Highlights
► Recombinant expression of a new type of Arabidopsis F-box protein. ► Arabidopsis F-box-Nictaba protein is a functional lectin. ► Arabidopsis F-box-Nictaba recognizes N-acetyllactosamine and Lewis structures.
Keywords: Carbohydrate-binding, Glycan array, Lewis structure, Plant lectin, Recombinant protein expression
Abbreviations: Fbs, sugar-binding F-box protein; LacNAc, N-acetyllactosamine; Nictaba, Nicotiana tabacum agglutinin
Abstract
The Arabidopsis thaliana genome contains a small group of bipartite F-box proteins, consisting of an N-terminal F-box domain and a C-terminal domain sharing sequence similarity with Nictaba, the jasmonate-induced glycan-binding protein (lectin) from tobacco. Based on the high sequence similarity between the C-terminal domain of these proteins and Nictaba, the hypothesis was put forward that the so-called F-box-Nictaba proteins possess carbohydrate-binding activity and accordingly can be considered functional homologs of the mammalian sugar-binding F-box or Fbs proteins which are involved in proteasomal degradation of glycoproteins. To obtain experimental evidence for the carbohydrate-binding activity and specificity of the A. thaliana F-box-Nictaba proteins, both the complete F-box-Nictaba sequence of one selected Arabidopsis F-box protein (in casu At2g02360) as well as the Nictaba-like domain only were expressed in Pichia pastoris and analyzed by affinity chromatography, agglutination assays and glycan micro-array binding assays. These results demonstrated that the C-terminal Nictaba-like domain provides the F-box-protein with a carbohydrate-binding activity that is specifically directed against N- and O-glycans containing N-acetyllactosamine (Galβ1-3GlcNAc and Galβ1-4GlcNAc) and poly-N-acetyllactosamine ([Galβ1-4GlcNAc]n) as well as Lewis A (Galβ1-3(Fucα1-4)GlcNAc), Lewis X (Galβ1-4(Fucα1-3)GlcNAc, Lewis Y (Fucα1-2Galβ1-4(Fucα1-3)GlcNAc) and blood type B (Galα1-3(Fucα1-2)Galβ1-3GlcNAc) motifs. Based on these findings one can reasonably conclude that at least the A. thaliana F-box-Nictaba protein encoded by At2g02360 can act as a carbohydrate-binding protein. The results from the glycan array assays revealed differences in sugar-binding specificity between the F-box protein and Nictaba, indicating that the same carbohydrate-binding motif can accommodate unrelated oligosaccharides.
1. Introduction
The discovery of Nictaba, a jasmonate-induced lectin in tobacco (Nicotiana tabacum) leaves, made an important contribution to plant glycobiology during the last decade [1,2]. For the first time evidence was presented that application of one plant hormone induced expression of a specific lectin in plants. In addition, the identification of Nictaba as a cytoplasmic/nuclear lectin preferentially interacting with N-acetylglucosamine (GlcNAc) oligomers put the search for the physiological role of plant lectins in a new perspective. Subsequent in silico analyses revealed that the 165 amino acid residue subunit of the Nictaba homodimer represents a structural (carbohydrate-binding) motif that might well be ubiquitous in terrestrial plants. Since all evidence indicates that this motif covers the entire tobacco lectin subunit it will further be referred to as the ‘Nictaba domain’.
True orthologs/homologs consisting of only Nictaba domain(s) are not widespread but numerous chimera proteins could be identified which possess a Nictaba domain fused to unrelated N- and C-terminal domains. Moreover, in silico analyses revealed that all currently sequenced plant genomes contain genes encoding proteins with a Nictaba-like domain either or not fused to an unrelated domain [3]. For some of the expressed proteins, evidence could be presented that they possess lectin activity owing to their Nictaba domain. For example, the well documented family of phloem lectins from Cucurbitaceae, also known as the PP2 proteins [4,5], groups proteins with an active GlcNAc-binding Nictaba-like domain linked to an undefined N-terminal domain and a short cysteine-rich C-terminal peptide. The PP2-A1 protein from Arabidopsis is composed of N-terminal domain followed by a Nictaba domain and has been shown to exhibit a carbohydrate-binding specificity similar to Nictaba [6].
A comprehensive analysis of the Arabidopsis genome/transcriptome revealed the occurrence of chimera proteins in which a Nictaba-like domain is C-terminally fused to an F-box domain [7–9]. F-box domains are conserved, structural motifs of 40–50 amino acid residues commonly used in F-box proteins as a protein–protein interaction domain involved in the direct binding of F-box proteins to the protein Skp1 [10]. Complexes consisting of Skp, Cullin and an F-box protein (SCF complexes) play a role in ubiquitin labeling of proteins destined for proteasomal degradation [11]. Within F-box proteins, F-box domains are mostly used in concert with other (C-terminal) protein–protein interaction domains such as leucine-rich repeats or WD repeats. Based on this bipartite structure, F-box proteins can assemble into functional SCF complexes and recruit proteins that are destined for degradation to the complex for ubiquitin labeling [12].
Although these F-box-Nictaba like sequences were first identified in Arabidopsis, homologs occur in all plant genomes sequenced thus far. Moreover, most plant transcriptomes comprise sequences encoding F-box-Nictaba chimers indicating that this group of proteins is widely expressed in plants [9]. Though not conclusive the apparently ubiquitous occurrence and expression of the genes indicate that the F-box-Nictaba proteins might fulfill a universal role in plants. Due to the presence of the F-box domain it is tempting to hypothesize that the F-box-Nictaba proteins are somehow involved in a proteasomal protein degradation process. This hypothesis follows the concept that the F-box-Nictaba proteins can be considered functional homologs of the mammalian Fbs proteins. These mammalian Fbs proteins also consist of an N-terminal F-box domain and a C-terminal carbohydrate-binding domain (unrelated to Nictaba), and are known to play a role in cytosolic proteasomal degradation of glycoproteins [13]. For the F-box-Nictaba protein under study, it has even been shown that it can interact with Skp1 homologs expressed in Arabidopsis [14].
To test the hypothesis of being functional homologs of the mammalian Fbs proteins the presumed carbohydrate-binding activity of the Nictaba domain of the plant F-box-lectin needs to be supported by experimental evidence. Sequence alignments indicated that the deduced amino acid sequences of the Nictaba domain of Arabidopsis thaliana F-box-Nictaba proteins share approximately 40% overall sequence similarity with Nictaba itself [9]. However, since this cannot guarantee that the carbohydrate-binding activity and specificity are conserved the lectin activity of the Arabidopsis F-box-Nictaba protein encoded by At2g02360 was investigated in detail. The At2g02360 sequence was selected because it shares the highest sequence similarity (64%) to Nictaba. To corroborate the carbohydrate-binding properties of this F-box-Nictaba protein the recombinant full length protein as well as the composing Nictaba domain were produced in Pichia pastoris and subsequently the recombinant proteins were purified and characterized. Here we show that both the entire F-box-Nictaba protein and its Nictaba domain possess carbohydrate-binding activity and exhibit virtually the same specificity towards N- and O-glycans containing N-acetyllactosamine (LacNAc) (Galβ1-3GlcNAc and Galβ1-4GlcNAc) and poly-LacNAc ([Galβ1-4GlcNAc]n) as well as Lewis A (Galβ1-3(Fucα1-4)GlcNAc), Lewis X (Galβ1-4(Fucα1-3)GlcNAc, Lewis Y (Fucα1-2Galβ1-4(Fucα1-3)GlcNAc) and blood type B (Galα1-3(Fucα1-2)Galβ1-3GlcNAc) motifs. This specificity differs from the preferential binding of the tobacco lectin to high-mannose N-glycans [15,16] which illustrates once more that the same structural motif can accommodate different oligosaccharides depending on the atomic structure of the lectin binding site(s).
2. Materials and methods
2.1. Construction of expression vectors
Cloning and heterologous expression of recombinant proteins were performed using the EasySelect™ Pichia Expression Kit from Invitrogen (Invitrogen, Carlsbad, CA, USA). The full-length cDNA template for At2g02360 (BX820545) was ordered from the INRA (Institut National de la Recherche Agronomique), Centre de Toulouse, Unité de Recherche 1258-CNRGV (Centre National de Ressources Génomiques Végétales) (Castanet-Tolosan Cedex, France). The full-length F-box-Nictaba cDNA sequence was amplified by PCR using the forward primer evd 553 (5′-GGACACGTGGGGGAGAAAACGCAGAGTTAAATCGG-3′) and a reverse primer evd 554 (5′-GCTTCCGCGGCGAGGATTTTAGCAGGTCGGATTTC-3′) under the following PCR conditions: 2 min denaturation at 95 °C, 30 cycles of 15 s −95 °C, 30 s −60 °C, 1 min 20 s −72 °C, additional 5 min elongation at 72 °C. Amplification of the Nictaba domain alone was achieved with forward primer evd 360 (5′-GGCGGAGAATTCAGCGTATGGTTAGAGAAAGCGAGTGGG-3′) and reverse primer evd 359 (5′-CCCGCTTGCGGCCGCGAGGATTTTAGCAGGTCGGATTTC-3′) using the following PCR conditions: 2 min denaturation at 94 °C, 25 cycles of 15 s – 94 °C, 30 s – 50 °C, 1 min – 72 °C, additional 5 min elongation at 72 °C. The amplified sequences were double digested with the restriction enzymes PmlI and SacII (Fermentas, St. Leon-Rot, Germany) and the resulting fragment was cloned in the shuttle vector pPICZαA. After transformation into E. coli Top10F cells using heat shock transformation E. coli transformants were selected on LB agar plates containing zeocin (100 μg/mL) and checked by colony PCR using 5′ and 3′-AOX1 specific primers (forward evd 21, 5′-GACTGGTTCCAATTGACAAGC-3′ and reverse evd 22, 5′-GCAAATGGCATTCTGACATCC-3′). PCR conditions were as follows: 12 min at 94 °C, 30 cycles of 15 s – 94 °C, 30 s –48 °C, 1 min – 72 °C, additional 5 min elongation at 72 °C. Plasmids from transformed E. coli colonies were purified using the E.Z.N.A. Plasmid Mini kit I (Omega Bio-Tek, Norcross, GA, USA). Afterwards, the sequences of the fusion constructs were verified by DNA sequencing (LGC Genomics, Berlin, Germany).
2.2. Pichia pastoris transformation and expression analysis
The plasmid DNA from selected E. coli cells was purified using the NucleoBond® Xtra Midi kit (Macherey-Nagel, Düren, Germany) and linearized using the restriction enzyme PmeI (plasmid construct for full-length F-box-Nictaba) or SacI (plasmid construct for Nictaba domain) (Fermentas) with overnight incubation at 37 °C. Subsequently, P. pastoris competent cells were transformed with the linearized expression vector using electroporation (Bio-Rad, Hercules, CA, USA) with the following pulse settings: 25 μF, 1.5 kV and 125 Ω. Transformants were selected on YPDS plates (1% yeast extract, 2% peptone, 2% dextrose, 1 M sorbitol, 2% agar) containing 300 μg/ml zeocin.
For expression analysis, several colonies were inoculated in 5 mL BMGY medium containing 1% yeast extract, 2% peptone, 1.34% yeast nitrogen base with ammonium sulfate and without amino acids, 4 × 10−5% biotin, 100 mM potassium phosphate (pH 6.0), 1% glycerol and 100 μg/mL zeocin. Cultures were grown at 22 °C in a shaker at 220 rpm. After 24 h of incubation Pichia cells were washed twice with sterilized water and transferred to 10 mL BMMY medium (BMGY medium containing 1% of methanol instead of 1% of glycerol). Cultures were grown for 4 days under the same conditions as before. Induction of recombinant protein expression was achieved by adding methanol twice a day (once in the morning and once in the evening) until a final concentration of 1%. Proteins in the culture medium were precipitated with trichloroacetic acid (10% final concentration) and analyzed by SDS–PAGE and Western blotting.
For expression of the recombinant proteins in large cultures transformed P. pastoris colonies were inoculated into 10 mL BMGY medium and grown for 24 h at 22 °C in a shaker at 220 rpm. Afterwards, cultures were transferred to 50 mL BMGY in 250 mL Erlenmeyer flasks and allowed to grow until the culture reached an optical density between 2 and 6 at 600 nm. Cells were then washed twice with sterilized water and resuspended in 250 mL of BMMY medium in 1L Erlenmeyer flasks. Cultures were grown for 3 days under the same conditions as before. Induction of recombinant protein expression was done by adding methanol twice a day until a final concentration of 1%. After 3 days of methanol induction, cultures were centrifuged for 10 min at 3000g and the culture medium was collected. Proteins from the culture medium were precipitated using ammonium sulfate at a final concentration of 80%. The culture medium with precipitated proteins was stored at 4 °C until use.
2.3. Purification of recombinant proteins
Purification of recombinant proteins was achieved by 5 chromatographic steps. The culture medium with precipitated proteins was centrifuged at 17 000g for 25 min and the resulting pellet was resuspended in phosphate buffered saline. After adjusting the ammonium sulfate concentration to 1 M and setting the pH to 7, the protein solution was loaded on a phenyl Sepharose column (GE Healthcare, Uppsala, Sweden) equilibrated with 1 M ammonium sulfate (pH 7). The column was washed with 1 M ammonium sulfate (pH 7) and eluted with 20 mM 1,3-diaminopropane. This eluted protein fraction was then loaded on a Q Fast Flow column equilibrated with 20 mM 1,3-diaminopropane. After washing with equilibration buffer bound proteins were eluted using 0.5 M NaCl/100 mM Tris–HCl (pH 8.7) buffer. After adding imidazole to a final concentration of 25 mM, the eluate from the Q Fast Flow column was applied on a Ni-Sepharose column (GE Healthcare) equilibrated with 0.5 M NaCl/25 mM imidazole/100 mM Tris–HCl (pH 8,7) buffer to purify the His6-tagged protein. The column was washed using the equilibration buffer and then stepwise elution of bound proteins was performed with 0.5 M NaCl/100 mM Tris–HCl (pH 8.7) buffer with increasing imidazole concentrations ranging from 50 to 250 mM imidazole. Fractions eluted from the Ni-Sepharose column were pooled and 1 M ammonium sulfate was added. After adjusting the solution to pH 7 the eluate was loaded on an ovomucoid-conjugated Sepharose 4B column equilibrated with 1 M ammonium sulfate (pH 7.0). After washing the ovomucoid-conjugated column with 1 M ammonium sulfate (pH 7.0), the bound proteins were eluted using 20 mM 1,3-diaminopropane. Finally this protein fraction was concentrated on a small Q Fast Flow column. The purified proteins were eluted into 0.5 M NaCl/20 mM 1,3-diaminopropane (pH 8.7) buffer.
The purity of the protein samples after each purification step was verified by SDS–PAGE and Western blot analysis. SDS–PAGE was performed using 15% polyacrylamide gels under reducing conditions [17]. Proteins were visualized by gel staining with Coomassie Brilliant Blue R-250. For Western blotting, samples separated by SDS–PAGE were electrotransferred to 0.45 μm polyvinylidene fluoride membranes (BiotraceTM PVDF, PALL, Gelman Laboratory, Ann Arbor, MI, USA). Membranes were blocked with 5% (w/v) milk powder in Tris-buffered saline (TBS: 150 mM NaCl, 10 mM Tris, 0.1% (v/v) Triton X-100, pH 7.6). Afterwards blots were incubated for 1 h with a mouse monoclonal anti-His (C-terminal) antibody (Invitrogen) diluted 1/5000 in TBS, washed 3 times in TBS and finally incubated with the 1/1000 diluted rabbit anti-mouse IgG secondary antibody labeled with horseradish peroxidase (Dako Cytomation, Glostrup, Denmark). Immunodetection was performed using a colorimetric assay with 3,3′-diaminobenzidine tetrahydrochloride (Sigma–Aldrich, St. Louis, MO, USA) as a substrate. All washes and incubations were conducted at room temperature on a platform with gentle shaking.
2.4. Agglutination assays
Trypsin-treated rabbit red blood cells (Bio-Mérieux, Marcy l’Etoile, France) were used to check the lectin activity of purified recombinant proteins. Assays using purified protein fractions (0.5 mg/mL) were performed as described by [18].
2.5. N-terminal sequence analysis
Samples of purified recombinant F-box-Nictaba protein and its Nictaba domain alone were analyzed by SDS-PAGE, electroblotted onto a BioTrace™ polyvinylidene fluoride membrane (Gelman Laboratory) and stained with a 1:1 mix of Coomassie Brilliant Blue and methanol. Protein bands were excised from the membrane and the N-terminal sequence was determined by Edman degradation on a capillary Procise 491cLC protein sequencer without alkylation of cysteines (Applied Biosystems).
2.6. Glycan array screening
The printed microarrays are described previously by [19]. Printed array version 5.0 was used for the analyses reported here (http://www.functionalglycomics.org/static/consortium/resources/resourcecoreh8.shtml). Analyses were performed as described by [18]. The complete primary data set for each protein is available on the website of the Consortium for Functional Glycomics (http://www.functionalglycomics.org).
3. Results
3.1. Trp residues essential for glycan binding are conserved in the Arabidopsis F-box-Nictaba protein
Sequence alignments revealed that the Nictaba domain of the Arabidopsis F-box-Nictaba protein At2g02360 (AtPP2-B10) exhibits 52% and 64% sequence identity and similarity at the amino acid level, respectively, with the Nictaba sequence deduced from the cDNA encoding the N. tabacum leaf lectin [1]. According to a recently reported three-dimensional model for Nictaba [16], an electronegatively charged region on the protein surface was predicted as the glycan-binding site. Mutational analyses of two tryptophan residues (Trp15 and Trp22) located in the N-terminal half of the Nictaba domain revealed that these two residues are essential for glycan binding activity of the tobacco lectin since replacement of these Trp residues by Leu completely abolished the interaction of the mutant protein with the glycan array. Interestingly, sequence alignments of the Nictaba domain of At2g02360 and the tobacco lectin itself (Fig. 1) revealed that these Trp residues (Trp118 and Trp125 in At2g02360) are highly conserved in the F-box protein suggesting potential lectin activity for the Arabidopsis F-box-Nictaba protein with a similar sugar specificity as the one reported for the lectin from tobacco.
Fig. 1.
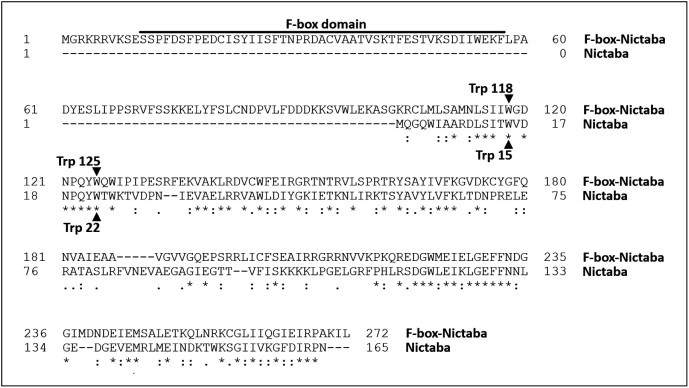
Sequence alignment of Nictaba and the Arabidopsis F-box-Nictaba protein.
3.2. Expression of recombinant Arabidopsis F-box-Nictaba protein in Pichia pastoris
To corroborate the glycan-binding activity and specificity of At2g02360 both the holoprotein (AA residues 1–272) and the composing Nictaba domain (AA residues 95–272) were expressed in the yeast P. pastoris. Sequences corresponding to the full-length protein and the Nictaba domain, respectively, were amplified by PCR and cloned in the pPICZαA vector, which was subsequently transferred into P. pastoris strain KM71H. SDS–PAGE analysis followed by Western blotting of crude protein extracts confirmed the successful expression of the His-tagged full-length protein as well as of the Nictaba domain in Pichia (Fig. 2A,B). The Pichia strains with the highest expression levels were selected and further used for the production of the recombinant proteins in large scale fed batch cultures. Both recombinant proteins were purified using a combination of ion exchange chromatography, metal affinity chromatography on a Ni-Sepharose column and affinity chromatography on an ovomucoid-Sepharose 4B matrix. SDS–PAGE and Western blot analysis confirmed the purity of the protein preparations (Fig. 2C,D). Yields amounted to approximately 250 μg and 500 μg per liter culture for the recombinant full-length F-box protein and recombinant Nictaba-like domain, respectively. The molecular mass of the recombinant F-box-Nictaba was estimated 34.9 kDa, which is in good agreement with the calculated molecular mass of 35 kDa of the recombinant protein (including a c-myc epitope and a His-tag). Unlike the recombinant F-box-Nictaba, which yielded a single polypeptide of approximately 35 kDa, the purified recombinant Nictaba domain yielded two major polypeptides of 26.4 and 24.3 kDa, respectively. N-terminal amino acid sequencing of the recombinant Nictaba domain yielded the sequence EAEAEFSVXLEEA with 85% sequence identity to the N-terminus of the recombinant protein expressed in Pichia and revealed that the higher molecular mass of the 26.4 kDa polypeptide is due to an incomplete removal of the N-terminal secretion sequence needed to direct the expressed proteins into the culture medium.
Fig. 2.
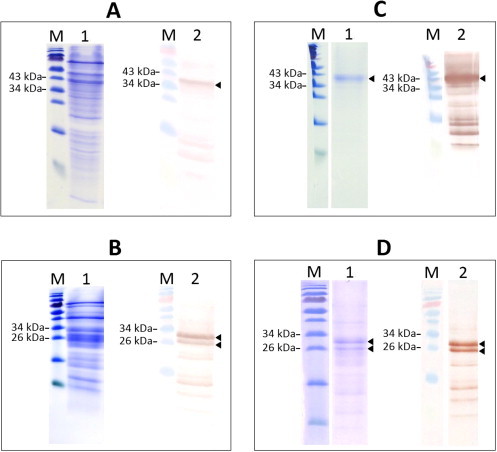
SDS/PAGE and Western blot pictures of unpurified and purified protein fractions M: protein marker (Fermentas, PageRuler, prestained); lane 1 for each panel: SDS/PAGE image; lane 2 for each panel: Western blot image. Panel A: Crude protein extract from Pichia pastoris culture medium containing recombinant F-box-Nictaba protein (50 μg/lane). Panel B: Crude protein extract from Pichia pastoris culture medium containing recombinant Nictaba domain from F-box-Nictaba protein (50 μg/lane). Panel C: Purified recombinant F-box-Nictaba protein (15 μg/lane). Panel D: Purified recombinant Nictaba domain from F-box-Nictaba protein (15 μg/lane).
The protein patterns shown in Fig. 2A–D suggest partial degradation of the recombinant proteins. Nevertheless these polypeptides could be detected using specific anti-His antibodies after purification on the ovomucoid column. Taken into account that the smaller protein polypeptides were also bound to the ovomucoid column these polypeptides also exhibit carbohydrate-binding activity.
3.3. Recombinant At2g02360 and its Nictaba domain bind N- and O-glycans carrying LacNAc structures
The fact that both recombinant proteins could be isolated by affinity chromatography on immobilized ovomucoid, a highly glycosylated protein carrying high-mannose N-glycans, already indicated that they possess carbohydrate-binding activity. Furthermore agglutination assays with the purified proteins also yielded a positive reaction in a test with rabbit erythrocytes (Results not shown). The glycan-binding activity of both recombinant proteins as well as of native Nictaba recombinantly expressed and purified before [16] was determined using the printed glycan array_v5.0 available from the Consortium for Functional Glycomics (USA) containing over 600 purified and synthesized glycan structures [19]. Fig. 3 summarizes the top 30 glycan structures most significantly (%CV < 40%) bound by the three proteins. A complete overview of the results for all tested glycan structures is available as Supplementary data (Table A). As shown in Fig. 3, the Nictaba domain as well as the full-length F-box-Nictaba protein showed interaction towards both N- and O-glycans containing type 1 and type 2 LacNAc (Galβ1-3GlcNAc and Galβ1-4GlcNAc) and poly-LacNAc type 2 ([Galβ1-4GlcNAc]n) structures as well as Lewis A (Galβ1-3(Fucα1-4)GlcNAc), Lewis X (Galβ1-4(Fucα1-3)GlcNAc), Lewis Y (Fucα1-2Galβ1-4(Fucα1-3)GlcNAc) and blood type B (Galα1-3(Fucα1-2)Galβ1-3GlcNAc) epitopes. It is apparent that the presence of the F-box domain does not hamper the glycan-binding capacity of the lectin domain since the glycan interaction profile of the full-length protein is very similar to the one obtained for the lectin domain. Therefore, the Nictaba domain present in the Arabidopsis At2g02360 protein can be considered a functional lectin domain which exhibits specificity for the Gal-GlcNAc sequence, both in a β1-3 and a β1-4 linkage. Previously the lectin from tobacco plants was shown to preferentially interact with GlcNAc oligomers and high-mannose N-glycans when tested on the first generation arrays (v2.1, v3 and v4) (Table A and [15,16]). A new screening using the latest available array v5 showed that Nictaba from tobacco is also able to recognize high-mannose glycans containing type 2 LacNAc structures, however at lower levels compared to the Nictaba domain from the Arabidopsis F-box-Nictaba protein under study.
Fig. 3.
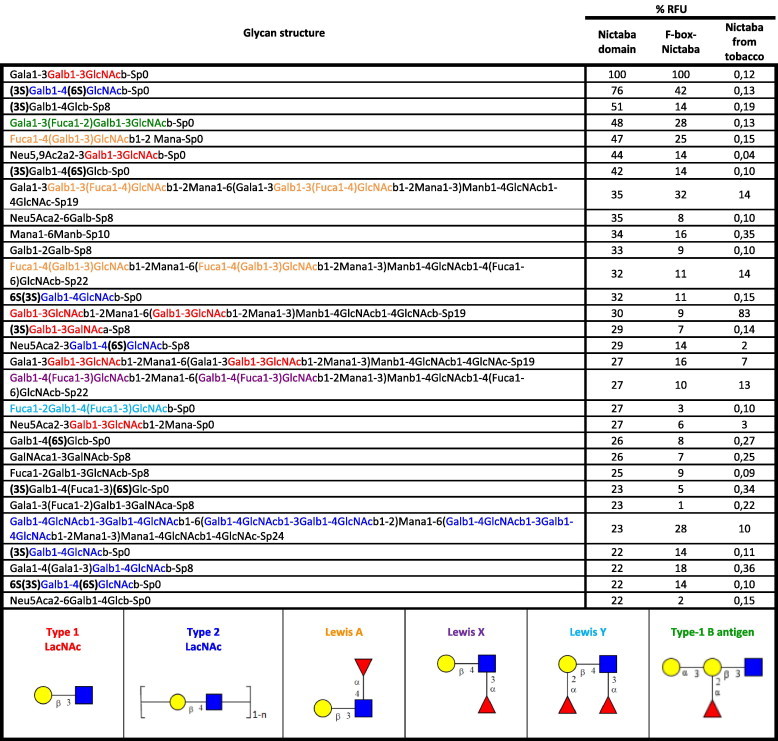
Overview of the top 30 glycan structures with highest reactivity on the glycan array for Nictaba domain of F-box-Nictaba from Arabidopsis thaliana. The most recurrent structures are colored as follows: type 1 LacNAc motifs – red, type 2 LacNAc motifs – dark blue, Lewis A structures – orange, Lewis X structures – violet, Lewis Y structures – blue and type-1 B antigen structures – green. The measurements on the right show the percentage relative fluorescence units (%RFU) of a glycan compared to the RFU of the glycan with the highest signal for each experiment. Nictaba from tobacco was analyzed using glycan array version 5.0.
4. Discussion
Judging from the results of the ovomucoid affinity chromatography, the agglutination assays and the glycan array analysis it can be concluded that the Arabidopsis F-box-Nictaba protein encoded by At2g02360 is a functional lectin. Parallel experiments with the Nictaba domain demonstrated that the lectin activity of At2g02360 resides in its C-terminal Nictaba domain. Based on the high sequence similarity between the C-terminal Nictaba domain of the Arabidopsis protein At2g02360 and Nictaba from tobacco leaves it was tempting to speculate that the F-box-Nictaba protein exhibited the same or at least similar glycan-binding properties as Nictaba and accordingly would interact preferentially with GlcNAc oligomers, and high-mannose and complex N-glycans. However, glycan array analysis clearly demonstrated that the Arabidopsis F-box-Nictaba protein exhibits a substantially different carbohydrate-binding specificity, recognizing type 1 and type 2 LacNAc, type 2 poly-LacNAc, Lewis A, Lewis X, Lewis Y, and type-1 B antigen motifs.
4.1. Occurrence of LacNAc motifs
In the last few years LacNAc structures have been studied most intensively in higher animals where they are responsible for blood group determination, cell-to-cell recognition and adhesion processes [20]. Different glycans containing LacNAc motifs have been also found in bacteria and viruses [21–23]. However, in plants only Lewis A motifs have been identified so far. These structures are localized at the cell surface (membrane-bound) or can be found in glycoproteins secreted by plant cells or in the Golgi apparatus where they are synthesized [24–26]. Although Lewis A structures are widespread within the plant kingdom including monocots, dicots and gymnosperms there has been some controversy to the presence of Lewis A motifs in A. thaliana and other members of the Brassicaceae family [24,25,27,28]. Nevertheless, it has been shown unambiguously that A. thaliana contains the indispensable enzymatic machinery and can synthesize Lewis A structures. Léonard et al. [29] demonstrated that A. thaliana possesses an active α1,4-fucosyltransferase (FUT13) capable of α1,4-fucosylation of the GlcNAc residue within the Galβ1-3GlcNAc structures. These Lewis A epitopes were detected in the plasma membrane and the Golgi vesicles of A. thaliana cells. Furthermore, Strasser et al. [30] reported the presence and activity of the β1,3-galactosyltransferase (GALT1) in A. thaliana responsible for the formation of Galβ1-3GlcNAc, a structure required for the synthesis of Lewis A structures. It has also been shown that the expression of Lewis A motifs in A. thaliana is tissue specific with relatively high levels in pedicels, stems and nodes, moderate levels in siliques and shoot apex, and relatively low levels in flowers and roots, whereas it was not detectable in leaves, which could explain previous problems in the detection of this glycan motif. Therefore, it seems evident that A. thaliana synthesizes Lewis A structures, however, most probably at substantially lower levels than other plants and/or the expression of the glycosyltransferases involved (i.e. β1,3-galactosyltransferase and α1,4-fucosyltransferase) is tissue and/or time specific. Unlike the type 1 LacNAc (Galβ1-3GlcNAc) and Lewis A motifs, the other structures recognized by the Arabidopsis F-box-Nictaba protein on the glycan array have not been reported in plants. However, this is not surprising since plants are missing the gene encoding β1,4-galactosyltransferase, the enzyme crucial for the biosynthesis of Galβ1-4GlcNAc structures [31]. As a consequence neither type 2 poly-LacNAc nor Lewis X epitopes can be synthesized by plants. Several research groups have shown that transgenic lines in which the human or rat β1,4-galactosyltransferase gene was introduced into the plant genome are capable of producing the mammalian-type Lewis structures [32,33]. Similarly, up till now no structures related to blood group B antigens could be identified in plants, which most probably also results from the lack of the necessary enzyme, the α1,3-galactosyltransferase. Clearly, no putative α1,3-galactosyltransferase gene has been identified in the genome of A. thaliana.
4.2. Relevance of plant lectin F-box-Nictaba recognizing LacNAc motifs
This is the first report of the characterization of a sugar-binding F-box protein in plants. Sugar-binding F-box proteins are already known since 2002, when they were first discovered in mammals [13]. Unlike the plant Fbs family, the mammalian Fbs protein family is a very small group consisting of only 5 homologous proteins, referred to as FBG1-FBG5. FBG1 and FBG2 exhibit carbohydrate-binding specificity towards high-mannose N-glycans similar to the glycan specificity reported for Nictaba [34,35]. Both FBG1 and FBG2 have been proposed to play a role in protein quality control by recognizing and targeting misfolded or incompletely assembled glycoproteins for degradation through the ER-associated degradation pathway [13]. Based on the striking similarities between the Fbs proteins and Nictaba for what concerns their localization pattern in the nucleus and cytoplasm of the cell, the three-dimensional conformation of the lectin domain and the carbohydrate-binding properties and the fact that F-box-Nictaba has been reported before to bind Skp1 [14], the hypothesis was put forward that the nucleocytoplasmic Arabidopsis F-box-Nictaba protein might be a functional homolog of the mammalian FBG1 and FBG2 proteins in plants [9]. However, the glycan arrays show that the plant F-box-Nictaba protein encoded by At2g02360 recognizes and binds type 1 LacNAc and Lewis A structures as well as type 2 poly-LacNAc, Lewis X and Y epitopes and blood group B antigens. As such, the sugar-binding specificity of the plant Fbs protein resembles better the one reported for the mammalian Fbs proteins FBG4 and FBG5, which exhibit strong affinity towards sulfated glycan structures and different glycans with type 2 LAcNAc (Galβ1-4GlcNAc) motifs (see Table A). Table A gives an overview of the glycan-binding properties of the tobacco lectin and all Fbs members published to date. Comparative analysis shows that within the mammalian Fbs protein family there is a wide divergence in carbohydrate-binding activity. While the mammalian Fbs protein FBG1 is highly specific to high-mannose N-glycans, FBG2 strongly binds not only high-mannose but also complex N-glycans and sulfated glycan structures. FBG3 does not exhibit carbohydrate-binding activity at all. Hence, although the mammalian Fbs proteins share high sequence similarity, they differ substantially in their glycan-binding properties. Due to the diversity in carbohydrate-binding specificity the different Fbs proteins are suggested to play divergent roles in the glycome regulation in mammals [35]. Likewise, it seems that the carbohydrate-binding site of F-box-Nictaba has developed a different specificity from the one of the Nictaba protein from tobacco, despite the similarity at the sequence level.
Taken into account that the Arabidopsis genome contains a whole family of homologous F-box-Nictaba proteins which slightly differ in the sequences of the Nictaba domains it is likely that these chimera proteins show broad differences in their fine specificities. Therefore it cannot be excluded that other plant F-box-Nictaba proteins might have carbohydrate-binding properties that are more similar to that of Nictaba.
4.3. Physiological significance
While it is obvious that type 1 LacNAc and Lewis A motifs can be synthesized by plants (including A. thaliana) a few questions still arise in view of the potential function of F-box-Nictaba in Arabidopsis. First, what is the role of these LAcNAc structures in plant physiology. In mammals Lewis A epitopes are involved in the cell-to-cell recognition, in selectin-dependent cell adhesion processes and in interactions with pathogens [20]. Thus, by analogy, it could be hypothesized that Lewis A motifs present at the plant cell surface might also be involved in cell-to-cell communication or in plant-pathogen interactions. Moreover, secretion of glycoproteins containing Lewis A epitopes could suggest a putative role in stress signaling [25,26,29]. Second, it remains to be elucidated what is the function and significance of the recognition of these structures by F-box-Nictaba protein in A. thaliana. In order to answer this question the spatial distribution and possibility of interaction between F-box-Nictaba and its putative glycosylated ligands, i.e. glycoproteins containing type 1 LAcNAc and/or Lewis A structures needs to be investigated in more detail. Microscopical analyses have shown that F-box-Nictabais located in the nucleocytoplasmic compartment of the plant cell [36]. At present Lewis A epitopes have been reported only at the cell surface or in glycoproteins secreted by plant cells or in the Golgi apparatus [25,26,29]. Since apart from Golgi stacks Lewis A structures have not been detected within any intracellular compartment of the plant cell further experiments are needed to investigate the location of F-box-Nictaba proteins and the presence of type 1 LacNAc structures in the plant cell. The difference in carbohydrate-binding specificity between Fbs proteins and F-box-Nictaba does not preclude that the sugar binding Arabidopsis protein can be considered a functional homolog of the mammalian Fbs proteins in ER-associated degradation, but indicates that the Arabidopsis protein will target another class of glycoproteins. Consequently, in order to elucidate the physiological role of F-box-Nictaba in Arabidopsis, future work will focus on the recognition of carbohydrate structures by F-box-Nictaba protein and the identification of putative interacting partners.
Our results also indicate that gene divergence within the family of Nictaba-related lectins lead to changes in carbohydrate binding specificity. The demonstration of these differences in specificity between Nictaba and F-box-Nictaba urges for extreme caution when making predictions regarding the specificity of lectins. Previously it was also shown that gene divergence within the legume lectin family [37], the jacalin-related lectins [38], the GNA-related lectins [39] and recently also within the EUL family [40] has resulted in changes in carbohydrate-binding specificity.
Acknowledgments
We thank Dr. David Smith and the Consortium for Functional Glycomicsfunded by the NIGMS GM62116 for the help and advice with the glycan array analysis. We thank Dr. Y. Kimura and Dr. M. Maeda for critical discussions.
Funding: This work was funded primarily by the Fund for Scientific Research – Flanders (FWO Grants G.0022.08 and KAN 1.5.069.09.N.), and the Research Council of Ghent University (projects BOF2005 /GOA/008 and BOF2007 /GOA/0017).
Appendix A. Supplementary data
Overview of glycans/glycoproteins bound by Nictaba and sugar-binding domains present in Arabidopsis and mammalian F-box proteins.
References
- 1.Chen Y., Peumans W.J., Hause B., Bras J., Kumar M., Proost P., Barre A., Rougé P., Van Damme E.J.M. Jasmonic acid methyl ester induces the synthesis of a cytoplasmic/nuclear chitooligosaccharide-binding lectin in tobacco leaves. FASEB J. 2002;16:905–907. doi: 10.1096/fj.01-0598fje. [DOI] [PubMed] [Google Scholar]
- 2.Van Damme E.J.M., Barre A., Rougé P., Peumans W.J. Cytoplasmic/nuclear plant lectins: a new story. Trends Plant Sci. 2004;9:484–489. doi: 10.1016/j.tplants.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Van Damme E.J.M., Lannoo N., Peumans W.J. Plant lectins. Adv. Bot. Res. 2009;48:107–209. [Google Scholar]
- 4.Read S.M., Northcote D.H. Subunit structure and interactions of the phloem proteins of Cucurbita maxima (pumpkin) Eur. J. Biochem. 1983;134:561–569. doi: 10.1111/j.1432-1033.1983.tb07603.x. [DOI] [PubMed] [Google Scholar]
- 5.Sabnis D.D., Hart J.W. The isolation and some properties of a lectin (haemagglutinin) from Cucurbita phloem exudate. Planta. 1978;142:97–101. doi: 10.1007/BF00385126. [DOI] [PubMed] [Google Scholar]
- 6.Beneteau J., Renard D., Marché L., Douville E., Lavenant L., Rahbé Y., Dupont D., Vilaine F., Dinant S. Binding properties of the N-acetylglucosamine and high-mannose N-glycan PP2-A1 phloem lectin in Arabidopsis. Plant Physiol. 2010;153:1345–1361. doi: 10.1104/pp.110.153882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinant S., Clark A.M., Zhu Y.M., Vilaine F., Palauqui J.C., Kusiak C., Thompson G.A. Diversity of the superfamily of phloem lectins (Phloem protein 2) in angiosperms. Plant Physiol. 2003;131:114–128. doi: 10.1104/pp.013086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gagne J.M., Downes B.P., Shiu S.H., Durski A.M., Vierstra R.D. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lannoo N., Peumans W.J., Van Damme E.J.M. Do F-box proteins with a C-terminal domain homologous with the tobacco lectin play a role in protein degradation in plants? Biochem. Soc. Trans. 2008;36:843–847. doi: 10.1042/BST0360843. [DOI] [PubMed] [Google Scholar]
- 10.Bai C., Sen P., Hofmann K., Ma L., Goebl M., Harper J.M., Elledge S.J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 11.Petroski M.D., Deshaies R.J. Function and regulation of cullin-ring ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;l6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 12.Lechner E., Achard P., Vansiri A., Potuschak T., Genschik P. F-box proteins everywhere. Curr. Opin. Plant Biol. 2006;9:1–8. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida Y., Tanaka K. Lectin-like ERAD players in ER and cytosol. Biochim. Biophys. Acta. 2010;1800:172–180. doi: 10.1016/j.bbagen.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi N., Kuroda H., Kuromori T., Hirayama T., Seki M., Shinozaki K., Shimada H., Matsui M. Expression and interaction analysis of Arabidopsis Skp1-related genes. Plant Cell Physiol. 2004;45:83–91. doi: 10.1093/pcp/pch009. [DOI] [PubMed] [Google Scholar]
- 15.Lannoo N., Peumans W.J., Van Pamel E., Alvarez R., Xiong T.-C., Hause G., Mazars C., Van Damme E.J.M. Localization and in vitro binding studies suggest that the cytoplasmic/nuclear tobacco lectin can interact in situ with high-mannose and complex N-glycans. FEBS Lett. 2006;580:6329–6337. doi: 10.1016/j.febslet.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 16.Schouppe D., Rougé P., Lasanajak Y., Barre A., Smith D.F., Proost P., Van Damme E.J.M. Mutational analysis of the carbohydrate binding activity of the tobacco lectin. Glycoconj. J. 2010;27:613–623. doi: 10.1007/s10719-010-9305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Al Atalah B., Fouquaert E., Vanderschaeghe D., Proost P., Balzarini J., Smith D.F., Rougé P., Lasanajak Y., Callewaert N., Van Damme E.J.M. Expression analysis of the nucleocytoplasmiclectin ‘Orysata’ from rice in Pichia pastoris. FEBS J. 2011;278:2064–2079. doi: 10.1111/j.1742-4658.2011.08122.x. [DOI] [PubMed] [Google Scholar]
- 19.Blixt O., Head S., Mondala T., Scanlan C., Huflejt M.E., Alvarez R., Bryan M.C., Fazio F., Calarese D., Stevens J., Razi N., Stevens D.J., Skehel J.J., van Die I., Burton D.R., Wilson I.A., Cummings R., Bovin N., Wong C.-H., Paulson J.C. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanley P., Cummings R.D. Structures common to different glycans. In: Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley P., Bertozzi C.R., Hart G.W., Etzler M.E., editors. Essentials of Glycobiology. 2nd ed. Cold Spring Harbor Laboratory Press; NY: 2009. pp. 175–198. Chapter 13. [Google Scholar]
- 21.Preston A., Mandrell R.E., Gibson B.W., Apicella M.A. The lipooligosaccharides of pathogenic gram-negative bacteria. Crit. Rev. Microbiol. 1996;22:139–180. doi: 10.3109/10408419609106458. [DOI] [PubMed] [Google Scholar]
- 22.Wang G., Ge Z., Rasko D.A., Taylor D.E. Lewis antigens in Helicobacter pylori: biosynthesis and phase variation. Mol. Microbiol. 2000;36:1187–1196. doi: 10.1046/j.1365-2958.2000.01934.x. [DOI] [PubMed] [Google Scholar]
- 23.Monzavi-Karbassi B., Luo P., Cunto-Amesty G., Jousheghany F., Pashov A., Weissman D., Kieber-Emmons T. Fucosylatedlactosamines participate in adhesion of HIV-1 envelope glycoprotein to dendritic cells. Arch. Virol. 2004;149:75–91. doi: 10.1007/s00705-003-0198-2. [DOI] [PubMed] [Google Scholar]
- 24.Fitchette-Lainé A.-C., Gomord V., Cabanes M., Michalski J.-C., Saint Macary M., Foucher B., Cavelier B., Hawes C., Lerouge P., Faye L. N-glycans harboring the Lewis a epitope are expressed at the surface of plant cells. Plant J. 1997;12:1411–1417. doi: 10.1046/j.1365-313x.1997.12061411.x. [DOI] [PubMed] [Google Scholar]
- 25.Fitchette A.-C., Cabanes-Macheteau M., Marvin L., Martin B., Satiat-Jeunemaitre B., Gomord V., Crooks K., Lerouge P., Faye L., Hawes C. Biosynthesis and immunolocalization of Lewis a-containing N-glycans in plant cells. Plant Physiol. 1999;121:333–343. doi: 10.1104/pp.121.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melo N.S., Nimtz M., Conradt H.S., Fevereiro P.S., Costa J. Identification of the human Lewisa carbohydrate motif in a secretory peroxidase from a plant cell suspension culture (Vacciniummyrtillus L.) FEBS Lett. 1997;415:186–191. doi: 10.1016/s0014-5793(97)01121-6. [DOI] [PubMed] [Google Scholar]
- 27.Rayon C., Cabanes-Macheteau M., Loutelier-Bourhis C., Salliot-Maire I., Lemoine J., Reiter W.D., Lerouge P., Faye L. Characterization of N-glycans from Arabidopsis. Application to a fucose-deficient mutant. Plant Physiol. 1999;119:725–734. doi: 10.1104/pp.119.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson I.B., Zeleny R., Kolarich D., Staudacher E., Stroop C.J., Kamerling J.P., Altmann F. Analysis of Asn-linked glycans from vegetable foodstuffs: widespread occurrence of Lewis a, core α1,3-linked fucose and xylose substitutions. Glycobiology. 2001;11:261–274. doi: 10.1093/glycob/11.4.261. [DOI] [PubMed] [Google Scholar]
- 29.Léonard R., Costa G., Darrambide E., Lhernould S., Fleurat-Lessard P., Carlué M., Gomord V., Faye L., Maftah A. The presence of Lewis a epitopes in Arabidopsis thaliana glycoconjugates depends on an active alpha4-fucosyltransferase gene. Glycobiology. 2002;12:299–306. doi: 10.1093/glycob/12.5.299. [DOI] [PubMed] [Google Scholar]
- 30.Strasser R., Singh Bondili J., Vavra U., Schoberer J., Svoboda B., Glössl J., Léonard R., Stadlmann J., Altmann F., Steinkellner H., Mach L. A unique β1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell. 2007;19:2278–2292. doi: 10.1105/tpc.107.052985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakker H., Bardor M., Molthoff J.W., Gomord V., Elbers I., Stevens L.H., Jordi W., Lommen A., Faye L., Lerouge P., Bosch D. Galactose-extended glycans of antibodies produced by transgenic plants. Proc. Natl. Acad. Sci. USA. 2001;98:2899–2904. doi: 10.1073/pnas.031419998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouwendal G.J.A., Florack D.E.A., Hesselink T., Cordewener J.H., Helsper J.P.F.G., Bosch D. Synthesis of Lewis X epitopes on plant N-glycans. Carbohydr. Res. 2009;344:1487–1493. doi: 10.1016/j.carres.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Karg S.R., Frey A.D., Kallio P.T. Reduction of N-linked xylose and fucose by expression of rat β1,4-N-acetylglucosaminyltransferase III in tobacco BY-2 cells depends on Golgi enzyme localization domain and genetic elements used for expression. J. Biotechnol. 2010;146:54–65. doi: 10.1016/j.jbiotec.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida Y., Adachi E., Fukiya K., Iwai K., Tanaka K. Glycoprotein-specific ubiquitin ligases recognize N-glycans in unfolded substrates. EMBO Rep. 2005;6:239–244. doi: 10.1038/sj.embor.7400351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glenn K.A., Nelson R.F., Wen H.M., Mallinger A.J., Paulson H.L. Diversity in tissue expression, substrate binding, and SCF complex formation for a lectin family of ubiquitin ligases. J. Biol. Chem. 2008;283:12717–12729. doi: 10.1074/jbc.M709508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lannoo, N., 2007. Functional characterization of a jasmonate-inducible carbohydrate-binding protein in tobacco plants. PhD thesis, Ghent University, p. 253.
- 37.Loris R., Hamelryck T., Bouckaert J., Wyns L. Legume lectin structure. Biochim. Biophys. Acta. 1998;1383:9–36. doi: 10.1016/s0167-4838(97)00182-9. [DOI] [PubMed] [Google Scholar]
- 38.Rougé P., Peumans W.J., Barre A., Van Damme E.J.M. A structural basis for the difference in specificity between the two jacalin-related lectins from mulberry (Morusnigra) bark. Biochem. Biophys. Res. Commun. 2003;304:91–97. doi: 10.1016/s0006-291x(03)00538-2. [DOI] [PubMed] [Google Scholar]
- 39.Fouquaert E., Smith D.F., Peumans W.J., Proost P., Balzarini J., Savvides S.N., Van Damme E.J.M. Related lectins from snowdrop and maize differ in their carbohydrate-binding specificity. Biochem. Biophys. Res. Commun. 2009;380:260–265. doi: 10.1016/j.bbrc.2009.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Hove J., Fouquaert E., Smith D.F., Proost P., Van Damme E.J.M. Lectin activity of the nucleocytoplasmic EUL protein from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2011;414:101–105. doi: 10.1016/j.bbrc.2011.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of glycans/glycoproteins bound by Nictaba and sugar-binding domains present in Arabidopsis and mammalian F-box proteins.


