalloidin increases the melting temperature of F-actin by ∼10 °C as a result of its strong stabilizing effect on the inter-subunit contacts in the actin filaments. In the sample with phalloidin we have also doubled the amount of F-actin to check if the actin effect on UTR261 conformation is saturated. It can be seen clearly that melting profiles of oxidised UTR261T36C/S242C in the presence and absence of phalloidin-F-actin are very similar (Table 1, Fig. 5D). Thus, oxidised UTR261T36C/S242C binds phalloidin-F-actin in the closed state. To verify that phalloidin does not alter the interaction of UTR261 with F-actin we also performed DSC on WT UTR261 in the presence of phalloidin-F-actin. Results showed that the effects of F-actin and phalloidin-F-actin on the conformation of WT UTR261 are similar (Table 1, Fig. 5A).
The melting profile of the reduced UTR261T36C/S242C in the complex with F-actin (Fig. 5C) was very similar to that of oxidised UTR261T36C/S242C (Fig. 5D). Again, to resolve reduced UTR261T36C/S242C and F-actin peaks we repeated the experiment with phalloidin. As one can see, in both scans reduced UTR261T36C/S242C melts as a single peak with Tm ∼68 °C (Table 1, Fig. 5C). Thus, the vast majority of the reduced UTR261T36C/S242C molecules adopt the closed conformation on F-actin, while in the absence of F-actin more molecules (∼60%) are in the open state (Fig. 5D). These results indicate that for UTR261T36C/S242C mutant F-actin favours the closed state. Whilst the vast majority of WT UTR261 binds F-actin in the open state, we cannot exclude that a small fraction may be in the closed state on F-actin.
In other tandem-CH domain ABD structures such as those for α-actinin or filamin, mutations in the inter-CH domain interface affect actin binding [16,17]. In all cases the mutations in the CH1:CH2 interface region do not alter the gross structural conformation, in that both α-actinin 4 and filamin B crystal structures adopt a compact structure whether or not the mutations are present. However the presence of the mutations does increase the affinity of both the α-actinin and filamin ABDs for F-actin [16,17]. The α-actinin mutants appeared to retain their compact shape as determined by analytical ultracentrifugation [16], whereas the filamin B mutants are also associated with a reduction in the melting temperatures for this ABD. This would argue at the very least in favour of inter-CH domain rearrangement on binding to F-actin, or even the possibility of the proteins adopting an open conformation as shown previously [18]. By contrast the cysteine mutants in utrophin increased the melting temperature and reduced actin binding suggesting that a loss of CH1–CH2 flexibility reduced their affinity for F-actin. The highest resolution cryo-EM reconstructions of F-actin and F-actin with a tandem-CH domain ABD – that of fimbrin, however, demonstrate unequivocally that the two CH domains remain in a closed conformation with very little rearrangement required to match the crystal structure [19] and reviewed in [20]. However the situation regarding the utrophin ABD is less clear.
A number of cryo-EM reconstructions of UTR261 with F-actin using different methods of analysis have arrived at different conclusions. The earliest models had been derived from helical reconstructions had proposed that utrophin bound to F-actin in an open conformation, but that there was an induced fit onto actin requiring some rearrangement of the orientation of the CH domains relative to their position in the crystal structure [9]. However using a different method of analysis – iterative helical real space reconstruction, the Egelman group arrived at an alternative model [12]. In this model, although again the utrophin ABD was fitted in an open conformation, it was able to interact with F-actin in two different states depending on whether one or both CH domains were in contact with actin. Furthermore, in the Egelman study, questions were raised over the validity of using helical averaging techniques to derive a reconstruction from heterogeneously decorated actin filaments, and also as to the polarity of the filament used in the reconstructions [12]. However a further reconstruction comprising the utrophin ABD and the first spectrin repeat bound to F-actin, arrived at a third model – that of a closed conformation for the utrophin (and dystrophin) ABD on F-actin [21]. A further reassessment of all the evidence by the Egelman lab provided convincing arguments for utrophin binding to actin in different modes but in an open conformation, see [22] and discussions therein. The actin binding and DSC data presented here indicate that utrophin ABD binds to actin in an open conformation and add further compelling weight to the open conformation hypothesis. More recently, and despite evidence from solution studies and crystal structures of a closed conformation for α-actinin CH domains [4,22], [16] a cryo-EM reconstruction of α-actinin bound to F-actin predicted an open conformation [18]. The use of cysteine mutagenesis has also been employed in a recent electron paramagnetic resonance study by Lin and colleagues [10] to examine the opening and closing of the utrophin CH domains in solution and on binding to actin. Interestingly, in solution they identified a conformation almost identical to that of a single utrophin ABD as seen in the crystal structure (as in Fig. 1C) but in apparent equilibrium with an equally abundant species with a more closed conformation. However on binding to actin, there is only one population evident and this has an even more open conformation [10]. Thus the authors also conclude that utrophin binds to actin in an open conformation, but via an induced fit mechanism, ironically a conclusion also reached from the earliest EM reconstructions a decade earlier [9].
Acknowledgements
This work was funded by an MRC studentship to M.J.F.B. and MRC Career Establishment Grant G0000104 to S.J.W. We gratefully acknowledge the assistance of Dr. Rosie Staniforth with CD and tryptophan fluorescence experiments.
References
- 1.Gimona M., Djinovic-Carugo K., Kranewitter W., Winder S. Functional plasticity of CH domains. FEBS Lett. 2002;513:98–106. doi: 10.1016/s0014-5793(01)03240-9. [DOI] [PubMed] [Google Scholar]
- 2.Winder S.J. Structural insights into actin-binding, branching and bundling proteins. Curr. Opin. Cell Biol. 2003;15:14–22. doi: 10.1016/s0955-0674(02)00002-9. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Alvarez B., Bobkov A., Sonnenberg A., Pereda J.M.D. Structural and functional analysis of the actin binding domain of plectin suggests alternative mechanisms for binding to F-actin and integrin β4. Structure. 2003;11:615–625. doi: 10.1016/s0969-2126(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 4.Franzot G., Sjoblom B., Gautel M., Carugo K.D. The crystal structure of the actin binding domain from a-actinin in its closed conformation: structural insight into phospholipid regulation of a-actinin. J. Mol. Biol. 2005;348:151–165. doi: 10.1016/j.jmb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Lehman W., Craig R., Kendrick-Jones J., Sutherland-Smith A.J. An open or closed case for the conformation of calponin homology domains on F-actin? J. Muscle Res. Cell Motil. 2004;25:351. doi: 10.1007/s10974-004-0690-7. [DOI] [PubMed] [Google Scholar]
- 6.Keep N.H., Winder S.J., Moores C.A., Walke S., Norwood F.L.M., Kendrick-Jones J. Crystal structure of the actin-binding region of utrophin reveals a head-to-tail dimer. Struct. Fold. Des. 1999;7:1539–1546. doi: 10.1016/s0969-2126(00)88344-6. [DOI] [PubMed] [Google Scholar]
- 7.Norwood F.L.M., Sutherland-Smith A.J., Keep N.H., Kendrick-Jones J. The structure of the N-terminal actin-binding domain of human dystrophin and how mutations in this domain may cause Duchenne or Becker muscular dystrophy. Struct. Fold. Design. 2000;8:481–491. doi: 10.1016/s0969-2126(00)00132-5. [DOI] [PubMed] [Google Scholar]
- 8.Winder S.J., Hemmings L., Maciver S.K., Bolton S.J., Tinsley J.M., Davies K.E., Critchley D.R., Kendrick-Jones J. Utrophin actin binding domain: analysis of actin binding and cellular targeting. J. Cell Sci. 1995;108:63–71. doi: 10.1242/jcs.108.1.63. [DOI] [PubMed] [Google Scholar]
- 9.Moores C.A., Keep N.H., Kendrick-Jones J. Structure of the utrophin actin-binding domain bound to F-actin reveals binding by an induced fit mechanism. J. Mol. Biol. 2000;297:465–480. doi: 10.1006/jmbi.2000.3583. [DOI] [PubMed] [Google Scholar]
- 10.Lin A.Y., Prochniewicz E., James Z.M., Svensson B., Thomas D.D. Large-scale opening of utrophin’s tandem calponin homology (CH) domains upon actin binding by an induced-fit mechanism. Proc. Natl. Acad. Sci. USA. 2011;108:12729–12733. doi: 10.1073/pnas.1106453108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winder S., Walsh M. Structural and functional characterization of calponin fragments. Biochem. Int. 1990;22:335–341. [PubMed] [Google Scholar]
- 12.Galkin V.E., Orlova A., VanLoock M.S., Rybakova I.N., Ervasti J.M., Egelman E.H. The utrophin actin-binding domain binds F-actin in two different modes: implications for the spectrin superfamily of proteins. J. Cell Biol. 2002;157:243–251. doi: 10.1083/jcb.200111097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winder S.J. Structure-function relationships in dystrophin and utrophin. Biochem. Soc. Trans. 1996;24:497–501. doi: 10.1042/bst0240497. [DOI] [PubMed] [Google Scholar]
- 14.Gonnelli M., Strambini G. Phosphorescence lifetime of tryptophan in proteins. Biochemistry. 1995;34:13847–13857. doi: 10.1021/bi00042a017. [DOI] [PubMed] [Google Scholar]
- 15.Levitsky D., Nikolaeva O., Orlov V., Pavlov D., Ponomarev M., Rostkova E. Differential scanning calorimetric studies on myosin and actin. Biochemistry (Mosc) 1998;63:322–333. [PubMed] [Google Scholar]
- 16.Lee S.H., Weins A., Hayes D.B., Pollak M.R., Dominguez R. Crystal structure of the actin-binding domain of [alpha]-actinin-4 Lys255Glu mutant implicated in focal segmental glomerulosclerosis. J. Mol. Biol. 2008;376:317–324. doi: 10.1016/j.jmb.2007.11.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawyer G.M., Clark A.R., Robertson S.P., Sutherland-Smith A.J. Disease-associated substitutions in the filamin B actin binding domain confer enhanced actin binding affinity in the absence of major structural disturbance. Insights from the crystal structures of filamin B actin binding domains. J. Mol. Biol. 2009;390:1030–1047. doi: 10.1016/j.jmb.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Galkin V.E., Orlova A., Salmazo A., Djinovic-Carugo K., Egelman E.H. Opening of tandem calponin homology domains regulates their affinity for F-actin. Nat. Struct. Mol. Biol. 2010;17:614–616. doi: 10.1038/nsmb.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galkin V.E., Orlova A., Cherepanova O., Lebart M.-C., Egelman E.H. High-resolution cryo-EM structure of the F-actin, fimbrin/plastin ABD2 complex. Proc. Natl. Acad. Sci. USA. 2008;105:1494–1498. doi: 10.1073/pnas.0708667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sjoblom B., Ylanne J., Djinovic-Carugo K. Novel structural insights into F-actin-binding and novel functions of calponin homology domains. Curr. Opin. Struct. Biol. 2008;18:702–708. doi: 10.1016/j.sbi.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Sutherland-Smith A.J., Moores C.A., Norwood F.L.M., Hatch V., Craig R., Kendrick-Jones J., Lehman W. An atomic model for actin binding by the CH domains and spectrin-repeat modules of utrophin and dystrophin. J. Mol. Biol. 2003;329:15–33. doi: 10.1016/s0022-2836(03)00422-4. [DOI] [PubMed] [Google Scholar]
- 22.Galkin V.E., Orlova A., VanLoock M.S., Egelman E.H. Do the utrophin tandem calponin homology domains bind F-actin in a compact or extended conformation? J. Mol. Biol. 2003;331:967–972. doi: 10.1016/s0022-2836(03)00842-8. [DOI] [PubMed] [Google Scholar]


