Abstract
The existence of G protein-coupled receptor (GPCR) dimers and/or oligomers has been demonstrated in heterologous systems using a variety of biochemical and biophysical assays. While these interactions are the subject of intense research because of their potential role in modulating signaling and altering pharmacology, evidence for the existence of receptor interactions in vivo is still elusive because of a lack of appropriate methods to detect them. Here, we adapted and optimized a proximity ligation assay (PLA) for the detection in brain slices of molecular proximity of two antigens located on either the same or two different GPCRs. Using this approach we were able to confirm the existence of dopamine D2 and adenosine A2A receptor complexes in the striatum of mice ex vivo.
Keywords: PLA, GPCR, oligomers
Introduction
GPCRs play critical roles in cell signaling. Their putative organization as dimers and/or oligomers in living cells has been proposed to fine-tune cell-cell communication by regulating receptor function and pharmacology (1). However, despite extensive evidence supporting the formation of GPCR oligomers in heterologous systems, the existence of such signaling complexes in their native environment remains controversial, mostly because of the lack of appropriate methodology (2). In the absence of a clear demonstration of receptor heteromerization in vivo, it has been difficult to differentiate downstream signaling crosstalk from heteromer-dependent signaling (Aburi et al., in press). Recently a time-resolved FRET approach (3) was used to demonstrate the existence of oligomers in native tissue, but this strategy requires high levels of native receptor expression and does not yet allow spatial or subcellular resolution. It has been suggested that GPCR heteromer levels can be regulated by drug exposure or by disease state (4), but this too is difficult to assess in vivo, especially in a manner that can be generally applicable to different receptor pairs. Here we have adapted a well-characterized Proximity Ligation Assay (PLA) to confirm the existence of GPCR complexes in striatal brain slices of mice ex vivo.
Material and methods
Animals
All procedures were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at Columbia University and the New York State Psychiatric Institute. WT, D2R KO (5) and A2AR KO (6) male congenic C57Bl/6j mice (postnatal ages 90–150 days) were used in this study. All animals were group-housed in a standard animal care facility with a 12-h light/dark cycle where they had free access to food and water.
Surgeries/viral injections
Viral injections were performed in a biohazard level 2 biochemical cabinet. Animals were anesthetized with a mixture of ketamine and xylazine via intraperitoneal injection and operated on using standard sterile conditions. The skull was drilled to allow the penetration of a fine glass pipette (8 to 15 μm inner diameter) containing a solution of GFP or D2L-R-mVenus expressing adeno-associated virus (GeneDetect, Auckland, New Zealand) or myc-CD8a-EGFP expressing lentivirus. The ejection of the virus was performed using a syringe under positive pressure. Each injection in the dorsal striatum consisted of a volume of 1 μl delivered bilaterally by a single injection at the following coordinates relative to Bregma (in mm): antero-posterior 1; ventro-dorsal 3; medial-lateral 1.7. The rate of injection was 0.2–0.3 μl per minute. Following the injections, the skull was covered with triple antibiotic ointment and the wound sutured. Animals were sacrificed 3–8 weeks later for histology and PLA analysis.
Brain tissue preparation
Fixed tissue: for most experiments, mice were anaesthetized and perfused intracardiacally with 50 ml of ice-cold 4% paraformaldehyde (PFA) in 0.1 M Na2HPO4/NaH2PO4 pH 7.4 buffer. Brains were post-fixed overnight in the same solution of PFA at 4 °C. Coronal sections (30 μm) were processed using a vibratome (Leica). Slices were collected in a cryoprotective solution (30% glycerol, 30% ethylene glycol in Tris 0.1 M pH 7.4) and kept at −20 °C until processing.
For experiments using fixed frozen tissue, brains were incubated 24 hours in 30% sucrose in 0.1 M Na2HPO4/NaH2PO4 pH 7.4 buffer following the postfixation step. Coronal sections (30 μm) were generated using a cryostat (Leica) and then mounted on slides. Slides were kept at −20 °C until processing.
Fresh frozen tissue
Mice were anesthetized and decapitated. Brains were quickly removed and frozen in isopentane (−20 °C). Coronal sections (30 μm) were generated using a crysotat and then mounted on slides that were kept at −80 °C until processing. Note that in the case of fresh frozen tissue, slices were fixed for 5 minutes in 4% PFA and then rinsed twice in TBS before staining.
Immunohistochemistry
Most of the experiments shown in this study were performed on floating sections. For sections mounted on slides, reagents were applied directly on the slides and covered with parafilm to prevent evaporation.
Sections were rinsed four times in Tris-buffered saline (TBS; 0.1 M Tris pH.7.4 and 0.9% (w/v) NaCl), and incubated with blocking solution (10% fetal bovine serum and 0.5% Triton X-100 in TBS) for 2 hours at room temperature. The slices were incubated with the primary antibodies overnight at 4 °C. After four rinses (30 minutes each) in TBS (0.2% Triton X-100), sections were incubated for 2 h at room temperature with the appropriate secondary antibodies conjugated with Alexa dyes (Invitrogen), then washed 6 more times (30 minutes each) with TBS (0.2% Triton X-100) and mounted on slides with Fluorsave (Calbiochem, Billerica, MA, USA) for observation.
In situ Proximity ligation assay (PLA)
PLA was performed in a similar manner as standard immunohistochemistry until the secondary antibody incubation step, and PLA probe dilution/incubation time, rolling circle amplification (RCA) times and polymerase concentrations were optimized for floating sections.
PLA secondary probes (see also below)
Slices (or slides when appropriate) were incubated for 2 hours at 37 °C in the blocking solution (same as for the primary antibodies) containing PLA secondary probes (Olink Bioscience, Uppsala, Sweden) diluted 1:5 under gentle agitation.
Hybridization and ligation steps
These steps were performed following the manufacturer’s protocol and under gentle agitation. For optimal labelling it is important to pre-warm the solutions at 37 °C before incubation with slices.
Amplification step
This step is crucial for the efficiency of the assay. As shown in the Results, a polymerase dilution of 1/80 was optimal for single recognition whereas the efficiency of the assay was significantly improved when diluted 1/40 for the dual recognition of D2R and A2AR. For both conditions incubation time was 2 hours under gentle agitation. Note that longer incubations increased non-specific diffuse signal (data not shown).
Detection step
This step was performed according to the manufacturer’s instructions except that the incubation time was prolonged to 90 minutes. Atto590 labelled oligonucleotide detection probes (Olink Bioscience) were used in all experiments.
After extensive washing in decreasing concentrations of SSC buffers (2x, 1x, 0.2x, 0.02x, 10 minutes each), slices were mounted on slides (in case of floating sections), dried and coverslips were applied with either Duolink Mounting Medium (Olink Bioscience) or FluorSave (Calbiochem).
PLA probes
For single detection of either D2R or A2AR, anti-rabbit PLUS/MINUS and anti-mouse PLUS/MINUS PLA probes were used respectively. For Dual recognition of D2R and mVenus or GFP, anti-rabbit PLUS and anti-chicken MINUS were used. For dual recognition of D2R and A2AR, most experiments were performed using anti-rabbit PLUS and anti-mouse MINUS. Anti-rabbit MINUS and anti-mouse PLUS were also used to control for efficiency (see SF 4).
Antibodies
Anti-D2R antibody (rabbit polyclonal; Millipore, AB5084P) was used at a concentration of 1/100 for immunohistochemistry, 1/300 for PLA single recognition, 1/200 for dual recognition with GFP or A2AR and 1/500 for immunoblots; these dilutions were independently optimized for each of the approaches.
Anti-A2AR (mouse monoclonal; Millipore, 05–717) was used at a concentration of 1/500 for immunohistochemistry and PLA single recognition, 1/300 for dual recognition with D2R. Anti-GFP antibody (chicken polyclonal; Abcam ab13970) was used at a concentration of 1/500 for PLA dual recognition with D2R.
Generation of the rabbit polyclonal anti-D2R antibody
Affinity-purified rabbit polyclonal sera directed against a peptide sequence corresponding to amino acids 246–305 (third intracellular loop) of the mouse D2R (antibody generated by GL Biochem, Shanghai, China) were produced. This antibody was used at a concentration of 1/500 for immunohistochemistry and PLA (both single and dual recognition).
Microscopy/quantification of PLA signal
Microscopy/images acquisition
Fluorescence images were acquired on an Olympus Fluoview FV1000 confocal laser scanning microscope using a 60X 1.42 NA Oil objective for high magnification images (with zoom when needed) and a 4X 0.16 NA objective for low magnification images. High-resolution images were acquired as a z-stack with a 0.2 μm z-interval. When necessary, sequential scanning was used to prevent crosstalk between channels for samples with multiple fluorophores.
Image Analysis/quantification of PLA signal
All images analyzed in this study were taken from the dorsal striatum of 2–3 mice for each genotype. For all experiments, quantifications were performed from at least 8 images (minimum 1 image per hemisphere; 2 slices for each animal; 2 animals). High-resolution (60X 1.4 NA) images from single scans were analyzed in ImageJ (NIH) to calculate the density of PLA puncta. Images were first smoothed and a threshold was selected manually to discriminate PLA puncta from background fluorescence. Once selected, this threshold was applied uniformly to all images in the sample set. The built in macro ‘Analyze Particles’ was then used to count and characterize all objects in the thresholded image. Objects larger than 5 μm2 were rejected, thereby effectively removing nuclei. The remaining objects were counted as PLA puncta.
Statistical analysis
Statistical analyses were performed with the StatView software (SAS institute). In the case of single recognitions of GPCRs (Figure 2), an unpaired t-test was used for the comparison of PLA signal density between WT and either D2R or A2AR KOs mice. For dual recognition (Figure 3), factorial ANOVA was first used to compare the three genotypes. Genotypes were then directly compared by post-hoc analyses (Fischer’s PLSD).
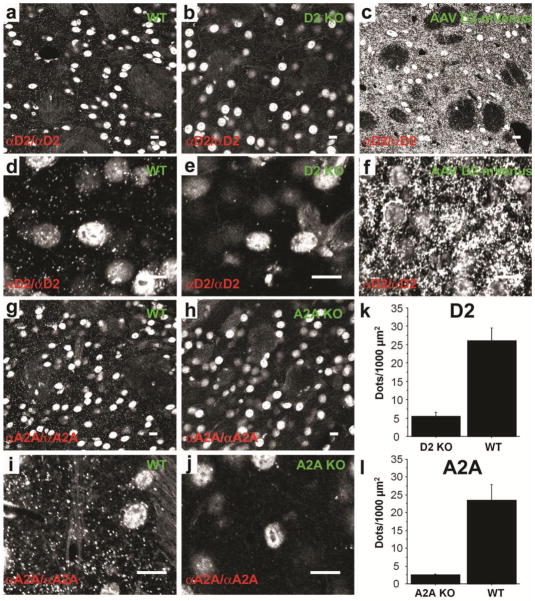
Figure 2. Detection of GPCRs ex vivo in striatal sections using PLA.
D2R was detected in the dorsal striatum of WT mice (a and d). PLA signal from single confocal slice was virtually absent in D2R KO mice (b and e) but was strongly increased when the receptor was overexpressed in the striatum by viral gene transfer of a D2L-R-mVenus (AAV D2-mVenus) (c and f). Similarly A2AR detection in the striatum of WT mice gave a strong PLA signal (g and i) that was absent in A2AR KO mice (h and j). Note that the non-specific nuclear signal (see also SF 3) is similar among genotypes and is unrelated to the presence of primary or secondary antibodies. (k;l) Quantification of PLA signals for D2R (k; unpaired t-test: t= −5.7; p<0.01) and A2AR (l; unpaired t-test: t=−4.2; p<0.01) demonstrates the difference of PLA signal density between WT and KO mice. Scale bars=10 μm.
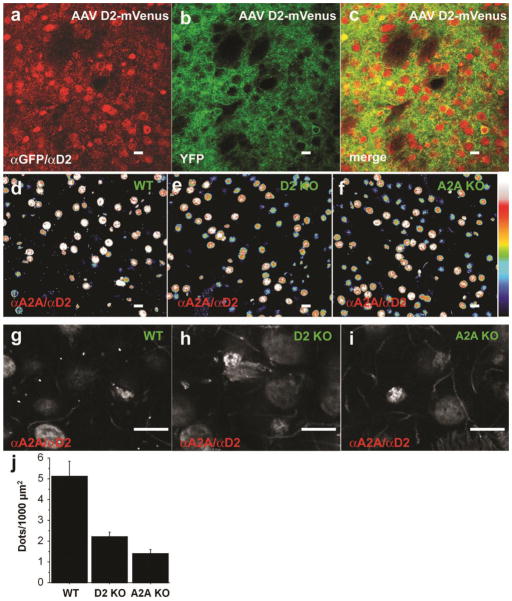
Figure 3. Detection of proximity ex vivo in striatal sections using PLA.
Proximity detection was performed on striatal slices overexpressing the D2L-R-mVenus (AAV D2-mVenus) using antibodies directed against D2-R and GFP (a–c). This can lead to intraprotomer as well as potential interprotomer proximity. (a) PLA signal. (b) Direct fluorescence of mVenus. (c) merge.
Proximity between D2R and A2AR was detected by PLA in the striatum of WT mice (d and g). The signal was virtually absent in both D2R (e and h) and A2AR (f and i) KO mice. (d–f are pseudocolors) (j) Quantification of PLA signals for D2R and A2AR proximity confirmed the significant difference of PLA signal density between WT and KO mice (ANOVA: p<0.01; post-hoc comparison: WT/A2A KO, p<0.01; WT/D2 KO, p=0.01; A2A KO/D2 KO, p=0.46). Scale bars=10 μm.
Immunoblots
Mice were anesthetized with isofluorane, decapitated and the brain was quickly removed and rapidly frozen in isopentane (−30 to −40 °C) and kept at −80° C until processing. Areas of interest were punched (Harris Unicore) from 150–300 μm slices generated with a cryostat (temperature −12 to −15 °C) and kept frozen until protein extraction. Samples were homogenized by application of 8 M urea 2% SDS in TB buffer and brief sonication. 10–40 μg proteins were separated by SDS-PAGE (8–12% acrylamide), transferred to PVDF membranes (BioRad) and immunoblotted. Anti-α-tubulin (Sigma; 1/10000) was used as control for loading. All the HRP-conjugated secondary antibodies were used at 1:5000 dilutions. Immunoblots were visualized with Supersignal West Pico (Pierce, Pittsburgh, PA, USA).
Results/discussion
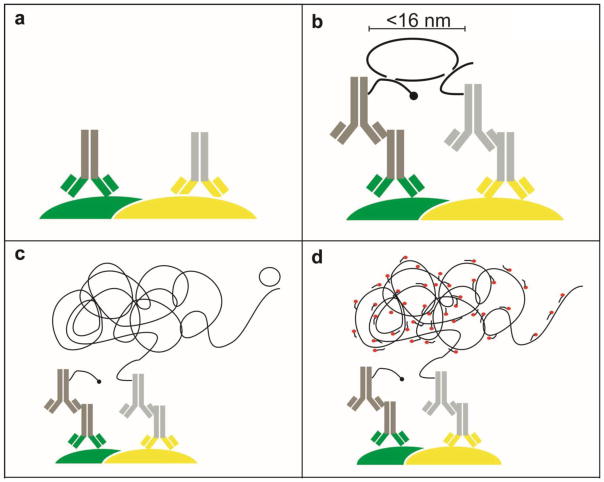
PLA is an antibody-based method in which either a single or two proteins (or antigens) are immunolabeled first with two primary antibodies and then with different species-specific secondary antibodies conjugated to complementary oligonucleotides (7,8). When two antibody molecules are in close proximity, the complementary DNA strands can be ligated, amplified and visualized with a fluorescent probe as distinct puncta. Each spot may represent a single complex containing each of two interacting proteins (or antigens). The maximal distance between the secondary antibodies in this assay is ~16 nm (Figure 1), only slightly larger than that for resonance energy transfer between fluorophores (~10 nm), the most common approach used to infer GPCR oligomerization. By measuring close proximity, PLA allows a validation in vivo of the molecular proximity of two endogenous proteins, something that cannot be established with simple colocalization studies, thereby making it possible to interrogate the existence and localization of interactions in vivo.
Figure 1. The Duolink assay principle.
(a) Two primary antibodies raised in different species are used to detect two target antigens of interest (here as examples, grey/green (left) against a target protein (green) and grey/yellow (right) against a second target protein (yellow)). (b) Each species-specific secondary antibody (dark grey and light grey, respectively) provided in the Duolink kit has a unique short DNA strand attached to it (black line). When the secondary antibodies are in close proximity, the DNA strands can interact through a subsequent addition of two other circle-forming DNA oligonucleotides (circular black line). The distance between the two secondary antibodies is a maximum of 16 nm as calculated from the number of nucleotides in the attached DNA arms. (c) After enzymatic ligation, the two added oligonucleotides are amplified via rolling circle amplification using a polymerase to yield a long concatemeric copy of the circle formed by ligation. (d) After the amplification reaction, labeled complementary oligonucleotide probes are added to highlight the product (red circles).
To establish the validity of such an approach for the detection of molecular proximity ex vivo, we focused on the proposed oligomerization of dopamine D2 and adenosine A2A receptors. There is extensive evidence for D2R-A2AR heteromerization in heterologous expression systems (9,10). In the striatum, an antagonistic functional interaction between these receptors has been shown both at the electrophysiological and behavioral levels using in vivo pharmacological approaches (11,12). The existence of such a heteromeric receptor complex in the striatum has also been recently inferred based on the effects of bivalent ligands (13).
The performance of the PLA system depends critically on the quality of the primary antibodies used, and GPCR antibodies are notoriously problematic (14). We first characterized the specificity of both anti-D2R and anti-A2AR antibodies used in this study by standard fluorescence immunohistochemistry and confocal microscopy (supplementary Figures (SF) 1 and 2). Multiple commercially available antibodies against the D2R showed no signal, or a strong unspecific staining that was still present in tissue from D2R knockout (KO) animals (data not shown). For one antibody tested staining was virtually absent in D2R KO mice, whereas in WT D2R staining was highly enhanced in the striatum compared to adjacent cortical areas (SF 1a), consistent with the pattern of D2R mRNA expression in the mouse brain (Allen brain Atlas). At high magnification some faint non-D2R-related residual staining (mostly somatic) was observed in D2R KO mice (SF 1b), consistent with the presence of a residual band in immunoblots of striatal extracts of D2R KO mice (SF 1c), but immunoblot analyses corroborated the dramatic D2R striatal enrichment (SF 1d). We also performed D2R immunostaining in mice that overexpressed a fusion protein in which the long form of D2R (D2LR) has been fused to the yellow fluorescent protein (mVenus) (striatal injection of D2LR-mVenus adeno-associated virus; AAV D2-mVenus) (SF 1e). D2LR-mVenus was stained robustly by the anti-D2R antibody in a pattern overlapping with mVenus fluorescence, further supporting the specificity of the anti-D2R antibody. The strong A2AR immunoreactivity was completely absent in the striatum of A2AR KO mice highlighting the high specificity of the anti-A2AR antibody (SF 2a). Using the same antibodies, we confirmed that D2R and A2AR partially colocalize in striatal neurons (SF 2b), as shown previously (15–17).
PLA has been used for analyzing protein-protein interactions in vitro. Several studies recently applied this approach to tissue, almost exclusively for the study of post-surgical tumors (18). One recent study used PLA on fresh frozen brain tissue for determining the vascular localization of membrane proteins (19) but to date PLA has not been used successfully for the study of protein-protein interactions in neurons ex vivo.
Tissue processing is a major factor related to subsequent antigen detection by antibodies (20); therefore, using a PLA single recognition paradigm (D2R only), we studied different protocols of fixation and tissue processing (SF 3). The PLA signal was significantly stronger when performed on fixed non-frozen floating sections compared to fixed frozen or fresh frozen sections treated on slides (SF 3). Intriguingly, fixation promoted a strong nuclear signal independent of the PLA signal. This is in accordance with the deleterious effect of aldehydes on DNA that might favor the binding of fluorescent oligonucleotides during the PLA reaction. This strong nuclear fluorescence in non-frozen floating sections was similar in both WT and D2R KO mice, whereas the specific PLA signal was greatly reduced in D2R KO mice (Figure 2a, b, d, e, k). The residual PLA signal in the D2R KO is likely accounted for by the residual staining observed with standard immunohistochemistry in the absence of the D2R (SF 1a,b). The PLA signal was dramatically enhanced in striatal slices in which the D2R was overexpressed (Figure 2c, f), demonstrating the D2R-specificity of the PLA signal. Similarly, there was virtually no PLA signal in A2AR KO mice compared to WT when single-recognition PLA was used to detect A2AR (Figure 2g–j, l). Altogether, these data validate PLA for detection of endogenous GPCRs in brain slices ex vivo. This method could be a reliable approach for the quantification of protein expression as an alternative to standard immunohistochemistry. Indeed, quantification of immunohistochemical signal relies on differences in intensity, a parameter that is extremely variable. In contrast, with the PLA approach, since each puncta represents a high concentration of fluorescence resulting from the single-molecule amplification resulting from several hundred–fold replication of the DNA circle formed as a result of the probe proximity, the puncta/dot number can be quantified independently of the intensity.
In vitro, PLA has been shown to be a reliable readout of molecular proximity of two antigens located on either the same or two distinct proteins (7,8,21). To adapt the approach for ex vivo studies, we first tested the ability to detect mVenus and D2R proximity in striatal slices of mice expressing D2LR-mVenus. This generated a strong PLA signal that colocalized with the direct mVenus fluorescence (Figure 3a–c). The signal was virtually absent in WT mice (SF 5b). Note that because the PLA assay is conducted on fixed tissue, the mobility of proteins in the membrane and/or cytoplasm and associated random collisions of the proteins are not expected to result in significant signal, in contrast to BRET and FRET assays in living cells. Consistent with this expectation, strong AAV-mediated overexpression of GFP in the cytoplasm did not result in an enhanced PLA signal between D2R and GFP (SF 5a). This finding confirms that close proximity – and not simply subcellular colocalization – is required to generate the specific PLA signal. Moreover, these data validate PLA for the detection of antigen proximity within a single protein ex vivo. Such an approach could also be suitable for the study of protein posttranslational modifications in vivo; for example, proximity between a phosphospecific antibody and another nonphosphospecific antibody to a different epitope of the same protein could be used to localize and quantitate phosphorylation ex vivo.
We next tested whether D2R-A2AR oligomers could be detected ex vivo. Performing PLA on striatal sections of WT mice gave a PLA signal that was significantly reduced in both D2R and A2AR KO mice (Figure 3d–j). In contrast to the single recognition of D2R (SF 4a), increasing the polymerase concentration during the amplification reaction improved the dynamic range for detection of interaction between D2R and A2AR (SF 4b). We confirmed the specificity of the D2R-A2AR PLA signal in WT mice by recapitulating the signal with PLA probes with reversed polarity (SF 5c–d). Moreover, the heteromer specific PLA signal – but not the unspecific nuclear one – was completely absent if either one PLA probe (SF 5e) or one primary antibody (SF 5f) was omitted.
To validate the specificity of the PLA approach for the detection of protein-protein interactions at the plasma membrane where random collision is likely to correlate with levels of protein expression, we overexpressed by viral gene transfer in the striatum of WT mice an exogenous non-neuronal protein, the transmembrane glycoprotein “cluster of differentiation 8” (CD8) fused to GFP. Single detection of GFP gave a strong PLA signal, confirming the high expression of CD8-GFP in the membrane of striatal neurons (SF 6a). CD8-GFP overexpression did not alter the formation and detection of D2R-A2AR heteromers by PLA (SF 6b, d). Despite the high expression level of CD8-GFP, the interaction between either D2R or A2AR with CD8-GFP was low (SF 6b′, b″, even if colocalization was quite extensive (SF 6c, c′). Since the level of expression of endogenous D2R and A2AR was much lower than that of CD8-GFP but their PLA signal with each other was substantially higher than with the overexpressed CD8-GFP, these data suggest that random collisions in the plasma membrane are much less effective in generating a PLA signal. Note that the CD8-GFP experiments were performed with an anti-D2R antibody we generated as an improvement over the commercially available antibody (see Methods). We verified its improved specificity by regular immunohistochemistry and single recognition PLA in both WT and D2R KO mice (data not shown). With this antibody we found a comparable amount of D2R-A2AR interaction in the striatum of WT mice as previously shown with the commercially available antibody, but the signal/noise was slightly improved respective to the residual PLA signal in D2R KO mice (compare Figure 3j with SF 6e), presumably due to a decrease in the nonspecific binding observed with the commercial antibody.
In summary, we have successfully translated PLA for the detection of single and interacting GPCRs ex vivo in brain tissue. By validating this approach in brain slices we provide evidence that PLA is a simple yet powerful tool for studying protein-protein interaction as well as antigen proximity in their native state ex vivo as initially described in vitro (8). Accordingly, ex vivo PLA can be used as a complementary approach to validate the existence of GPCR oligomers characterized in vitro, as we have recently demonstrated for dopamine D2 and delta-opioid receptors (Aburi et al. in preparation). Moreover, the fact that this assay is performed on intact fixed tissue offers the opportunity of directly studying the localization and regulation of endogenous oligomers.
Supplementary Material
Acknowledgments
We thank H. Thomas Lee for the generous gift of the A2A KO mice and Eleanor Simpson and Mahalaxmi Aburi for their helpful comments on the manuscript. This work was supported in part by US National Institutes of Health grants DA022413, MH054137 (to J.A.J.) and F32NS064732 (to R.A.P), by the Research Associate Award from the Research Foundation for Mental Hygiene (to P.T.), by the Lieber Center for Schizophrenia Research and Treatment (to J.A.J), an EMBO Long-Term fellowship and the Basque Country Government (to E.U.), and by a post-doctoral fellowship from the Fondation pour la Recherche Médicale (to M-L.R.).
Footnotes
Authors contributions:
P.T., M-L.R., E.U. and J.A.J conceived the study and designed the experiments; P.T., J.C. performed the experiments; R.A.P generated the CD8-GFP construct and lentivirus and tested its efficiency for intracerebral injection. H.D.V. supervised and helped with imaging; C.S. raised and provided the D2R KO mice; M.S. and M.G. provided reagents; P.T. analyzed the data; P.T. and J.A.J wrote the manuscript; all the authors participated to the interpretation of the data and edited the manuscript.
Competing interests statement:
Maria Slättman and Mats Gullberg work for the company that developed the technique that has been adapted in this study.
References
- 1.Kenakin T, Miller LJ. Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol Rev. 2010;62:265–304. doi: 10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pin JP, Neubig R, Bouvier M, Devi L, Filizola M, Javitch JA, Lohse MJ, Milligan G, et al. International Union of Basic and Clinical Pharmacology. LXVII. Recommendations for the recognition and nomenclature of G protein-coupled receptor heteromultimers. Pharmacol Rev. 2007;59:5–13. doi: 10.1124/pr.59.1.5. [DOI] [PubMed] [Google Scholar]
- 3.Albizu L, Cottet M, Kralikova M, Stoev S, Seyer R, Brabet I, Roux T, Bazin H, et al. Time-resolved FRET between GPCR ligands reveals oligomers in native tissues. Nat Chem Biol. 2010;6:587–594. doi: 10.1038/nchembio.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta A, Mulder J, Gomes I, Rozenfeld R, Bushlin I, Ong E, Lim M, Maillet E, et al. Increased abundance of opioid receptor heteromers after chronic morphine administration. Sci Signal. 2010;3:ra54. doi: 10.1126/scisignal.2000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung MY, Skryabin BV, Arai M, Abbondanzo S, Fu D, Brosius J, Robakis NK, Polites HG, et al. Potentiation of the D2 mutant motor phenotype in mice lacking dopamine D2 and D3 receptors. Neuroscience. 1999;91:911–924. doi: 10.1016/s0306-4522(98)00705-2. [DOI] [PubMed] [Google Scholar]
- 6.Chen JF, Huang Z, Ma J, Zhu J, Moratalla R, Standaert D, Moskowitz MA, Fink JS, Schwarzschild MA. A(2A) adenosine receptor deficiency attenuates brain injury induced by transient focal ischemia in mice. J Neurosci. 1999;19:9192–9200. doi: 10.1523/JNEUROSCI.19-21-09192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 8.Soderberg O, Leuchowius KJ, Gullberg M, Jarvius M, Weibrecht I, Larsson LG, Landegren U. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods. 2008;45:227–232. doi: 10.1016/j.ymeth.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Fuxe K, Ferre S, Canals M, Torvinen M, Terasmaa A, Marcellino D, Goldberg SR, Staines W, et al. Adenosine A2A and dopamine D2 heteromeric receptor complexes and their function. J Mol Neurosci. 2005;26:209–220. doi: 10.1385/JMN:26:2-3:209. [DOI] [PubMed] [Google Scholar]
- 10.Agnati LF, Guidolin D, Leo G, Carone C, Genedani S, Fuxe K. Receptor-receptor interactions: A novel concept in brain integration. Prog Neurobiol. 2010;90:157–175. doi: 10.1016/j.pneurobio.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Agnati LF, Leo G, Vergoni AV, Martinez E, Hockemeyer J, Lluis C, Franco R, Fuxe K, Ferre S. Neuroprotective effect of L-DOPA co-administered with the adenosine A2A receptor agonist CGS 21680 in an animal model of Parkinson’s disease. Brain Res Bull. 2004;64:155–164. doi: 10.1016/j.brainresbull.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Ferre S, Ciruela F, Canals M, Marcellino D, Burgueno J, Casado V, Hillion J, Torvinen M, et al. Adenosine A2A-dopamine D2 receptor-receptor heteromers. Targets for neuro-psychiatric disorders. Parkinsonism Relat Disord. 2004;10:265–271. doi: 10.1016/j.parkreldis.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Soriano A, Ventura R, Molero A, Hoen R, Casado V, Cortes A, Fanelli F, Albericio F, et al. Adenosine A2A receptor-antagonist/dopamine D2 receptor-agonist bivalent ligands as pharmacological tools to detect A2A-D2 receptor heteromers. J Med Chem. 2009;52:5590–5602. doi: 10.1021/jm900298c. [DOI] [PubMed] [Google Scholar]
- 14.Michel MC, Wieland T, Tsujimoto G. How reliable are G-protein-coupled receptor antibodies? Naunyn Schmiedebergs Arch Pharmacol. 2009;379:385–388. doi: 10.1007/s00210-009-0395-y. [DOI] [PubMed] [Google Scholar]
- 15.Fink JS, Weaver DR, Rivkees SA, Peterfreund RA, Pollack AE, Adler EM, Reppert SM. Molecular cloning of the rat A2 adenosine receptor: selective co-expression with D2 dopamine receptors in rat striatum. Brain Res Mol Brain Res. 1992;14:186–195. doi: 10.1016/0169-328x(92)90173-9. [DOI] [PubMed] [Google Scholar]
- 16.Augood SJ, Emson PC. Adenosine A2a receptor mRNA is expressed by enkephalin cells but not by somatostatin cells in rat striatum: a co-expression study. Brain Res Mol Brain Res. 1994;22:204–210. doi: 10.1016/0169-328x(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 17.Svenningsson P, Le Moine C, Kull B, Sunahara R, Bloch B, Fredholm BB. Cellular expression of adenosine A2A receptor messenger RNA in the rat central nervous system with special reference to dopamine innervated areas. Neuroscience. 1997;80:1171–1185. doi: 10.1016/s0306-4522(97)00180-2. [DOI] [PubMed] [Google Scholar]
- 18.Weibrecht I, Leuchowius KJ, Clausson CM, Conze T, Jarvius M, Howell WM, Kamali-Moghaddam M, Soderberg O. Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev Proteomics. 2010;7:401–409. doi: 10.1586/epr.10.10. [DOI] [PubMed] [Google Scholar]
- 19.Winkler EA, Bell RD, Zlokovic BV. Pericyte-specific expression of PDGF beta receptor in mouse models with normal and deficient PDGF beta receptor signaling. Mol Neurodegener. 2010;5:32. doi: 10.1186/1750-1326-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paavilainen L, Edvinsson A, Asplund A, Hober S, Kampf C, Ponten F, Wester K. The impact of tissue fixatives on morphology and antibody-based protein profiling in tissues and cells. J Histochem Cytochem. 2010;58:237–246. doi: 10.1369/jhc.2009.954321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mocanu MM, Varadi T, Szollosi J, Nagy P. Comparative analysis of fluorescence resonance energy transfer (FRET) and proximity ligation assay (PLA) Proteomics. 2011;11:2063–2070. doi: 10.1002/pmic.201100028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.