Highlights
► We analyzed the effects of cyclin D1 on the migration of cancer cell lines. ► The Wnt pathway promotes cellular migration via its target gene cyclin D1. ► Cyclin D1 rearranges the actin cytoskeleton and destabilizes adherens junctions.
Keywords: Wnt pathway, Cell migration, β-Catenin, Cyclin D1, Adherens junction
Abstract
The Wnt pathway regulates cell proliferation, mobility and differentiation. Among the many Wnt target genes is CCND1 which codes for cyclin D1. Cyclin D1, in complex with cdk4 and cdk6, regulates G1/S phase transition during cell cycle. Independently of CDK, cyclin D1 also regulates the migration of macrophages. Here we analyzed the effects of cyclin D1 on the migration of cancer cell lines using the transwell migration and scratch assays. We also tested the effect of cyclin D1 and β-catenin on E-cadherin-mediated cell–cell contacts. Our results show that the Wnt pathway promotes cellular migration via its target gene cyclin D1. Moreover we show that cyclin D1 influences the actin cytoskeleton and destabilizes adherens junctions.
1. Introduction
The canonical Wnt pathway plays an important role in the cellular proliferation and differentiation and leads to tumor formation when aberrantly activated [1]. If no Wnt signal is present, the proto-oncoprotein β-catenin is bound by a multiprotein complex consisting of the proteins adenomatous polyposis Coli (APC), axin or conductin/axin2, the serine-threonine kinases casein kinase 1 (CK1) and glycogen synthase kinase-3β (GSK-3β). In this so-called destruction complex β-catenin is phosphorylated and therefore marked for ubiquitination via β-transducin repeat containing protein (β-TrCP) and the subsequent proteasomal degradation, so the level of free β-catenin in the cytosol is kept low. In cells with active pathway the destruction complex is destabilized. As a consequence β-catenin is no longer bound and phosphorylated. So β-catenin accumulates in the cytoplasm and translocates into the nucleus where it binds to transcription factors of the T-cell factor/lymphoid enhancer factor (TCF/LEF) family. The binding of β-catenin replaces co-repressors and recruits other co-activators to the promoter [2].
Beside its function in the activation of the Wnt pathway, there is a second pool of β-catenin at the plasma membrane. Here it plays an important role in the formation of E-cadherin-mediated cell–cell contacts, as it connects proteins of the adherens junctions with the actin cytoskeleton [3]. On the one hand β-catenin interacts with the cytoplasmic domain of E-cadherin and on the other hand it also binds α-catenin, an actin-binding protein. Recent studies have shown that this complex is rather dynamic, because α-catenin cannot bind E-cadherin and β-catenin at the same time [4–6]. Adherens junctions are involved in the epithelial–mesenchymal transition (EMT) during tumor formation. One hallmark of EMT is the loss of function of E-cadherin, which results in the dissociation of the E-cadherin–β-catenin–α-catenin complex from the membrane [7–11]. The loss of E-cadherin-mediated cellular adhesion leads to an increased β-catenin dependent transcription [12–14] and is associated with a poor prognosis [15–17].
CCND1, which encodes the cyclin dependent kinase (cdk) activator cyclin D1 was found as a Wnt target gene in several systems, e.g. in human colorectal cancer cells [18,19] and human teratocarcinoma cells [20]. In other studies, however, these results were not confirmed [21–23].
The central role of cyclin D1 is the regulation of G1/S phase transition in the cell cycle [24–27]. In complex with cdk4 and cdk6 cyclin D1 integrates extracellular signals and mitogens into cell cycle progression [28]. There are also some cdk4- and cdk6-independent functions of cyclin D1. Among these are the inhibition of transcription factors like SP1 [29], cyclin D1-binding myb-like protein 1 (DMPI) [30] and nuclear receptors like the androgen receptor [31,32], the peroxisome proliferator-activated receptor γ (PPARγ) [33] and the thyroid hormone receptor [34]. Moreover cyclin D1 seems to influence the migration of cells in a cdk-independent manner. Macrophages of Ccnd1−/− mice show an increased cellular adhesion compared to macrophages from wild type mice. As a consequence the migration of these macrophages is impaired [35]. The pro-migratory effect of cyclin D1 is due to the inhibition of the rho-associated, coiled-coil containing protein kinase 2 (ROCK2) and thrombospondin-1 (TSP1), which both inhibit the migration of cells [36]. Among the substrates of ROCK2 and TSP1 are the kinases MLC and LIM, which both lead to the formation of stress fibers and thus to a decreased migration rate.
2. Materials and methods
2.1. Cell lines and cell cultivation
The colorectal carcinoma cell lines HCT116, SW480 and the cervix carcinoma cell line HeLa were purchased from ATCC-LGC (Wesel, Germany). HCT116 cells harbor activating mutations in the Wnt and the Ras pathway. Due to the deletion of the serine coding codon 45 in the β-catenin coding gene CTNNB1 the phosphorylation of β-catenin by the kinase CKIα/ε is not possible [37]. Furthermore the proto-oncogene KRAS is mutated at codon 13 leading to the constitutive activation of the proliferation activating Ras pathway in HCT116 cells. SW480 cells have no functional APC protein [38] based on a non-sense mutation at codon 1338 of the first allele leading to a truncated APC protein and the LOH of the second allele. HeLa cells were originally established from a HPV-18 infected cervical carcinoma. The viral infection led to the inactivation of the p53/Rb tumor suppressor network. Cell lines were cultured at 37 °C and 7.5% CO2 in a humidified atmosphere in Dulbecco’s Modified Eagle Medium (DMEM). The medium was supplemented with 10% (v/v) fetal calf serum (FCS), penicillin (2.85 U/ml) and streptomycin (2.85 μg/ml), l-glutamine (11.4 mM), non-essential amino acids (0.57 mM) and sodium pyruvate (11 nM). Cell growth was controlled by light microscopy, and cell number was determined with a hemocytometer.
2.2. Transient siRNA transfection
siRNA was purchased from Dharmacon (distributed by Perbio, Bonn, Germany). Among these siRNAs were cyclin D1 siGENOME SMARTpool, c-myc siGENOME SMARTpool and the β-catenin siGENOME SMARTpool. As a control siCONTROL non-targeting #1 was used. Cells were transfected using the transfection reagent DharmaFECT 1 (Dharmacon) by following the protocol of the manufacturer. DharmaFECT contains non-ionic and cationic lipids, which form liposome complexes with the negative charged siRNA. After fusion with the cell membrane these complexes are ingested by endocytosis.
2.3. Transwell migration assay
The migration rate of cells was determined with the QCM chemotaxis 96-well cell migration assay (Millipore, Schwalbach, Germany) by following the supplied protocol. The migration assay was performed 48 h after siRNA transfection. Cells were incubated with serum-free medium 12 h previously. Medium with 10% FCS was used as attractant.
2.4. Scratch assay
Migration of HeLa cells was also tested with a scratch assay. Cells were cultured in 3.5 cm plates, transfected with siRNA and 48 hours after transfection the confluent monolayer was scratched with a tip to produce a 100–150 μm cell free scratch. After washing the cells the scratch was documented every hour over a period of 14 hours (Leitz Labovert).
2.5. Immunocytochemistry and fluorescence microscopy
The actin cytoskeleton was stained with phalloidin/TRITC (Sigma Aldrich, München, Germany). Cells were washed with phosphate buffered saline (PBS) and fixed by incubation for 20 min in formaldehyde (3.7% in PBS). After this the cells were washed with PBS and permeabilized in cold acetone/methanol (1:1) for 5 min at −20 °C and air-dried for 10 min at room temperature (RT). Unspecific antigens were blocked by incubation in blocking solution (1% milk powder in PBS) for 15 min at RT. After washing the cells were incubated with phalloidin/TRITC (1:1000 in PBS with 0.5% Tween20, PBS/T) for 1 h at RT. Cells were again washed with PBS and the cell nuclei were counterstained in 0.05 μg/ml DAPI (4,6-diamidin-2-phenylindol) in PBS for 5 min. After washing with PBS the cells were mounted on slides with fluorescent mounting medium (Dako, Glostrup, Denmark). Analysis and evaluation were performed with the microscope Axiophot (Zeiss, Oberkochen, Germany) and the software Axiovision Release 4.6.3.
E-cadherin-mediated cell–cell junctions were visualized with an anti-E-cadherin antibody (Dianova, Hamburg). Cells were washed with PBS and fixed as described above. After fixation in formaldehyde cells were permeabilized in 1% Triton x-100 in PBS for 10 min at RT. Unspecific antigens were blocked by incubation in blocking solution (2% BSA in PBS) for 30 min. Primary anti-E-cadherin antibody was added at a 1:100 dilution in blocking solution and incubated for 1 h at RT. Cells were washed twice with PBS and incubated with the secondary anti-mouse-Cy3 antibody (GE Healthcare, München, Germany) at a 1:100 dilution in blocking solution again for 1 h at RT. After washing with PBS cells were treated as indicated above.
2.6. Flow cytometry
Cells were cultured in 10 cm plates, transfected with siRNA and harvested 48 h after transfection. Cells were centrifuged for 5 min at 1500 rpm and resuspended in PBS. Fixation and permeabilization of 106 cells were performed with commercial reagents (BD Cytofix/Cytoperm Fixation/Permeabilization Kit from Becton Dickinson, Heidelberg, Germany) by following the protocol of the manufacturer. Staining of the actin cytoskeleton with phalloidin/FITC was performed as described above for phalloidin/TRITC in fluorescence microscopy. Cells were analyzed by FACS (LSRII from Becton Dickinson) and data were evaluated with the software FlowJo (TreeStar Inc., Ashland, USA).
3. Results
3.1. Cyclin D1 and β-catenin promote cellular migration
The influence of the Wnt pathway and cyclin D1 on cellular migration was tested using the cell lines HCT116, SW480 and HeLa. The cells were transfected with cyclin D1 and β-catenin siRNA and a transwell migration assay was performed (Fig. 1).
Fig. 1.
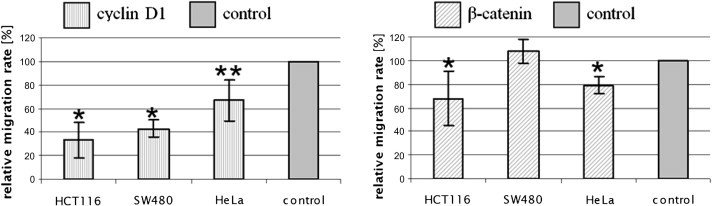
Knock-down of cyclin D1 and β-catenin lowers relative migration rate. Bars show relative migration rates after siRNA transfection. The migration rate of control cells, which were treated with control siRNA, was set as 100%. The statistical analysis was performed with the Student’s t-test. ∗ indicates P < 0.005, ∗∗ indicates P < 0.05.
In this assay cyclin D1 siRNA treated cells showed a significant decrease up to 67% in the migration rate. Transfection with β-catenin siRNA led to an inhibition of at least 20%. In SW480 cells β-catenin siRNA treatment showed no effect on cellular migration.
A scratch assay with HeLa cells was performed (Fig. 2). In this assay the results of the transwell migration assay could be confirmed, as the migration was inhibited after cyclin D1 siRNA and β-catenin siRNA transfection. Control cells showed again a confluent monolayer after 6 h, in cyclin D1 and β-catenin siRNA transfected cells the scratch was closed after 10 and 13 h, respectively. These results indicate that the Wnt pathway promotes cellular migration via its target gene cyclin D1.
Fig. 2.
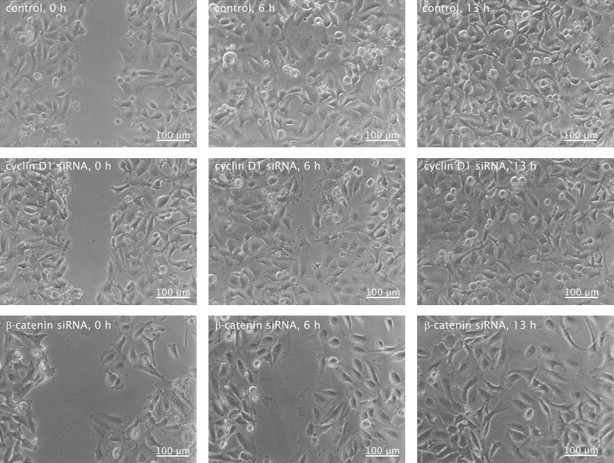
HeLa cells are impaired in their migration after treatment with siRNA against β-catenin or cyclin D1. Representative photographs of the cellular migration in the scratch assay after 6 and 13 h. Control cells were transfected with control siRNA.
3.2. Cyclin D1 and β-catenin influence the actin cytoskeleton
To investigate the effect on cellular migration in more detail the influence of the Wnt pathway and cyclin D1 on the actin cytoskeleton was analyzed. Cells were transfected with siRNA against β-catenin or cyclin D1 and stained with TRITC- or FITC-coupled phalloidin. Subsequently, the cells were investigated with the help of immunofluorescence microscopy (Fig. 3) or flow cytometry (Fig. 4), respectively.
Fig. 3.
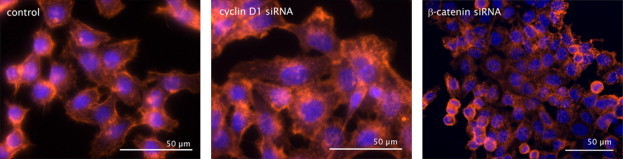
The actin cytoskeleton rearranges after knock-down of cyclin D1 or β-catenin. Immunofluorescences of siRNA treated and phalloidin/TRITC (red) stained HCT116 cells. Control cells were transfected with control siRNA. Nuclei are counterstained with DAPI (blue).
Fig. 4.
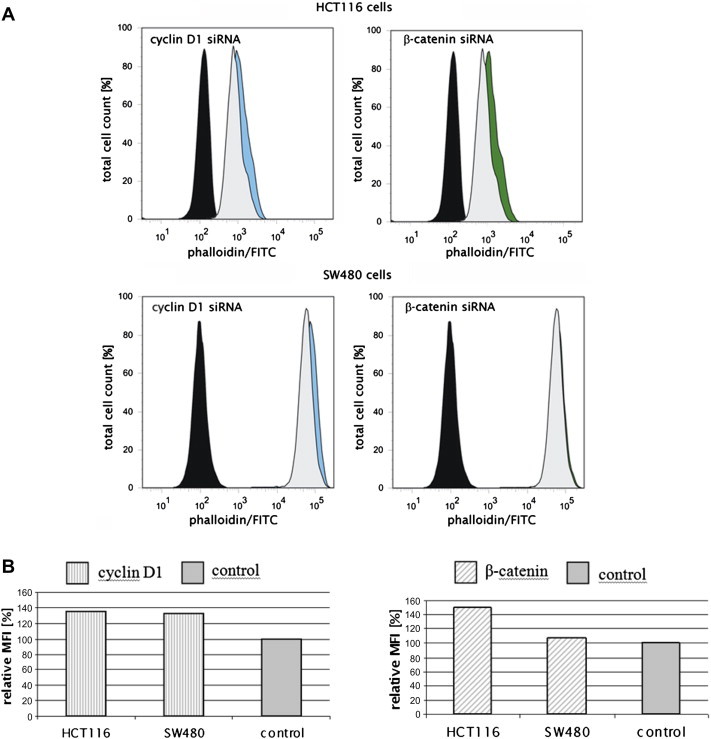
Cyclin D1 influences polymerization of the actin cytoskeleton in colorectal cancer cells. Flow cytometry analysis of phalloidin/FITC stained and siRNA treated HCT116 and SW480 cells. Representative histograms for cyclin D1 (blue) and β-catenin siRNA (green) transfected cells are shown (A). As a control the cells were either treated with control siRNA (gray) or were left untreated and unstained (black). (B) Quantification of the relative mean fluorescence intensity (MFI).
The fluorescence microscopic analysis showed a rearrangement of the actin cytoskeleton after cyclin D1 siRNA transfection in all tested cell lines. Treatment with β-catenin siRNA influenced the actin cytoskeleton in HCT116 and HeLa cells, but not in SW480 cells (Fig. 3 and unshown data).
Next, the fluorescence intensity of phalloidin/FITC was quantified using flow cytometry. Results showed that cyclin D1 siRNA and β-catenin siRNA treatment of HCT116 cells and cyclin D1 siRNA transfection of SW480 cells lead to an increased mean fluorescence intensity (MFI) (Fig. 4). As a conclusion cyclin D1 either inhibits actin polymerization or promotes actin depolymerization in colorectal cancer cells.
3.3. Cyclin D1 and β-catenin destabilize E-cadherin-mediated adherens junctions
Furthermore the effects of cyclin D1 and β-catenin siRNA transfection on the formation of E-cadherin-mediated adherens junctions were analyzed via fluorescence microscopy. Therefore cells were stained with a Cy3-coupled anti-E-cadherin antibody. HCT116 and SW480 cells showed an increase in the number of adherens junctions after cyclin D1 and β-catenin siRNA treatment (Fig. 5). In HeLa cells no effect could be observed (data not shown).
Fig. 5.
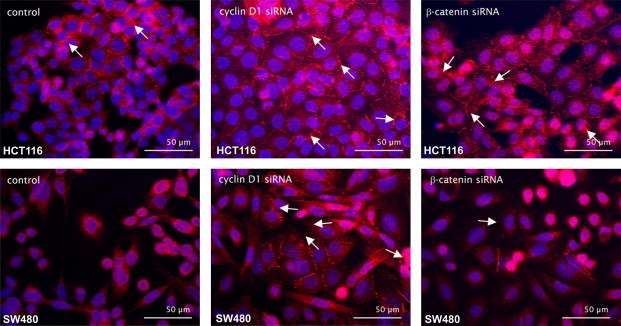
Formation of E-cadherin-mediated adherens junctions in HCT116 and SW480 cells after transfection with siRNA against cyclin D1 or β-catenin. Immunofluorescences of siRNA treated and E-cadherin/Cy3 (red) stained HCT cells and SW480 cells. As a control the cells were transfected with control siRNA. Nuclei are marked with DAPI (blue). White arrows show E-cadherin-mediated cellular contacts.
4. Discussion
Increased migration and decreased adhesion of cells are important hallmarks in advanced carcinogenesis, as both contribute to the ability of tumor cells to destabilize the intercellular context and to metastasize. The actin cytoskeleton is directly located under the cell membrane and is important for a number of biological functions in eukaryotic cells. It determines the form and polarity of cells, regulates cell division and plays a role in migration [39]. The movement of cells can be divided into three phases. First, the protrusion of the leading edge when the cell reorganizes the actin network at this side. Second, the adhesion of the cell to the substrate at the leading edge and the de-adhesion at the cell body and rear of the cell. Finally, the movement of the cell body, when contractile forces of the acto-myosin network pull the cell forward [40]. The actin cytoskeleton is regulated by different members of the family of Rho GTPases. The best characterized are RhoA, Rac1 and Cdc42. RhoA induces stress fibers, which play an important role in cell contraction, and is also involved in the formation of adhesion plaques, which are essential for cell–substrate contacts. Rac1 is associated with the formation of lamellipodias and Cdc42 mediates the polymerization of actin to filopodias [39].
The study presented here shows a significant influence of the Wnt pathway and namely cyclin D1 on cellular migration. Cyclin D1 promotes the migration of both colorectal and cervical carcinoma cell lines, whereas an effect of β-catenin was only visible in HCT116 and HeLa cells but not in the second tested colorectal cancer cell line SW480. The same was observed in the polymerization of the actin cytoskeleton, where β-catenin has no influence in SW480 cells but in HCT116 and HeLa cells. So the influence of the Wnt signaling pathway via cyclin D1 on cellular migration might be due to effects on the polymerization or depolymerization of the actin cytoskeleton. It is known, that macrophages and MEFs (mouse embryonic fibroblasts) of cyclin D1 deficient mice show an increased cellular adhesion and an impaired migration compared to those of wild type mice [35]. Li et al. [36] could show that the influence of cyclin D1 on cellular migration is due to the transcriptional inhibition of ROCK2 and TSP1, which are involved in the formation of stress fibers and therefore inhibit migration. These results could not be confirmed in our study via quantitative real-time RT-PCR (data not shown). There was no influence of cyclin D1 on the transcription of ROCK2 and TSP1 found in the tested colorectal and cervical cancer cell lines. Therefore cyclin D1 promotes cellular migration via an alternative mechanism. The cdk inhibitor p27Kip1 is involved in the inhibition of cellular proliferation of normal cells. In cancer cells p27Kip1 often translocates from the nucleus to the cytoplasm. Here it is not degraded like other tumor suppressors, but acts as a regulator of the cytoskeleton and cell migration [41–43]. p27Kip1 binds to RhoA and inhibits the interaction with the activating GEF [44]. Moreover it has been shown that p21 binds and inhibits the RhoA effectors ROCK1 and -2 in the cytoplasm [45]. Furthermore another cdk inhibitor, p57Kip2, binds to the kinase LIMK1 without affecting the activity directly. Instead, the interaction leads to the nuclear localization of the kinase and consequently to its inhibition [46]. These studies suggest that proteins of the Cip/Kip family inhibit the Rho signaling pathway at different sites. As a consequence there is a reduction in the formation of Rho-induced stress fibers and focal adhesions. In this model cyclin D1 promotes cellular migration by rescuing p27Kip1 from degradation, increasing the amount of this protein. Future studies should measure the activity of the actin cytoskeleton regulating proteins RhoA, Rac1 and Cdc42 to elucidate the role of cyclin D1 on cell migration.
E-cadherin mediates cellular adhesion and the formation of cell–cell contacts, as it connects proteins of adherens junctions with the actin cytoskeleton [3]. During carcinogenesis the cells undergo the epithelial–mesenchymal transition, which is characterized by the loss of these cell–cell contacts resulting in an increased motility [15,16]. The results of this study show that an aberrantly activated Wnt pathway destabilizes E-cadherin-mediated cell–cell contacts via cyclin D1. As a consequence the integrity of the cell layer is disturbed and the invasion of tumor cells facilitated.
In conclusion, the results presented here indicate that cyclin D1 influences the movement of cancer cells by promoting cell migration and destabilizing adherens junctions.
References
- 1.Giles R.H., van Es J.H., Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta. 2003;1653(1):1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 2.Daniels D.L., Weis W.I. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 2005;12(4):364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 3.Brembeck F.H., Rosario M., Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr. Opin. Genet. Dev. 2006;16(1):51–59. doi: 10.1016/j.gde.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Yamada S., Pokutta S., Drees F., Weis W.I., Nelson W.J. Deconstructing the cadherin–catenin–actin complex. Cell. 2005;123(5):889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drees F., Pokutta S., Yamada S., Nelson W.J., Weis W.I. Alpha-catenin is a molecular switch that binds E-cadherin–beta-catenin and regulates actin-filament assembly. Cell. 2005;123(5):903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pokutta S., Weis W.I. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu. Rev. Cell Dev. Biol. 2007;23:237–261. doi: 10.1146/annurev.cellbio.22.010305.104241. [DOI] [PubMed] [Google Scholar]
- 7.Brabletz T., Jung A., Reu S. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. USA. 2001;98(18):10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behrens J., Mareel M.M., Van Roy F.M., Birchmeier W. Dissecting tumor cell invasion: epithelial cells acquire invasive properties after the loss of uvomorulin-mediated cell–cell adhesion. J. Cell Biol. 1989;108(6):2435–2447. doi: 10.1083/jcb.108.6.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vleminckx K., Vakaet L., Jr., Mareel M., Fiers W., van Roy F. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991;66(1):107–119. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- 10.Behrens J. The role of cell adhesion molecules in cancer invasion and metastasis. Breast Cancer Res. Treat. 1993;24(3):175–184. doi: 10.1007/BF01833258. [DOI] [PubMed] [Google Scholar]
- 11.Perl A.K., Wilgenbus P., Dahl U., Semb H., Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392(6672):190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 12.Orsulic S., Huber O., Aberle H., Arnold S., Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J. Cell Sci. 1999;112(Pt 8):1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- 13.Stockinger A., Eger A., Wolf J., Beug H., Foisner R. E-cadherin regulates cell growth by modulating proliferation-dependent beta-catenin transcriptional activity. J. Cell Biol. 2001;154(6):1185–1196. doi: 10.1083/jcb.200104036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottardi C.J., Wong E., Gumbiner B.M. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J. Cell Biol. 2001;153(5):1049–1060. doi: 10.1083/jcb.153.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thiery J.P. Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 16.Birchmeier C., Birchmeier W., Gherardi E., Vande Woude G.F. Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol. 2003;4(12):915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 17.Lilien J., Balsamo J. The regulation of cadherin-mediated adhesion by tyrosine phosphorylation/dephosphorylation of beta-catenin. Curr. Opin. Cell Biol. 2005;17(5):459–465. doi: 10.1016/j.ceb.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Tetsu O., McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398(6726):422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 19.Shtutman M., Zhurinsky J., Simcha I. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA. 1999;96(10):5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willert J., Epping M., Pollack J.R., Brown P.O., Nusse R. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev. Biol. 2002;2(1):8. doi: 10.1186/1471-213x-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van De W.M., Sancho E., Verweij C. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111(2):241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 22.Sansom O.J., Reed K.R., Hayes A.J. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18(12):1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreu P., Colnot S., Godard C. Crypt-restricted proliferation and commitment to the Paneth cell lineage following Apc loss in the mouse intestine. Development. 2005;132(6):1443–1451. doi: 10.1242/dev.01700. [DOI] [PubMed] [Google Scholar]
- 24.Morgan D.O. Principles of CDK regulation. Nature. 1995;374(6518):131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 25.Matsushime H., Roussel M.F., Sherr C.J. Novel mammalian cyclins (CYL genes) expressed during G1. Cold Spring Harb. Symp. Quant. Biol. 1991;56:69–74. doi: 10.1101/sqb.1991.056.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Motokura T., Bloom T., Kim H.G. A novel cyclin encoded by a bcl1-linked candidate oncogene. Nature. 1991;350(6318):512–515. doi: 10.1038/350512a0. [DOI] [PubMed] [Google Scholar]
- 27.Xiong Y., Beach D. Population explosion in the cyclin family. Curr. Biol. 1991;1(6):362–364. doi: 10.1016/0960-9822(91)90193-z. [DOI] [PubMed] [Google Scholar]
- 28.Coqueret O. Linking cyclins to transcriptional control. Gene. 2002;299(1–2):35–55. doi: 10.1016/s0378-1119(02)01055-7. [DOI] [PubMed] [Google Scholar]
- 29.Opitz O.G., Rustgi A.K. Interaction between Sp1 and cell cycle regulatory proteins is important in transactivation of a differentiation-related gene. Cancer Res. 2000;60(11):2825–2830. [PubMed] [Google Scholar]
- 30.Inoue K., Sherr C.J. Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent-kinase-independent mechanism. Mol. Cell Biol. 1998;18(3):1590–1600. doi: 10.1128/mcb.18.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knudsen K.E., Cavenee W.K., Arden K.C. D-type cyclins complex with the androgen receptor and inhibit its transcriptional transactivation ability. Cancer Res. 1999;59(10):2297–2301. [PubMed] [Google Scholar]
- 32.Petre C.E., Wetherill Y.B., Danielsen M., Knudsen K.E. Cyclin D1: mechanism and consequence of androgen receptor co-repressor activity. J. Biol. Chem. 2002;277(3):2207–2215. doi: 10.1074/jbc.M106399200. [DOI] [PubMed] [Google Scholar]
- 33.Wang C., Pattabiraman N., Zhou J.N. Cyclin D1 repression of peroxisome proliferator-activated receptor gamma expression and transactivation. Mol. Cell Biol. 2003;23(17):6159–6173. doi: 10.1128/MCB.23.17.6159-6173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin H.M., Zhao L., Cheng S.Y. Cyclin D1 is a ligand-independent co-repressor for thyroid hormone receptors. J. Biol. Chem. 2002;277(32):28733–28741. doi: 10.1074/jbc.M203380200. [DOI] [PubMed] [Google Scholar]
- 35.Neumeister P., Pixley F.J., Xiong Y. Cyclin D1 governs adhesion and motility of macrophages. Mol. Biol. Cell. 2003;14(5):2005–2015. doi: 10.1091/mbc.02-07-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z., Wang C., Prendergast G.C., Pestell R.G. Cyclin D1 functions in cell migration. Cell Cycle. 2006;5(21):2440–2442. doi: 10.4161/cc.5.21.3428. [DOI] [PubMed] [Google Scholar]
- 37.Rubinfeld B., Robbins P., El Gamil M. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997;275(5307):1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 38.Munemitsu S., Albert I., Souza B., Rubinfeld B., Polakis P. Regulation of intracellular beta-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc. Natl. Acad. Sci. USA. 1995;92(7):3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279(5350):509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 40.Ananthakrishnan R., Ehrlicher A. The forces behind cell movement. Int. J. Biol. Sci. 2007;3(5):303–317. doi: 10.7150/ijbs.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bloom J., Pagano M. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin. Cancer Biol. 2003;13(1):41–47. doi: 10.1016/s1044-579x(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 42.Slingerland J., Pagano M. Regulation of the cdk inhibitor p27 and its deregulation in cancer. J. Cell Physiol. 2000;183(1):10–17. doi: 10.1002/(SICI)1097-4652(200004)183:1<10::AID-JCP2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 43.Philipp-Staheli J., Payne S.R., Kemp C.J. P27(Kip1): regulation and function of a haplo insufficient tumor suppressor and its misregulation in cancer. Exp. Cell Res. 2001;264(1):148–168. doi: 10.1006/excr.2000.5143. [DOI] [PubMed] [Google Scholar]
- 44.Besson A., Gurian-West M., Schmidt A., Hall A., Roberts J.M. P27Kip1 modulates cell migration through the regulation of RhoA activation. Genes Dev. 2004;18(8):862–876. doi: 10.1101/gad.1185504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S., Helfman D.M. Cytoplasmic p21Cip1 is involved in Ras-induced inhibition of the ROCK/LIMK/cofilin pathway. J. Biol. Chem. 2004;279(3):1885–1891. doi: 10.1074/jbc.M306968200. [DOI] [PubMed] [Google Scholar]
- 46.Yokoo T., Toyoshima H., Miura M. P57Kip2 regulates actin dynamics by binding and translocating LIM-kinase 1 to the nucleus. J. Biol. Chem. 2003;278(52):52919–52923. doi: 10.1074/jbc.M309334200. [DOI] [PubMed] [Google Scholar]


