Abstract
Seagrass beds are the foundation species of functionally important coastal ecosystems worldwide. The world’s largest losses of the widespread seagrass Zostera marina (eelgrass) have been reported as a consequence of wasting disease, an infection with the endophytic protist Labyrinthula zosterae. During one of the most extended epidemics in the marine realm, ∼90% of East and Western Atlantic eelgrass beds died-off between 1932 and 1934. Today, small outbreaks continue to be reported, but the current extent of L. zosterae in European meadows is completely unknown. In this study we quantify the abundance and prevalence of the wasting disease pathogen among 19 Z. marina populations in northern European coastal waters, using quantitative PCR (QPCR) with primers targeting a species specific portion of the internally transcribed spacer (ITS1) of L. zosterae. Spatially, we found marked variation among sites with abundances varying between 0 and 126 cells mg−1 Z. marina dry weight (mean: 5.7 L. zosterae cells mg−1 Z. marina dry weight ±1.9 SE) and prevalences ranged from 0–88.9%. Temporarily, abundances varied between 0 and 271 cells mg−1 Z. marina dry weight (mean: 8.5±2.6 SE), while prevalences ranged from zero in winter and early spring to 96% in summer. Field concentrations accessed via bulk DNA extraction and subsequent QPCR correlated well with prevalence data estimated via isolation and cultivation from live plant tissue. L. zosterae was not only detectable in black lesions, a sign of Labyrinthula-induced necrosis, but also occurred in green, apparently healthy tissue. We conclude that L. zosterae infection is common (84% infected populations) in (northern) European eelgrass populations with highest abundances during the summer months. In the light of global climate change and increasing rate of marine diseases our data provide a baseline for further studies on the causes of pathogenic outbreaks of L. zosterae.
Introduction
Seagrass beds are among the most threatened coastal ecosystems worldwide [1] while at the same time, they provide very important ecological functions as nursery habitat, sediment stabilizer, and via carbon and nutrient fixation [2]. We are now witnessing a century of accelerated seagrass decline driven by growing human populations, coastal development, ecological degradation and climate change [1], [3], [4]. However, the world’s largest and fastest losses of Zostera marina occurred in the 1930’s and were attributed to eelgrass wasting disease, caused by the net-slime mold Labyrinthula zosterae (Straminopiles, an endophytic protist reviewed by [5]). Among the many other known factors causing eelgrass decline, the role of pathogens has so far largely been neglected, although diseases are already noticeably on the increase not only in marine ecosystem [6], [7]. The main objective of this study was to obtain first quantitative data on the prevalence and abundance of the wasting disease pathogen Labyrinthula zosterae in contemporary Z. marina populations of Northern Europe.
Although detailed data are scarce, it is generally accepted that Z. marina beds were very common before the disease struck throughout the North Atlantic (see e.g. [8] for the Wadden Sea, [9] for the Netherlands, [10] for Danmark, [11] for the German Baltic and [12] 2008 for France). Historical records of a large eelgrass industry producing insulation and mattresses suggest high abundances of extended eelgrass beds in France, The Netherlands and Canada [13], [14]. This changed dramatically when in the 1930’s, a pandemic caused by the net-slime mold L. zosterae struck eelgrass beds on both sides of the North Atlantic. Beginning in 1930, eelgrass beds disappeared from large areas ranging from New Brunswick to north-west Carolina at the Atlantic West Coast within only two years [15], [16]. In 1931, similar die-offs were reported from Brittany and the Norman-Breton Gulf in France [17], and in the subsequent year from sublitoral eelgrass beds in the Dutch Wadden Sea [18]. In 1933, the epidemic reached southeast England [19], the northern German Wadden Sea [13] and the Danish west coast, while it arrived in Norway and the Baltic in 1934 [13], [20]. Eelgrass bed recolonization was slow and accompanied by new outbreaks until 1965 [12], [21]–[24]. In many regions, only intertidal meadows have recovered [8], [25], while subtidal Z. marina beds have never recovered and are today restricted to remnant patches within tidal creeks [26], [27]. In the 1980s new outbreaks of wasting disease were reported from the Atlantic coast of Nova Scotia and New England [24] and the Pacific northwest coast of North America [28], Brittany (France) and Grevelingen lagoon (The Netherlands, [24]), demonstrating that pathogenic strains of L. zosterae are still present in contemporary eelgrass beds.
During pathogenic outbreaks of L. zosterae, eelgrass plants exhibit a fast spread of black lesions on all leaves within hours, followed by leaf abscission, rhizome discoloration and mortality [29]–[31]. Even in the 1930’s, Labyrinthula was microscopically identified in diseased plants and experimental inoculation of healthy plants with infected leaves was reproducible [29]. In 1991, after the recurrence of wasting disease on the Atlantic and Pacific US coasts, Muehlstein et al. [30] identified Labyrinthula zosterae as the causative agent of wasting disease according to Kochs postulates. A recent survey of Labyrinthula-isolates (N = 53) from six northern European sites and one southern location (Adriatic Sea) identified three species based on a 1400 bp region of the 18S small subunit rDNA, all isolated from apparently healthy Z. marina beds. While the most common isolate was L. zosterae, two additional culturable Labyrinthula species were also found [32].
In order to quantify infection, Burdick et al. [33] introduced the “wasting index”, which estimates the percentage of necrotic tissue for each leaf on a vegetative shoot. Although valuable as a first step towards quantification, this indirect method has several disadvantages. First, not all lesions are caused by Labyrinthula spp. and second, not all Labyrinthula spp. result in observable lesions (see also [23]). Most importantly, we still do not know what triggers the pathogenic outbreaks of L. zosterae, given that the endophyte has been and remains omnipresent in eelgrass beds ([31], pre-wasting disease; [34], post-wasting disease in the 1930s; and [32] contemporary eelgrass beds).
Thus, a method was needed that allows the determination of L. zosterae abundance independent of the presence or absence of lesions. To this end, we previously developed a quantitative PCR (QPCR) assay based on species specific ITS primers [35], using DNA extraction from live or dried plant tissue.
In the present study, we (1) surveyed Z. marina tissue with our QPCR assay across 19 locations of its European range including Portugal, Germany, Denmark, southern Norway and western Sweden; (2) we compared our assay against the presence of lesions and success in isolating L. zosterae (for a subset of five locations) and (3) followed L. zosterae concentration over time at one western Baltic and one in the Wadden Sea location. The goal of the study was to establish a baseline of prevalence of the endophyte including temporal variation in infection.
Materials and Methods
Sampling
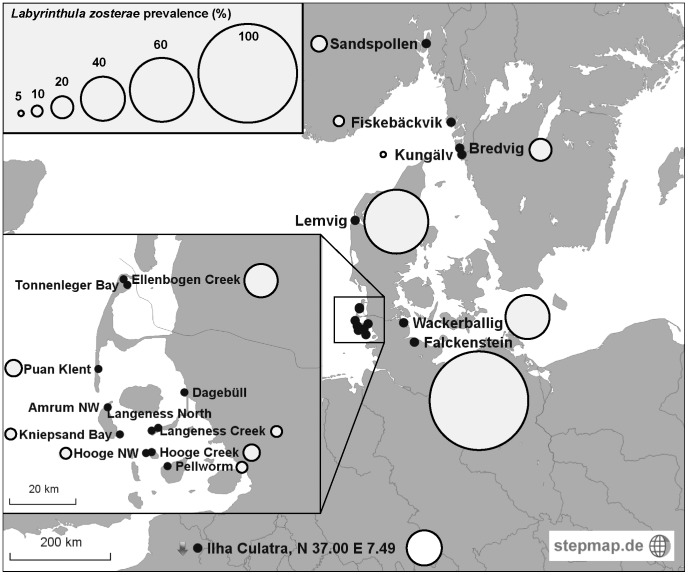
In total, we sampled 19 coastal sited in a water depth of 0.5–3 m (Fig. 1). Eighteen of the 19 sites were situated within the affected region of the 1930’s wasting disease epidemic, while they presently show no signs of decline due to wasting disease (Fig. 1). We were particularly interested to analyze the few remnant permanently submerged Zostera marina populations in Wadden Sea tidal creeks, because they are the only subtidal sites that recovered after the wasting disease. These subtidal populations consist of vegetation patches of 0.5–5 m width, distributed along creek banks (33% cover, ±5.5 SE). The intertidal populations sampled in the Wadden Sea are continuous but show sparse eelgrass coverage (mean of all sample sites: 13.4% ±0.5 SE) with low shoot densities (71 shoots m−2±1.8 SE). Although intertidal plants are phenotypically distinct from subtidal Z. marina (e.g. shoot lengthSylt_intertidal_september_2012∶24.7 cm ±0.9 SE, shoot length Sylt_subtidal_september_2012∶63.3 cm ±2.9 SE). Microsatellite analysis confirmed low but significant genetic differentiation (F ST = 0.009, P = 0.067) between Wadden Sea populations, resulting from divergent selection detected on genes linked with three of 25 microsatellite loci tested [36]. All other populations in this study were continuous eelgrass beds in 0.5–3 m water depth, extending over several 100 of m2 (Table 1).
Figure 1. Prevalences of Labyrinthula zosterae in Zostera marina populations.
Circle size proportional to percent prevalence, n = 18–21.
Table 1. Sampling locations, salinity and sample size for assessing spatial variance in abundance and prevalence.
| Area | Location | Geograph.coordinates | Sampling date | Salinity (psu) | N | % leaves with lesions | % pos. cult. from leaves with lesions | ||||||||
| Langeness, Wadden Sea, Germany | Langeness North | N 54.6396 E 08.5781 | 22.07.12 | >30 | 20 | 0 | No data | ||||||||
| Langeness Creek* | N 54.6320 E 08.5440 | 28.06.11 | >30 | 20 | 13 | No data | |||||||||
| Hooge, Wadden Sea, Germany | Hooge-NW | N 54.5700 E 08.5180 | 23.07.12 | >30 | 20 | 0 | No data | ||||||||
| Hooge Creek | N 54.5723 E 08.5240 | 23.07.12 | >30 | 20 | 0 | No data | |||||||||
| Amrum, Wadden Sea Germany | Kniepsand Bay | N 54.6203 E 08.3970 | 20.07.12 | >30 | 20 | 0 | No data | ||||||||
| Amrum-NW | N 54.6960 E 08.3400 | 19.07.12 | >30 | 20 | 0 | No data | |||||||||
| Pellworm, Wadden Sea, Germany | Pellworm | N 54.5504 E 08.5990 | 26.07.12 | >30 | 20 | 0 | No data | ||||||||
| Dagebüll, Wadden Sea, Germany | Dagebüll | N 54.7212 E 08.7051 | 24.07.12 | >30 | 20 | 0 | No data | ||||||||
| Sylt, Wadden Sea, Germany | Puan Klent | N 54.0798 E 08.2960 | 18.07.12 | >30 | 20 | 2 | No data | ||||||||
| Tonnenleger Bay | N 55.0258 E 08.4323 | 17.07.12 | >30 | 20 | 0 | No data | |||||||||
| Ellenbogen Creek | N 55.0410 E 08.4130 | 04.07.11 | >30 | 20 | 40 | 42.86 | |||||||||
| Limfjord, Denmark | Lemvik* | N 56.6300 E 08.2961 | 28.05.11 | >30 | 19 | 58 | 100.00 | ||||||||
| Skagerrak, Norway | Sandspollen* | N 59.6657 E 10.5869 | 10.05.10 | 20–25 | 21 | 57 | 100.00 | ||||||||
| Åbyfjord, Sweden | Fiskebäckvik* | N 58.3362 E 11.4078 | 01.06.11 | 20–25 | 18 | No data | 11.11 | ||||||||
| Gullmarsfjord, Sweden | Bredvik+Snäckebacke-bukten | N 58.1987 E 11.3244 | 03.07.11 | 20–30 | 20 | No data | No data | ||||||||
| Kungälv | N 57.5405 E 11.4083 | 04.07.11 | 6–14 | 20 | No data | No data | |||||||||
| Flensburg Fjord, Germany | Wackerballig* | N 54.7557 E 09.8668 | 12.07.11 | 15–17 | 19 | 80 | 100.00 | ||||||||
| Kiel Fjord, Germany | Falckenstein | N 54.3954 E 10.1935 | 15.07.11 | 15–17 | 20 | 95 | 100.00 | ||||||||
| Faro lagoon, Portugal | Ilha Culatra | N 37.0005 W 07.4921 | 05.08.11 | 36 | 20 | No data | No data | ||||||||
The percentage of Zostera marina plants with lesions and successful Labyrinthula isolations are shown where available,
= subset of 5 populations chosen for methods comparison.
At each site, fresh leaves of at least twenty Z. marina-shoots were collected between May and August of the years 2010 (1 site), 2011 (8 sites) and 2012 (10 sites, Table 1), separately stored in Zip-lock bags with ambient sea water and kept cool until return to the lab 1–3 days later. Sampling at Ellenbogen Creek was permitted by nature conservation authority and Mr. Diedrichsen, the owner of this private property. We took care that by picking a leaf piece the entire plant was kept alive in situ and/or sampled outside areas not open to public. Therefore no special permission was necessary at all other sites.
Before starting the spatial survey, we wanted to address within-plant variation in Labyrinthula zosterae abundance. To this end DNA was extracted from all leaves of eight individual plants of two sites (Lemvig and Wackerballig), dividing each leaf in three sections (top, middle, basis). Initial QPCR-assay results revealed that the highest L. zosterae prevalences and/or abundances were found in the middle part of the 3rd oldest leaf (for means and statistical tests see Tables 2 and 3); therefore, we analyzed the 3rd leave in all subsequent samples.
Table 2. Mean Labyrinthula zosterae abundance and prevalence in different leaf parts.
| Leaf part | N | L. zosterae cells×mgplant DW−1 | Std. Err | Prevalence (%) |
| Top | 27 | 0.30 | 0.14 | 18.92 |
| Middle | 22 | 71.31 | 67.54 | 38.71 |
| Basis | 19 | 9.37 | 3.08 | 31.03 |
Abundance per g Zostera marina dry weight (DW) with standard errors: Wilcoxon-Kruskal-Wallis Testleaf part: df = 2, X2 = 6.05, p = 0.05, planned comparisonabundance: top< middle **, basis<middle**. Prevalence (%): Nominal logistic regressionleaf parts: df = 2, deviance = 14.47, p = 0.001, planned comparisonprevalence: top< middle*, * = significantly different at p>0.05, ** = p<0.02.
Table 3. Mean Labyrinthula zosterae abundance and prevalence among different Zostera marina leaves.
| Leaf number | N | L. zosterae cells×mg plant DW−1 | Std. Err. | Prevalence (%) |
| 1 | 16 | 6.00 | 2.52 | 12.50 |
| 2 | 19 | 5.22 | 2.01 | 10.53 |
| 3 | 18 | 6.00 | 80.84 | 50.00 |
| 4 | 12 | 0.33 | 0.09 | 33.33 |
| 5 | 3 | 2.48 | 0.00 | One data point only |
Abundance per g Zostera marina weight (DW) with standard errors: Wilcoxon-Kruskal-Wallis-testleaf number: df = 4, X2 = 5.37, p = 0.25). Prevalence (%): Nominal logistic regressionleaf number: df = 4, deviance = 9.71, p = 0.05, planned comparisonprevalence: leaf 2<3**, ** = significantly different at p>0.02.
After sampling, leaves from all populations were air dried. Leaves from five of these populations (Table 1) were additionally examined for black lesions on the leaves. Then all leaves were cut in half, longitudinally. One half was dried for later DNA extraction, the other half served as inoculum for cultivation of Labyrinthula zosterae on seawater-agar medium.
To assess temporal variation in L. zosterae prevalence and abundance, the same population was sampled 14x in Falckenstein (7.4., 21.4., 5.5., 19.5., 9.6., 23.6., 7.7., 15.7., 5.8., 28.9., 1.11. and 28.1.2011, 23.2. and 25.3.2012) and 6x at Ellenbogen creek (18.5., 9.6., 4.7., 4.8., 5.9. und 10.11.2011).
DNA Extraction
Ca. 2–4 mg of dried leaf material was first ground in a ball mill (Retsch, Germany) at maximal speed setting for 5 min. DNA extractions of L. zosterae were performed with an Invisorb spin tissue mini kit (Invitek, Berlin, Germany) following the manufacturer’s instructions. To enhance extraction efficiency and to ensure that even low amounts of target DNA were carried through the filter absorption steps, 1 µL (containing ∼500 ng) of UltraPure™ salmon sperm DNA solution (Invitrogen, life technologies, USA) was added to each extraction to saturate silica columns with DNA. Target DNA was purified using a one-step PCR inhibitor removal kit (Zymo Research, USA).
Quantitative PCR (QPCR)-assay
Following on the original assay protocol of Bergmann [35] we modified the method to enhance specificity and sensitivity by developing a novel, TaqMan based assay with the consensus sequence of Labyrinthula zosterae ([35]; accession numbers JN121409-13). Using the software PrimerXpress (Applied Biosystems) the forward primer Laby_ITS_Taq_f: TTGAACGTAACATTCGACTTTCGT and the reverse primer Laby_ITS_Taq_r: ACGCATGAAGCGGTCTTCTT were identified, along with the probe Laby_ITS_Taq_pr: TGGACGAGTGTGTTTTG that carried the fluorescence label 6-Fam at the 5′ end and the dark quencher BHQ-1 at the 3′ end. Reactions were carried out using standard conditions recommended by the manufacturer using the 10 µL TaqMan universal Master Mix (Applied Biosystems, now Life Technologies) in a 20 µL reaction volume: 2 µL 1∶10 diluted template DNA, 2.4 µL (40.8 nM) of the two primers, 2.4 µL Milli-Q H2O and 0.8 µL probe (50 nM), respectively. The thermo-cycling program on a Step-One QPCR machine was 2 min at 50°C and 10 min at 95°C, followed by 48 cycles at 95°C for 15 s and 1 min at 60°C. All samples were tested in triplicate and the standard deviation of triplicates never exceeded 0.3 units of cycle threshold (Ct). Only CT values <39 were considered. Standard curves using preparations of Labyrinthula zosterae with known cell numbers attained correlation coefficients between r2 = 0.97 and 0.99 and a detection limit of ∼0.01 cells. Abundance as the number of L. zosterae cells in each milligram (dry weight) Zostera marina sample was calculated from the linear regression of the standard curve (standard cell number against mean standard Ct calculated from all QPCR reactions; 150 cells 22.493 Ct ±0.060 SE, 15 cells = 27.080 Ct ±0.080 SE, 0.5cells = 32.215 Ct ±0.125 SE).
where a = intercept, b = slope and w = sample dry weight. Cell numbers were multiplied by 10 because the samples were diluted 1∶10 prior QPCR.
Prevalence was calculated as the percentage of samples of each site with a Ct<39.
Cultures
Seawater-agar medium
For one liter of seawater-agar medium (for 50 Petri-dishes 10 cm in diameter) : 12 g agar (bacteriological grade, Roth, Germany ), 1 g glucose, 0.1 g yeast extract (Roth, Germany), 0.1 g peptone (Fluka, Germany) in 1 L Milli-Q water were mixed and autoclaved 30 min at 121°C. Immediately following the autoclave step, 25 g Instant Ocean (Instant Ocean, Spectrum Brands, USA) artificial sea salt was added (salinity: 25 psu). After cooling to 50°C, 25 mL Penicillin-Streptomycin (MP Biomedicals, USA) and 10 mL horse serum (Invitrogen, USA) were added, mixed, and the medium poured immediately.
Labyrinthula-isolation
Ca. 2 cm-long leaf pieces taken from the middle part of each 3rd leaf were dipped in 0.5% hypochlorite (bleach) solution in seawater for 20 s of surface sterilization, rinsed with Milli-Q water for 10 s and soaked in artificial seawater for 1 min. Washed leaf samples were separately placed on the agar plates and incubated at 25°C in a climate cabinet without light. Cultures were checked under the dissecting scope after three, five and eight days for growing L. zosterae.
Statistical Analysis
To compare mean Labyrinthula zosterae-abundances (cell numbers obtained by QPCR-assay) we used non-parametric tests because data were markedly non-normally distributed. L. zosterae abundance in different positions of the leaf/in leaves of different age was compared by Wilcoxon/Kruskal-Wallis-tests (implemented in software JMP 9, SAS Institute, USA) followed by planned contrasts to identify which leaf parts/leaves were different. Likewise, spatial and temporal patterns in L. zosterae abundance were tested for statistically significant variation using Wilcoxon/Kruskal-Wallis-tests (implemented in software JMP 9, SAS Institute, USA). Nominal logistic regression was applied to nominal data, i.g. the presence/absence of lesions on leaves and prevalence measurements. Prevalence was defined depending on the method used. Using culturing, it was defined as successful/unsuccessful isolation of L. zosterae from fresh leaf material. When using QPCR on dried leaf material, positive prevalence was defined as a PCR reaction with a Ct-value <39, from dried leaf material.
Results
Prevalence and Abundance of Labyrinthula Zosterae
Using the QPCR assay, L. zosterae was present in 16 of 19 populations tested, with a statistically significant variation of prevalence among sites (Nominal logistic regressionsite: df = 18, deviance = 116.06, p = 0.0001). Because we had no a priori expectations about site-specific abundances, we did not perform any post-hoc tests. The highest prevalence of 88.9% was found in Falckenstein, the population in Kiel Fjord. Lemvig plants were ranked second in terms of prevalence (58%, Fig. 1). The Swedish Kungälv population showed the lowest prevalence (5%). No L. zosterae was found in Tonnenleger Bay, Amrum NW, Dagebüll and Langeness North (intertidal populations, Table 1).
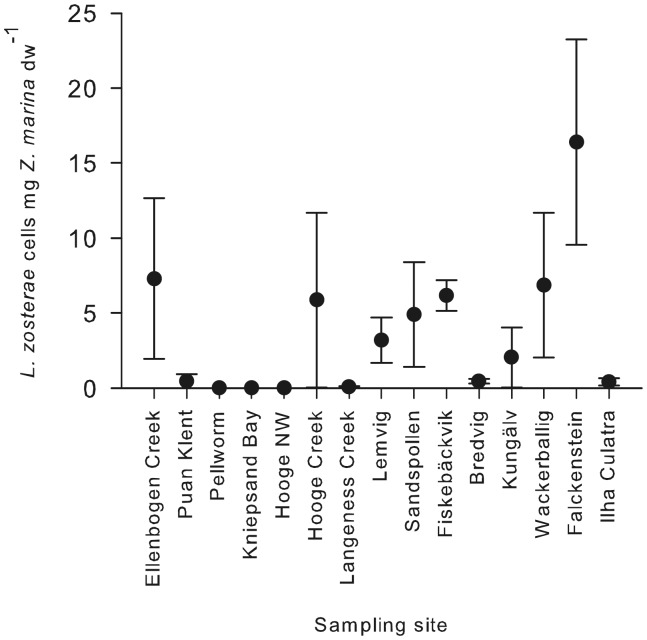
The abundance of L. zosterae was standardized relative to eelgrass dry weight (DW) and revealed high variation within and among sites (minimum: 0.01 L. zosterae cells mg−1 plant DW, maximum: 504 L. zosterae cells mg−1 plant DW, Fig. 2). Note that cell numbers <1 are possible because the amplified ITS-region belongs to the multi-copy rDNA gene and the detection limit per PCR-reaction was 0.01 cells. Similar to prevalence, abundance was highest in Falckenstein (16.40 cells mg−1 plant DW ±6.84 SE), followed by Fiskebäckvik (6.17 cells mg−1 plant DW ±1.03 SE) as shown in Fig. 3. The lowest abundances were found in the positive samples from Hooge NW and Pellworm Creek (0.01 cells mg−1 plant DW). Site differences were significant (Wilkoxon/Kruskal-Wallis-testsite: χ2 = 25.27, df = 14, p = 0.032; note that only positive values were included into the analysis resulting in an exclusion of sites without L. zosterae).
Figure 2. Differences in the abundance of Labyrinthula zosterae in infected Zostera marina plants from 15 sites.
Means with standard error bars, N = 18–21.
Figure 3. Temporal variation in the abundance and prevalence of Labyrinthula zosterae in infected Zostera marina plants.
Means with standard error bars, N = 10–25, Falckenstein = Baltic Sea, Ellenbogen Creek = Wadden Sea (sublitoral).
Lesion, Isolation and Prevalence of Labyrinthula Zosterae
For a subset of five sites, we investigated the presence of lesions and the isolation success of Labyrinthula zosterae in addition to QPCR-assay analysis. Prevalences of Labyrinthula zosterae assessed as isolation success via cultivation did not differ significantly from obtained via the QPCR assay. We analyzed the method applied together with site differences in prevalence in one model. Differences were only found for site and not for the method used (Fig. 4, Nominal logistic regressionmethod and site: method: df = 1, deviance = 0.04, p = 0. 850, site: df = 2, deviance = 20.28, p = 0.0004, method×site: df: 4, deviance 3.245, p = 0.5177, ns). The mean prevalence across all sites was 26% for the QPCR-approach and 30% for the isolation approach.
Figure 4. Differences in the prevalence of Labyrinthula zosterae in Zostera marina-samples detected by QPCR-assay versus isolation.
Samples from five sites, isolation on seawater-agar-culture plates, N = 18–21.
The percentage of leaves with lesions (small black or brown spots, between 1 mm and 2 cm in diameter) differed markedly among populations ranging from 11% in Fiskebäckvik to 80% in Wackerballig (Table 1, Nominal logistic regressionlesion for site: df = 4, deviance = 17.81, p = 0.0013). Across all sites, the probability of obtaining a positive L. zosterae culture or a positive QPCR result was significantly higher in leaves with lesions that without, although the protist was also found in plants without lesions. 48.8% of the leaves where L. zosterae has been detected by QPCR showed lesions, whereas the protist was only found in 10.4% leaves without lesions (Nominal logistic regressionlesion: df = 1, deviance = 15.87, p = 0.001, log odds ratio = 1.39, SE = 0.585). Using isolation, L. zosterae could be detected in 56.5% leaves with lesions but only in 8.3% without (Nominal logistic regressionlesion: df = 1, deviance = 32.37, p = 0.0001, log odds ratio = 2.88, SE = 0.616).
Interestingly, isolates of L. zosterae were easily obtained from lesions on the leaves at Sandspollen, Fiskebäckvik, Wackerballig and Lemvig, whereas this was not the case with the leaves from sublitoral eelgrass plants in Ellenbogen Creek. Here, 57% of the Labyrinthula isolated came from green leaves without any lesions.
Temporal Variation in Abundance and Prevalence of Labyrinthula zosterae
At two selected sites, prevalence and abundance of L. zosterae were monitored throughout one year. Overall the temporal patterns were congruent. Prevalence data varied strongly and ranged between 0 and 25% between April and June, 67–95% between the end of June and September. At the western Baltic Sea site of Falckenstein (Table 1 and Fig. 3) L. zosterae occurred at very low abundances between April and June (0.01–0.09 cells mg−1 Z. marina dry weight), increasing from the end of June and September (4.4–24.3 cells mg−1 Z. marina dry weight) and declining from October until March (ca. 1 cell/mg Z. marina dry weight (Wilkoxon/Kruskal-Wallis-testsampling date: df = 12, χ2 = 141.40, p<0.0001). The Wadden Sea site at Ellenbogen Creek (Table 1 and Fig. 3) revealed much lower prevalences and abundances than the Baltic Sea Falckenstein location. Here, only about 20% of plants were infected during the July-August period and abundances also remained low (0.6–0.9 cells mg−1 Z. marina dry weight, Wilkoxon/Kruskal-Wallis-testsampling date: df = 4, χ2 = 28.256, p<0.0001).
Discussion
After nearly a century of investigations on Labyrinthula zosterae as putative agent of eelgrass wasting disease there is still no conclusive picture of what triggers pathogenic outbreaks. We show here that background prevalence is extremely high in contemporary eelgrass beds in northern Europe, with up to 89% of the plants carrying L. zosterae. Using a specific QPCR assay, we show that Labyrinthula zosterae is present in almost all populations assessed even though most plants showed few lesions, let alone signs of an epidemic outbreak. The QPCR assay thus provides a valuable tool to assess background levels (∼0.01 cells mg−1 DW) of L. zosterae independent of lesions. Prevalence, as determined by either QPCR data or isolation and culture were comparable. Since the latter is far more laborious and slow, preference should be given to a QPCR assay which also works with dried samples. A direct comparison of QPCR values with the “wasting disease index” [33] has to be interpreted with caution, as the QPCR and the wasting index measure different processes. The wasting disease index reflects the cumulative pathogenic effects of a L. zosterae infection (including e.g. defense reactions of the plant), whereas the QPCR value reflects abundance only. The two should be seen as complementary.
Currently we do not know whether the very low background concentrations of the L. zosterae endophyte in winter and spring are to the only inoculum that gives rise to high abundances during summer, or whether eelgrass leaves are secondarily infected every year from L. zosterae spores the environment. Although a number of life history studies on L. zosterae have been conducted earlier [37]–[40], the details of zoospore formation as well as the emergence and location of resting stages (cysts) in the environment remain unknown. While have not yet searched for resting stages in the sediments and/or water column our QPCR approach may be the suitable tool to do so. Equally unknown is how the endophyte disperses which could take place via the drift of decaying infected leaves. L. zosterae can be transmitted rapidly by direct contact of leaves (AC Bockelmann, personal observation).
With a mean value of 5.7 L. zosterae cells mg−1 Z. marina dry weight (±1.9 SE), abundances of L. zosterae seem low on an absolute scale but are consistent with a scenario of chronic, non-pathogenic infection, while the variation across and among sites is very high. Z. marina plants from four intertidal sites in the Wadden Sea were completely uninfected, even in summer. High intra- as well as inter-population variability may be due to stochastic infection dynamics [41], [42], genotypic resistance effects of the host, as shown for other pathogen-host associations [43], [44] or due to differential physiological activity among leaves and among individuals. For example, a single eelgrass shoot from one individual can harbor 20,000 times as many L. zosterae cells as a shoot from another individual just a few meters away (this study). Rapid changes in abundance of Labyrinthula spp. have been shown in culture where cells can spread 10 mm hr−1 [30]; spread has also been shown to correlate with reduced photosynthetic capacity across an infected area of the leaf at a velocity of 0.8 mm hr−1 [45]. Thus infection of the physiologically most active parts of the plants undoubtedly contributes to high intra-individual variation. The extremely low abundance of L. zosterae in subtidal Wadden Sea populations may be a result of high resistance to infection, resulting from the 1930s epidemic which destroyed almost all subtidal eelgrass beds.
Experimental investigations of L. zosterae and lesion development revealed that neither high temperatures, nor high salinity or low light availability could be identified as variables that satisfactory explain the 1930’s pandemic [5], [33], [46]–[48]. Next to environmental factors, interactions with biotic effects such as herbivory [49] and competition with epiphytes and bacteria on the leaf surface [50] are likely to impact infection dynamics. Our QPCR assay also provides the opportunity to study historical museum material (AC Bockelmann unpublished) in order to determine whether the L. zosterae present in today eelgrass meadows is the same strain that caused the 1930’s wasting disease epidemic and thus provide a clue about the endophyte’s possible origins.
A commensalistic or even mutualistic relationship [43], [44] for Labyrinthula species is also worthy of further investigation, as has been shown for many terrestrial plant-endophyte associations [43], [44], [51]–[53]. Several other Labyrinthula species have now been identified in the Baltic [32], suggesting that a commensally association may be more likely than previously supposed. It is conceivable that the presence of the endophyte in low concentration confers some sort of chemical protection against other infections like known from bacteria or fungi [53], [54]. Schmoller [55] found that in culture Labyrinthula coenocystis can actually be nourished by a bacterial film. Furthermore, the rapid decay and mineralization of senescent leaves [50] could alleviate nutrient limitation for eelgrass plants. Switches between pathogenic and mutualistic relationships are common in plant-endophyte symbiosis [56], [57], which could also be the case here. There is thus a pressing need to experimentally disentangle the role of different environmental and biotic factors as well as the mechanism of host defense [58].
In culture, morphological differences in colony growth form, cell morphology, and in pathogenicity and infectiousness have been observed, which suggests different genetic backgrounds [59], [60], (AC Bockelmann, personal observation). However, there is currently no genetic or definitive experimental data available. Whereas species differences have been documented using 18S ribosomal rDNA sequence analysis [32], there are currently no genetic markers to distinguish among specific strains that are of commensalistic vs. pathogenic nature.
With climate change resulting in a multitude of altered environmental conditions, for example warmer temperatures and ocean acidification, marine diseases in several taxonomic groups are already noticeably increasing [6], [61]–[63]. Given that endophytes such as Labyrinthula species are diverse and that only very few have been studied thus far (as L. zosterae for Z. marina), it may be useful to other endophytes in addition to Labyrinthula zosterae in future studies on eelgrass health and performance [64].
Acknowledgments
Samples from Fiskebekvik were collected by Susanne Landis and Olivia Roth; other Swedish samples by Per Moksnes. Sandspollen samples were kindly provided by Stein Fredriksen and Frithjof Moy. Eylem Elma, Petra Kadel and Janina Brakel helped with sampling in the Wadden Sea. Ashwin Engelen kindly sent samples from Portugal. Thanks to all of them.
Funding Statement
Funding was provided by Excellence Cluster ‘The Future Ocean’ (GEOMAR, Kiel, Germany), and the Ministry for Agriculture, Nature Conservation and Rural Development of Schleswig-Holstein (LLUR, Germany). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Orth RJ, Carruthers TJB, Dennison WC, Duarte CM, Fourqurean JW, et al. (2006) A Global Crisis for Seagrass Ecosystems. BioScience 56: 987–996. [Google Scholar]
- 2. Costanza R, d'Arge R, de Groot R, Farber S, Grasso M, et al. (1997) The value of the world's ecosystem services and natural capital. Nature 387: 253–260. [Google Scholar]
- 3. Duffy JE (2006) Biodiversity and the functioning of seagrass ecosystems. Mar Ecol-Prog Ser 311: 233–250. [Google Scholar]
- 4. Waycott M, Duarte CM, Carruthers TJB, Orth RJ, Dennison WC, et al. (2009) Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proc Natl Acad Sci USA 106: 12377–12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Muehlstein LK (1989) Perspectives on the wasting disease of eelgrass Zostera marina . Dis Aquat Organ 7: 211–221. [Google Scholar]
- 6. Harvell CD, Kim K, Burkholder JM, Colwell RR, Epstein PR, et al. (1999) Emerging Marine Diseases–Climate Links and Anthropogenic Factors. Science 285: 1505–1510. [DOI] [PubMed] [Google Scholar]
- 7. Jones KE, Patel GP, Levy MA, Storeygard A, Balk D, et al. (2008) Global trends in emerging infectious diseases. Nature 451: 990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reise K, Herre E, Sturm M (1989) Historical changes in the benthos of the Wadden Sea around the island of Sylt in the North Sea. Helgoland Mar Res 43: 417–433. [Google Scholar]
- 9. de Jonge V, de Jong D, van den Bergs J (1996) Reintroduction of eelgrass (Zostera marina) in the Dutch Wadden Sea; Review of research and suggestions for management measures. J Coastal Cons 2: 149–158. [Google Scholar]
- 10.Boström C, Baden SP, Krause-Jensen D (2003) The seagrasses of Scandinavia and the Baltic Sea. In: Green EP, Short FT (eds) World Atlas of Seagrasses. University of California Press, Berkeley, 27–37.
- 11. Meyer T, Nehring S (2006) Anpflanzung von Seegraswiesen (Zostera marina L.) als interne Maßnahme zur Restaurierung der Ostsee - Plantation of seagrass beds (Zostera marina L.) as internal measure for restoration of the Baltic Sea. Rostock. Meeresbiolog Beitr 15: 105–119. [Google Scholar]
- 12. Godet L, Fournier J, van Katwijk M, Olivier F, Le Mao P, et al. (2008) Before and after wasting disease in common eelgrass Zostera marina along the French Atlantic coasts: a general overview and first accurate mapping. Dis Aquat Organ 79: 249–255. [DOI] [PubMed] [Google Scholar]
- 13. Wohlenberg E (1935) Beobachtungen über das Seegras, Zostera marina L., und seine Erkrankung im nordfriesischen Wattenmeer. Beiträge zur Heimatforschung in Schleswig-Holstein, Hamburg und Lübeck, Sonderdruck aus Nordelbingen 11: 1–19. [Google Scholar]
- 14. Wyllie-Echeverria S, Cox PA (1999) The seagrass (Zostera marina, Zosteraceae) industry of Nova Scotia (1907–1960). Econom Bot 53: 419–426. [Google Scholar]
- 15. Cottam C (1933) Disappearance of eelgrass along the Atlantic Coast. Plant Dis Rep 17: 46–53. [Google Scholar]
- 16. Cotton AD (1933) Disappearance of Zostera marina . Nature 132: 483–483. [Google Scholar]
- 17. Fischer-Piette E, Heim R, Larni R (1932) Note preliminaire sur une maladie bacterienne des Zosteres . Compt Rend Acad Sc Paris 195: 1420. [Google Scholar]
- 18. van der Werff A (1938) A new parasitic organism in Zostera marina . Chron Bot 4: 498–499. [Google Scholar]
- 19.Butcher RW (1934) Zostera, Report on the present condition of Eel Grass on the coasts of England, based on a survey during August to October 1933. J du Conseil 9.
- 20.Blegvad H (1935) En epidemisk sygdom i bændeltangen (Zostera marina L.). In: Blegvad H (ed) Beretning til Mineteriet for Søfart og Fisken fra den Danske Biologiske Station. C.A. Reitzels Forlag, København, 1–8.
- 21. Addy CE, Aylward DA (1944) Status of Eelgrass in Massachusetts during 1943. J Wildlife Manage 8: 269–275. [Google Scholar]
- 22. Blois JC, Francaz JM, Gaudichon M, Gaudichon S, Le Bris L (1961) Observations sur les herbiers à Zostères de la région de Roscoff. Cah Biol Mar 2: 223–262. [Google Scholar]
- 23.Rasmussen E (1977) The wasting disease of eelgrass (Zostera marina) and its effects on environmental factors and fauna. In: McRoy CH, Hellferish C (eds) Seagrass ecosystems, a scientific perspective. Marcel Dekker, New York, 1–51.
- 24. Short FT, Ibelings BW, Den Hartog C (1988) Comparison of a current eelgrass disease to the wasting disease in the 1930s. Aquatic Botany 30: 295–304. [Google Scholar]
- 25. Reise K, Kohlus J (2008) Seagrass recovery in the Northern Wadden Sea? Helgoland Marine Research 62: 77–84. [Google Scholar]
- 26. Kastler T, Michaelis H (1997) Der Rückgang der Seegrasbestände im niedersächsischen Wattenmeer. Berichte der Forschungsstelle Küste 41: 119–139. [Google Scholar]
- 27.Dolch T, Buschbaum C, Reise K (2009) Seegras-Monitoring im Schleswig-Holsteinischen Wattenmeer 2008. Landesamtes für Landwirtschaft, Umwelt und ländliche Räume des Landes Schleswig-Holstein, Flintbek.
- 28. Short FT, Muehlstein LK, Porter D (1987) Eelgrass wasting disease: Cause and recurrence of a marine epidemic. Biol Bull 173: 557–562. [DOI] [PubMed] [Google Scholar]
- 29.Renn CE (1936) The Wasting Disease of Zostera marina. I. A Phytological Investigation of the Diseased Plant. Biol Bull 70 148–158.
- 30. Muehlstein LK, Porter D, Short FT (1991) Labyrinthula zosterae sp. nov., the causative agent of wasting disease of eelgrass, Zostera marina . Mycologia 83: 180–191. [Google Scholar]
- 31. den Hartog C (1989) Early records of wasting-disease-like damage patterns in eelgrass Zostera marina . Dis Aquat Organ 7: 223–226. [Google Scholar]
- 32. Bockelmann AC, Beining K, Reusch TBH (2012) Widespread occurrence of endophytic Labyrinthula spp. in northern European eelgrass Zostera marina beds. Mar Ecol Prog Ser 445: 109–116. [Google Scholar]
- 33. Burdick DM, Short FT, Wolf J (1993) An Index to Assess and Monitor the Progression of Wasting Disease in Eelgrass Zostera marina . Mar Ecol Prog Ser 94: 83–90. [Google Scholar]
- 34. Hily C, Raffin C, Brun A, den Hartog C (2002) Spatio-temporal variability of wasting disease symptoms in eelgrass meadows of Brittany (France). Aquat Bot 72: 37–53. [Google Scholar]
- 35. Bergmann N, Fricke B, Schmidt MC, Tams V, Beining K, et al. (2011) A quantitative real-time PCR assay for the seagrass pathogen Labyrinthula zosterae . Mol Ecol Res 11: 1076–1081. [DOI] [PubMed] [Google Scholar]
- 36. Oetjen K, Reusch TBH (2007) Genome scans detect consistent divergent selection among subtidal vs. intertidal populations of the marine angiosperm Zostera marina . Mol Ecol 16: 5156–5157. [DOI] [PubMed] [Google Scholar]
- 37. Pokorny KS (1967) Labyrinthula . J Eukaryot Microbiol 14: 697–708. [Google Scholar]
- 38. Amon JP, Perkins FO (1968) Structure of Labyrinthula sp. Zoospores. J Protozool 15: 543–546. [Google Scholar]
- 39. Bartsch G (1971) Cytologische Beobachtungen an Labyrinthula coenocystis Schmoller bei verschiedenen Kulturbedingungen. Zeitschrift für Allg. Mikrobiologie 11: 79–90. [DOI] [PubMed] [Google Scholar]
- 40.Porter D (1990) Phylum Labyrinthulomycota. In: Margulis L,Corliss JO, Melkonian M, Chapman DJ (eds) Handbook of Protoctista. Jones and Bartlett Publishers, Boston.
- 41. Anderson RM, May RM (1978) Regulation of stability of host-parasite population interactions. I. Regulatory Processes, J Anim Ecol 47: 219–247. [Google Scholar]
- 42. Thrall PH, Burdon JJ (2003) Evolution of virulence in a plant host-pathogen metapopulation. Science 299: 1735–1737. [DOI] [PubMed] [Google Scholar]
- 43. Gilbert GS (2002) Evolutionary ecology of plant diseases in natural ecosystems. Annu Rev Phytopathol 40: 13–43. [DOI] [PubMed] [Google Scholar]
- 44.Cheplick GP, Faeth SH (2009) Ecology and evolution of the grass-endophyte symbiosis. Oxford University Press, Oxford.
- 45. Ralph PJ, Short FT (2002) Impact of the wasting disease pathogen, Labyrinthula zosterae, on the photobiology of eelgrass Zostera marina . Mar Ecol Prog Ser 226: 265–271. [Google Scholar]
- 46. Giesen WBJT, Van Katwijk MM, Den Hartog C (1990) Temperature, salinity, insolation and wasting disease of eelgrass (Zostera marina L.) in the Dutch Wadden Sea in the 1930's. Netherlands J Sea Res 25: 395–404. [Google Scholar]
- 47. Vergeer LHT, Aarts TL, de Groot JD (1995) The `wasting disease' and the effect of abiotic factors (light intensity, temperature, salinity) and infection with Labyrinthula zosterae on the phenolic content of Zostera marina shoots. Aquat Bot 52: 35–44. [Google Scholar]
- 48. McKone KL, Tanner CE (2009) Role of salinity in the susceptibility of eelgrass Zostera marina to the wasting disease pathogen Labyrinthula zosterae . Mar Ecol Prog Ser 377: 123–130. [Google Scholar]
- 49. Bowles JW, Bell SS (2004) Simulated herbivory and the dynamics of disease in Thalassia testudinum . Mar Ecol Prog Ser 283: 127–132. [Google Scholar]
- 50. Raghukumar S, Damare VS (2011) Increasing evidence for the important role of Labyrinthulomycetes in marine ecosystems. Bot Mar 54: 3–11. [Google Scholar]
- 51. Saikkonen K, Wäli P, Helander M (2004) Faeth SH (2004) Evolution of endophyte plant symbioses. Trends Plant Sci 9: 275–280. [DOI] [PubMed] [Google Scholar]
- 52. Paszkowski U (2006) Mutualism and parasitism: the yin and yang of plant symbioses. Curr Opin Plant Biol 9: 364–370. [DOI] [PubMed] [Google Scholar]
- 53. van Loon LC, Bakker PA, Pieterse CM (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36: 453–483. [DOI] [PubMed] [Google Scholar]
- 54. Ramamoorthy V, Viswanathan R, Raguchander T, Prakasam V, Samiyappan R (2001) Induction of systemic resistance by plant growth promoting rhizobacteria in crop plants against pests and diseases. Crop Prot 20: 1–11. [Google Scholar]
- 55. Schmoller H (1960) Kultur und Entwicklung von Labyrinthula coenocystis n. sp. Arch f Mikrobiol 36: 365–372. [PubMed] [Google Scholar]
- 56. Johnson NC, Graham JH, Smith FA (1997) Functioning of mycorrhizal associations along the mutualism-parasitism continuum. New Phytol 135: 575–585. [Google Scholar]
- 57. Kniskern JM, Rausher MD (2006) Environmental variation mediates the deleterious effects of Coleosporium ipomoeae on Ipomoea pupurea . Ecology 87: 675–685. [DOI] [PubMed] [Google Scholar]
- 58. Steele L, Caldwell M, Boettcher AA, Arnold T (2005) Seagrass-pathogen interactions : 'pseudo-induction' of turtlegrass phenolics near wasting disease lesions. Mar Ecol Prog Ser 303: 123–131. [Google Scholar]
- 59. Muehlstein LK, Porter D, Short FT (1988) Labyrinthula sp., a marine slime mold producing the symptoms of wasting disease in eelgrass, Zostera marina . Mar Biol 99: 465–472. [Google Scholar]
- 60. Martin DL, Boone E, Caldwell MM, Major KM, Boettcher AA (2009) Liquid culture and growth quantification of the seagrass pathogen, Labyrinthula spp. Mycologia 101: 632–635. [DOI] [PubMed] [Google Scholar]
- 61. Cook T, Folli M, Klinck J, Ford S, Miller J (1998) The Relationship between Increasing Sea-surface Temperature and the Northward Spread of Perkinsus marinus (Dermo) Disease Epizootics in Oysters. Estuar Coast Shelf Sci 46: 587–597. [Google Scholar]
- 62. Ward JR, Lafferty KD (2004) The Elusive Baseline of Marine Disease: Are Diseases in Ocean Ecosystems Increasing? PLoS Biol 2: 0542–0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Karvonen A, Rintamäki Pi, Jokela J, Valtonen ET (2010) Increasing water temperature and disease risks in aquatic systems: Climate change increases the risk of some, but not all, diseases. Int J Parasitol 40: 1483–1488. [DOI] [PubMed] [Google Scholar]
- 64. Garrett KA, Dendy SP, Frank EE, Rouse MN, Travers SE (2006) Climate Change Effects on Plant Disease: Genomes to Ecosystems. Annu Rev Phytopathol 44: 489–509. [DOI] [PubMed] [Google Scholar]