Abstract
GTPases are important regulatory proteins that hydrolyze GTP to GDP. A novel GTP-hydrolysis mechanism is employed by MnmE, YqeH and FeoB, where a potassium ion plays a role analogous to the Arginine finger of the Ras-RasGAP system, to accelerate otherwise slow GTP hydrolysis rates. In these proteins, two conserved asparagines and a ‘K-loop’ present in switch-I, were suggested as attributes of GTPases employing a K+-mediated mechanism. Based on their conservation, a similar mechanism was suggested for TEES family GTPases. Recently, in Dynamin, Fzo1 and RbgA, which also conserve these attributes, a similar mechanism was shown to be operative. Here, we probe K+-activated GTP hydrolysis in TEES (TrmE-Era-EngA-YihA-Septin) GTPases – Era, EngB and the two contiguous G-domains, GD1 and GD2 of YphC (EngA homologue) – and also in HflX, another GTPase that also conserves the same attributes. While GD1-YphC and Era exhibit a K+-mediated activation of GTP hydrolysis, surprisingly GD2-YphC, EngB and HflX do not. Therefore, the attributes identified thus far, do not necessarily predict a K+-mechanism in GTPases and hence warrant extensive structural investigations.
Keywords: GTPases, Hydrolysis mechanism, K+ stimulated hydrolysis, K-loop, HAS-GTPases
Abbreviations: TEES, TrmE-Era-EngA-YihA-Septin; HAS-GTPases, Hydrophobic Amino acid Substituted for catalytic glutamine-GTPases; Era, E. coli ras; EngA, Essential Neisseria gonorrhoeae GTPase
Highlights
▸ An emerging alternative mechanism of GTP hydrolysis is mediated by K+ ions. ▸ Features characteristic of the K+-mediated mechanism were suggested. ▸ As TEES family GTPases possess these, they were suggested to employ this mechanism. ▸ Not all GTPases that possess these features utilize the K+-mediated mechanism. ▸ Unambiguously identifying determinants of K+ mechanism requires extensive studies.
1. Introduction
GTPases or G-proteins are ubiquitously found in all the domains of life and regulate several key biological processes. This regulation is provided by a G-domain that facilitates GTP binding and its subsequent hydrolysis to GDP. Although, GTPases are divided into several families based on the associated function, the G-domain is widely conserved [1–3]. The residues required for GTP binding and hydrolysis are provided by conserved motifs G1 (GxxxxGKS/T or P-loop), G2 (T), G3 (DxxG) and G4 (NKxD) [4]. In accordance to their biological activity, G-proteins could be grouped into two classes: conventional G-proteins and G-proteins activated by nucleotide dependent dimerization. The first class comprises the Ras superfamily GTPases, while the second class includes a number of GTPases with varied biological functions: The latter are characterized by low affinity for nucleotides and distinct modes of dimerization [5].
GTP hydrolysis mechanisms have been well studied in classical GTPases such as Ras and hetero-trimeric Gα-proteins. In these, the intrinsic hydrolysis rates are usually slow. Therefore, GAPs (GTPase Activiating Proteins), by providing a positively charged residue such as an arginine, accelerate hydrolysis rates; this arginine, termed ‘Arginine finger’ together with a conserved Gln (from Switch-II/G3) in the G-domain, form a part of the catalytic machinery to stabilize the transition state and hence lower the activation energy barrier [6,7].
In the last several years, variations to this canonical GTP hydrolysis mechanism have been noted. These encompass variations in the Arginine finger and the catalytic Gln; the latter is found in HAS-GTPases (Hydrophobic Amino acid Substituted for catalytic glutamine-GTPases) [8]. A noteworthy difference in catalytic machinery was brought out by structural and biochemical studies on MnmE, wherein the role of the catalytic Gln is taken over by a Glu (present in a helix α2 following the G3 motif) and a water molecule. Surprisingly, MnmE does not require a GAP and instead a K+ substitutes for the role of an Arginine finger in stabilizing the transition state of the reaction (see supplementary Fig. S1) [9]. A K+-mediated stimulation of GTP hydrolysis was subsequently shown for GTPases, YqeH [10], FeoB and RbgA [11,12]. Besides GTPases, ATPases like GroEL, Hsc70 and YchF too, display a K+-mediated stimulation of ATP hydrolysis; of these, YchF is unique in bearing a high sequence similarity to GTPases [13–15]. In dynamin, a member of large GTPase family, the role of the catalytic glutamine is taken over by the carbonyl group of Thr65 and the backbone nitrogen of Gly139. The catalytic water is further oriented by a secondary water molecule, which acts as a bridge between the Gln40 side chain and the carbonyl oxygen of Gly139. Surprisingly, in case of dynamin, not only K+ but also Na+ was shown to substitute for the Arginine finger. In these, K+/Na+ are stabilized by the carbonyl groups of two glycine residues in the switch 1 region and Ser41 in the P-loop [16].
Structural and biochemical studies on MnmE and later on FeoB identified the determinants of K+-mediated mechanism as two conserved Asn residues – one in the P-loop (G1) and another following it (GxxN xGKSxLxN); and the presence of an insertion termed K-loop, in Switch-I (G2). Similarly a conserved Asn in yeast Fzo1 [17] and Ser [16] in dynamin present in the P-loop, were found to be stabilize the metal ion (K+/Na+) stimulating GTP hydrolysis. Based on the conservation of these features, it was further suggested that GTPases of the TEES family, i.e. TrmE/MnmE, FeoB, EngA, Era, YihA/EngB and Septins, would also follow a similar K+-mediated mechanism (see Fig. 1) [4]. Members of the TEES family mediate diverse cellular functions. MnmE regulates tRNA modification, FeoB a transmembrane protein is responsible for ferrous ion uptake, EngA, Era and EngB are involved in ribosome assembly, while Septins are structural proteins required for cell division and compartmentalization.
Fig. 1.
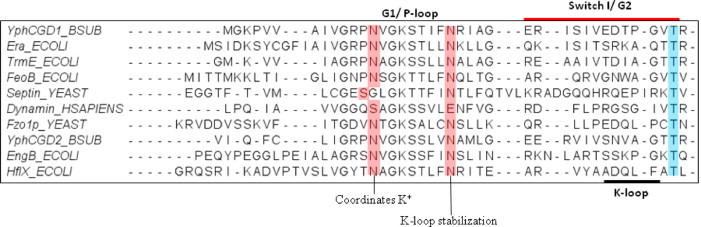
Multiple sequence alignment of TEES family GTPases. Primary sequences of respective GTPases were retrieved from NCBI database and were subjected to multiple sequence alignment (MSA) using clustalX [2]. The region spanning G1 and G2 is shown. Residues suggested to coordinate the potassium ion and stabilizing the K-loop, are highlighted in red. K-loop (indicated) adopts a conformation optimal to coordinate the K+ ion. Switch I region is indicated by a red bar above the sequences and a conserved Thr from G2 is highlighted in blue. Conserved residues suggested to confer K+ dependent stimulation of GTP hydrolysis in TEES family of proteins were identified based on this alignment (For interpretation of reference to colour in this figure legend, the reader is referred to the web version of this article.).
To verify if these features are the hallmark of K+ utilizing GTPases, we investigated GTP hydrolysis in TEES family GTPases Era, EngB and YphC (Bacillus subtilis homologue of EngA). YphC is a unique GTPase with two contiguous G-domains GD1 and GD2 (termed GD1-YphC and GD2-YphC, from here on). Employing these, GTP hydrolysis was probed in GD1-YphC and GD2-YphC, independently. Surprisingly, only Era and GD1-YphC, but not EngB and GD2-YphC showed stimulation of GTP hydrolysis in presence of K+. Additionally, HflX, which is not a member of the TEES family, but conserves the aforesaid attributes, was also investigated. It did not show a K+-mediated stimulation of GTP hydrolysis, either. This study allows us to conclude that currently identified attributes are not accurate indicators of K+-mediated GTP hydrolysis, and detailed structural and mechanistic investigations would be needed to comprehensively identify attributes characteristic to GTPases employing a K+-mediated mechanism.
2. Materials and methods
2.1. Site directed mutagenesis
Asn to Ala point mutants YphC-N13A, YphC-N185A and Era-N18A, were generated to abrogate K+ binding in GD1-YphC, GD2-YphC and Era, respectively. Site directed mutagenesis was performed using the overlapping PCR technique. Amplicons were synthesized by Pfu DNA polymerase (Fermentas) using appropriate primers containing the desired mutation along with the full length primer (Table S1) of the corresponding genes. Wild type genes coding for the proteins were used as templates. Amplicons were then digested with NdeI and HindIII restriction endonucleases and cloned into pQE2 expression vector. Mutations were confirmed by DNA sequencing.
2.2. Cloning, expression and purification of proteins
DNA sequences corresponding to wild type YphC, GD1-YphC, GD2-YphC and EngB were PCR amplified from the Bacillus subtilis genome (Table S1) using pfu DNA polymerase (Fermentas). Similarly, wild type era was amplified using the E. coli genome. Amplified products were digested with NdeI and HindIII restriction endonucleases and were further cloned into pQE2 expression vector (Qiagen). E. coli hflx was cloned and purified as described earlier [18].
E. coli DH5α competent cells were transformed with the recombinant plasmids for overexpressing the proteins. Cells were grown in LB medium containing 100 μg of ampicillin per ml at 37 °C until OD600 reached to ∼0.6, induced with 0.2 mM isopropyl-β-D-thiogalactopyranosid (IPTG) and incubated for 8–10 h at 22 °C. The cells were harvested by centrifugation at 5000 x g and resuspended in lysis buffer (50 mM Tris–Cl buffer (pH 7.5)), 0.3 M NaCl, 5% (v/v) glycerol, 0.5 mg/ml Lysozyme and 100 μg/ml of Hiscocktail (sigma) and lysed by 5 cycles of freeze–thaw procedure. This was followed by DNase and RNase treatment (5 μg/ml) for 1 h at 4 °C and lysates thus obtained were centrifuged at 35,000 x g at 4 °C for 1 h in 50 mL oak-ridge tubes (Sorvall Evolution SS-34 rotor). Clarified supernatant was loaded on a 5 mL Global His-trap affinity column (Amersham) pre-equilibrated with 5 CV of binding buffer B (50 mM Tris–HCl (pH 8.0) at 4 °C, 300 mM NaCl, 5% glycerol 2 mM β-mercaptoethanol). The column was washed with 10 CV of washing buffer containing 50 mM Tris–Cl (pH 8.0), 300 mM NaCl, 10 mM Imidazole and 1 mM DTT. Proteins were then eluted using an imidazole linear gradient (0–500 mM in buffer B). Fractions containing the protein were analyzed by SDS–PAGE and further subjected to size exclusion chromatography using superdex 200 column (Amersham). The proteins were eluted with elution buffer (20 mM Tris–Cl (pH 8.0), 150 mM NaCl, 1 mM DTT). Nucleotide free proteins were then prepared as described earlier [19]. Briefly, purified proteins were subjected to extensive dialysis in 50 mM Tris–HCl buffer (pH 7.5), 150 mM NaCl, 1 mM DTT and 5 mM EDTA. The buffer was exchanged with 20 mM Tris–Cl (pH 8.0), 150 mM NaCl, 1 mM DTT and the concentration was estimated using BCA assay (Sigma). Aliquots were made, flash frozen in liquid nitrogen and stored at −80 °C until required.
All Asn to Ala mutants (YphC-N13A, YphC-N185A and Era-N13A) were overexpressed and purified as above, except that size exclusion chromatography was not performed and fractions containing the protein after imidazole gradient elution, were collected and buffer was exchanged with 20 mM Tris-Cl (pH 8.0), 150 mM NaCl, 1 mM DTT using (Millipore Amicon) ultra centrifugation filter tubes.
2.3. Nucleotide binding assays
Nucleotide binding assays were conducted with a fluorescently labelled nucleotide analogue (Jena Biosciences). Typically, an N-methyl-2′/3′-O-anthraniloyl (mant) fluorophore group attached to the nucleotide was excited at 355 nm (slit width of 5 nm) and the emission at 448 nm was monitored between 400 nm and 600 nm (slit width of 10 nm) using a spectrofluorimeter (Perkin Elmer). To measure the emission spectra for protein mant-nucleotide complexes, reactions were carried out in 150 μl reaction mixtures in buffer F containing 50 mM Tris–Cl (pH 7.5), 150 mM NaCl, 5 mM MgCl2 and 5 mM DTT. Protein complexes were prepared by incubating 0.8 μM of the respective proteins with 0.4 μM mant–nucleotides in Buffer F at room temperature for 10 min. For studying protein mantGDP·AlFx·M+ complex (M = metal), 200 mM of M+ (i.e. NaCl or KCl) and 5 μM of NaF and 500 nM of AlCl3 were added to the reaction mixture.
2.4. GTP hydrolysis assays
Catalytic efficiency of the wild type and mutant proteins were probed by performing GTP hydrolysis using radioactive [α32P]-GTP. The assays were performed in a buffer containing 50 mM Tris–Cl (pH 7.5), 5 mM MgCl2, 1 mM DTT and traces of radiolabelled GTP with 200 μM GTP and 10 μM of protein. For stimulation of GTP hydrolysis, 200 mM of M+ (NaCl, KCl, NH4Cl, RbCl and CsCl) were used. Reaction mixture (10 μl) was incubated at 30 °C for 1 h and the reaction was stopped by the addition of 1 M formic acid. Separation of hydrolyzed GDP was performed on thin layer chromatography (TLC). For this, samples were centrifuged and 5 μl of reaction mixture was spotted on a polyethyleneimine-coated TLC plate (Merck), resolved in 1.5 M KH2PO4 (pH 3.4) buffer. PEI-TLC plates were subjected to autoradiography to detect the formation of [α32P]-GDP. Autoradiograms were obtained and spots corresponding to GDP were scraped from the TLC plate, and the counts (cpm) were measured using a scintillation counter (Perkin Elmer).
3. Results
3.1. K+ dependent acceleration of GTP hydrolysis by Era and GD1 of YphC
An alternative GTP hydrolysis mechanism driven by potassium ions was found operative in GTPases MnmE, YqeH and FeoB, [9–11]. More recently in RbgA, Fzo1 and Dynamin, either K+ or Na+ ions were found to stimulate GTP hydrolysis [12,16,17]. This mechanism is in contrast to the well known mechanism employed by classical GTPases like Ras, which utilize an ‘Arginine finger’ from an interacting GAP protein to stabilize the transition state and facilitate GTP hydrolysis [6]. Interestingly, Scrima and Wittinghofer elucidated that in MnmE, a potassium ion performs a role analogous to the Arginine finger. The crystal structure of MnmE bound to the transition state analogue, GDP·AlF3, showed that the K+ occupies a position equivalent to the terminal atoms of the Arginine finger. It is held in place by interactions with backbone atoms of residues contributed by a loop termed the ‘K-loop’, which is present between conserved motifs G1 and G2 or in switch-I of the G-domain. Later, YqeH was also found to utilize a K+ dependent mechanism [10]. Concurrently, structural studies on FeoB revealed that it also utilizes a similar mechanism for GTP hydrolysis [11]. In this study, the authors proposed that the K+ mediated mechanism is conferred by the K-loop and two conserved asparagines (GxxNxGKSxLxN; x-represents any residue) in or near the P-loop; this Asn is substituted by a Ser in case of dynamin [16]. These Asn residues and the K-loop were suggested as features characteristic of K+ mediated activation in TEES family GTPases.
Therefore, presence of the two conserved Asn residues and K-loop (Fig. 1) in TEES family GTPases, YphC (EngA homologue in B. subtilis), Era and EngB prompted us to investigate whether a similar mechanism is operative in these. GTP hydrolysis was assayed using radiolabelled GTP, in presence of various metal ions (Section 2.4). As anticipated, YphC shows a significantly stimulated GTP hydrolysis in presence of KCl, RbCl and NH4Cl (Fig. 2A).
Fig. 2.
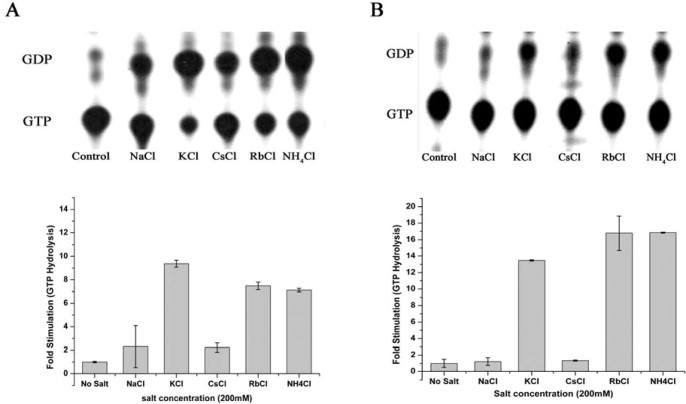
Potassium stimulates GTP hydrolysis of YphC and Era. KCl, RbCl and NH4Cl stimulate GTP hydrolysis significantly, whereas NaCl and CsCl do not. A) An autoradiogram showing the hydrolysis of GTP to GDP by YphC, in presence of the indicated salts, along with a control that lacks any protein is shown. Amount of GTP hydrolysed in presence of salts was quantified and represented as fold stimulation (GTP hydrolysis by the protein in the absence of any salt represents 1-fold). The experiments were repeated thrice and error bars are shown. (B) GTP hydrolysis by Era was monitored similarly as in (A).
We further probed whether both GD1 and GD2 or only one of these invoke K+ dependent stimulation of GTP hydrolysis. For this, individual G-domains GD1-YphC and GD2-YphC were generated and were similarly employed in GTP hydrolysis assays. Interestingly, only GD1-YphC, but not GD2-YphC, shows a K+ dependent stimulation (Fig. 3A). Incidentally, GD1 is also known to exhibit a higher GTP hydrolysis rate than GD2 [20]. A similar K+ dependent acceleration of GTP hydrolysis was also observed for Era; like with EngA, NH4Cl and RbCl, but not NaCl or CsCl, accelerate GTP hydrolysis (Fig. 2B). In contrast, GTP hydrolysis by EngB, is not influenced by metal ions (Fig. 3B). These results indicate that despite the conservation of elements, regarded necessary for K+ mediated mechanism, in the primary sequences, not all TEES family GTPases exhibit the anticipated stimulation in presence of K+. To verify this, another GTPase, HflX that conserves these elements but is not a member of TEES family was also employed for GTP hydrolysis assays. HflX, does not show K+ dependent stimulation of GTP hydrolysis, either (Fig. 3C).
Fig. 3.
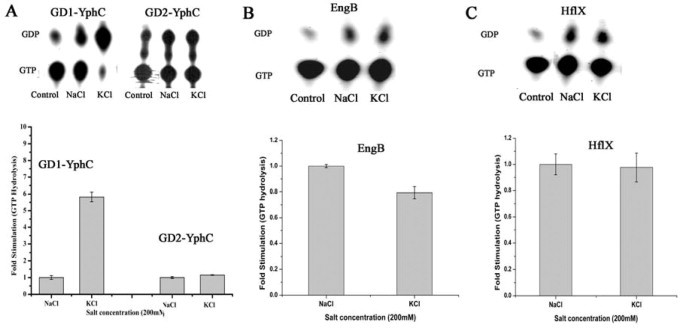
Potassium dependent GTPase activity in EngB, HflX and the two G domains GD1 and GD2 of YphC. Individual G domains, GD1-YphC and GD2-YphC were generated and used to determine the effect of K+ and Na+ ions on GTP hydrolysis by the two domains, like in Fig. 2. (A) An autoradiogram showing the hydrolysis of GTP to GDP by GD1-YphC (left panel) and GD2-YphC-(right panel) in presence of 200 mM NaCl and 200 mM KCl, along with a control devoid of protein. Amount of GTP hydrolyzed in presence of salts was quantified (from the above autoradiograms) and represented as fold stimulation considering GTP hydrolysis in presence of Na+ to be 1-fold. The experiments were repeated thrice and error bars are shown. GTP hydrolysis assays were conducted as above, for (B) EngB and (C) HflX.
3.2. Role of K+ in stabilizing the transition state
Crystal structures of Ras and Gα-proteins elucidated that the transition state is stabilized through an Arginine finger supplied either in cis or in trans. Also, AlF3 in complex with GDP (GDP·AlF3) mimics the transition state. Here, AlF3 occupies the position of the γ-phosphate of GTP [7]. Interestingly, in MnmE, which utilizes K+ to accelerate GTPase activity, it has been shown that K+ stabilizes and facilitates stronger binding of GDP·AlF3 complex and acts as a GTPase Activating Element (GAE), analogous to the Arginine finger, to neutralize the developing negative charge developed during catalysis [9]. Encouraged by the finding that YphC and Era also utilize K+ to stimulate GTP hydrolysis (Fig. 2), we investigated whether K+ acts as a GAE and similarly enhances GDP·AlFx binding in these proteins. Fluorescently labelled N-methyl-2′/3′-O-anthraniloyl nucleotides-GDP (mGDP) in presence of AlFx and different monovalent salts was assayed for binding. Proteins were first incubated with mGDP and an increased fluorescence (compared to that with mGDP alone) was observed. Addition of AlCl3 and NaF, that form AlFx and in turn the mGDP·AlFx transition state mimic, did not show a further increase in fluorescence. However, when K+ were added to this mixture, fluorescence emission increased substantially, indicating the formation of a stable transition state complex in YphC-WT. The same was not seen in presence of Na+ (Fig. 4A). Similarly, GD1-YphC showed increased mGDP fluorescence in the presence of AlFx and K+, but not in presence of Na+ (Fig. 4B). In contrast, but in concurrence with the GTP hydrolysis assays, where K+ accelerates GTP hydrolysis only in GD1-YphC, but not in GD2-YphC, increase in fluorescence in presence of KCl/NaCl and AlFx was not observed when GD2-YphC was employed (Fig. 4C).
Fig. 4.
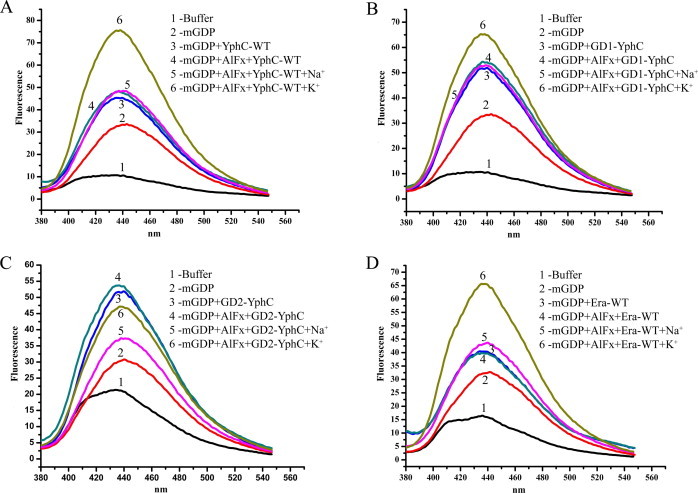
K+ ion stabilizes the transition state. Nucleotide binding assays, employing fluorescent N-methyl-2’/3’-O-anthraniloyl nucleotides GDP (mGDP) were carried out to assay stabilization of the transition state mimic mGDP·AlFx in presence of monovalent ions KCl and NaCl. (A) The emission spectra, monitored at 380–600 nm, show that the presence of K+ ions enhances the binding of the transition state analogue (mGDP·AlFx) in full length YphC-WT (spectra 6), while Na+ does not show a similar effect (spectra 5). (B) GD1-YphC shows a similar trend in enhancing the binding of the transition state analog only in the presence of K+, (C) while GD2-YphC does not display a similar trend. (D) Era too shows significant binding of mGDP·AlFx in presence of K+, but not with Na+.
Along the same lines, mGDP binding assays with Era also illustrate an increase in fluorescence, reflecting the formation of a stabilized transition state in presence of K+ and AlFx, but not Na+ and AlFx (Fig. 4D). Overall, these results reveal a K+ dependent stimulation of GTPase activity due to the stabilization of the transition state by potassium ions, for YphC and Era proteins of the TEES family.
3.3. P- loop Asn is essential for K+ mediated stimulation of GTP hydrolysis
The catalytically important K+ is stabilized by number of interactions, as seen in the crystal structure of MnmE. In this structure, the K+ is held in place due to interactions from three different regions: first is the α and β phosphates of GTP/GDP, interactions from backbone atoms of residues in the K-loop, and an important interaction mediated by the side chain carbonyl oxygen of a conserved Asn226 of the P-loop (GxxNxGKS/T) [9]. Similarly, analogous residues, Asn11, Asn169 and Ser41 from the P-loops of FeoB,YqeH and dynamin were inferred to stabilize the K+. Mutational studies involving mutants N226K in MnmE, the equivalent N11A in FeoB [11], or N169L in YqeH [10] and S41A [16] resulted in either a significantly compromised (72% in MnmE) or completely abolished K+ dependent GTPase activity (for FeoB and YqeH). A similar Asn is present in the P-loops of YphC and Era, too. In order to ascertain whether it is critical for K+ dependent stimulation of GTPase activity in these proteins, we generated Asn to Ala mutants, YphC-N13A (i.e. in the P-loop of GD1), YphC-N185A (i.e. in the P-loop of GD2) and Era-N18A, and assayed for their ability to bind and hydrolyze GTP. As anticipated, for YphC-N13A and Era-N18A mutant, potassium dependent acceleration of GTPase activity was completely abolished (Fig. 5A and C). Whereas, in line with the observation that GD2 does not utilize potassium mediated mechanism, the YphC-N185A mutant displayed a K+ stimulated GTPase activity like the wild type YphC, which is contributed by GD1-YphC (Fig. 5B). Furthermore, fluorescent nucleotide binding assays, with the transition state mimic mGDP·AlFx in the presence of Na+ and K+, suggest that YphC-N13A and Era-N18A mutants do not show an increase in nucleotide binding but show an unexpected decrease in binding (Fig. 5D and F); as anticipated, YphC-N185A mutant behaves like the wild type protein (Fig. 5E) since GD1-YphC continues to bind the transition state analogue. This suggests that the mutation of the conserved P-loop Asn to Ala, compromises K+ binding and hence abolishes the stimulation of GTP hydrolysis, indicating an important role for this residue in coordinating the K+ at the active site of these proteins, like for MnmE, YqeH, FeoB and dynamin.
Fig. 5.
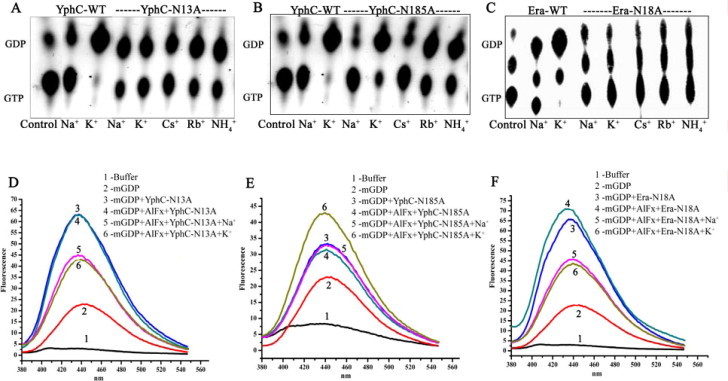
Mutating P-loop asparagines in YphC and Era, abolishes K+ ion dependent stimulation of GTP hydrolysis and transition state stabilization. A conserved Asn in the P-loop, critical for coordinating the K+ ion, was mutated to alanine in YphC and Era to create YphC-N13A (of GD1), YphC-N185A (of GD2) and Era-N18A mutants. GTP hydrolysis in presence of various ions (indicated in the insets) were measured using [α32P]-GTP. Autoradiograms show the hydrolysis of GTP to GDP, in presence of various salts (as indicated), by (A) YphC-N13A, (B) by YphC-N185A and (C) by Era-N18A. In all cases, a control devoid of protein was included. Fluorescent mGDP binding assays, as in Fig. 4, were carried out for (D) YphC-N13A (E) YphC-N185A and (F) Era-N18A. Unlike with wild type proteins (Fig. 4), in these, enhanced binding of the transition state mimic mGDP·AlFx in the presence of KCl is not observed for YphC-N13A and Era-N18A. This is in line with the observations that Yphc-GD1 and Era use a K+ dependent stimulation of GTP hydrolysis.
4. Discussion
The emergence of a novel class of GTPases, like the HAS-GTPases that carry a hydrophobic residue in lieu of the Gln, suggested that newer mechanisms, other than that seen for classical GTPases such as Ras and Gα-like-proteins, to hydrolyze GTP must exist; a few possibilities were proposed [8] and indeed, recent studies on a variety of HAS-GTPases verified these. The K+ mediated GTP hydrolysis mechanism, exhibited by GTPases MnmE, YqeH, FeoB, RbgA, dynamin and more recently YchF (an ATPase of the Obg family GTPases) [9–12,15,16] is an emerging alternative theme by which GTP may be hydrolyzed. Structural and biochemical studies on MnmE, FeoB, and YqeH revealed that a K+, hexacoordinated by a K-loop and two conserved Asn residues around the P-loop, performs a role akin to the Arginine finger, by stabilizing the transition state complex (Fig. S1) [9,11]. It was specualted that a similar mechanism may exist in other members of the TEES family. Here, we set out to investigate the same using GD1-YphC, GD2-YphC, Era and EngB as candidate GTPases that conserve the aforesaid attributes suggested for GTPases employing a K+ mediated mechanism (Fig. 1). Surprisingly, such a mechanism was observed only for GD1-YphC (and therefore for full length YphC protein) and for Era. In contrast, GD2-YphC and EngB do not seem to exhibit a similar response in spite of the conserved Asn in the P-loop and the presence of a K-loop. Recent reports on dynamin, Fzo1 and RbgA that do not belong to the TEES family, but conserve these attributes, reiterate that K+ stimulates GTP hydrolysis in these [12,16,17]: An exception being, that in dynamins, Na+ also stimulate GTP hydrolysis, and a Ser in the P-loop substitutes for the conserved Asn of the TEES family GTPases (Fig. 1). Intrigued by all these observations, we probed HflX – another GTPase that conserves the aforesaid attributes, but does not belong to the TEES family. HflX does not exhibit a K+ mediated stimulation of GTP hydrolysis, either (Fig. 3C). These findings bring forth the view that mere conservation of a P-loop Asn/Ser and a K-loop are not reliable indicators of K+ stimulated GTP hydrolysis.
In line with the observations for MnmE, YqeH, FeoB and YchF, Rb+ and NH4+ having a similar ionic radii as K+ (K+ 138 pm, NH4+ 144 pm, and Rb+ 152 pm) also efficiently accelerate GTPase activity in GD1-YphC and Era. Whereas, Na+ with smaller radius (Na+ 102 pm) and Cs+ with a larger radius (Cs+ 167 pm) were unable to accelerate GTP hydrolysis (Fig. 2A and B). Nucleotide binding assays are also in agreement with these observations. YphC-WT and GD1-YphC show appreciable increase in binding the transition analogue GDP·AlFx in presence of K+ (Fig. 4). In addition, and similarly as in MnmE, FeoB, YqeH and (Ser in) dynamin, mutational analysis underscores the importance of the conserved P-loop Asn in coordinating the K+ in YphC and Era GTPases (Fig. 5).
It is noteworthy that for EngA, GD1 undergoes a drastic conformational change depending upon the nucleotide bound state; it was inferred that this conformational change is due to GTP hydrolysis by GD1 [21]. Also, GTP hydrolysis at GD1 is a key factor that distinguishes two distinct ribosome bound states of EngA, i.e. bound to 50S alone or to 30S, 50S and 70S [22]. Together with the critical role of GD1 in regulating the activity of the protein, and an exclusive requirement of K+ for GTP hydrolysis in GD1 alone (but not in GD2), suggests an important regulation mediated by K+ towards EngA's role in ribosome biogenesis. Further, since GD2 does not employ a K+ dependent mechanism, it may require a positively charged residue contributed by a GAP like regulatory protein to drive GTP hydrolysis: Alternatively, a novel mechanism may be employed in GD2. Also, realizing the two ribosome bound states of EngA requires GD2 to be GTP bound [22]. Given the low GTP hydrolysis rate of GD2, the likely employment of a mechanism distinct from that employed by GD1 (i.e. the K+ mechanism) appears meaningful. Therefore, it may be that triggering GTP hydrolysis at GD2 is an event that occurs when ribosome biogenesis is completed.
Although suggested before [9,11], this is the first experimental demonstration of a K+ mediated mechanism in GD1-YphC and Era. Together with the recent reports on Fzo1, Dynamin and RbgA, K+ mediated mechanism continues to emerge as an alternative theme. Given this, primary sequence based prediction of GTPases employing such a mechanism becomes important. In this context, an important understanding brought out by this work is the finding that GD2-YphC, EngB and HflX do not use K+ to accelerate GTP hydrolysis, although they possess the suggested attributes of K+ mediated GTP hydrolysis. Therefore, extensive biochemical and structural investigations are necessary to unambiguously identify attributes unique to GTPases employing a K+ mediated mechanism. Until then, it is difficult to rule out K+/Na+ mediated GTP hydrolysis in GTPases, regardless of the conserved Asn/Ser residues and/or the presence of K-loop in these proteins.
Acknowledgements
We acknowledge Dr. Sushil Kumar Tomar for several helpful discussions during the course of this work. AR acknowledges Indian Council of Medical Research (ICMR) India for fellowship. SM acknowledges CSIR for fellowship. We thank the Departments of Biotechnology (DBT), Science and Technology (DST), Indian Council of Medical Research (ICMR), India, for financial support.
Supplementary Material
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fob.2012.07.008.
Appendix. Supplementary material
Note added in Proof
While this manuscript was under revision, another work speculating GTPases likely employing a potassium dependent mechanism, had appeared (Ash et.al. FEBS Lett. Volume 586, Pages 2218-2224, July, 2012). Our study contradicts some of these conclusions and indicates that the current indicators for the employment of potassium (or cation) dependent mechanism in GTPases, are insufficient.
References
- 1.Bourne H.R., Sanders D.A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 2.Leipe D.D., Wolf Y.I., Koonin E.V., Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 3.Bourne H.R., Sanders D.A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 4.Wittinghofer A., Vetter I.R. Structure–function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- 5.Gasper R., Meyer S., Gotthardt K., Sirajuddin M., Wittinghofer A. It takes two to tango: regulation of G proteins by dimerization. Nat. Rev. Mol. Cell. Biol. 2009;10:423–429. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 6.Mittal R., Ahmadian M.R., Goody R.S., Wittinghofer A. Formation of a transition-state analog of the Ras GTPase reaction by Ras-GDP, tetrafluoroaluminate, and GTPase-activating proteins. Science. 1996;273:115–117. doi: 10.1126/science.273.5271.115. [DOI] [PubMed] [Google Scholar]
- 7.Scheffzek K., Ahmadian M.R., Wittinghofer A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem. Sci. 1998;23:257–262. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- 8.Mishra R., Gara S.K., Mishra S., Prakash B. Analysis of GTPases carrying hydrophobic amino acid substitutions in lieu of the catalytic glutamine: Implications for GTP hydrolysis. Proteins-Struct. Funct. Bioinformat. 2005;59:332–338. doi: 10.1002/prot.20413. [DOI] [PubMed] [Google Scholar]
- 9.Scrima A., Wittinghofer A. Dimerisation-dependent GTPase reaction of MnmE: how potassium acts as GTPase-activating element. EMBO J. 2006;25:2940–2951. doi: 10.1038/sj.emboj.7601171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anand B., Surana P., Prakash B. Deciphering the catalytic machinery in 30S ribosome assembly GTPase YqeH. PLoS One. 2010;5:e9944. doi: 10.1371/journal.pone.0009944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ash M.R., Guilfoyle A., Clarke R.J., Guss J.M., Maher M.J., Jormakka M. Potassium-activated GTPase reaction in the G protein-coupled ferrous iron transporter B. J. Biol. Chem. 2010;285:14594–14602. doi: 10.1074/jbc.M110.111914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Achila D., Gulati M., Jain N., Britton R.A. Biochemical characterization of the ribosome assembly GTPase RbgA in Bacillus subtilis. J. Biol. Chem. 2012;287:8417–8423. doi: 10.1074/jbc.M111.331322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J., Boisvert D.C. Structural basis for GroEL-assisted protein folding from the crystal structure of (GroEL-KMgATP)(14) at 2.0 angstrom resolution. J. Mol. Biol. 2003;327:843–855. doi: 10.1016/s0022-2836(03)00184-0. [DOI] [PubMed] [Google Scholar]
- 14.Wilbanks S.M., McKay D.B. How potassium affects the activity of the molecular chaperone Hsc70. II. Potassium binds specifically in the ATPase active site. J. Biol. Chem. 1995;270:2251–2257. doi: 10.1074/jbc.270.5.2251. [DOI] [PubMed] [Google Scholar]
- 15.Tomar S.K., Kumar P., Prakash B. Deciphering the catalytic machinery in a universally conserved ribosome binding ATPase YchF. Biochem. Biophys. Res. Commun. 2011;408:459–464. doi: 10.1016/j.bbrc.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 16.Chappie J.S., Acharya S., Leonard M., Schmid S.L., Dyda F. G domain dimerization controls dynamin's assembly-stimulated GTPase activity. Nature. 2010;465:435–440. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anton F., Fres J.M., Schauss A., Pinson B., Praefcke G.J.K., Langer T., Escobar-Henriques M. Ugo1 and Mdm30 act sequentially during Fzo1-mediated mitochondrial outer membrane fusion. J. Cell Sci. 2011;124:1126–1135. doi: 10.1242/jcs.073080. [DOI] [PubMed] [Google Scholar]
- 18.Jain N., Dhimole N., Khan A.R., De D., Tomar S.K., Sajish M., Dutta D., Parrack P., Prakash B. E. coli HflX interacts with 50S ribosomal subunits in presence of nucleotides. Biochem. Biophys. Res. Commun. 2009;379:201–205. doi: 10.1016/j.bbrc.2008.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan K.-H., Li T., Wong C.-O., Wong K.-B. Structural basis for GTP-dependent dimerization of hydrogenase maturation factor HypB. Plos ONE. 2012;7:e30547. doi: 10.1371/journal.pone.0030547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson V.L., Hwang J., Fox E., Inouye M., Stock A.M. Domain arrangement of Der, a switch protein containing two GTPase domains. Structure. 2002;10:1649–1658. doi: 10.1016/s0969-2126(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 21.Muench S.P., Xu L., Sedelnikova S.E., Rice D.W. The essential GTPase YphC displays a major domain rearrangement associated with nucleotide binding. Proc. Natl. Acad. Sci. U S A. 2006;103:12359–12364. doi: 10.1073/pnas.0602585103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomar S.K., Dhimole N., Chatterjee M., Prakash B. Distinct GDP/GTP bound states of the tandem G-domains of EngA regulate ribosome binding. Nucleic Acids Res. 2009;37:2359–2370. doi: 10.1093/nar/gkp107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


