Abstract
Chronic eosinophilic leukemia is a clonal disease characterized by hypereosinophilia and eosinophilia-related pathologic manifestations. Recently, the fusion gene FIP1L1/PDGFRA was found in the long arm of chromosome 4 and its expression has been shown to be associated with development of a clinical hypereosinophilic syndrome (HES) in a significant proportion of patients. FIP1L1/PDGFRα, the product of the gene FIP1L1/PDGFRA, is a constitutively activated tyrosine kinase and can be inhibited by imatinib mesylate. Several investigations have tried to dissect the mechanism of leukemogenesis and signaling induced by FIP1L1/PDGFRα in cell lines, primary human eosinophils and in murine myeloproliferative models. In this review, we analyzed the current knowledge on the relationship between FIP1L1/PDGFRα-induced signaling and eosinophil proliferation, survival and activation, specially focusing on its possible role in the modulation of cytokine and chemoattractant signaling pathways.
Keywords: FIP1L1/PDGFRα, chronic eosinophilic leukemia, hypereosinophilic syndrome eosinophils, mast cells
Introduction
Hypereosinophilic syndrome (HES) was first described by Hardy and Anderson in 1968 (Hardy and Anderson, 1968). The diagnostic criteria of this hematological disorder were proposed by Chusid et al. in 1975 (Chusid et al. 1975) and have been almost invariably accepted until recently. These criteria include unexplained severe peripheral blood eosinophilia (higher than 1500 eosinophils/mm3) sustained for over 6 months and accompanied by end-organ damage resulting from direct organ infiltration by eosinophils. The generic term “unexplained eosinophilia” has been used to exclude any allergic, inflammatory, infectious or neoplastic diseases, including specific eosinophilia induced by chronic or acute myelogenous leukemia, myelodysplastic syndromes or other myeloproliferative disorders.
In 2001, the World Health Organization (WHO) proposed a set of criteria that distinguish chronic eosinophilic leukemia (CEL) from HES (Bain et al. 2001). These criteria were based on the exclusion of the diseases mentioned above, together with the absence of a T-cell population with an aberrant phenotype and abnormal cytokine production, and the presence of a clonal cytogenetic abnormality or clonality, or a blast content in the peripheral blood (higher than 2%) or marrow (more than 5% but less than 19%). However, there was still the general consensus to use HES as a broad category to define heterogeneous conditions having hypereosinophilia and eosinophilic tissue infiltrations (Klion et al. 2006). Two types of HES/CEL, in particular have been described; Intrinsic: those with clonal expansion of a myeloid progenitor population with primary eosinophil differentiation including FIP1L1/PDGFRα+ CEL and CEL demonstrating with cytogenetic abnormalities; and Extrinsic: eosinophil expansion responding to a clonal expansion of T-cells, expressing high levels of the eosinophil-differentiating cytokine interleukin-5 (IL-5), including a subgroup of HES/CEL patients who present clonal T-cell populations, expressing aberrant phenotypes and producing Th2 cytokines such as IL-5 (Bank et al. 2001, Cogan et al. 1994, Raghavachar et al. 1987; Simon et al. 1999).
To examine the role of genetic abnormalities in cases of hypereosinophilia, multiple methods have been used to diagnose clonal disorders, including classical cytogenetical analysis of purified eosinophils, fluorescent in situ hybridization, cytogenetic analysis of purified eosinophils and X-chromosome inactivation analysis through the human androgen receptor gene analysis (HUMARA) (Chang et al. 1999). However, the analysis of clonality in HES may have been limited in these studies by a low frequency of chromosomal anomalies and a dramatic male predominance (Gilliland et al. 2004; Pardanani et al. 2006) that prevented X-inactivation studies. More recent analysis of other cohorts has not shown this male predominance (M.E. Rothenberg, personal communication). The chromosomal abnormalities include trisomy 8 (Guitard et al. 1994; Ma et al. 1995; Quiquandon et al. 1995; Weinfeld et al. 1977), deletion of chromosome Y (Needleman et al. 1990); t(8;9)(p21–23;p23–24)(Reiter et al. 2005); del(6)(q24) and ins(9;4) (q34;q12q31) (Schoch et al. 2004). Interestingly, many of the cases of clonal eosinophilia are associated with lymphoproliferative disorders (T-cell lymphomas, Hodgkin’s lymphoma or acute lymphoblastic leukemia), mastocytosis or other chronic myeloproliferative disorders. Also, it includes some specific subtypes of acute myeloid leukemia affecting the chromosome 16 (M4Eo inv(16)(p13q22) or t(16;16)(p13;q22)) (Le Beau et al. 1985; Marlton et al. 1995), resulting in chimeric fusion of the CBFb and MYH11 genes; t(5;16)(q33;q22) (Bhambhani et al. 1986), and t(16;21)(p11;q22) (Mecucci et al. 1985); the leukemia M2 with t(8;21)(q22;q22) (Swirsky et al. 1984) which links the acute myeloid leukemia-1 (AML1) and eight-twenty-one (ETO) genes; the presence of monosomy 7 (Song and Park 1987) or trisomy 1 (Harrington et al. 1988); t(10;11)(p14;q21) (Broustet et al. 1986), and rare eosinophil myelodysplastic syndromes (t(1;7) or dic(1;7)) (Matsushima et al. 1995).
These findings only accounted for a small fraction of patients with HES. In fact, until recently, the absence of significant specific markers of clonality made the diagnosis of CEL to be based on indirect clinical or laboratory data, such as the presence of hepatosplenomegaly, morphological dysplasia of eosinophils or other cell lineages, myeloid immaturity, bone marrow (BM) fibrosis, elevated serum vitamin B12 or mastocytosis-associated elevated serum tryptase levels (Roufosse et al. 2004). Since chronic myelogenous leukemia (CML) shares many of these clinicobiological features, there was often confusion about when to apply the diagnosis of CEL. Therefore, most of patients with the criteria defined by Chusid et al. remained diagnosed as HES.
The finding that a novel fusion gene, called FIP1 like 1/platelet derived growth factor receptor alpha (FIP1L1/PDGFRA) is responsible for a significant proportion of HES/CEL cases has changed the expectations of diagnosis and therapy in this disease, and has modified the view about incidence of clonal disorders in HES (Cools et al. 2003a).
In this review, we focus on the new advances in the pathogenesis and molecular mechanisms involved in CEL and the development of animal models that can explain the pathogenesis, clinical presentation and therapeutic targets for FIP1L1/PDGFRα-induced CEL.
Pathogenesis of FIP1L1/PDGFRα-induced HES/CEL
In 2001, a group reported a successful response to administration of imatinib mesylate in one patient with HES, that had been unsuccessfully treated with interferon-alpha and hydroxyurea (Schaller and Burkland, 2001). The rationale of its use was based on the efficacy demonstrated of imatinib in another myeloproliferative disease, BCR (breakpoint cluster region)/ABL (Abelson leukemia virus gene)-positive CML (Deininger and Druker, 2003; Druker et al. 1996). Imatinib was used and led to a rapid remission without any significant side effects. Subsequently, several groups reported more HES individuals who responded to imatinib (Ault et al. 2002; Cortes et al. 2003; Gleich et al. 2002; Pardanani et al. 2003b). When these studies were combined, 40–80% patients showed complete remission after imatinib therapy (Cortes et al. 2003; Gleich et al. 2002; Pardanani et al. 2003b). However, the molecular basis of the imatinib response remained unclear. In 2003, two independent groups used different approaches to identify the molecular mechanism of imatinib responsiveness. The first group found an imatinib-responder patient who presented the translocation t(1;4) (q44;q12) (Cools et al. 2003a). The combination of this patient’s response to imatinib (able to inhibit not only BCR/ABL-induced proliferation but also c-kit and platelet-derived-growth factor receptor beta), and the finding of a translocation at the chromosomal region 4q12, which contained the genes PDGFRA and cKIT, led to the discovery of a chimeric PDGFRA transcript with the novel gene FIP1L1 (from chromosomal region 4q12 and homologue to the FIP1 gene from Saccharomyces cerevisiae) translating the FIP1L1/PDGFRα fusion protein. A second group identified an eosinophil cell line (Eol-1) which proliferation was inhibited by imatinib. The cell line expressed a novel 110 kDa phosphorylated fusion protein, composed of an N-terminal region, encoded by a gene of unknown function (that corresponded to the same gene that had just been named FIP1L1), and the C-terminal region derived from the intracellular domain of PDGFRα (Griffin et al. 2003).
That a tyrosine kinase domain is responsible to develop CEL has been emphasized by the finding of a significant number of cases of myeloproliferative disorders with eosinophilia, where there were other fusion genes affecting the PDGFRA gene, or other genes that coded for tyrosine kinase receptors like PDGFRB and fibroblast growth factor receptor 1 (FGFR1). An excellent review of these fusion genes and others that induce eosinophilia or hypereosinophilia, usually associated with neutrophilic myeloproliferation has been recently published (Tefferi et al. 2006).
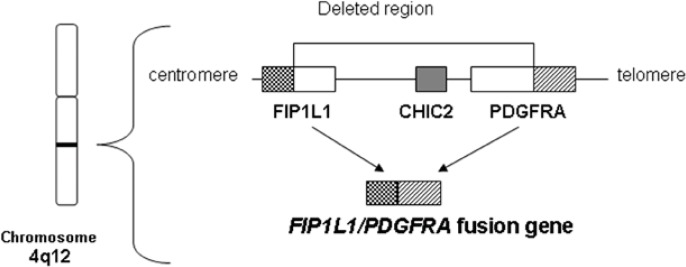
The FIP1L1/PDGFRA fusion gene is generated by an approximately 800 Kb interstitial chromosomal deletion on 4q12 (Figure 1). Sequencing of the resulting FIP1L1/PDGFRA fusion gene revealed breakpoints scattered in FIP1L1, between exons 7 and 13 that affected coding and non-coding sequences, whereas breakpoints in PDGFRA gene are exclusively restricted to exon 12 of chromosome 4, affecting the coding sequence of its juxta-membrane (JM) region, which is known to have an autoinhibitory function (Cools et al. 2003a; Roche-Lestienne et al. 2005; Vandenberghe et al. 2004). The translated protein also includes the full tyrosine kinase domain which becomes constitutively activated (see below).
Figure 1.
Interstitial deletion of 4q12 resulting in generating FIP1L1/PDGFRA fusion gene.
The FIP1L1/PDGFRA fusion gene is generated from the fusion of FIP1L1 gene and PDGFRA gene by approximately 800kbp deletion(Cools et al. 2003a). This chromosomal deletion includes the cysteine-rich hydrophobic domain 2 (CHIC2) locus. Deletion of the CHIC2 locus at 4q12 in fluorescence in situ hybridization (FISH) is a surrogate marker for FIP1L1/PDGFRA fusion gene(Pardanani et al. 2003a).
For detection of the FIP1L1/PDGFRA fusion gene, FISH analysis and/or RT-PCR are necessary. The interstitial chromosomal deletion on 4q12 of FIP1L1/PDGFRα+ patients includes the cysteine-rich hydrophobic domain 2 (CHIC2) locus. Nested or real-time RT-PCR of the FIP1L1/PDGFRA mRNA, on purified peripheral blood leukocytes or purified eosinophils, and analysis of the loss of CHIC2 locus on 4q12 by FISH analysis as a surrogate marker (Pardanani et al. 2003a) (Fig. 1), have been used to diagnose the presence of such an interstitial deletion. Expression of the FIP1L1/PDGFRA fusion gene or deletion of the surrogate marker CHIC2 have been detected in non-eosinophilic cells, including neutrophils, monocytes, mast cells, BM CD34+ cells and even lymphoid cells from a fraction of patients, suggesting that the fusion of the FIP1L1 and PDGFRA genes may occur in hematopoietic stem cells or early progenitors (HSC/P) (Pardanani et al; 2003a, Robyn et al. 2006; Tefferi et al. 2004).
Overall, the incidence of FIP1L1/PDGFRα+ HES/CEL patients ranged from 4% to 60% (Table 1), becoming the most frequent clonal defect demonstrated in HES/CEL. The expression of the FIP1L1/PDGFRA gene especially correlated with a severe subtype of HES, called myeloproliferative variant. The wide range in the frequency of expression of the FIP1L1/PDGFRA gene may derive from the use of different diagnostic techniques, as well as biased patient selection. Notably, a relatively-frequent subgroup of FIP1L1/PDGFRα+ patients also have elevated serum tryptase levels, BM infiltration of mast cells (which presented an abnormal spindle shape), and/or expression of the low-affinity IL-2 receptor (CD25). These are parameters associated with systemic mastocytosis (Klion et al. 2003; Pardanani et al. 2003a; Tefferi et al. 2004). While a physiological relationship between mast cells and eosinophils is well documented (e.g. mast cells produce eosinophil-active cytokines and mediators including IL-5, GM-CSF, histamine, and leukotriene LTD4, and eosinophils stimulate mast cells with their products such as major basic protein), there is a strong possibility that FIP1L1/PDGFRα directly affects both eosinophil and mast cell proliferation and function, or induces closer crosstalk between them. Consistent with a direct effect of FIP1L1/PDGFRα on mast cells, mast cells have been shown to be FIP1L1/PDGFRα+ in these patients (Robyn et al. 2006).
Table 1.
Reports of patients with FIP1L1/PDGFRA fusion gene in primary hypereosinophilia patients
| Year | Diagnosis | % F/P+ # | Diagnosis of F/P+ patients (No.) | Comments | references |
|---|---|---|---|---|---|
| 2003 | HES, AML-EoMPD | 56 (9/16) | CEL (8), AML-Eos (1)** | A patient with F/P T674I mutation relapsed | (Cools et al. 2003a) |
| 2003 | HES | 56 (5/9) | CEL (5) | Elevated serum tryptase (5/5) | (Klion et al. 2003) |
| 2003 | SM+Eos | 60 (3/5) | SM+Eos (3) | D816V mutation (2/5) | (Pardanani et al. 2003a) |
| 2004 | M-HES | 100 (7/7) | CEL (7) | Serum IL-5 levels were low or undetectable | (Klion et al. 2004b) |
| 2004 | HES/CEL | 47 (8/17) | CEL (8) | Splenomegaly (5/8) Elevated serum vitamin B12 (8/8) |
(Vandenberghe; et al. 2004). |
| 2004 | HES** | 50 (2/4) | CEL (2) | Serum IL-5 levels were low | (Klion et al. 2004a) |
| 2004 | HES, c-Eos SM-Eos |
14 (11/81) | CEL (1), SM+Eos (10) | Imatinib partial responder in non-F/P(4/17) | (Pardanani et al. 2004) |
| 2004 | HES | 67 (2/3) | CEL (2) | Both patients had pruritus and pulmonary infiltrates | (Smith et al. 2004) |
| 2005 | HES | 17 (6/35) | CEL (6) | TCRg rearrangement (11/35), Elevated tryptase of F/P+ (3/3) |
(Roche-Lestienne et al. 2005) |
| 2005 | HES, un-Eos | 38 (10/26) | CEL (10) | Significantly more frequent hepatosplenomegaly in F/P+ patients | (La Starza et al. 2005) |
| 2006 | un-Eos | 10 (4/40) | CEL (3), AML-CEL (1)** | 4/40 clonal eosinophilia including ins(9;4)(q34;q12q31), AML following CEL | (Bacher et al. 2006) |
| 2006 | HES/CEL | 25 (2/8) | CEL (2) | MHES (1), F/P+CEL (2) and Ly-HES (1) | (Helbig et al. 2006) |
| 2006 | HES, un-Eos | 11 (31/270) | CEL (31) | 9/217 in non-F/P including KIF5B-PDGFRA fusion had PDGFRA overexpression | (Score et al. 2006) |
| 2006 | HES | 4 (32/830) | CEL (32) | Update on previous report on 2004. | (Pardanani et al. 2006) |
HES, hypereosinophilic syndrome; AML-EoMPD, acute myeloid leukemia following eosinophilic myeloproliferative disorder; SM-Eos, systemic mastocytosis with persistent eosinophilia; M-HES, myeloproliferative variant of HES; CEL, chronic eosinophilic leukemia; c-Eos,clonal eosinophilia; un-Eos, persistent unexplained eosinophilia, F/P, FIP1L1/PDGFRA fusion gene
patients with HES refractory to or intolerant of therapy with corticosteroids, hydroxyurea, and interferon-α
both AML patients following persistant hypereosinophilia had trisomy 8.
Between brackets, numerator: number of patients with FIP1L1/PDGFRα expression; denominator: number of patients with primary eosinophilia.
Molecular mechanism of FIP1L1/PDGFRα activation and signaling
The FIP1L1/PDGFRα fusion protein is a dysregulated tyrosine kinase (Cools et al. 2003a). The physiological and pathological role of one of the fused genes, FIP1L1, remains unknown. A recent report has demonstrated that the activation of FIP1L1/PDGFRα tyrosine kinase domain depends on the integrity of the JM domain kinase of PDGFRα while the FIP1L1 is dispensable for FIP1L1/PDGFRα tyrosine kinase activation (Stover et al. 2006). The JM domain has been previously shown to play a crucial role in the autoregulation of other receptor tyrosine kinases (RTKs) (Hubbard, 2004). When the JM domain is disrupted by a mutation, the PDGFR tyrosine kinase becomes constitutively activated. Two conserved tryptophan residues in the JM domain are required for its normal inhibitory activity (Stover et al. 2006). Indeed, all the reported breakpoints of the PDGFRA gene in HES/CEL patients induce truncations of the JM-containing exon 12 (Cools et al. 2003a; Roche-Lestienne et al. 2005; Vandenberghe et al. 2004). Interestingly, similar to what has been described in BCR/ABL-induced CML (Li et al. 2001), an imatinib-resistant mutation in the FIP1L1/PDGFRA fusion gene has been identified that results in relapse after an initial response to imatinib (Cools et al. 2003a). A mutation of the threonine at position 674, (analogous to the T315I mutation of BCR/ABL), located in the ATP-binding region of the PDGFRα protein, confers FIP1L1/PDGFRα resistance to imatinib in in vitro and in vivo models of myelo- and lympho-proliferation (Cools et al. 2003a; Stover et al. 2005). Intracellular signaling of FIP1L1/PDGFRα has been investigated in transfected Ba/F3 cells, an IL-3-dependent hematopoietic cell line. After transfection with FIP1L1/PDGFRα, these cells demonstrate IL-3 independent growth and intense activation of the transcriptional factor STAT5 (Cools et al. 2003a; Stover et al. 2006). A similar process occurring in EoL-1 cells (Cools et al. 2003a; Verstovsek et al. 2006) which endogenously express the FIP1L1/PDGFRA fusion gene (Cools et al. 2004; Griffin et al. 2003). STAT5-phosphorylation is inhibited by imatinib in a dose-dependent manner, whereas it was resistant to imatinib in FIP1L1/PDGFRα (T674I)-expressing Ba/F3 cells (Cools et al. 2003a). STAT3 and STAT5 transcriptional factors would appear to be activated either directly by FIP1L1/PDGFRα or through interaction with Janus activated kinase (JAK) (Li et al. 2005; Zhang et al. 2004). Notably, it has been recently shown that JAK2 and STAT3/STAT5 are upregulated in FIP1L1/PDGFRα-expressing primary granulocytes and rapidly down-regulated by imatinib treatment (Li et al. 2005) (Fig. 2). In contrast to the STAT5 pathway, the involvement of MAPK pathway in FIP1L1/PDGFRα fusion protein signaling is still controversial. Early studies on the downstream signaling of FIP1L1/PDGFRα showed that, unlike other activated tyrosine kinases, FIP1L1/PDGFRα does not appear to induce ERK1/2 activation (Cools et al. 2003a). However, a more recent study showed that ERK1/2 is indeed a downstream target of FIP1L1/PDGFRα in both FIP1L1/PDGFRα expressing Ba/F3 and EoL-1 cell lines (Lierman et al. 2006).
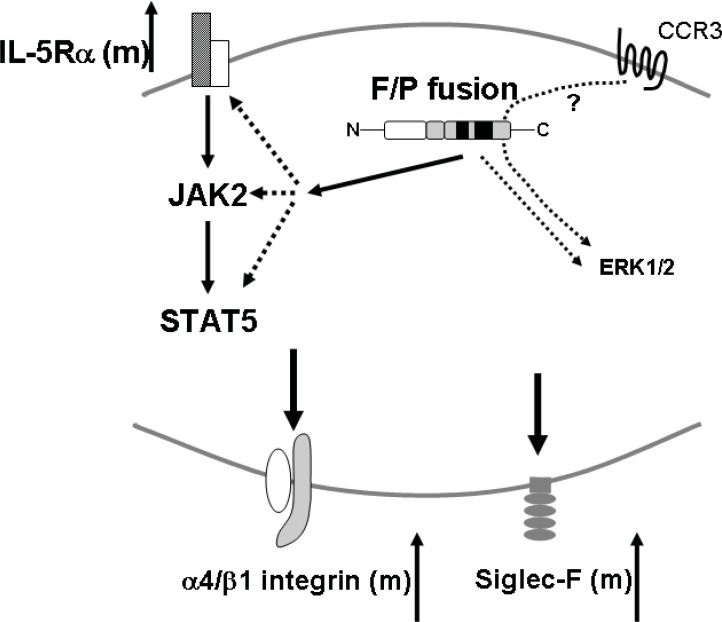
Figure 2.
FIP1L1/PDGFRα+ eosinophil signaling.
FIP1L1/PDGFRα+ primary eosinophils express upregulated IL-5 receptor (IL-5Rα) and JAK2/STAT5 pathway may be involved in FIP1L1/PDGFRα induced disease development(Li et al. 2005; Yamada et al. 2006; Zhang et al. 2004). FIP1L1/PDGFRα may phosphorylate ERK1/2 though its direct signal or indirect signal such as transactivation(Adachi et al. 2006; Lierman et al. 2006). There is a possibility that CCR3/ERK1/2 pathway is amplified by FIP1L1/PDGFRα+(Adachi et al. 2006). It is speculated that the integrated signaling may upregulate the expressions α4 integrin and Siglec-F (Yamada et al. 2006).
RTKs such as PDGFR and epidermal cell growth factor receptor (EGFR) can be transactivated through some G-protein-coupled receptors, without ligand-RTK interaction (Daub et al. 1996; Herrlich et al. 1998). In the same way, the transactivation of EGFR through the chemokine receptor 3 (CCR3), a receptor for eosinophil selective chemokines (eotaxins) has been recently reported in epithelial cells (Adachi et al. 2004). An intriguing possibility is that the CCR3-MAP kinase activation pathway in eosinophils may be modulated through RTK activation, specifically PDGFR. It has been shown that ERK phosphorylation and eotaxin-induced chemotaxis of primary eosinophils are specifically inhibited by the PDGFR-inhibitor AG1295 (Adachi et al. 2006). These authors also demonstrated that, although at low levels, both PDGFRα and PDGFRβ are endogenously expressed in eosinophils (Adachi et al. 2006). Taken together, these findings imply that FIP1L1/PDGFRα fusion may also modify the CCR3-MAP kinase pathway and the eosinophil functions dependent on that pathway (Fig. 2).
FIP1L1/PDGFRα-induced myeloproliferative disease models
Cools and colleagues reported a murine model of disease induced after BM transplantation of FIP1L1/PDGFRA-transduced HSC/P (Cools et al. 2003b). The introduction of the FIP1L1/PDGFRA fusion gene by itself into BM HSC/P induced a myeloproliferative disorder. The disease was characterized by severe leukocytosis with mild eosinophilia (5–20%) in the peripheral blood and myeloid tissue infiltration, mostly neutrophil infiltration in multiple organs similar to that found in p210-BCR/ABL induced CML-like disease. The disease development was completely inhibited by imatinib. Several groups have also determined the response of FIP1L1/PDGFRα-induced myeloproliferative disease to the tyrosine kinase inhibitors, PKC412 (Cools et al. 2003b) and nilotinib (Stover et al. 2005; von Bubnoff et al. 2006) in this murine model, demonstrating that both inhibitors were efficient in preventing disease development. This was clinically interesting, as the possibility of imatinib-resistant mutations has been reported (Cools et al. 2003a; Ohnishi et al. 2006). FIP1L1/PDGFRα expression alone induced a moderate eosinophilia, which did not fully resemble human HES/CEL. Indeed, we have also found that FIP1L1/PDGFRα expression preferentially induces eosinophilia when compared to proliferation of other myeloid lineages, but is not sufficient to develop result in severe blood and tissue eosinophilia resembling human HES/CEL in the mouse (Yamada et al. 2006). This might be due to specific differences between human and murine hematopoiesis, or to the presence in HES/CEL of secondary events that are needed to facilitate the development of hypereosinophilia (Figure 3).
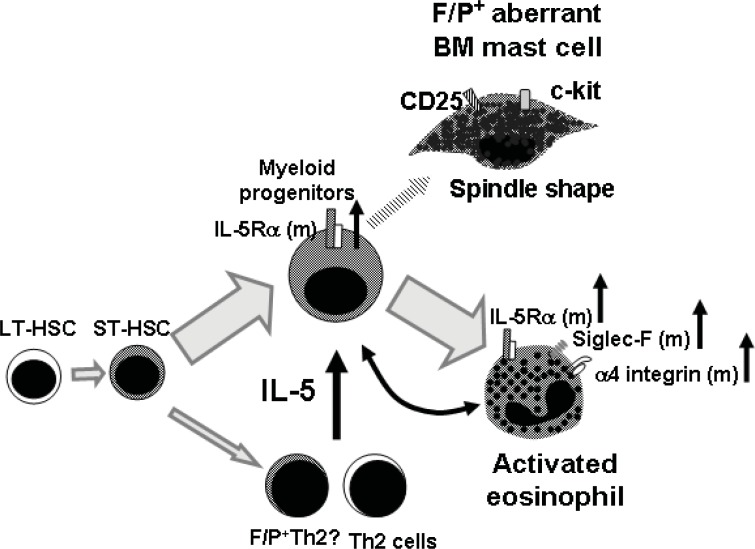
Figure 3.
FIP1L1/PDGFRα fusion promotes aberrant eosinophil and mast cell development synergized with IL-5 signaling.
FIP1L1/PDGFRα fusion would occur in short term repopulating stem cells or early progenitors and then differentiated into not only FIP1L1/PDGFRα+ myeloid cells but also FIP1L1/PDGFRα+ lymphoid cells (Robyn et al. 2006; Tefferi et al. 2004; Yamada et al. 2006). The characterization of FIP1L1/PDGFRα+ lymphocytes has not been performed but there are possibilities that FIP1L1/PDGFRα preferentially differentiated lymphoid cells into IL-5- producing Th2 cells, or other highly IL-5 producing Th2 cells may be involved. IL-5Rα expression and frequency of IL5Ra-expressing cells are specifically increased in FIP1L1/PDGFRα+ cells (Yamada et al. 2006), suggesting that IL-5 response in FIP1L1/PDGFRα+ cells may be upregulated. Mature FIP1L1/PDGFRα+ eosinophils demonstrate upregulation of α4 integrin and Siglec-F indicating the eosinophilis would be activated (Yamada et al. 2006). In addition to eosinophils, FIP1L1/PDGFRα+ aberrant mast cells, which have spindle shape and express CD25/ckit, have been observed in bone marrow of FIP1L1/PDGFRα+ patients (Klion et al. 2003).
A subgroup of patients with HES displays aberrant, sometimes clonal, CD3−/CD4+ Th2 lymphocytes which secrete large amounts of IL-5 (Bank et al. 2001) where anti-IL-5 treatment has been shown to be effective (Garrett et al. 2004; Plotz et al. 2003). In some patients, elevated IL-5 levels are not detected in circulation, even in patients with HES that respond to anti-IL-5 treatment (Garrett et al. 2004; Klion et al. 2004a; Owen et al. 1989), indicating that paracrine (Simon et al. 1999) and autocrine (Lamkhioued et al. 1996) effects of IL-5 produced by local T-cells and/or eosinophils may have critical roles in HES. Serum IL-5 levels have also been reported to be elevated in imatinib-responder HES patients including FIP1L1/PDGFRα+ HES/CEL patients (Pardanani and Tefferi, 2004). In addition to IL-5 levels, there is a possibility that FIP1L1/PDGFRα+ lymphocytes may affect the disease development in humans since it has been demonstrated that a subpopulation of CD3+ cells in patients with FIP1L1/PDGFRα+-expressing HES/CEL also express FIP1L1/PDGFRα (Robyn et al. 2006; Tefferi et al. 2004). Interestingly, a T-cell-dependent murine model of IL-5 overexpression (induced by the CD2-IL-5 transgenic) is associated with blood eosinophilia but not tissue eosinophilia (Dent et al. 1990). Since neither IL-5 nor FIP1L1/PDGFRα overexpression alone induce substantial tissue eosinophilia, is possible that the combination of two or more events may be needed to induce the development of HES/CEL-like disease (Fig. 3). Clinical studies that combine imatinib and anti-IL-5 therapies have not been reported, but may represent an attractive approach to the therapy of FIP1L1/PDGFRα-positive CEL.
These collective findings motivated us to develop a FIP1L1/PDGFRα-induced disease model in the presence of T-cell-dependent IL-5 overexpression. In order to determine if the expression of the FIP1L1/PDGFRA fusion gene in the presence of T-cell IL-5 overexpression induces HES-like disease in mice, lethally irradiated wild-type mice were transplanted with FIP1L1/PDGFRα-transduced HSC/P derived from CD2-IL-5 Tg mouse BM (Yamada et al. 2006). These mice developed a rapidly progressive phenotype featuring intense leukocytosis, hepatosplenomegaly, strikingly high eosinophilia and eosinophilic infiltration of non-hematopoietic as well as hematopoietic tissues, resembling human HES (CEL-like mice). The eosinophils in FIP1L1/PDGFRα induced CEL-like mice expressed increased levels of surface alpha 4-integrin and Siglec-F, two molecules that are involved in eosinophil activation (Fig. 2). In addition, the level of expression and frequency of IL-5Rα+ cells was increased in FIP1L1/PDGFRα+ splenocytes. IL-5Rα expression was not upregulated in p210-BCR/ABL fusion positive cells, demonstrating FIP1L1/PDGFRα specificity for the IL-5 pathway (Yamada et al. 2006). Since both IL-5R and FIP1L1/PDGFRα fusion protein can activate the JAK2/STAT5 pathway (Buitenhuis et al. 2003; Cools et al. 2003a; de Groot et al. 1998), this may be the mechanism by which the combination of FIP1L1/PDGFRα expression and the overexpression of IL-5 converge at the JAK/STAT signaling pathway and triggering a CEL-like disease. This effect would be noteworthy since FIP1L1/PDGFRα may amplify the IL-5 signaling through upregulation of receptor expression (Fig. 2).
In order to determine if the murine CEL-like disease model represents a stem cell or early progenitor-derived proliferative disease, secondary transplantation from diseased mice was performed (Yamada et al. 2006). When a high number of splenocytes from primary CEL-like mice was transplanted into lethally-irradiated secondary recipients, mice developed a similar CEL-like disease, and at the same time, demonstrated FIP1L1/PDGFRα expression in peripheral blood B- and T-cells for up to 7 weeks after secondary transplantation. These data strongly suggest the involvement of a short-term repopulating stem cell or an early myeloid progenitor (Fig. 3).
A major question is whether the combination of IL-5 overexpression together with other fusion genes may induce the same type of hypereosinophilia. This is underscored by the fact that human p210-BCR/ABL-induced CML is sometimes associated with hypereosinophilia in the context of a general chronic myeloproliferative disorder (eosinophilic variant of CML) (Bennett et al. 1994; Gotlib et al. 2000). Therefore, we introduced the exogenous expression of the p210-BCR/ABL fusion gene in HSC/P and tested it against the in vivo effect of FIP1L1/PDGFRα fusion protein in the presence of IL-5 overexpression. Notably, p210-BCR/ABL, in the presence of IL-5 overexpression induced eosinophilia at significantly lower levels than that induced by FIP1L1/PDGFRα. Moreover, unlike FIP1L1/PDGFRα-expressing splenocytes (Yamada et al. 2006); p210-BCR/ABL+ cells had no upregulation of IL-5Rα expression (Yamada et al. 2006).
Summary
The FIP1L1/PDGFRα fusion protein has provoked great attention since it was described as a cause of HES/CEL. The role of FIP1L1/PDGFRα in leukemogenesis, through its constitutively active tyrosine kinase activity, has been demonstrated in in vitro and in vivo models. However, in terms of the role of specific hypereosinophilia, it has only recently been shown that FIP1L1/PDGFRα is specifically associated with the development of hypereosinophilia in vivo when combined with IL-5 overexpression (Figures 2 and 3). Although, the mechanism behind the aberrant eosinophilopoiesis induced by FIP1L1/PDGFRα in patients remains elusive, this finding suggests that the blocking of IL-5 pathway such as anti-IL-5 treatment would be effective for intrinsic HES including FIP1L1/PDGFRα+ CEL as well as IL-5-producing T-lymphocyte-associated HES/CEL. Further elucidation of the specific mechanisms by which FIP1L1/PDGFRα induces CEL will shed light on the development of new molecular therapies for CEL/HES and probably, other eosinophilic disorders.
Acknowledgments
We are grateful to David A. Williams for his help in the development of the CEL murine model project. The work presented in this review was also supported by the Akita Medical School Fund for International Cooperation and Exchange (Y.Y.), Japan Allergy Foundation Fund for International Cooperation and Exchange (Y.Y.), American Heart Association Ohio Valley Affiliate Postdoctoral Fellowship (Y.Y.), Campaign Urging Research for Eosinophilic Disease (C.U.R.E.D, M.E.R), the National Blood Foundation (J.A.C.) and University of Cincinnati Cancer Center (J.A.C., M.E.R.). We also want to thank Margaret O’Leary for editing assistance.
References
- Adachi T, Cui CH, Kanda A, Kayaba H, Ohta K, Chihara J. Activation of epidermal growth factor receptor via CCR3 in bronchial epithelial cells. Biochem. Biophys. Res. Commun. 2004;320:292–296. doi: 10.1016/j.bbrc.2004.05.172. [DOI] [PubMed] [Google Scholar]
- Adachi T, Hanaka S, Yano T, Yamamura K, Yoshihara H, Nagase H, Chihara J, Ohta K. The role of platelet-derived growth factor receptor in eotaxin signaling of eosinophils. Int. Arch. Allergy Immunol. 2006;140(Suppl 1):28–34. doi: 10.1159/000092708. [DOI] [PubMed] [Google Scholar]
- Ault P, Cortes J, Koller C, Kaled ES, Kantarjian H. Response of idiopathic hypereosinophilic syndrome to treatment with imatinib mesylate. Leuk. Res. 2002;26:881–884. doi: 10.1016/s0145-2126(02)00046-2. [DOI] [PubMed] [Google Scholar]
- Bacher U, Reiter A, Haferlach T, Mueller L, Schnittger S, Kern W, Schoch C. A combination of cytomorphology, cytogenetic analysis, fluorescence in situ hybridization and reverse transcriptase polymerase chain reaction for establishing clonality in cases of persisting hypereosinophilia. Haematologica. 2006;91:817–820. [PubMed] [Google Scholar]
- Bain B, Pierre R, Imbert M, Vardiman JW, Brunning RD, Flandrin G. Chronic eosinophilic leukaemia and the hypereosinophilic syndrome. IARC Press; Lyon, France: 2001. [Google Scholar]
- Bank I, Amariglio N, Reshef A, Hardan I, Confino Y, Trau H, Shtrasburg S, Langevitz P, Monselise Y, Shalit M, Rechavi G. The hypereosinophilic syndrome associated with CD4+CD3− helper type 2 (Th2) lymphocytes. Leuk. Lymphoma. 2001;42:123–133. doi: 10.3109/10428190109097684. [DOI] [PubMed] [Google Scholar]
- Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick H, Sultan C, Cox C. The chronic myeloid leukaemias: guidelines for distinguishing chronic granulocytic, atypical chronic myeloid, and chronic myelomonocytic leukaemia. Proposals by the French-American-British Cooperative Leukaemia Group. Br. J. Haematol. 1994;87:746–754. doi: 10.1111/j.1365-2141.1994.tb06734.x. [DOI] [PubMed] [Google Scholar]
- Bhambhani K, Inoue S, Tyrkus M, Gohle N. Acute myelomonocytic leukemia type M4 with bone marrow eosinophilia and t(5;16)(q33;q22) Cancer Genet. Cytogenet. 1986;20:187–188. doi: 10.1016/0165-4608(86)90128-7. [DOI] [PubMed] [Google Scholar]
- Broustet A, Bernard P, Dachary D, David B, Marit G, Lacombe F, Issanchou AM, Reiffers J. Acute eosinophilic leukemia with a translocation (10p+;11q−) Cancer Genet. Cytogenet. 1986;21:327–333. doi: 10.1016/0165-4608(86)90213-x. [DOI] [PubMed] [Google Scholar]
- Buitenhuis M, Baltus B, Lammers JW, Coffer PJ, Koenderman L. Signal transducer and activator of transcription 5a (STAT5a) is required for eosinophil differentiation of human cord blood-derived CD34+ cells. Blood. 2003;101:134–142. doi: 10.1182/blood-2002-03-0740. [DOI] [PubMed] [Google Scholar]
- Chang HW, Leong KH, Koh DR, Lee SH. Clonality of isolated eosinophils in the hypereosinophilic syndrome. Blood. 1999;93:1651–1657. [PubMed] [Google Scholar]
- Chusid MJ, Dale DC, West BC, Wolff SM. The hypereosinophilic syndrome: analysis of fourteen cases with review of the literature. Medicine (Baltimore) 1975;54:1–27. [PubMed] [Google Scholar]
- Cogan E, Schandene L, Crusiaux A, Cochaux P, Velu T, Goldman M. Brief report: clonal proliferation of type 2 helper T cells in a man with the hypereosinophilic syndrome. N. Engl. J. Med. 1994;330:535–538. doi: 10.1056/NEJM199402243300804. [DOI] [PubMed] [Google Scholar]
- Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, Kutok J, Clark J, Galinsky I, Griffin JD, Cross NC, Tefferi A, Malone J, Alam R, Schrier SL, Schmid J, Rose M, Vandenberghe P, Verhoef G, Boogaerts M, Wlodarska I, Kantarjian H, Marynen P, Coutre SE, Stone R, Gilliland DG. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N. Engl. J. Med. 2003a;348:1201–1214. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- Cools J, Quentmeier H, Huntly BJ, Marynen P, Griffin JD, Drexler HG, Gilliland DG. The EOL-1 cell line as an in vitro model for the study of FIP1L1-PDGFRA-positive chronic eosinophilic leukemia. Blood. 2004;103:2802–2805. doi: 10.1182/blood-2003-07-2479. [DOI] [PubMed] [Google Scholar]
- Cools J, Stover EH, Boulton CL, Gotlib J, Legare RD, Amaral SM, Curley DP, Duclos N, Rowan R, Kutok JL, Lee BH, Williams IR, Coutre SE, Stone RM, DeAngelo DJ, Marynen P, Manley PW, Meyer T, Fabbro D, Neuberg D, Weisberg E, Griffin JD, Gilliland DG. PKC412 overcomes resistance to imatinib in a murine model of FIP1L1-PDGFRalpha-induced myeloproliferative disease. Cancer Cell. 2003b;3:459–469. doi: 10.1016/s1535-6108(03)00108-9. [DOI] [PubMed] [Google Scholar]
- Cortes J, Ault P, Koller C, Thomas D, Ferrajoli A, Wierda W, Rios MB, Letvak L, Kaled ES, Kantarjian H. Efficacy of imatinib mesylate in the treatment of idiopathic hypereosinophilic syndrome. Blood. 2003;101:4714–4716. doi: 10.1182/blood-2003-01-0081. [DOI] [PubMed] [Google Scholar]
- Daub H, Weiss FU, Wallasch C, Ullrich A. Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature. 1996;379:557–560. doi: 10.1038/379557a0. [DOI] [PubMed] [Google Scholar]
- de Groot RP, Coffer PJ, Koenderman L. Regulation of proliferation, differentiation and survival by the IL-3/IL-5/GM-CSF receptor family. Cell Signal. 1998;10:619–628. doi: 10.1016/s0898-6568(98)00023-0. [DOI] [PubMed] [Google Scholar]
- Deininger MW, Druker BJ. Specific targeted therapy of chronic myelogenous leukemia with imatinib. Pharmacol. Rev. 2003;55:401–423. doi: 10.1124/pr.55.3.4. [DOI] [PubMed] [Google Scholar]
- Dent LA, Strath M, Mellor AL, Sanderson CJ. Eosinophilia in transgenic mice expressing interleukin 5. J. Exp. Med. 1990;172:1425–1431. doi: 10.1084/jem.172.5.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- Garrett JK, Jameson SC, Thomson B, Collins MH, Wagoner LE, Freese DK, Beck LA, Boyce JA, Filipovich AH, Villanueva JM, Sutton SA, Assa’ad AH, Rothenberg ME. Anti-interleukin-5 (mepolizumab) therapy for hypereosinophilic syndromes. J. Allergy Clin. Immunol. 2004;113:115–119. doi: 10.1016/j.jaci.2003.10.049. [DOI] [PubMed] [Google Scholar]
- Gilliland G, Cools J, Stover EH, Wlodarska I, Marynen P. FIP1L1-PDGFRalpha in hypereosinophilic syndrome and mastocytosis. Hematol. J. 2004;5(Suppl 3):S133–137. doi: 10.1038/sj.thj.6200439. [DOI] [PubMed] [Google Scholar]
- Gleich GJ, Leiferman KM, Pardanani A, Tefferi A, Butterfield JH. Treatment of hypereosinophilic syndrome with imatinib mesilate. Lancet. 2002;359:1577–1578. doi: 10.1016/S0140-6736(02)08505-7. [DOI] [PubMed] [Google Scholar]
- Gotlib V, Darji J, Bloomfield K, Chadburn A, Patel A, Braunschweig I. Eosinophilic variant of chronic myeloid leukemia with vascular complications. Leuk. Lymphoma. 2003;44:1609–1613. doi: 10.3109/10428190309178786. [DOI] [PubMed] [Google Scholar]
- Griffin JH, Leung J, Bruner RJ, Caligiuri MA, Briesewitz R. Discovery of a fusion kinase in EOL-1 cells and idiopathic hypereosinophilic syndrome. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7830–7835. doi: 10.1073/pnas.0932698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitard AM, Horschowski N, Mozziconacci MJ, Michel G, George F, Capodano AM, Perrimond H. Hypereosinophilic syndrome in childhood: trisomy 8 and transformation to mixed acute leukaemia. Nouv. Rev. Fr. Hematol. 1994;35:555–559. [PubMed] [Google Scholar]
- Hardy WR, Anderson RE. The hypereosinophilic syndromes. Ann. Intern. Med. 1968;68:1220–1229. doi: 10.7326/0003-4819-68-6-1220. [DOI] [PubMed] [Google Scholar]
- Harrington DS, Peterson C, Ness M, Sanger W, Smith DM, Vaughan W. Acute myelogenous leukemia with eosinophilic differentiation and trisomy-1. Am. J. Clin. Pathol. 1988;90:464–469. doi: 10.1093/ajcp/90.4.464. [DOI] [PubMed] [Google Scholar]
- Helbig G, Stella-Holowiecka B, Grosicki S, Bober G, Krawczyk M, Wojnar J, Reiter A, Hochhaus A, Holowiecki J. The results of imatinib therapy for patients with primary eosinophilic disorders. Eur. J. Haematol. 2006;76:535–536. doi: 10.1111/j.1600-0609.2006.00652.x. [DOI] [PubMed] [Google Scholar]
- Herrlich A, Daub H, Knebel A, Herrlich P, Ullrich A, Schultz G, Gudermann T. Ligand-independent activation of platelet-derived growth factor receptor is a necessary intermediate in lysophosphatidic, acid-stimulated mitogenic activity in L cells. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8985–8990. doi: 10.1073/pnas.95.15.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2004;5:464–471. doi: 10.1038/nrm1399. [DOI] [PubMed] [Google Scholar]
- Klion AD, Bochner BS, Gleich GJ, Nutman TB, Rothenberg ME, Simon HU, Wechsler ME, Weller PF, The Hypereosinophilic Syndromes Working, G Approaches to the treatment of hypereosinophilic syndromes: a workshop summary report. J. Allergy Clin. Immunol. 2006;117:1292–1302. doi: 10.1016/j.jaci.2006.02.042. [DOI] [PubMed] [Google Scholar]
- Klion AD, Law MA, Noel P, Kim YJ, Haverty TP, Nutman TB. Safety and efficacy of the monoclonal anti-interleukin-5 antibody SCH55700 in the treatment of patients with hypereosinophilic syndrome. Blood. 2004a;103:2939–2941. doi: 10.1182/blood-2003-10-3620. [DOI] [PubMed] [Google Scholar]
- Klion AD, Noel P, Akin C, Law MA, Gilliland DG, Cools J, Metcalfe DD, Nutman TB. Elevated serum tryptase levels identify a subset of patients with a myeloproliferative variant of idiopathic hypereosinophilic syndrome associated with tissue fibrosis, poor prognosis, and imatinib responsiveness. Blood. 2003;101:4660–4666. doi: 10.1182/blood-2003-01-0006. [DOI] [PubMed] [Google Scholar]
- Klion AD, Robyn J, Akin C, Noel P, Brown M, Law M, Metcalfe DD, Dunbar C, Nutman TB. Molecular remission and reversal of myelofibrosis in response to imatinib mesylate treatment in patients with the myeloproliferative variant of hypereosinophilic syndrome. Blood. 2004b;103:473–478. doi: 10.1182/blood-2003-08-2798. [DOI] [PubMed] [Google Scholar]
- La Starza R, Specchia G, Cuneo A, Beacci D, Nozzoli C, Luciano L, Aventin A, Sambani C, Testoni N, Foppoli M, Invernizzi R, Marynen P, Martelli MF, Mecucci C. The hypereosinophilic syndrome: fluorescence in situ hybridization detects the del(4)(q12)-FIP1L1/PDGFRA but not genomic rearrangements of other tyrosine kinases. Haematologica. 2005;90:596–601. [PubMed] [Google Scholar]
- Lamkhioued B, Gounni AS, Aldebert D, Delaporte E, Prin L, Capron A, Capron M. Synthesis of type 1 (IFN gamma) and type 2 (IL-4, IL-5, and IL-10) cytokines by human eosinophils. Ann. NY Acad. Sci. 1996;796:203–208. doi: 10.1111/j.1749-6632.1996.tb32582.x. [DOI] [PubMed] [Google Scholar]
- Le Beau MM, Diaz MO, Karin M, Rowley JD. Metallothionein gene cluster is split by chromosome 16 rearrangements in myelomonocytic leukaemia. Nature. 1985;313:709–711. doi: 10.1038/313709a0. [DOI] [PubMed] [Google Scholar]
- Li B, Zhang GS, Dai CW, Pei MF. [The activation of JAK/STAT signal pathway in hypereosinophilic syndrome and the patients therapeutic response to imatinib] Zhonghua Yi Xue Za Zhi. 2005;85:448–452. [PubMed] [Google Scholar]
- Li S, Gillessen S, Tomasson MH, Dranoff G, Gilliland DG, Van Etten RA. Interleukin 3 and granulocyte-macrophage colony-stimulating factor are not required for induction of chronic myeloid leukemia-like myeloproliferative disease in mice by BCR/ABL. Blood. 2001;97:1442–1450. doi: 10.1182/blood.v97.5.1442. [DOI] [PubMed] [Google Scholar]
- Lierman E, Folens C, Stover EH, Mentens N, Van Miegroet H, Scheers W, Boogaerts M, Vandenberghe P, Marynen P, Cools J. Sorafenib is a potent inhibitor of FIP1L1-PDGFRalpha and the imatinib-resistant FIP1L1-PDGFRalpha T674I mutant. Blood. 2006;108:1374–1376. doi: 10.1182/blood-2006-02-004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SK, Wong KF, Chan JK, Kwong YL. Refractory cytopenia with t(1;7),+8 abnormality and dysplastic eosinophils showing intranuclear Charcot-Leyden crystals: a fluorescence in situ hybridization study. Br. J. Haematol. 1995;90:216–218. doi: 10.1111/j.1365-2141.1995.tb03405.x. [DOI] [PubMed] [Google Scholar]
- Marlton P, Keating M, Kantarjian H, Pierce S, O’Brien S, Freireich EJ, Estey E. Cytogenetic and clinical correlates in AML patients with abnormalities of chromosome 16. Leukemia. 1995;9:965–971. [PubMed] [Google Scholar]
- Matsushima T, Murakami H, Kim K, Uchiumi H, Murata N, Tamura J, Sawamura M, Karasawa M, Naruse T, Tsuchiya J. Steroid-responsive pulmonary disorders associated with myelodysplastic syndromes with der(1q;7p) chromosomal abnormality. Am. J. Hematol. 1995;50:110–115. doi: 10.1002/ajh.2830500207. [DOI] [PubMed] [Google Scholar]
- Mecucci C, Bosly A, Michaux JL, Broeckaert-Van Orshoven A, Van den Berghe H. Acute nonlymphoblastic leukemia with bone marrow eosinophilia and structural anomaly of chromosome 16. Cancer Genet. Cytogenet. 1985;17:359–363. doi: 10.1016/0165-4608(85)90120-7. [DOI] [PubMed] [Google Scholar]
- Needleman SW, Mane SM, Gutheil JC, Kapil V, Heyman MR, Testa JR. Hypereosinophilic syndrome with evolution to myeloproliferative disorder: temporal relationship to loss of Y chromosome and c-N-ras activation. Hematol. Pathol. 1990;4:149–155. [PubMed] [Google Scholar]
- Ohnishi H, Kandabashi K, Maeda Y, Kawamura M, Watanabe T. Chronic eosinophilic leukaemia with FIP1L1-PDGFRA fusion and T674I mutation that evolved from Langerhans cell histiocytosis with eosinophilia after chemotherapy. Br J Haematol. 2006 doi: 10.1111/j.1365-2141.2006.06221.x. [DOI] [PubMed] [Google Scholar]
- Owen WF, Rothenberg ME, Petersen J, Weller PF, Silberstein D, Sheffer AL, Stevens RL, Soberman RJ, Austen KF. Interleukin 5 and phenotypically altered eosinophils in the blood of patients with the idiopathic hypereosinophilic syndrome. J. Exp. Med. 1989;170:343–348. doi: 10.1084/jem.170.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardanani A, Brockman SR, Paternoster SF, Flynn HC, Ketterling RP, Lasho TL, Ho CL, Li CY, Dewald GW, Tefferi A. FIP1L1-PDGFRA fusion: prevalence and clinicopathologic correlates in 89 consecutive patients with moderate to severe eosinophilia. Blood. 2004;104:3038–3045. doi: 10.1182/blood-2004-03-0787. [DOI] [PubMed] [Google Scholar]
- Pardanani A, Ketterling RP, Brockman SR, Flynn HC, Paternoster SF, Shearer BM, Reeder TL, Li CY, Cross NC, Cools J, Gilliland DG, Dewald GW, Tefferi A. CHIC2 deletion, a surrogate for FIP1L1-PDGFRA fusion, occurs in systemic mastocytosis associated with eosinophilia and predicts response to imatinib mesylate therapy. Blood. 2003a;102:3093–3096. doi: 10.1182/blood-2003-05-1627. [DOI] [PubMed] [Google Scholar]
- Pardanani A, Ketterling RP, Li CY, Patnaik MM, Wolanskyj AP, Elliott MA, Camoriano JK, Butterfield JH, Dewald GW, Tefferi A. FIP1L1-PDGFRA in eosinophilic disorders: prevalence in routine clinical practice, long-term experience with imatinib therapy, and a critical review of the literature. Leuk. Res. 2006;30:965–970. doi: 10.1016/j.leukres.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Pardanani A, Reeder T, Porrata LF, Li CY, Tazelaar HD, Baxter EJ, Witzig TE, Cross NC, Tefferi A. Imatinib therapy for hypereosinophilic syndrome and other eosinophilic disorders. Blood. 2003b;101:3391–3397. doi: 10.1182/blood-2002-10-3103. [DOI] [PubMed] [Google Scholar]
- Pardanani A, Tefferi A. Imatinib therapy for hypereosinophilic syndrome and eosinophilia-associated myeloproliferative disorders. Leuk. Res. 2004;28(Suppl 1):S47–52. doi: 10.1016/j.leukres.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Plotz SG, Simon HU, Darsow U, Simon D, Vassina E, Yousefi S, Hein R, Smith T, Behrendt H, Ring J. Use of an anti-interleukin-5 antibody in the hypereosinophilic syndrome with eosinophilic dermatitis. N. Engl. J. Med. 2003;349:2334–2339. doi: 10.1056/NEJMoa031261. [DOI] [PubMed] [Google Scholar]
- Quiquandon I, Claisse JF, Capiod JC, Delobel J, Prin L. alpha-Interferon and hypereosinophilic syndrome with trisomy 8: karyotypic remission. Blood. 1995;85:2284–2285. [PubMed] [Google Scholar]
- Raghavachar A, Fleischer S, Frickhofen N, Heimpel H, Fleischer B. Tlymphocyte control of human eosinophilic granulopoiesis. Clonal analysis in an idiopathic hypereosinophilic syndrome. J. Immunol. 1987;139:3753–3758. [PubMed] [Google Scholar]
- Reiter A, Walz C, Watmore A, Schoch C, Blau I, Schlegelberger B, Berger U, Telford N, Aruliah S, Yin JA, Vanstraelen D, Barker HF, Taylor PC, O’Driscoll A, Benedetti F, Rudolph C, Kolb HJ, Hochhaus A, Hehlmann R, Chase A, Cross NC. The t(8;9)(p22;p24) is a recurrent abnormality in chronic and acute leukemia that fuses PCM1 to JAK2. Cancer Res. 2005;65:2662–2667. doi: 10.1158/0008-5472.CAN-04-4263. [DOI] [PubMed] [Google Scholar]
- Robyn J, Lemery S, McCoy JP, Kubofcik J, Kim YJ, Pack S, Nutman TB, Dunbar C, Klion AD. Multilineage involvement of the fusion gene in patients with FIP1L1/PDGFRA-positive hypereosinophilic syndrome. Br. J. Haematol. 2006;132:286–292. doi: 10.1111/j.1365-2141.2005.05863.x. [DOI] [PubMed] [Google Scholar]
- Roche-Lestienne C, Lepers S, Soenen-Cornu V, Kahn JE, Lai JL, Hachulla E, Drupt F, Demarty AL, Roumier AS, Gardembas M, Dib M, Philippe N, Cambier N, Barete S, Libersa C, Bletry O, Hatron PY, Quesnel B, Rose C, Maloum K, Blanchet O, Fenaux P, Prin L, Preudhomme C. Molecular characterization of the idiopathic hypereosinophilic syndrome (HES) in 35 French patients with normal conventional cytogenetics. Leukemia. 2005;19:792–798. doi: 10.1038/sj.leu.2403722. [DOI] [PubMed] [Google Scholar]
- Roufosse F, Cogan E, Goldman M. Recent advances in pathogenesis and management of hypereosinophilic syndromes. Allergy. 2004;59:673–689. doi: 10.1111/j.1398-9995.2004.00465.x. [DOI] [PubMed] [Google Scholar]
- Schaller JL, Burkland GA. Case report: rapid and complete control of idiopathic hypereosinophilia with imatinib mesylate. Med. Gen. Med. 2001;3:9. [PubMed] [Google Scholar]
- Schoch C, Reiter A, Bursch S, Schnittger S, Hiddemann W, Kern W, Haferlach T. Chromosome Banding Analysis, FISH and RT-PCR Performed in Parallel in Hypereosinophilic Syndrome Establishes the Diagnosis of Chronic Eosinophilic Leukemia in 22% of Cases: A Study on 40 Patients. Blood. 2004;104:2444. [Google Scholar]
- Score J, Curtis C, Waghorn K, Stalder M, Jotterand M, Grand FH, Cross NC. Identification of a novel imatinib responsive KIF5B-PDGFRA fusion gene following screening for PDGFRA overexpression in patients with hypereosinophilia. Leukemia. 2006;20:827–832. doi: 10.1038/sj.leu.2404154. [DOI] [PubMed] [Google Scholar]
- Simon HU, Plotz SG, Dummer R, Blaser K. Abnormal clones of T cells producing interleukin-5 in idiopathic eosinophilia. N. Engl. J. Med. 1999;341:1112–1120. doi: 10.1056/NEJM199910073411503. [DOI] [PubMed] [Google Scholar]
- Smith KJ, Jacobson E, Hamza S, Skelton H. Unexplained hypereosinophilia and the need for cytogenetic and molecular genetic analyses. Arch. Dermatol. 2004;140:584–588. doi: 10.1001/archderm.140.5.584. [DOI] [PubMed] [Google Scholar]
- Song HS, Park SK. A case of monosomy-7 eosinophilic leukemia and neurofibromatosis, terminated with disseminated cryptococcosis. Korean J. Intern. Med. 1987;2:131–134. doi: 10.3904/kjim.1987.2.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover EH, Chen J, Folens C, Lee BH, Mentens N, Marynen P, Williams IR, Gilliland DG, Cools J. Activation of FIP1L1-PDGFRalpha requires disruption of the juxtamembrane domain of PDGFRalpha and is FIP1L1-independent. Proc Natl Acad Sci USA, 2006;103:8078–8083. doi: 10.1073/pnas.0601192103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover EH, Chen J, Lee BH, Cools J, McDowell E, Adelsperger J, Cullen D, Coburn A, Moore SA, Okabe R, Fabbro D, Manley PW, Griffin JD, Gilliland DG. The small molecule tyrosine kinase inhibitor AMN107 inhibits TEL-PDGFR{beta} and FIP1L1-PDGFR{alpha} in vitroand in vivo. Blood. 2005 doi: 10.1182/blood-2005-05-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirsky DM, Li YS, Matthews JG, Flemans RJ, Rees JK, Hayhoe FG. 8;21 translocation in acute granulocytic leukaemia: cytological, cytochemical and clinical features. Br. J. Haematol. 1984;56:199–213. doi: 10.1111/j.1365-2141.1984.tb03948.x. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Lasho TL, Brockman SR, Elliott MA, Dispenzieri A, Pardanani A. FIP1L1-PDGFRA and c-kit D816V mutation-based clonality studies in systemic mast cell disease associated with eosinophilia. Haematologica. 2004;89:871–873. [PubMed] [Google Scholar]
- Tefferi A, Patnaik MM, Pardanani A. Eosinophilia: secondary, clonal and idiopathic. Br. J. Haematol. 2006;133:468–492. doi: 10.1111/j.1365-2141.2006.06038.x. [DOI] [PubMed] [Google Scholar]
- Vandenberghe P, Wlodarska I, Michaux L, Zachee P, Boogaerts M, Vanstraelen D, Herregods MC, Van Hoof A, Selleslag D, Roufosse F, Maerevoet M, Verhoef G, Cools J, Gilliland DG, Hagemeijer A, Marynen P. Clinical and molecular features of FIP1L1-PDFGRA (+) chronic eosinophilic leukemias. Leukemia. 2004;18:734–742. doi: 10.1038/sj.leu.2403313. [DOI] [PubMed] [Google Scholar]
- Verstovsek S, Giles FJ, Quintas-Cardama A, Manshouri T, Huynh L, Manley P, Cortes J, Tefferi A, Kantarjian H. Activity of AMN107, a novel aminopyrimidine tyrosine kinase inhibitor, against human FIP1L1-PDGFR-alpha-expressing cells. Leuk Res. 2006 doi: 10.1016/j.leukres.2006.03.012. [DOI] [PubMed] [Google Scholar]
- von Bubnoff N, Gorantla SP, Thone S, Peschel C, Duyster J. The FIP1L1-PDGFRA T674I mutation can be inhibited by the tyrosine kinase inhibitor AMN107 (nilotinib) Blood. 2006;107:4970–4971. doi: 10.1182/blood-2006-01-0285. author reply 4972. [DOI] [PubMed] [Google Scholar]
- Weinfeld A, Westin J, Swolin B. Ph1-negative eosinophilic leukaemia with trisomy 8. Case report and review of cytogenetic studies. Scand J Haematol. 1977;18:413–420. doi: 10.1111/j.1600-0609.1977.tb02095.x. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Rothenberg ME, Lee AW, Akei HS, Brandt EB, Williams DA, Cancelas JA. The FIP1L1-PDGFRA fusion gene cooperates with IL-5 to induce murine hypereosinophilic syndrome (HES)/chronic eosinophilic leukemia (CEL)-like disease. Blood. 2006;107:4071–4079. doi: 10.1182/blood-2005-08-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GS, Li B, Pei MF, Dai CW, Zheng WL, Shen JK. [Identification of FIP1L1-PDGFRA fusion, and expression of signal transducer and activator of transcription 5 in hypereosinophilic syndrome] Zhonghua Yi Xue Za Zhi. 2004;84:1541–1544. [PubMed] [Google Scholar]