Abstract
Increasing evidence that several drug compounds exert their effects through interactions with multiple targets is boosting the development of research fields that challenge the data reductionism approach. In this article, we review and discuss the concepts of drug repurposing, polypharmacology, chemogenomics, phenotypic screening and highthroughput in vivo testing of mixture-based libraries in an integrated manner. These research fields offer alternatives to the current paradigm of drug discovery, from a one target–one drug model to a multiple-target approach. Furthermore, the goals of lead identification are being expanded accordingly to identify not only ‘key’ compounds that fit with a single-target ‘lock’, but also ‘master key’ compounds that favorably interact with multiple targets (i.e. operate a set of desired locks to gain access to the expected clinical effects).
Keywords: chemogenomics, drug repurposing, in vivo testing, phenotypic screening, polypharmacology, target fishing
Drug discovery has undergone transformations over the years, moving from in vivo models to a single targetselective drug for a single mechanism. Following the lock and key model proposed by Erlich more than a century ago [1], over the previous decades, drug discovery efforts have focused on identifying single selective drugs that target a single mechanism; that is, identifying ligands (‘keys’) that fit into specific targets (‘locks’) (Figure 1). This strategy has been largely motivated by the reductionist view of systems biology [2], aided by the ever-increasing understanding of biological processes at the molecular level (e.g. molecular interactions of a chemical compound with a target protein). Thus, there has been augmented interest in identifying drugs that interact with specific targets. This pursuit, to identify suitable ligands for single targets, has been aided further by in vitro high-throughput screening (HTS) with a validated target. This approach has been coupled with advances in the design and synthesis of extensive combinatorial libraries [3] and has enabled scientists to screen vast regions of chemical space [4] for biologically active compounds in a relatively short amount of time. Despite the several successful applications of in vitro HTS [5], the process can be inefficient because the resulting hits often lack efficacy in vivo. Drug design at the single molecular target level ‘is blind’ to other processes that are inevitably connected through complex networks with higher levels of the hierarchical nature of biological systems [2]. In addition to the technical advances of HTS, computational structure-based approaches have also evolved around the single-target approach. The overall goal of such methods is to help identify compound candidates for screening against a particular target in a faster and more economical manner. The constantly increasing availability of high resolution three-dimensional (3D) structures of therapeutically relevant targets has motivated the development and application of computer structure-based strategies [6].
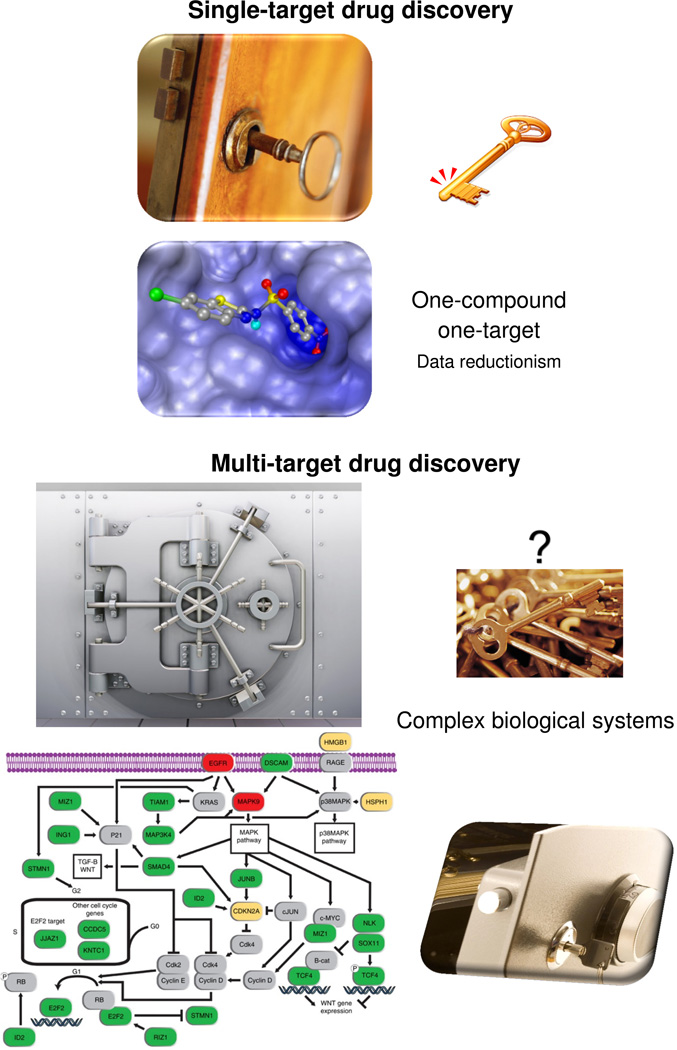
Figure 1.
[LM1]Schematic representation of the single-target and multitarget drug discovery paradigms. Although one ligand (key) compound might fit and operate a single target (lock), diseases are associated with complex biological processes and, in several cases, with multiple targets, which are more difficult to ‘unlock’.
It is currently evident that the concept that one drug acts on a single receptor is not as effective as expected from the reductionism view of the lock and key model [2]. The growing evidence for polypharmacology (i.e. that clinical effects are often because of the interaction of single or multiple drugs with multiple targets [7]) is encouraging the shift to experimental and computational multitarget approaches (Figure 1) [8]. For example, the methodologies for understanding structure–activity relations (SAR) for single biological endpoints are being adapted to model structure–multiple-activity relations [9]. By analogy with the lock and key model, one of the current challenges would be to find the ‘master keys’ that operate a set of several locks to gain access to the desirable clinical effect. This is a general concept reminiscent of the ‘molecular master keys’ proposed by Müller, who extensively discussed the use of privileged structures, frequently employed in medicinal chemistry, to hit targets from a gene family [10]. Of course the ‘master key’ should not operate on any possible lock to avoid adverse effects (e.g. ‘promiscuous keys’). In this context, the challenge starts with finding the set of multiple targets (multiple locks) that are associated with a desired clinical effect and that might not be from the same target family. As discussed by Merino et al., it is important to emphasize that there are different levels of polypharmacology (or promiscuity) that could lead to positive or negative effects [11]. Such levels will largely depend on the dose of the drug. It might be that, at therapeutic doses, a drug has a positive clinical effect owing to its interaction with multiple targets. However, depending on the dose, interactions of the drug with antitargets will lead to adverse effects [11].
This review is divided into three main sections. Drug repurposing is discussed first as a primary application of polypharmacology to speed up the drug discovery process. This section covers not only the repurposing of drugs, but also the repositioning of chemical databases initially designed, by human or nature, for other purposes. A discussion then follows on chemogenomics, a growing field that aims to integrate systematically the chemical and target spaces. The third section, high-throughput in vivo testing of mixture-based libraries, presents a general strategy to identify new lead compounds that directly address the polypharmacology in drug discovery efforts.
Drug repurposing
A direct application of polypharmacology is drug repurposing (also called drug repositioning), which is an increasingly growing approach to speed up the drug discovery process by identifying a new clinical use for an existing approved drug [12,13]. A closely related concept is drug rescue, in which a new indication is pursued for compounds that have failed to reach the clinic or failed in human clinical trials for lack of efficacy against the indication they were originally directed [13]. Groups from academia and other research environments working with cancer-related, neglected, rare and other diseases, are actively looking at repositioning compounds that are already approved for other indications [14,15]. Drug repurposing has occurred in many instances by serendipity [12]. However, there are ongoing efforts to conduct drug repurposing systematically. To this end, at least three general strategies have been envisioned, namely; chemical, biological and text data mining [16]. One of the several notable examples of drug repurposing is illustrated by the ‘high throughput’ in vivo pharmacology platform theraTRACE® [16]. Of note, Oprea and Mestres recently emphasized that a successful drug repurposing campaign involves not only identifying new targets for old drugs, but also factors such as adverse effect tolerance and analysis of the intellectual property landscape [17]. Current progress in drug repurposing, including experimental and computational strategies conducted in several companies and research groups in academia, successful applications and legal aspects, for example, have been the subject of extensive reviews, and the special issue of one journal [12,15,16,18].
The successful application of drug repurposing, which is primarily a retrospective approach, is encouraging the prospective search of potential alternative targets to compounds developed for a particular disease. Furthermore, approaches such as phenotypic screening aim to identify hit compounds considering broader areas of the ‘target space’ (e.g. signaling pathways, metabolic networks, or the entire expressed proteomes) to which small molecules are exposed [19,20]. One of the major challenges of phenotypic screening is the identification of the target(s) of a hit molecule, (i.e. target deconvolution) and the elucidation of the mode of action [21–23]. Nonetheless, phenotypic screening is valuable for identifying hit compounds for complex processes, such as epigenetic modulation [21].
Repurposing of chemicals that differ from drugs
Experimental and/or computational target fishing approaches can be applied not only to approved drugs (e.g. for drug repurposing), but also to other types of compound library. This is facilitated by the availability of large compound collections in the public domain [24], including databases of small molecules [25], natural products [26] and virtual compounds synthetically available [27]. Research groups in industry or academia can look for potential targets for existing inhouse compound libraries, for example, combinatorial libraries or focused libraries initially designed for different targets (e.g. repurposing of focused libraries [28]). In addition, there is increased interest in the scientific community to search systematically for potential targets of natural products [29,30]. For example, it has been known for centuries that herbal remedies or compounds used in traditional Chinese medicine (TCM) are effective for the treatment of many diseases. The advent of large compound collections, such as the TCM database [31], has opened up the possibility to search the targets of the active components using computational approaches [32]. Recent analysis of the molecular complexity, structural diversity and molecular properties of the TCM database reveals that this collection is a rich source of molecules to expand the traditional medicinally relevant chemical space [33]. Another relevant example is increased interest in investigating the role of dietary components in regulating epigenetic events leading to the research area of nutritional epigenomics [34].
Food materials designated as ‘generally recognized as safe’ (GRAS) [35] are attractive sources from which to identify molecules with health-promoting effects and that complement the chemical space of drugs [36,37]. Flavoring substances in the GRAS list (i.e. those that comprise discrete chemical entities) are beginning to appeal as a source to uncover bioactive compounds with potential health-related benefits. In particular, there is currently much interest in exploring possible secondary benefits of flavor ingredients, such as those relating to health and wellness [38]. Using computational approaches to uncover potential applications of food chemicals has led to the research area of foodinformatics [37] (perhaps a particular case of cheminformatics).
Chemogenomics: Integrating chemical and target spaces
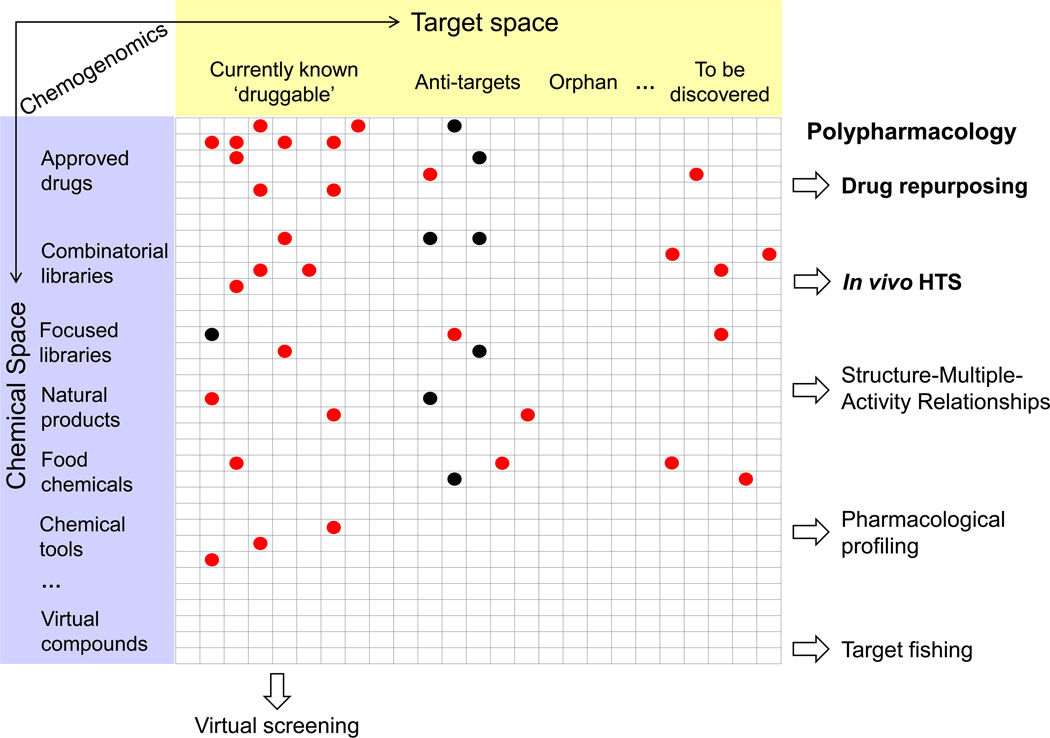
Ideally, polypharmacology can be fully explored if there was readily available information linking the relation between the entire chemical and target spaces. In the pursuit of this goal, chemogenomics has emerged as a research field that aims to identify all possible ligands for all possible targets [39,40]. Chemogenomics and related concepts, reviewed here, are schematically illustrated in Figure 2.
Figure 2.
[LM2]Schematic representation of the relations between the concepts reviewed in this article. The table represents the association between all possible molecules in chemical space (rows) (organized in different types of chemical library), and targets in the ‘target space’ (columns) (collected in different types of target). The red circles indicate that there exists a compound–target interaction, whereas the black circles denote that there is no interaction. Empty cells indicate that the intersecting compound–target interaction is not known. Abbreviation: HTS, high throughput screening.
Databases annotated with biological activity
Despite the fact that the full knowledge of the integration of chemical and target spaces is a goal far from being completed soon (if ever), the availability of experimental activity data of chemical compounds screened across different biological endpoints represents an important step in this direction. Although large databases are stored internally by pharmaceutical and biotech companies, there are several compound databases annotated with biological activity available in the public domain. PubChem, ChEMBL and Binding Database are examples of major databases recently reviewed [41], and represent an important source for mining complex ligand–target relations and unveil potential new biological activities for molecules screened in a particular biological assay. Recent advances in the integration and mining of chemical databases annotated with biological activity are represented by the Open Pharmacological Concept Triple Store (Open PHACTS) project [42] and the PharmaTrek web explorer (http://cgl.imim.es/pharmatrek) [43]. These types of major initiative aim to create and systematically explore an integrated pharmacological space, and represent an effort to facilitate open innovation in drug discovery research, including multitarget approaches [42,43].
Target fishing and ligand profiling
Despite the rich chemical databases available to date annotated with biological activity, current experimental data are not sufficient to fill in all possible relations between chemical and target spaces [40]. Given that a considerably large and perhaps prohibitive investment of time and resources would be required to generate experimental data to cover all possible chemical–target associations, computational approaches are actively being pursued to identify either new ligands for known targets (computational or virtual screening) or putative targets for known ligands (target fishing) (Figure 2). Computational approaches, such as docking, similarity searching and pharmacophore modeling, are a few examples of well-established virtual screening techniques that can be used to select compounds for further experimental investigation. Of course, despite several successful applications of virtual screening methods, these approaches are not free from pitfalls [44], and the improvement of computational screening methods is actively being pursued [45–47].
Target fishing leads to the generation of the global pharmacological profile or ligand profiling [40]. In addition, computational approaches are being developed to visualize and mine the SAR of chemogenomic data sets [48,49]. Similar to virtual screening, the prediction of the polypharmacological profile of bioactive compounds can be performed based on the chemical structure of the bioactive compounds themselves (ligand-based methods) or on the structure of the targets (structure-based methods). Specific methodologies and successful applications of both approaches have been the subject of extensive reviews [39,40,50,51]. More recently, computational and experimental approaches are emerging that investigate systematically potential additional targets of chemical probes used in chemical biology [52,53].
High-throughput in vivo testing of mixture-based libraries
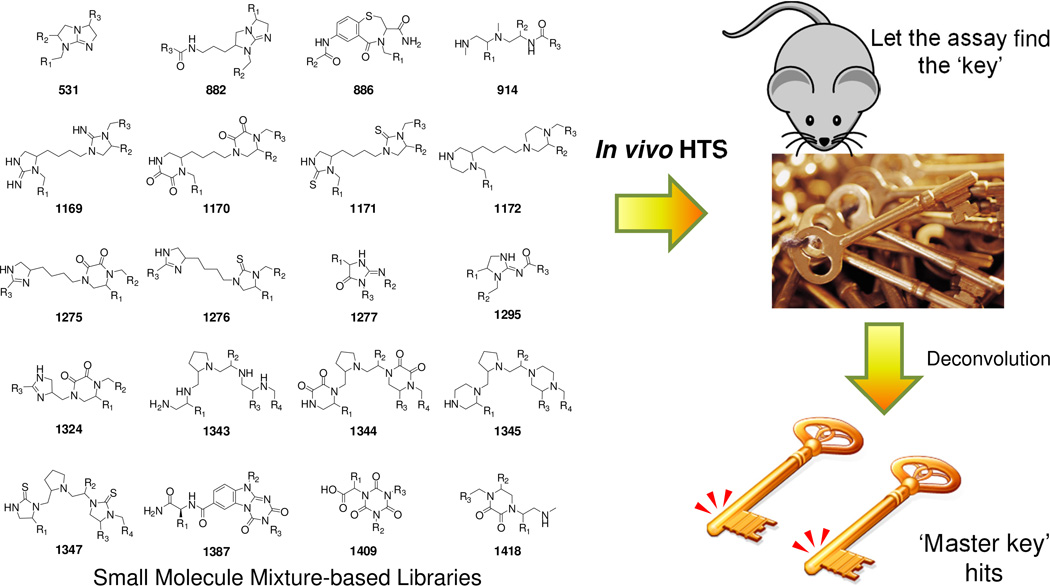
Owing to the frequently observed lack of efficacy of hits identified from in vitro HTS discussed above, it has been recognized that an ideal process would include early demonstration that individual compounds are active in vivo in disease-relevant models before further development. In fact, many drugs have been identified through the initial in vivo testing of either natural product mixtures or individual compounds. Interestingly, several approved drugs were developed based on favorable animal model data without a clear understanding of the mechanism of action of the drug at the molecular level [11]. In fact, as pointed out by Merino et al., ‘despite the broad scientific knowledge around human drug targets, today’s pharmacopoeia still includes many drugs that are being prescribed with unknown Mechanism of Action’ [11]. Houghten et al. have largely emphasized that one approach to circumvent the high attrition rate in current drug discovery endeavors is to use in vivo models directly in the discovery phase to identify candidates with desirable biological profiles while simultaneously eliminating those compounds with poor absorption, distribution, metabolism and elimination (ADME) and/or pharmacokinetic properties [54,55]. The idea of testing an entire library in vivo containing thousands to millions of individual compounds one at a time is simply not practical owing to the extreme economic and time requirements. To address this impracticality, in vivo testing of mixture-based libraries has emerged as a successful approach that is both rapid and efficient, because only those compounds displaying efficacy in disease models are selected for further pharmacological and chemical development (Figure 3). The screening of the highly dense mixture-based libraries [3,56–58] explores uncovered regions of the traditional medicinally relevant chemical space [33], increases the potential of identifying activity cliffs (chemical compounds with highly similar structures but significantly different biological activities) and provides a rapid understanding of the SAR associated with novel leads and targets. Different approaches for the use of mixture-based libraries, namely positional scanning libraries, have been developed over the past 20 years. The use of these systematically arranged mixtures enables one to utilize a set of samples containing exponentially more compounds than samples to identify a single active compound [54]. Additionally, these mixture libraries are formatted in such a way that one can identify potentially synergistic effects and then use an iterative deconvolution [59] to identify the compounds responsible. In fact, these systematically formatted mixture-based libraries are ideally suited to identify synergistic effects. Thus, the outcome of this process is the identification of lead compounds with in vivo activity early in the drug discovery effort, providing more advanced lead compounds than the traditional in vitro HTS process.
Figure 3.
Schematic representation of in vivo high throughput screening (HTS) of mixture-based libraries. A representative core scaffold of mixture-based libraries is shown. The assay directly points to the hit compounds that might act through the interaction with multiple targets (‘master key’ compounds), which are readily identified after deconvolution.
In addition to identifying lead compounds with mechanisms of action involving a single known target or multiple known targets, in vivo HTS also enables the discovery of lead molecules with novel mechanism of actions. This, in turn, opens up the possibility of identifying new biological targets and/or pathways for therapeutic intervention. Therefore, in vivo HTS is led completely by the overall activity of the compounds in the animal model, with no preconceived bias for the method of action. Several examples of lead compounds identified using in vivo HTS have been reviewed elsewhere [54,55,60].
Concluding remarks
The fact that several drugs exert their effect through the interaction with multiple targets is shifting the drug discovery paradigm from the one target-one drug model to a multiple-target approach. In this context, instead of pursuing highly selective compounds for unique targets, that is, ‘single keys for specific locks,’ the goal is shifting towards identifying ideal ‘master keys’ that selectively operate a set of ‘multiple locks’ to gain access to a clinical benefit that is usually associated with a complex biological process. Of course, the ‘master key’ should not open any lock (antitargets) to avoid adverse effects. In this scenario, the challenge is to also identify the set of targets that are associated with a desired clinical effect. Experimental and computational chemogenomic approaches are actively being developed and applied to identify systematically potential additional targets of existing or virtual chemical compounds. Drug repurposing is a primary example of the beneficial application of polypharmacology. The basic idea of ‘repositioning’ for health benefits can be extended to other chemical libraries initially designed for other applications, such as focused libraries, GRAS chemicals or natural products. In vivo testing of mixture-based libraries represents an efficient approach to identify compounds with the desirable effect and rapidly identify chemical compounds that might act as selective or master keys to gain access to novel and effective therapeutic treatments. In vivo HTS technology also opens up the possibility to uncover compounds with novel mechanisms of action and identify therapeutically relevant synergistic combinations of compounds.
Research Highlights.
Drug discovery is changing from a one target-one drug to a multiple-target model
Drug repurposing is a positive application of polypharmacology
“Repurposing” can be extended to chemical libraries other than known drugs
Chemogenomics and in vivo testing may uncover ‘master key’ leads
‘Master key’ compounds may operate a set of desired ‘locks’ (multiple-targets)
Acknowledgments
We thank the Institute of Chemistry of the National Autonomous University of Mexico (UNAM) and the State of Florida, Executive Office of the Governor’s Office of Tourism, Trade, and Economic Development. This work was supported by NIDA grant R01 DA031370-02 (to R.A.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fischer E. Einfluss der Configuration auf die Wirkung der Enzyme. Ber. Dtsch. Chem. Ges. 1894;27:2985–2993. [Google Scholar]
- 2.Maggiora G. The reductionist paradox: are the laws of chemistry and physics sufficient for the discovery of new drugs? J. Comput. Aided Mol. Des. 2011;25:699–708. doi: 10.1007/s10822-011-9447-8. [DOI] [PubMed] [Google Scholar]
- 3.Houghten RA, et al. Mixture-based synthetic combinatorial libraries. J. Med. Chem. 1999;42:3743–3778. doi: 10.1021/jm990174v. [DOI] [PubMed] [Google Scholar]
- 4.Medina-Franco JL, et al. Visualization of the chemical space in drug discovery. Curr. Comput. Aided Drug Des. 2008;4:322–333. [Google Scholar]
- 5.Roy A, et al. Open access high throughput drug discovery in the public domain: a Mount Everest in the making. Curr. Pharm. Biotechnol. 2010;11:764–778. doi: 10.2174/138920110792927757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Congreve M, et al. Structural biology and drug discovery. Drug Discov. Today. 2005;10:895–907. doi: 10.1016/S1359-6446(05)03484-7. [DOI] [PubMed] [Google Scholar]
- 7.Paolini GV, et al. Global mapping of pharmacological space. Nat. Biotechnol. 2006;24:805–815. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 8.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 9.Medina-Franco JL, et al. Multitarget structure–activity relationships characterized by activity-difference maps and consensus similarity measure. J. Chem. Inf. Model. 2011;51:2427–2439. doi: 10.1021/ci200281v. [DOI] [PubMed] [Google Scholar]
- 10.Müller G. Medicinal chemistry of target family-directed masterkeys. Drug Discov. Today. 2003;8:681–691. doi: 10.1016/s1359-6446(03)02781-8. [DOI] [PubMed] [Google Scholar]
- 11.Merino A, et al. Drug profiling: knowing where it hits. Drug Discov. Today. 2010;15:749–756. doi: 10.1016/j.drudis.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 13.Aubé J. Drug repurposing and the medicinal chemist. ACS Med. Chem. Lett. 2012;3:442–444. doi: 10.1021/ml300114c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dueñas–González A, et al. The prince and the pauper. A tale of anticancer targeted agents. Mol. Cancer. 2008;7:33. doi: 10.1186/1476-4598-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekins S, et al. In silico repositioning of approved drugs for rare and neglected diseases. Drug Discov. Today. 2011;16:298–310. doi: 10.1016/j.drudis.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Loging W, et al. Cheminformatic/bioinformatic analysis of large corporate databases: application to drug repurposing. Drug Discov. Today Ther. Strateg. 2011;8:109–116. [Google Scholar]
- 17.Oprea TI, Mestres J. Drug repurposing: far beyond new targets for old drugs. AAPS J. 2012;14:759–763. doi: 10.1208/s12248-012-9390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipinski CA. Drug repurposing. Drug Discov. Today Ther. Strateg. 2011;8:57–59. doi: 10.1016/j.ddstr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clemons PA. Complex phenotypic assays in high-throughput screening. Curr. Opin. Chem. Biol. 2004;8:334–338. doi: 10.1016/j.cbpa.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert IH, et al. Finding new hits in neglected disease projects: target or phenotypic based screening? Curr. Top. Med. Chem. 2011;11:1284–1291. doi: 10.2174/156802611795429176. [DOI] [PubMed] [Google Scholar]
- 21.Lu Q, et al. Perspectives on the discovery of small-molecule modulators for epigenetic processes. J. Biomol. Screening. 2012;17:555–571. doi: 10.1177/1087057112437763. [DOI] [PubMed] [Google Scholar]
- 22.Montolio M, et al. Identification of small molecule inhibitors of amyloid β-induced neuronal apoptosis acting through the imidazoline I2 receptor. J. Med. Chem. 2012;55:9838–9846. doi: 10.1021/jm301055g. [DOI] [PubMed] [Google Scholar]
- 23.Laggner C, et al. Chemical informatics and target identification in a zebrafish phenotypic screen. Nat. Chem. Biol. 2012;8:144–146. doi: 10.1038/nchembio.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbosa AJM, Del Rio A. Freely accessible databases of commercial compounds for high-throughput virtual screenings. Curr. Top. Med. Chem. 2012;12:866–877. doi: 10.2174/156802612800166710. [DOI] [PubMed] [Google Scholar]
- 25.Irwin JJ, Shoichet BK. ZINC – a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yongye AB, et al. Molecular scaffold analysis of natural products databases in the public domain. Chem. Biol. Drug Des. 2012;80:717–724. doi: 10.1111/cbdd.12011. [DOI] [PubMed] [Google Scholar]
- 27.Reymond J-L, et al. Chemical space as a source for new drugs. MedChemComm. 2010;1:30–38. [Google Scholar]
- 28.López-Vallejo F, et al. Integrating virtual screening and combinatorial chemistry for accelerated drug discovery. Comb. Chem. High Throughput Screen. 2011;14:475–487. doi: 10.2174/138620711795767866. [DOI] [PubMed] [Google Scholar]
- 29.Rollinger JM. Accessing target information by virtual parallel screening – the impact on natural product research. Phytochem. Lett. 2009;2:53–58. [Google Scholar]
- 30.Lauro G, et al. Inverse virtual screening of antitumor targets: pilot study on a small database of natural bioactive compounds. J. Nat. Prod. 2011;74:1401–1407. doi: 10.1021/np100935s. [DOI] [PubMed] [Google Scholar]
- 31.Chen CY-C. TCM Database@Taiwan: the world’s largest traditional chinese medicine database for drug screening in silico. PLoS ONE. 2011;6:e15939. doi: 10.1371/journal.pone.0015939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai T-Y, et al. iScreen: world’s first cloud-computing web server for virtual screening and de novo drug design based on TCM database@Taiwan. J. Comput. Aided Mol. Des. 2011;25:525–531. doi: 10.1007/s10822-011-9438-9. [DOI] [PubMed] [Google Scholar]
- 33.López-Vallejo F, et al. Expanding the medicinally relevant chemical space with compound libraries. Drug Discov. Today. 2012;17:718–726. doi: 10.1016/j.drudis.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Szarc vel Szic K, et al. Nature or nurture: let food be your epigenetic medicine in chronic inflammatory disorders. Biochem. Pharmacol. 2010;80:1816–1832. doi: 10.1016/j.bcp.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 35.Burdock GA, et al. The importance of GRAS to the functional food and nutraceutical industries. Toxicology. 2006;221:17–27. doi: 10.1016/j.tox.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 36.Scalbert A, et al. Databases on food phytochemicals and their health-promoting effects. J. Agric. Food Chem. 2011;59:4331–4348. doi: 10.1021/jf200591d. [DOI] [PubMed] [Google Scholar]
- 37.Medina-Franco JL, et al. Chemoinformatic analysis of GRAS (Generally Recognized as Safe) flavor chemicals and natural products. PLoS ONE. 2012;7:e50798. doi: 10.1371/journal.pone.0050798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.López-Vallejo F, et al. Computational methods for the discovery of mood disorder therapies. Expert Opin. Drug Discov. 2011;6:1227–1245. doi: 10.1517/17460441.2011.637106. [DOI] [PubMed] [Google Scholar]
- 39.Jacoby E. Computational chemogenomics. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2011;1:57–67. [Google Scholar]
- 40.Rognan D. Structure-based approaches to target fishing and ligand profiling. Mol. Inf. 2010;29:176–187. doi: 10.1002/minf.200900081. [DOI] [PubMed] [Google Scholar]
- 41.Nicola G, et al. Public domain databases for medicinal chemistry. J. Med. Chem. 2012;55:6987–7002. doi: 10.1021/jm300501t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams AJ, et al. Open PHACTS: semantic interoperability for drug discovery. Drug Discov. Today. 2012;17:1188–1198. doi: 10.1016/j.drudis.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 43.Carrascosa MC, et al. PharmaTrek: a semantic web explorer for open innovation in multitarget drug discovery. Mol. Inf. 2012;31:537–541. doi: 10.1002/minf.201200070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scior T, et al. Recognizing pitfalls in virtual screening: a critical review. J. Chem. Inf. Model. 2012;52:867–881. doi: 10.1021/ci200528d. [DOI] [PubMed] [Google Scholar]
- 45.Ripphausen P, et al. State-of-the-art in ligand-based virtual screening. Drug Discov. Today. 2011;16:372–376. doi: 10.1016/j.drudis.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 46.Sanders MPA, et al. Comparative analysis of pharmacophore screening tools. J. Chem. Inf. Model. 2012;52:1607–1620. doi: 10.1021/ci2005274. [DOI] [PubMed] [Google Scholar]
- 47.Cheng TJ, et al. Structure-based virtual screening for drug discovery: a problem-centric review. AAPS J. 2012;14:133–141. doi: 10.1208/s12248-012-9322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lounkine E, et al. Chemotography for multi-target sar analysis in the context of biological pathways. Bioorg. Med. Chem. 2012;20:5416–5427. doi: 10.1016/j.bmc.2012.02.034. [DOI] [PubMed] [Google Scholar]
- 49.Yongye AB, Medina-Franco JL. Data mining of protein-binding profiling data identifies structural modifications that distinguish selective and promiscuous compounds. J. Chem. Inf. Model. 2012;52:2454–2461. doi: 10.1021/ci3002606. [DOI] [PubMed] [Google Scholar]
- 50.Koutsoukas A, et al. From in silico target prediction to multi-target drug design: current databases, methods and applications. J. Proteomics. 2011;74:2554–2574. doi: 10.1016/j.jprot.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 51.Jenkins JL. Large-scale QSAR in target prediction and phenotypic HTS assessment. Mol. Inf. 2012;31:508–514. doi: 10.1002/minf.201200002. [DOI] [PubMed] [Google Scholar]
- 52.Gregori-Puigjané E, et al. Identifying mechanism-of-action targets for drugs and probes. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11178–11183. doi: 10.1073/pnas.1204524109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antolín AA, et al. Identification of Pim kinases as novel targets for PJ34 with confounding effects in PARP biology. ACS Chem. Biol. 2012;7:1962–1967. doi: 10.1021/cb300317y. [DOI] [PubMed] [Google Scholar]
- 54.Houghten R, et al. In vitro and direct in vivo testing of mixture-based combinatorial libraries for the identification of highly active and specific opiate ligands. AAPS J. 2006;8:E371–E382. doi: 10.1007/BF02854908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carroll FI, Houghten RA. From rapid in vitro screening to rapid in vivo screening in the drug discovery process. Neuropsychopharmacology. 2009;34:251–252. doi: 10.1038/npp.2008.160. [DOI] [PubMed] [Google Scholar]
- 56.Pinilla C, et al. Advances in the use of synthetic combinatorial chemistry: mixture-based libraries. Nat. Med. 2003;9:118–122. doi: 10.1038/nm0103-118. [DOI] [PubMed] [Google Scholar]
- 57.Houghten RA, et al. Strategies for the use of mixture-based synthetic combinatorial libraries: scaffold ranking, direct testing, in vivo and enhanced deconvolution by computational methods. J. Comb. Chem. 2008;10:3–19. doi: 10.1021/cc7001205. [DOI] [PubMed] [Google Scholar]
- 58.Armishaw CJ, et al. A synthetic combinatorial strategy for developing α-conotoxin analogs as potent α7 nicotinic acetylcholine receptor antagonists. J. Biol. Chem. 2010;285:1809–1821. doi: 10.1074/jbc.M109.071183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pinilla C, et al. Rapid identification of high-affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. Biotechniques. 1992;13:901–905. [PubMed] [Google Scholar]
- 60.Reilley K, et al. Identification of two novel, potent, low-liability antinociceptive compounds from the direct in vivo screening of a large mixture-based combinatorial library. AAPS J. 2010;12:318–329. doi: 10.1208/s12248-010-9191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]