Abstract
Multiple lines of evidence implicate over-expression and activation of the androgen receptor (AR) in the progression of prostate cancer (PC) to androgen-independence (AI) and resistance to therapy. The mechanisms leading to AR over-expression are not fully understood but binding of Sp1 to specific Sp1-binding sites in the AR promoter and 5′-untranslated region (5′-UTR) was shown to up-regulate AR transcription. In this work, we further characterized the role of Sp1 in the control of AR transcription and explored its potential as a therapeutic target in androgen-dependent (AD) and independent (AI) LNCaP cells. We identified a pair of new Sp1-binding site in the 5′-UTR of AR which we named ARSp1-3. ARSp1-3 binds Sp1 with higher affinity than other known Sp1-binding sites in the promoter/5′-UTR and in transfection experiments, the ARSp1-3 reporter showed higher transcriptional activity in AI than in AD cells. Treatment of these cells with nanomolar concentrations of Mithramycin inhibited binding of Sp1 to its binding sites in the promoter/5′-UTR of the AR gene but more specifically the binding of ARSp1-3 while other regulatory elements of the AR promoter were not affected. Inhibition of Sp1 binding by Mithramycin decreased the AR transcription and transactivation of PSA reporter constructs. At the lowest concentrations, Mithramycin decreased endogenous AR protein and proliferation of AD and AI LNCaP cells. The combinations of Mithramycin with either paclitaxel or bicalutamide were highly synergistic.
Conclusion:
Sp1 binding induces AR transcription in LNCaP cells. The higher affinity of ARSp1-3 for Sp1 may support higher AR mRNA levels in AI than AD LNCaP cells. Mithramycin is a potent and specific inhibitor of Sp1 and AR transcription with potential, at very low concentrations, to enhance the efficacy of hormones or taxane based therapy in patients with recurrent or androgen-independent progression that sustain AR expression.
Keywords: Mithramycin, prostate cancer, Sp1, chemo-hormonal therapy, androgen receptor
Introduction
Approximately 30% of men treated for localized prostate cancer (PC) will experience systemic progression requiring androgen-ablative hormone therapy. Over 70% of these men will develop resistance to hormone therapy and disease progression due to selection or adaptation of androgen-independent clones (Isaacs and Coffey, 1981; Bruchovsky, Rennie et al. 1990). For these patients, the number of active agents is small and the duration of response is limited (Ferrari, Chachoua et al. 2001). Therefore, an understanding of the mechanisms leading to resistance and new approaches to design more disease specific treatments is warranted.
A common denominator in PC progression is sustained or increased expression and activity of the AR even under androgen-deprived conditions. A variety of mechanisms have been implicated in this process: 1) amplification of wild type AR gene or 2) mutations that render the receptor protein hypersensitive to low levels of androgen or non-androgenic ligands (Newmark, Hardy et al. 1992; Gaddipati, McLeod et al. 1994; Visakorpi, Hyytinen et al. 1995; Koivisto, Kononen et al. 1997; Linja, Savinainen et al. 2001), 3) activation by cross-talk with growth factor (GF) receptor signaling pathways (Hobisch, Eder et al. 1998; Craft, Shostak et al. 1999; Gioeli, Mandell et al. 1999; Sadar, 1999; Yoshida, Dubauskas et al. 1999; Signoretti, Montironi et al. 2000; Wen, Hu et al. 2000; Matsuda, Junicho et al. 2001; Shi, Brands et al. 2001), 4) excessive recruitment of transcriptional co-activators (Horwitz, Jackson et al. 1996; Anzick, Kononen et al. 1997; Miyamoto, Yeh et al. 1998), or loss of AR suppressor proteins (Wang, Ossowski et al. 2004). Altered expression of the AR has been demonstrated both in human tissues and animal model systems (Visakorpi, Hyytinen et al. 1995; Koivisto, Kononen et al. 1997; Linja, Savinainen et al. 2001; Chen, Welsbie et al. 2004). Real-time RT-PCR studies of human samples identified AR expression in all hormone-refractory tumors analyzed and the level of expression was on average 6-fold higher than in AD tumors or benign prostate hyperplasias (Linja, Savinainen et al. 2001). Amplification of the wild type AR is found in one-third of hormone-refractory cases (Koivisto, Hyytinen et al. 1995; Visakorpi, Hyytinen et al. 1995). Using microarray-based profiling of isogenic PC xenograft models, Chen et al, identified a modest increases in AR mRNA and protein as the only change consistently associated with the development of resistance to hormone therapy (Chen, Welsbie et al. 2004). Increased AR transcription has also been observed in PC that respond to endocrine therapy for more than 12 months suggesting that it is an early event in progression (Koivisto, Kononen et al. 1997). It is also apparent that AR over-expression is associated with abnormal expression of AR-target genes (Gregory, Hamil et al. 1998), which support androgen independent cell proliferation and survival in very low levels of androgens after castration (Kallioniemi and Visakorpi, 1996).
The significance of increased expression of AR in androgen-independent (AI) PC cells was demonstrated by the remarkable inhibition of cell proliferation and induction of apoptosis following specific down-regulation of AR mRNA by siRNA or antisense strategies (Eder, Culig et al. 2000; Ofelia L, 2002).
Thus, treatments aimed at suppression of AR transcription in PC cells have the potential to be effective, and to reduce resistance to androgen deprivation and chemotherapy irrespective of whether mutations, amplification, or aberrant cross-talk with other co-regulatory molecules support AR activation.
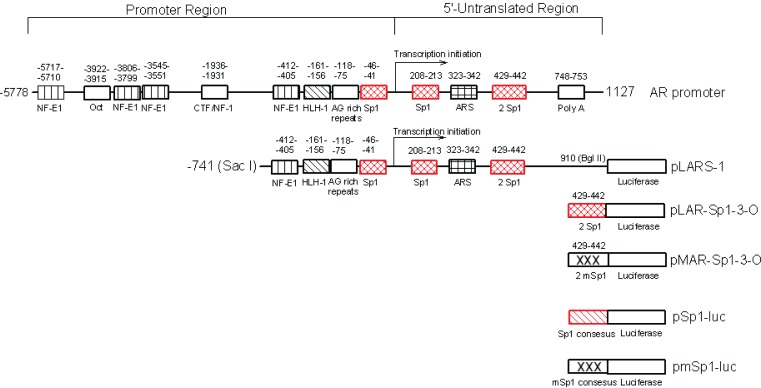
Unlike most genes, the promoter of the AR lacks the TATA and CCAAT box motifs that regulate its transcription. Instead, sequences scattered throughout the promoter and 5′-untranslated region (5′-UTR) of AR bind transcription factors with positive (Sp1) and negative (neurofibromatosis 1, NFkappa B and ARS regulatory activity (Mizokami and Chang, 1994; Grossmann and Tindall, 1995; Supakar, Jung et al. 1995; Takane and McPhaul, 1996; Song, Jung et al. 1999; Wang, Ossowski et al. 2004). Multiple functional Sp1 elements have been identified (Husmann, Wilson et al. 1990; Tilley, Marcelli et al. 1990; Tilley, Marcelli et al. 1990; Tilley, Wilson et al. 1990; Faber, King et al. 1991; Faber, van Rooij et al. 1991) but more specifically two of them located near the transcriptional initiation site of AR (Figure 1) at −46 to −41 (Sp1-1) and 208 to 213 (Sp1-2) (Mizokami and Chang, 1994; Takane and McPhaul, 1996; Ryu, Zhou et al. 1999) have been shown to up-regulate AR transcription (Mizokami and Chang, 1994; Takane and McPhaul, 1996).
Figure 1.
Diagrammatic representation of the regulatory sites surrounding the human AR promoter and 5′-UTR identified by computer program Gene-Runner. pLARS-1, pAR-Sp1-3-O and its mutant pMAR-Sp1-3-O are AR-luciferase reporters constructed in our laboratory and were used for in this study. pSp1-luc and pmSp1-luc used in this study are plasmids containing three tandem repeats of consensus Sp1 sites and their corresponding mutant that drive luciferase gene (kindly provided by Dr. Yoshihiro Sowa, University of Medicine, Kamigyo-ku, Kyoto, 602, Japan.).
Mithramycin (plicamycin) is an antitumor antibiotic used in the past for treatment of germ cell tumors and cancer related hypercalcemia (Bonadonna, 1971; Calabresi P, 1991). Mithramycin has been shown to interfere with Sp1 binding to GC rich Sp1 regulatory sequences (Mir and Dasgupta, 2001; Mir and Dasgupta, 2003) and to inhibit gene transcription (Ryu, Zhou et al. 1999; Yoo, Jeong et al. 2002).
In this study we explored further the role of Sp1 in the transcription and regulation of the AR gene promoter in PC cells that increase the levels of AR transcription with AI progression, and tested a potential therapeutic role for Sp1 suppression in the treatment of advanced PC.
We utilized the androgen-dependent (AD) LNCaP cells and its AI LNCaP derivative which has a genetically distinct profile with 4 fold higher level of AR mRNA and phosphorylated protein (Gao, Ossowski et al. 1999; Wang, Ossowski et al. 2001; Liu, Xia et al. 2003). In this model, down-regulation of AR can reverse the growth and apoptosis response of AI cells to that of AD cells (Wang, Ossowski et al. 2001).
The results of this work indicate that we identified a new pair of Sp1 binding sites, in the 5′UTR of AR (ARSp1-3) which may contribute to the higher levels of AR expressed by the AI LNCaP cells. We confirmed that Mithramycin is a specific inhibitor of Sp1 binding including to ARSP1-3 and a strong suppressor of AR transcription and transactivation activity. In doing so, Mithramycin reduced AR protein expression and inhibited growth of AD and AI LNCaP cells and the combinations of Mithramycin with either paclitaxel or bicalutamide were highly synergistic. The data supports Mithramycin targets AR transcription and has potential to enhance the response to chemotherapy and reverse the resistance to anti-androgens of PC patients with androgen-independent progression.
Materials and Methods
Reagents
PSA reporter-plasmid GRE-tk-LUC (GRE) was kindly provided by Dr. Albert O. Brinkmann (de Ruiter, Teuwen et al. 1995). The full length of p21 promoter reporter pWWP, its mutant containing Sp1 sites but not p53 regulator sequences pWP101, and pSp1-luc containing three tandem repeats of consensus Sp1 sites driving the luciferase gene as well as its mutant pmSp1-luc (Sowa, Orita et al. 1997; Liu, Jiang et al. 2003) were kindly provided by Dr. Yoshihiro Sowa (University of Medicine, Kamigyo-ku, Kyoto, 602, Japan.). The pLARS-1 reporter that contains the Sac I - Bgl II region of the human AR promoter and 5″-UTR from −741 to 910 covering 3 Sp1 sites located at −46, 208 and 429, respectively, was constructed as previous described (Wang, Ossowski et al. 2004). The reporter pAR-Sp1-3-O and its mutant (Fig. 1) were constructed by inserting into the basic luciferase-reporter pGL3, a series of three repeats of the Sp1 sequence, +429 to +442, found in the 5′-UTR of AR gene and the DNA sequence were verified. Antibodies against PSA, AR, p21, and β-actin as well as secondary antibodies were purchased from DAKO Corporation (Carpinteria, CA) and Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), respectively. Effectene transfection reagent and luciferase assay kit were acquired from Qiagen (Valencia, CA) and Promega (Madison, WI). ECL Western detection kit and films were purchased from Amersham Biosciences (Buckingamshire, England). Mithramycin, paclitaxel and other chemicals were purchased from Sigma (St. Louis, MO). Bicaluta-mide (Casodex, AstraZeneca) was extracted from commercially available tablets.
Cell Culture
LNCaP cells were purchased from the American Type Culture Collection (Rockville, MD) and were maintained in RPMI 1640 (GIBCO, Gaithersburg, MD) with 10% heat-inactivated bovine serum (FBS). The androgen independent (AI) LNCaP subline derived from LNCaP cells, was maintained in RPMI 1640 medium containing 10% charcoal-stripped, heat-inactivated FBS (CSFBS) (Hyclone Laboratories, Inc., Logan, UT) and 5 μg/ml of insulin as described previously (Gao, Ossowski et al. 1999).
Cell Transfection and Luciferase Assay
AD or AI LNCaP cells grown exponentially, were seeded into 60 mm dishes at a density of 105 per ml. After incubation for 24 hr at 5% CO2 and 37° C, the cells were transfected with 1 μg/dish of luciferase reporter pWWP, pWP101, pSp1-luc, pmSp1-luc, or pLARS-1 using Effectene (Qiagen, Valencia, CA). 0.5 μg of pSV-β- galactosidase control vector (Progema, Madison, WI) was co-transfected as an internal control. Twenty-four hrs after transfection, the cells were exposed for an additional 24 hrs to increasing concentrations of Mithramycin, or other agents, as indicated. The cells were washed, lysed, and the lysates were used for luciferase activity assay using Promega luciferase assay system. The β-galactosidase activity was determined using β-galactosidase Assay System (Promega, Madison, WI) according to manufacturer’s instructions. The luciferase activities were normalized on the basis of galactosidase activity, and were expressed as units per mg of proteins.
MTT and Combination Analysis
MTT was used to estimate cell growth as described previously (Wang, Liu et al. 1997). Briefly, AD or AI LNCaP cells grown exponentially were aliquoted into 96-well plates at a density of 5000 cells/200 μl per well in RPMI 1640 medium containing 10% FBS for AD and 10% CSFBS for AI. Twenty-four hrs after the incubation, the cells were exposed for three days to serial dilution of mithramycin, paclitaxel or bicalutamide, or mithramycin plus bicalutamide (1:3300) or plus paclitaxel (1:1). After the incubation, 100 μl of the medium was removed from each of the wells and 50 μl of a 1mg/ml solution of 3-(4, 5-dimethyl-thiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) was added, the cells were incubated for an additional 4 hrs and then 200 μl of 0.04 N HCl-isopropanol was added to each well to dissolve the black formazan precipitates, The absorbance was measured on a 96- well micro-plate reader (Dynex Technologies, Chantilly, VA). The percentages of growth inhibition (1-T/C%) and combination indexes (CI) were calculated, and analyzed using the PC program, CalcuSyn, of Biosoft edited by T.-C. Chou, Memorial Sloan-Kettering Cancer Center, New York, and M.P. Hayball, at Cambridge, U.K., 1996 (Chou, Motzer et al. 1994). The combination index (CI) was used to evaluate the outcome of the combinations. A CI greater than 1, indicates the combination is antagonistic, CI equal to 1, indicates the combination is additive, and CI smaller than 1 that the combination is synergistic (Chou, Motzer et al. 1994).
Mobility Gel Shift Assay (EMSA)
Was performed as described previously (Wang, Liu et al. 1994). Briefly, Five μg of nuclear proteins extracted from AD and AI cells was reacted for 30 minutes at room temperature with the respective [32P]-labeled double-stranded oligos of ARSp1-1: 5′-CGG CCC GGT GGG GGC GGG AC-3′, ARSp1-2: 5′-AGC CGC CGC CCG GAG CTG CC-3′, and ARSp1-3: 5′-CCA CCC GCC CCC CCG CCC CC-3′, or ARNF1: 5′-CGC TCT TAT CAG TCC T-3′, in binding buffer composed of 10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 0.5% (v/v) glycerol and 2.0 μg poly(dI)(dC) in a final volume of 25 μl. The reaction mixtures were then subjected to 8% native low ionic strength PAGE (Wang, Liu et al. 1994). The binding complexes were visualized by exposing the dried gel to X-ray film at −80° C overnight.
Southwestern (DNA-protein) Blot Analysis
Southwestern blot was performed essentially according to a method described by Yang et al., (Yang, Tsai et al. 1994) with minor modifications. Four aliquots of 50 μg of nuclear extracts from AD and AI cells were subjected to SDS-PAGE, and electro-transferred to nitrocellulose membrane. The membrane was then blocked with 5% fat free dry milk in a binding buffer as described in EMSA for 1 hr at room temperature. After 3 washes with the binding buffer, the membranes were hybridized at 4° C for 1 hr with [32P]- labeled oligos of AR-Sp1-1 or AR-Sp1-3, in the presence of 1 μg/ml oligos competitor wt-PSA-Sp1 (Wang, Liu et al. 1997). After 3 washes with the binding buffer, the membranes were exposed to X-ray film at −80° C.
Western Blotting of Cellular Proteins
Sp1, AR, PSA, and p21WAF1 (p21) protein levels were determined by Western blot analysis as described previously (Wang, Liu et al. 1997). Total cellular proteins were extracted under conditions described previously (Wang, Liu et al. 1997) from AD or AI LNCaP cells under various treatments conditions as indicated. Fifty μg cellular extracts were separated on a 10% SDS-PAGE, electro-transferred to nitrocellulose filters, and immuno blotted initially with antibodies against p21WAF1 or AR or PSA or other indicated target. The same membranes were stripped and re-probed with β-actin for loading control. Quantitation by densitometry of the X-ray films was done using an Imaging Densitometer Model GS-720 (Bio-Rad Lab. Hercules, CA).
Results
Identification of ARSp1-3 and its functionality analysis
By means of a computer-aided analysis of the 5′-UTR of the AR gene, we identified two new Sp1 binding sites adjacent to each other at 429 and 442 in close proximity to the initiation site of AR transcription (Fig. 1). We named this previously unidentified pair ARSp1-3.
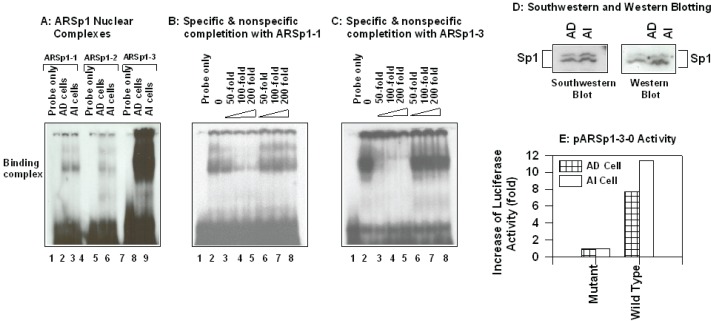
To study the role of the known and new Sp1-binding sites in the regulation of AR transcription in PC cells, we analyzed the binding affinity of nuclear proteins extracts from AD and AI LNCaP cells by EMSA using 32P-lableled oligos for ARSp1-1, ARSp1-2 and ARSp1-3 as probes. As expected, similar nuclear binding complex formed with each oligo but the binding affinity for ARSp1-3 was significantly stronger and similar in both AD and AI cells (Figure 2, panel A). The specificity of the binding of the nuclear extracts to these Sp1 sites was confirmed in competition assays shown in Fig. 2B & C. In the presence of unlabeled ARSp1-1 or ARSp1-3 oligos (lane 3–5) there was a progressive concentration-dependent reduction in the complex formation that was not observed when a non-specific Sp1 oligo sequence corresponding to the PSA promoter was used (lane 6–8) (Wang, Liu et al. 1997).
Figure 2.
Role of Sp1 elements present in AR promoter and/or 5′-UTR of the gene in the regulation of AR transcription. Panel A–C: EMSA: 5 μg nuclear extracts from AD and AI cells were reacted with 32P-labled oligo probe corresponding to the sequence to indicated Sp1 element in a binding buffer containing 1 μg Poly (dIdC) in the absence (panel A) or presence (panel B&C) of specific (panel B&C, lane 3 – 5) or nonspecific (panel B&C, lane 6 – 8) oligo competitors. The reaction was performed at room temperature for 30 min in a final volume of 20 μl, and the reaction mixtures were subjected to 8% native polyacrylmide gel as described in the “Materials and Methods.” Panel D: Southwestern and Western blot analysis: Nuclear proteins from AD and AI cells were subjected to SDS-PAGE, electrotransfered to nitrocellulose membrane and hybridized to 32P-labled ARSp1 oligo (Southwestern blot), the same membrane stripped and re-probed with Sp1 monoclonal antibody (Western blot). The two protein bands with appear molecular weight 95 and 105 kd represent phosphorylated and dephosphorylated Sp1 protein. Panel E: Luciferase activity of pARSp1-3-0 in AD and AI cells: AD and AI cells at exponential growth phase were transfected with 1 μg per 6- mm dish of pARSp1-3-0 or its mutant pMARSp1-3-0 and pSV-β-galactosidase (internal control) in serum free RPMI 1640 medium for 24 hrs. The medium was replaced with fresh serum containing medium, and the cells were incubated for another 24 hrs. The cells were then lysed and luciferase/β-galactosidase activities determined.
To confirm that Sp1-protein was involved in the complex formation with ARSp1-3, we performed a Southwestern using ARSp1-3 as probe. As shown in Fig. 2 D, a phosphorylated- and dephosphorylated- form of Sp1 were indeed the major protein components of the nuclear binding complex formed with ARSp1-3 in both AD and AI LNCaP cells. The specificity of the Sp1 signal was further verified by Western blot using an Sp1 monoclonal antibody (Jackson, MacDonald et al. 1990) which also showed a stronger signal in AI cells suggesting a relation to their higher levels of AR.
To investigate the transcriptional regulation of AR under ARSp1-3, we compared the luciferase activity of the wild type pARSp1-3-O reporter construct to its mutant (Fig. 1) after transfection into AD and AI LNCaP cells. As shown in Fig.2, panel E, an over 7-fold increase in AD cells and 11-fold in AI cells of the wide type reporter activity vs. its mutant were observed. These differences correlate with a 4 fold higher level of AR mRNA that is constitutively expressed by AI cells (Wang, Ossowski et al. 2001). Overall, these results support a role for ARSp1-3 in the regulation of AR transcription in PC cells and as a contributing factor to the higher levels of AR mRNA constitutively expressed by AI LNCaP cells.
Mithramycin targets ARSp1 sites and AR transcription
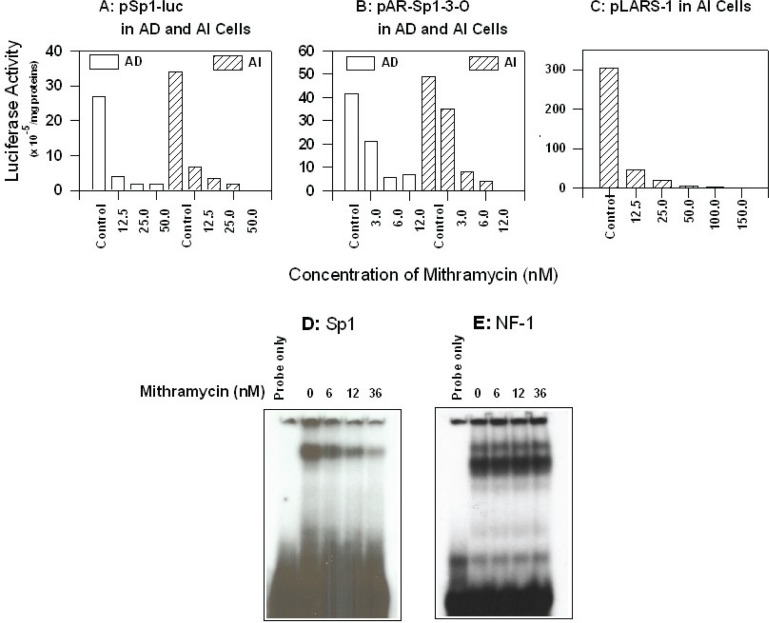
To determine whether Mithramycin inhibits Sp1 transcriptional regulation activity of AR in AD and AI LNCaP cells, the pSp1-luc reporter containing three tandem consensus Sp1 sites or the mutant, pmtSp1-luc (Fig. 1), were transfected into these cells and the luciferase reporter activity measured with and without treatment with Mithramycin at concentrations in the range of 12–50 nM. As shown in Figure 3A, 12 nM Mithramycin was sufficient to inhibit luciferase activity of pSp1-luc in both AD and AI cells but had no effect on the same cells transfected with the mutant pmtSp1-luc (data not shown).
Figure 3.
Panel A–C: Inhibitory effect of Mithramycin on Sp1-driven luciferase reporter activity: AD and AI cells in exponential growth phase were co-transfected with pSp1-luc, pAR-Sp1-1-3-O, or pLARS-1, and pSV-β-galactosidase (internal control) in serum-free RPMI 1640 medium. After twenty-four hours, the medium was replaced with full serum medium (10 %, FBS for AD or 10% charcoal-stripped FBS for AI cells), and the cells were exposed to different concentrations of Mithramycin, and incubated for an additional 24 hours. The cells were washed, lysed, and used to assay the activities of luciferase and β-galactosidase, respectively, using Promega assay kits (No activity was observed using mutant forms of the Sp1 segments). Panel D–E: Effects of mithramycin on the formation of DNA-nuclear binding complexes. 5 μg of nuclear extracts from AD cells exposed to different concentrations of mithramycin were reacted with [32P]-labeled ARSp1-3 (panel D) or ARNF-1 oligos (panel E) in binding buffer for 30 min, the EMSA was then performed as described in the “Materials and Methods.”
As shown in Fig. 3B, Mithramycin also had a strong concentration-dependent inhibition of ARSp1-3-O in AD and AI LNCaP cells. A 50% reduction of pAR-Sp1-3-O luciferase activity (Fig. 1) was seen in AD cells at 3 nM and reached 80% reduction in both AD and AI cells at 6 nM. To determine whether the presence of other regulatory elements in the AR promoter, including suppressors of AR such as NF1 could account for the inhibition of AR transcription in AI cells, we transfected these cells with pLARS-1 (Fig.1) and measured luciferase activity after treatment with Mithramycin. The luciferase activity of pLARS-1 at 12 nM Mithramycin (IC50 of 1.87 ± 0.02 nM), was suppressed compared to untreated control AI transfected cells (Fig. 3C) to a similar extent as ARSp1-3-O, suggesting that Mithramycin does not affect other regulatory elements in the AR promoter that could abrogate its suppressive activity.
Mithramycin does not interfere with other regulatory binding sites on the AR promoter
To further assess the specificity of Mithramycin to inhibit ARSp-1 binding without increasing the binding of negative regulatory elements such as NF1, we treated AD cells with increasing concentrations of Mithramycin and performed EMSA of the nuclear extract using ARSp1-3 and ARNF-1 oligos as probes. As shown in Fig. 3D–E, there was a concentration-dependent decrease in Sp1-nuclear binding complexes starting at 6 nM Mithramycin that did not affect in any way the ARNF-1/nuclear binding complex formation while level of Sp1 proteins was not affected (data not shown). Thus, decrease of AR transcription by Mithramycin is specifically through its interference with Sp1 protein binding to the AR promoter.
Mithramycin down-regulates endogenous AR levels and inhibits AR transactivation of down stream targets
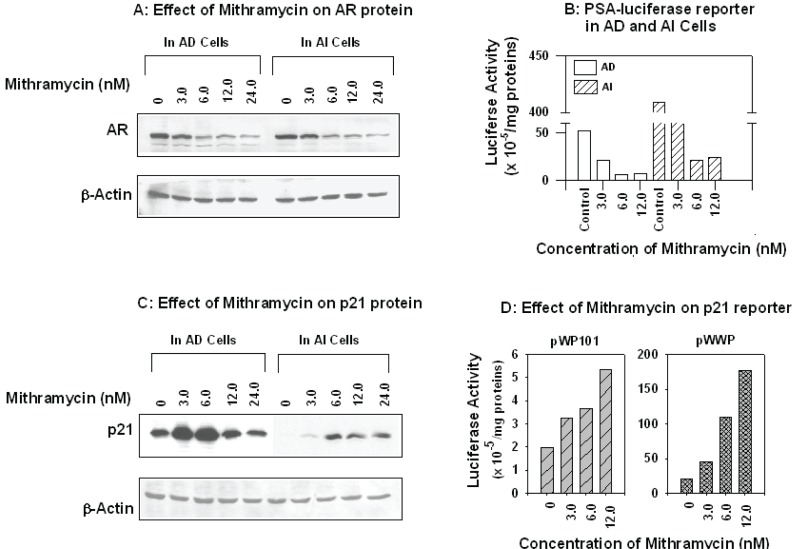
To determine whether Mithramycin could modulate endogenous AR levels and transactivation activity through inhibition Sp1, AD and AI LNCaP cells were treated with Mithramycin at the same nanomolar concentrations used to down regulate AR transcription. As shown in Figure 4A, there was a marked reduction in AR protein levels by Western blot and the response peaked to an equal extent in both AD and AI cells at 6.0 nM. Using the same concentrations of Mithramycin we then measured the effect on PSA reporter activation. As shown in Fig. 4B, PSA expression was markedly reduced in AD cells indicating that the down-regulation of endogenous AR levels is sufficient to decrease its transcriptional activation of down stream targets.
Figure 4.
Effects of Mithramycin on the expression of AR, AR down-stream genes PSA and p21. Panel A & C: AD and AI cells grown exponentially were exposed to indicated concentrations of Mithramycin for 24 hrs, the cells were harvested, washed, and total proteins extracted. Fifty μg were subjected to SDS-PAGE, electro-transferred to nitrocellulose membrane, and immuno-blotted with antibodies against AR (Santa Cruz Biotechnology, panel A) or p21 (DAKO, panel C). The same membrane was stripped and re-probed with β-Actin antibody for loading control. Panel B & D: AD and/or AI cells in exponential growth phase were co-transfected with PSA-luciferase reporter (B) or p21 reporters pWP101, pWWP (D) and pSV-β-galactosidase (internal control) in serum-free RPMI 1640 medium. After twenty-four hours, the medium was replaced with full serum medium (10 %, FBS for AD or 10% charcoal-stripped FBS for AI cells), and the cells were exposed to indicated concentrations of Mithramycin and incubated for an additional 24 hours. The cells were washed, lysed, and used to assay the activities of luciferase and β-galactosidase, respectively, using Promega assay kits. The activity of luciferase was normalized to β-galactosidase, and expressed as units per mg proteins.
In previous work, we determined that the expression of the cycle inhibitor p21 could be restored in AI LNCaP cells through activation of Sp1 or inhibition of AR mRNA by treatment with the histone deacetylase inhibitor, trichstatin A (Wang, Ossowski et al. 2001). Other agents, such as sodium butyrate, alkyl phospholipids were also reported to induce p21 through Sp1 activation (Nakano, Mizuno et al. 1997; De Siervi, Marinissen et al. 2004). To examine whether Mithramycin-mediated Sp1 inhibition would counteract the induction of p21 in AD and AI cells following AR down-regulation, we tested the effect on p21 expression in these cells after 24 hours treatment with 3–12 nM Mithramycin. Surprisingly, at 6 nM Mithramycin, a strong stimulation of p21 expression above baseline levels was observed in AD cells and the expression of p21 was restored from undetectable to detectable levels in AI cells (Fig. 4C).
The induction of p21 by Mithramycin was further supported by gene transfection assay in AD cells where full-length of p21 promoter reporter pWWP (wild type) and pWP101 that contains Sp1 but not p53 regulatory sites were used. As shown in Fig. 4D, a strong induction of the wild type p21 reporter activity in a concentration-dependent manner was observed after the transfeced cells were exposed to Mithramycin. Interestingly, a moderate induction by Mithramycin of pWP101 reporter that contains Sp1, but not p53 binding sites also obtained under the same experimental conditions. These results suggest that in AD and AI LNCaP cells Mithramycin does not inhibit binding of Sp1 activity to the p21 promoter to the same extent that it affects binding of Sp1 to the AR promoter. These results also suggest that transcription of p21 is less dependent on Sp1 binding than AR (Wang, Ossowski et al. 2001).
Mithramycin Inhibits Growth of AD and AI LNCaP Cells
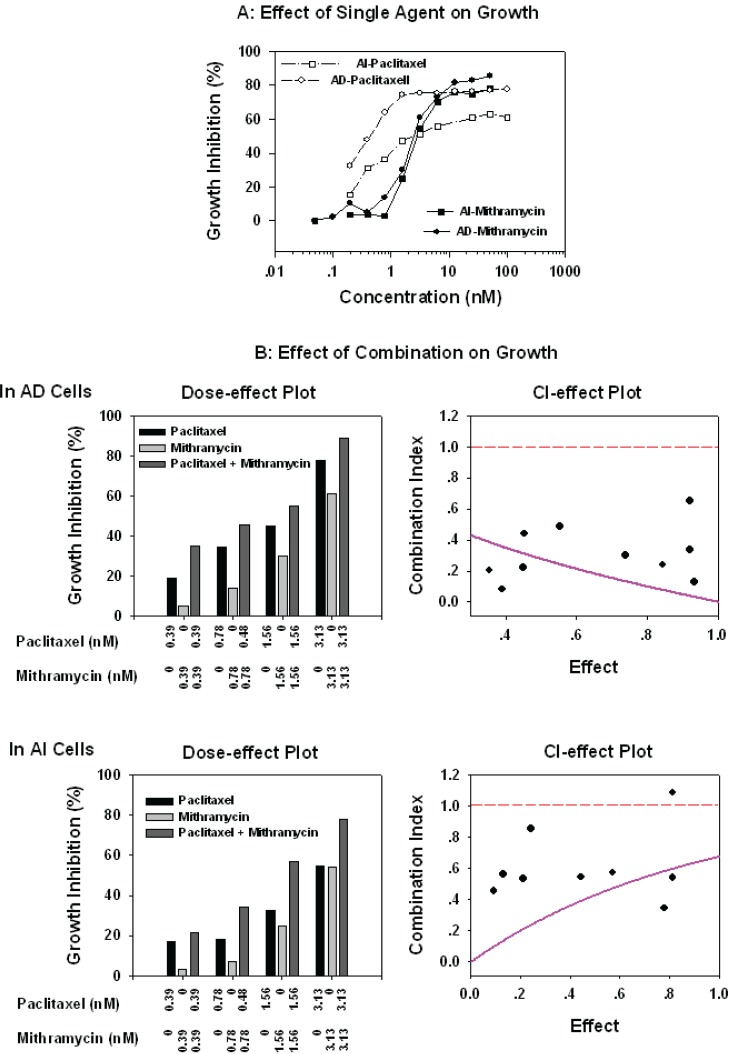
Given the effect of Mithramycin on AR and p21 expression we investigated the effect on proliferation of AD and AI LNCaP cells either alone or in combination with taxanes or the anti-androgen bicalutamide which is frequently used as second line hormonal therapy of patients with progressive disease. As shown in Figure 5, nanomolar concentrations of Mithramycin had an equally strong, growth inhibitory effect on both AD (IC50 3.603 ± 0.837 nM) and AI (IC50 4.660 ± 2.190 nM) cells. In contrast, the IC50 of paclitaxel was approximately 50-fold higher in AI than AD cells (Gao, Ossowski et al. 1999; Wang, Ossowski et al. 2001; Liu, Jiang et al. 2003) but the combinations of Mithramycin:paclitaxel were highly synergistic with a marked reduction in the Dm (medium effect) of both drugs. For paclitaxel the Dm for AD cells decreased from 1.7 nM to 0.234 nM and for AI cells from 6.300 nM to 2.491 nM. For Mithramycin, the Dm for AD cells decreased from 3.604 nM to 0.234 nM and for AI cells decreased from 6.281 nM to 2.491 nM.
Figure 5.
Inhibitory of Mithramycin and paclitaxel alone (panel A) or in combination on cell growth (panel B): AD and AI cells grown exponentially in 96-well dishes were exposed to a series dilution of Mithramycin or paciltaxel or Mithramycin plus paciltaxel at ratio of 1:1 for 3 days. The cells growth was measured by MTT using a standard protocol, percentages of growth inhibition (1-T/C)% calculated, and the combination index (CI) analyzed by a computer program, CalcuSyn, of Biosoft as described in the “Materials and Methods.” The read dash-line in Effect-CI plot represents the CI value equal to 1 (additive). The values above the line indicate antagonism, and below the line are synergistic. The pink line stimulated by the computer program represents the trend of the combination.
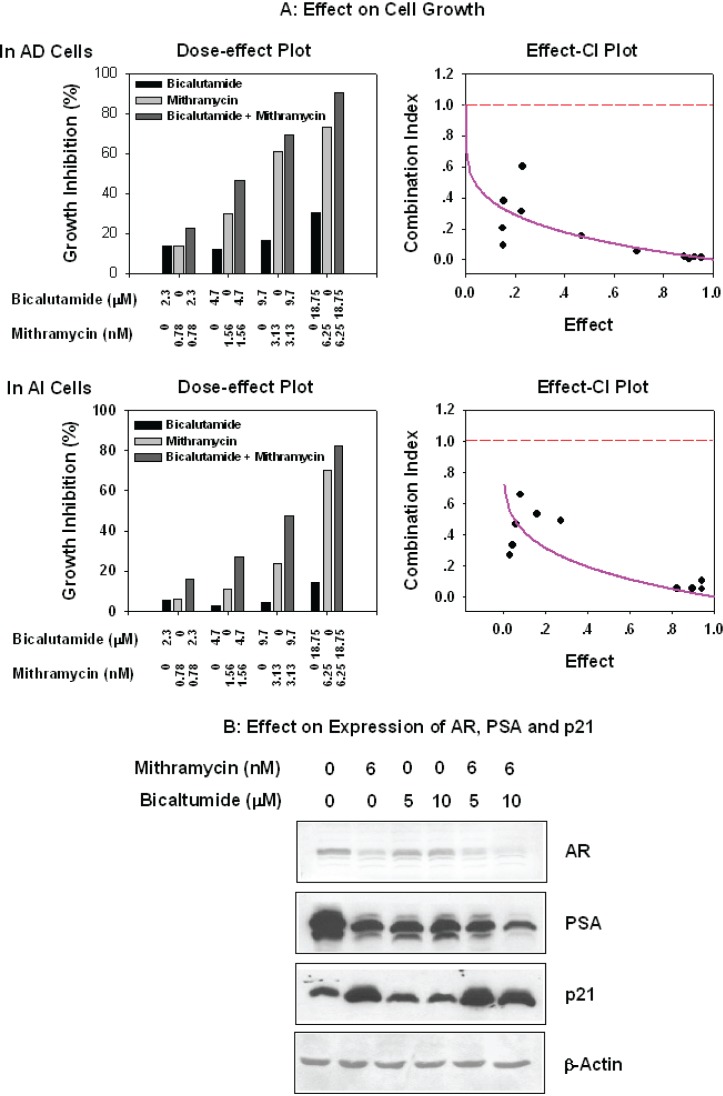
To assess the proliferative response of AD and AI cells to bicalutamide in the presence of Mithramycin, the cells were treated with combinations of these drugs in ratios of 1:3300. As shown in Figure 6A, Mithramycin alone had an equal inhibitory effect on proliferation of AD and AI LNCaP cells and was more pronounced than bicalutamide even in AI cells that are resistant to this agent. In contrast, the Mithramycin:bicalutamide combinations were strongly synergistic. The combination indexes (CI) were less that 1 (Effect-CI plot) and there was a marked reduction in the Dm (medium effect) of both drugs. In the case of bicalutamide, the Dm for AD cells decreased from 38.7 to 3.330 μM and for AI cells from 59.4 to 11.9 μM, In the case of Mithramycin the Dm for AD decreased from 3.604 to 0.0011 nM cells and for AI cells from 6.281 nM to 0.032 nM.
Figure 6.
Effect of combination of Mithramycin and bicalutamide on cell growth and expression of AR, PSA, and p21. Panel A: Synergistic combination of Mithramycin and bicalutamide by MTT. Exponentially growing AD and AI cells in 96-well dishes were were exposed for three days to serial dilution of Mithramycin or bicalutamide, or Mithramycin plus bicalutamide at ratio of 1:3300. The cell growth was measured by MTT, and the percentages of inhibition (1-T/C%) were calculated, and combination index (CI) analyzed by a PC program, CalcuSyn, of Biosoft (see ligand for Figure 5). Panel B: Effects of the combination of mithramycin with bicalutamide on expressions of AR, PSA, and p21. AD and AI cells at exponential growth were exposed to indicated concentration of Mithramycin, bicalutamide alone or their combination for 24 hrs. The cells were harvested, washed, and total proteins extracted. Fifty μg were subjected to SDS-PAGE, electro-transferred to nitrocellulose membrane, and immuno-blotted with antibodies against AR, PSA, and p21 (Santa Cruz and DAKO).
As shown in Fig. 6B, the growth inhibition induced by Mithramycin:biclutamide combinations coincided with reduction of AR and increase of p21 levels suggesting that these molecular events mediated the growth inhibitory response and loss of resistance of AI cells to bicalutamide (Wang, Ossowski et al. 2001).
Discussion
We have evaluated the role of Sp1 in the regulation of AR transcription in AD and AI LNCaP cells and the potential therapeutic value of inhibiting Sp1 to control AR overexpression and the androgen-independent phenotype of PC progression.
The results of this work show that Sp1 binds to known Sp1 binding sites in the promoter/5′-UTR of the AR gene and that the affinity is greater for binding ARSp1-3, a newly identified pair of Sp1 binding sites in the same region (Fig. 1, and 2A). The binding of Sp1 to these sites is specific (Fig. 2B–D) and serves to up-regulate transcription of AR in both AD and AI LNCaP cells but preferentially of the latter which displayed higher level of AR transcription activity than AD cells (Fig. 2D). This finding suggests that consistent with our previous observation that Sp1 binds to ARSp1-3 in vivo as measured by chromatin immunoprecipitation assay (ChIP), suggesting that ARSp1-3 might contribute to sustain the 4 fold higher levels of AR mRNA expression constitutively observed in the AI LNCaP cells (Wang, Ossowski et al. 2001). Overall, these results link deregulation of Sp1 activity as a mechanism of sustaining AR expression in PC cells and promoting over-expression of AR with androgen-independent progression. Given the critical role of AR in PC progression (Burnstein 2005; Yuan, Li et al. 2006), these findings suggest that Sp1 might be a therapeutic target that could be exploited to downregulate AR transcription, decrease proliferation and development of progressive resistance of AI cells to hormones and chemotherapy (Ren, Zhang et al. 2000; Yang, Fung et al. 2005; Yuan, Gong et al. 2005).
We confirmed that in PC cells, very low concentrations of Mithramycin are required to specifically inhibit binding of Sp1 to Sp1 binding sites in the AR promoter (Ryu, Zhou et al. 1999; Yoo, Jeong et al. 2002) and that it does not affect the binding of negative regulatory element such as NF1 (Fig.1) (Fig. 3D–E). Concordant with the loss of Sp1 binding, nanomolar concentration of Mithramycin reduced the transcription of AR in AI cells to the level of AD cells. The inhibition of transcription of AR was not abrogated by the presence of multiple regulatory elements in the promoter of AR (Fig. 3 A–C) and the levels of AR protein expression were also decreased (Fig. 4A). Consequently, the transactivation of downstream target genes like PSA was shut-off (Fig. 4B) and the expression of p21 which is completely repressed in AI LNCaP cells was released (Wang, Ossowski et al. 2001). This was a surprising observation since the promoter of p21 also contains active Sp1 binding sites which would be expected to be inhibited rather than induced by Mithramycin (Nakano, Mizuno et al. 1997; De Siervi, Marinissen et al. 2004).
The sensitivity of AI cells to suppression of AR transcription and AR protein expression and function by Mithramycin provides additional support to the tight link between AR and Sp1 activation making these cells especially vulnerable to inhibition of Sp1. We speculate that the remarkable efficacy of Mithramycin in suppressing AR transcription in these cells may also be due to the imbalance generated by the activity of the unaffected negative regulatory elements in the promoter of AR in the absence of Sp1 (Supakar, Jung et al. 1995). Therefore it is likely that tumor cells that do not express AR may not be equally responsive to Mithramycin.
Finally, we observed that the low concentrations of Mithramycin required for inhibiting Sp1 binding had an equivalent growth suppressive effect on AD and AI LNCaP cells. Moreover, Mithramycin was more effective than paclitaxel for AI cells and the combination was highly synergistic. Given the 7-fold reduction in the Dm of paclitaxel (from 1.7 nM to 0.234 nM), it is possible that the activity and duration of response to taxanes could be enhanced without increasing toxicity. Another remarkable observation was the equivalent and synergistic growth inhibition achieved with the combination of Mithramycin and bicalutamide on AI LNCaP cells compared to AD cells. This observation indicates that Mithramycin can reverse resistance of AI cells to anti-androgens and to prolong the response to hormonal therapy.
These preclinical studies provide support to a new therapeutic strategy of PC based on targeting Sp1 activity to reduce AR transcription and reverse resistance to hormones and/or enhance response to taxane based therapy. It provides the rationale for testing combinations of these agents in clinical trials of patients with androgen-independent, hormone refractory PC.
Acknowledgments
This work was supported by a Chemotherapy Foundation, New York.
References
- Anzick SL, Kononen J, et al. “AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer”. Science. 1997;277(5328):965–8. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Bonadonna G. “Chemotherapy of testicular tumors.”. Acta. Chir. Belg. 1971;70(4):393–400. [PubMed] [Google Scholar]
- Bruchovsky N, Rennie PS, et al. “Effects of androgen withdrawal on the stem cell composition of the Shionogi carcinoma.”. Cancer Res. 1990;50(8):2275–82. [PubMed] [Google Scholar]
- Burnstein KL. “Regulation of androgen receptor levels: implications for prostate cancer progression and therapy.”. J. Cell. Biochem. 2005;95(4):657–69. doi: 10.1002/jcb.20460. [DOI] [PubMed] [Google Scholar]
- Calabresi P, a. C.B.A. In: The Pharmacological basis of therapeutics. Goodman, Gilmann, editors. Macmillan; NY: 1991. pp. 1209–1263. [Google Scholar]
- Chen CD, Welsbie DS, et al. “Molecular determinants of resistance to antiandrogen therapy.”. Nat. Med. 2004;10(1):33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- Chou TC, Motzer RJ, et al. “Computerized quantitation of synergism and antagonism of taxol, topotecan, and cisplatin against human teratocarcinoma cell growth: a rational approach to clinical protocol design.”. J. Natl. Cancer Inst. 1994;86(20):1517–24. doi: 10.1093/jnci/86.20.1517. [DOI] [PubMed] [Google Scholar]
- Craft N, Shostak Y, et al. “A mechanism for hormone-independent prostate cancer through modulation of androgen receptor signaling by the HER-2/neu tyrosine kinase.”. Nat. Med. 1999;5(3):280–5. doi: 10.1038/6495. [DOI] [PubMed] [Google Scholar]
- de Ruiter PE, Teuwen R, et al. “Synergism between androgens and protein kinase-C on androgen-regulated gene expression.”. Mol. Cell. Endocrinol. 1995;110(1–2):R1–6. doi: 10.1016/0303-7207(95)03534-e. [DOI] [PubMed] [Google Scholar]
- De Siervi A, Marinissen M, et al. “Transcriptional Activation of p21(waf1/cip1) by Alkylphospholipids: Role of the Mitogen-Activated Protein Kinase Pathway in the Transactivation of the Human p21(waf1/cip1) Promoter by Sp1.”. Cancer Res. 2004;64(2):743–750. doi: 10.1158/0008-5472.can-03-2505. [DOI] [PubMed] [Google Scholar]
- Eder IE, Culig Z, et al. “Inhibition of LncaP prostate cancer cells by means of androgen receptor antisense oligonucleotides.”. Cancer Gene. Ther. 2000;7(7):997–1007. doi: 10.1038/sj.cgt.7700202. [DOI] [PubMed] [Google Scholar]
- Faber PW, King A, et al. “The mouse androgen receptor. Functional analysis of the protein and characterization of the gene.”. Biochem. J. 1991;278(Pt 1):269–78. doi: 10.1042/bj2780269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber PW, van Rooij HC, et al. “Characterization of the human androgen receptor transcription unit.”. J. Biol. Chem. 1991;266(17):10743–9. [PubMed] [Google Scholar]
- Ferrari AC, Chachoua A, et al. “A Phase I/II study of weekly paclitaxel and 3 days of high dose oral estramustine in patients with hormone-refractory prostate carcinoma”. Cancer. 2001;91(11):2039–45. [PubMed] [Google Scholar]
- Gaddipati JP, McLeod DG, et al. “Frequent detection of codon 877 mutation in the androgen receptor gene in advanced prostate cancers.”. Cancer Res. 1994;54(11):2861–4. [PubMed] [Google Scholar]
- Gao M, Ossowski L, et al. “Activation of Rb and decline in androgen receptor protein precede retinoic acid-induced apoptosis in androgen-dependent LNCaP cells and their androgen-independent derivative.”. J. Cell. Physiol. 1999;179(3):336–46. doi: 10.1002/(SICI)1097-4652(199906)179:3<336::AID-JCP11>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Gioeli D, Mandell JW, et al. “Activation of mitogen-activated protein kinase associated with prostate cancer progression.”. Cancer Res. 1999;59(2):279–84. [PubMed] [Google Scholar]
- Gregory CW, Hamil KG, et al. “Androgen receptor expression in androgen-independent prostate cancer is associated with increased expression of androgen-regulated genes.”. Cancer Res. 1998;58(24):5718–24. [PubMed] [Google Scholar]
- Grossmann ME, Tindall DJ. “The androgen receptor is transcriptionally suppressed by proteins that bind single-stranded DNA.”. J. Biol. Chem. 1995;270(18):10968–75. doi: 10.1074/jbc.270.18.10968. [DOI] [PubMed] [Google Scholar]
- Hobisch A, Eder IE, et al. “Interleukin-6 regulates prostate-specific protein expression in prostate carcinoma cells by activation of the androgen receptor.”. Cancer Res. 1998;58(20):4640–5. [PubMed] [Google Scholar]
- Horwitz KB, Jackson TA, et al. “Nuclear receptor coactivators and corepressors.”. Mol. Endocrinol. 1996;10(10):1167–77. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- Husmann DA, Wilson CM, et al. “Antipeptide antibodies to two distinct regions of the androgen receptor localize the receptor protein to the nuclei of target cells in the rat and human prostate”. Endocrinology. 1990;126(5):2359–68. doi: 10.1210/endo-126-5-2359. [DOI] [PubMed] [Google Scholar]
- Isaacs JT, Coffey DS. “Adaptation versus selection as the mechanism responsible for the relapse of prostatic cancer to androgen ablation therapy as studied in the Dunning R-3327-H adenocarcinoma.”. Cancer Res. 1981;41(12 Pt 1):5070–5. [PubMed] [Google Scholar]
- Jackson SP, MacDonald JJ, et al. “GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase”. Cell. 1990;63(1):155–65. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- Kallioniemi OP, Visakorpi T. “Genetic basis and clonal evolution of human prostate cancer.”. Adv. Cancer Res. 1996;68:225–55. doi: 10.1016/s0065-230x(08)60355-3. [DOI] [PubMed] [Google Scholar]
- Koivisto P, Hyytinen E, et al. “Analysis of genetic changes underlying local recurrence of prostate carcinoma during androgen deprivation therapy.”. Am. J. Pathol. 1995;147(6):1608–14. [PMC free article] [PubMed] [Google Scholar]
- Koivisto P, Kononen J, et al. “Androgen receptor gene amplification: a possible molecular mechanism for androgen deprivation therapy failure in prostate cancer.”. Cancer Res. 1997;57(2):314–9. [PubMed] [Google Scholar]
- Linja MJ, Savinainen KJ, et al. “Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer.”. Cancer Res. 2001;61(9):3550–5. [PubMed] [Google Scholar]
- Liu G, Xia T, et al. “The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CBP.”. J Biol Chem. 2003 doi: 10.1074/jbc.M210696200. [DOI] [PubMed] [Google Scholar]
- Liu XM, Jiang JD, et al. “Unique induction of p21(WAF1/CIP1)expression by vinorelbine in androgen-independent prostate cancer cells.”. Br. J. Cancer. 2003;89(8):1566–73. doi: 10.1038/sj.bjc.6601317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Junicho A, et al. “Cross-talk between signal transducer and activator of transcription 3 and androgen receptor signaling in prostate carcinoma cells.”. Biochem. Biophys. Res. Commun. 2001;283(1):179–87. doi: 10.1006/bbrc.2001.4758. [DOI] [PubMed] [Google Scholar]
- Mir MA, Dasgupta D. “Interaction of antitumor drug, mithramycin, with chromatin.”. Biochem. Biophys. Res. Commun. 2001;280(1):68–74. doi: 10.1006/bbrc.2000.4075. [DOI] [PubMed] [Google Scholar]
- Mir MA, Dasgupta D. “Association of anticancer drug mithramycin with H1-depleted chromatin: a comparison with native chromatin.”. J. Inorg. Biochem. 2003;94(1–2):72–7. doi: 10.1016/s0162-0134(02)00576-7. [DOI] [PubMed] [Google Scholar]
- Miyamoto H, Yeh S, et al. “Promotion of agonist activity of antiandrogens by the androgen receptor coactivator, ARA70, in human prostate cancer DU145 cells.”. Proc. Natl. Acad. Sci. U.S.A. 1998;95(13):7379–84. doi: 10.1073/pnas.95.13.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizokami A, Chang C. “Induction of translation by the 5′-untranslated region of human androgen receptor mRNA.”. J. Biol. Chem. 1994;269(41):25655–9. [PubMed] [Google Scholar]
- Nakano K, Mizuno T, et al. “Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line.”. J. Biol. Chem. 1997;272(35):22199–206. doi: 10.1074/jbc.272.35.22199. [DOI] [PubMed] [Google Scholar]
- Newmark JR, Hardy DO, et al. “Androgen receptor gene mutations in human prostate cancer.”. Proc. Natl. Acad. Sci. U.S.A. 1992;89(14):6319–23. doi: 10.1073/pnas.89.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofelia LZ-M, Schmidt LJ, Huang H, Tindall DJ. “Disruption of androgen receptor function inhibits proliferation of androgen refractory prostate cancer cells.”. Cancer Res. 2002;62:1008–1013. [PubMed] [Google Scholar]
- Ren F, Zhang S, et al. “Tea polyphenols down-regulate the expression of the androgen receptor in LNCaP prostate cancer cells”. Oncogene. 2000;19(15):1924–32. doi: 10.1038/sj.onc.1203511. [DOI] [PubMed] [Google Scholar]
- Ryu S, Zhou S, et al. “The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1”. Nature. 1999;397(6718):446–50. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- Sadar MD. “Androgen-independent induction of prostate-specific antigen gene expression via cross-talk between the androgen receptor and protein kinase A signal transduction pathways.”. J. Biol. Chem. 1999;274(12):7777–83. doi: 10.1074/jbc.274.12.7777. [DOI] [PubMed] [Google Scholar]
- Shi Y, Brands FH, et al. “Her-2/neu expression in prostate cancer: high level of expression associated with exposure to hormone therapy and androgen independent disease.”. J. Urol. 2001;166(4):1514–9. doi: 10.1016/s0022-5347(05)65822-3. [DOI] [PubMed] [Google Scholar]
- Signoretti S, Montironi R, et al. “Her-2-neu expression and progression toward androgen independence in human prostate cancer.”. J. Natl. Cancer Inst. 2000;92(23):1918–25. doi: 10.1093/jnci/92.23.1918. [DOI] [PubMed] [Google Scholar]
- Song CS, Jung MH, et al. “Negative regulation of the androgen receptor gene promoter by NFI and an adjacently located multiprotein-binding site.”. Mol. Endocrinol. 1999;13(9):1487–96. doi: 10.1210/mend.13.9.0350. [DOI] [PubMed] [Google Scholar]
- Sowa Y, Orita T, et al. “Histone deacetylase inhibitor activates the WAF1/Cip1 gene promoter through the Sp1 sites.”. Biochem. Biophys. Res. Commun. 1997;241(1):142–50. doi: 10.1006/bbrc.1997.7786. [DOI] [PubMed] [Google Scholar]
- Supakar PC, Jung MH, et al. “Nuclear factor kappa B functions as a negative regulator for the rat androgen receptor gene and NF-kappa B activity increases during the age-dependent desensitization of the liver.”. J. Biol. Chem. 1995;270(2):837–42. doi: 10.1074/jbc.270.2.837. [DOI] [PubMed] [Google Scholar]
- Takane KK, McPhaul MJ. “Functional analysis of the human androgen receptor promoter.”. Mol. Cell Endocrinol. 1996;119(1):83–93. doi: 10.1016/0303-7207(96)03800-2. [DOI] [PubMed] [Google Scholar]
- Tilley WD, Marcelli M, et al. “Expression of the human androgen receptor gene utilizes a common promoter in diverse human tissues and cell lines.”. J. Biol. Chem. 1990;265(23):13776–81. [PubMed] [Google Scholar]
- Tilley WD, Marcelli M, et al. “Recent studies of the androgen receptor: new insights into old questions.”. Mol. Cell. Endocrinol. 1990;68(2–3):C7–10. doi: 10.1016/0303-7207(90)90178-b. [DOI] [PubMed] [Google Scholar]
- Tilley WD, Wilson CM, et al. “Androgen receptor gene expression in human prostate carcinoma cell lines.”. Cancer Res. 1990;50(17):5382–6. [PubMed] [Google Scholar]
- Visakorpi T, Hyytinen E, et al. “In vivo amplification of the androgen receptor gene and progression of human prostate cancer.”. Nat. Genet. 1995;9(4):401–6. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- Wang LG, Liu XM, et al. “Differential binding of nuclear c-ets-1 protein to an intron I fragment of the c-myb gene in growth versus differentiation.”. Cell Growth Differ. 1994;5(11):1243–51. [PubMed] [Google Scholar]
- Wang LG, Liu XM, et al. “Down-regulation of prostate-specific antigen expression by finasteride through inhibition of complex formation between androgen receptor and steroid receptor-binding consensus in the promoter of the PSA gene in LNCaP cells.”. Cancer Res. 1997;57(4):714–9. [PubMed] [Google Scholar]
- Wang LG, Ossowski L, et al. “Overexpressed androgen receptor linked to p21WAF1 silencing may be responsible for androgen independence and resistance to apoptosis of a prostate cancer cell line.”. Cancer Res. 2001;61(20):7544–51. [PubMed] [Google Scholar]
- Wang LG, Ossowski L, et al. “Androgen receptor level controlled by a suppressor complex lost in an androgen-independent prostate cancer cell line”. Oncogene. 2004;23(30):5175–84. doi: 10.1038/sj.onc.1207654. [DOI] [PubMed] [Google Scholar]
- Wen Y, Hu MC, et al. “HER-2/neu promotes androgen-independent survival and growth of prostate cancer cells through the Akt pathway.”. Cancer Res. 2000;60(24):6841–5. [PubMed] [Google Scholar]
- Yang Q, Fung KM, et al. “Androgen receptor signaling is required for androgen-sensitive human prostate cancer cell proliferation and survival.”. Cancer Cell. Int. 2005;5(1):8. doi: 10.1186/1475-2867-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TH, Tsai WH, et al. “Purification and characterization of nucleolin and its identification as a transcription repressor.”. Mol. Cell Biol. 1994;14(9):6068–74. doi: 10.1128/mcb.14.9.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J, Jeong MJ, et al. “Activation of dynamin I gene expression by Sp1 and Sp3 is required for neuronal differentiation of N1E-115 cells.”. J. Biol. Chem. 2002;277(14):11904–9. doi: 10.1074/jbc.M111788200. [DOI] [PubMed] [Google Scholar]
- Yoshida BA, Dubauskas Z, et al. “Mitogen-activated protein kinase kinase 4/stress-activated protein/Erk kinase 1 (MKK4/SEK1), a prostate cancer metastasis suppressor gene encoded by human chromosome 17.”. Cancer Res. 1999;59(21):5483–7. [PubMed] [Google Scholar]
- Yuan H, Gong A, et al. “Involvement of transcription factor Sp1 in quercetin-mediated inhibitory effect on the androgen receptor in human prostate cancer cells”. Carcinogenesis. 2005;26(4):793–801. doi: 10.1093/carcin/bgi021. [DOI] [PubMed] [Google Scholar]
- Yuan X, Li T, et al. “Androgen Receptor Remains Critical for Cell-Cycle Progression in Androgen-Independent CWR22 Prostate Cancer Cells.”. Am. J. Pathol. 2006;169(2):682–96. doi: 10.2353/ajpath.2006.051047. [DOI] [PMC free article] [PubMed] [Google Scholar]