Abstract
Regulators of G protein Signaling (RGS) proteins negatively modulate G protein coupled receptor (GPCR) signaling activity by accelerating G protein hydrolysis of GTP, hastening pathway shutoff. A wealth of data from cell culture experiments using exogenously expressed proteins indicates that RGS9 and other RGS proteins have the potential to down-regulate a significant number of pathways. We have used an array of biochemical and tissue staining techniques to examine the subcellular localization and membrane binding characteristics of endogenous RGS9-2 and known binding partners in rodent striatum and tissue homogenates. A small fraction of RGS9-2 is present in the soluble cytoplasmic fraction, whereas the majority is present primarily associated with the plasma membrane and structures insoluble in non-ionic detergents that efficiently extract the vast majority of its binding partners, R7BP and Gβ5. It is specifically excluded from the cell nucleus in mouse striatal tissue. In cultured striatal neurons, RGS9-2 is found at extrasynaptic sites primarily along the dendritic shaft near the spine neck. Heterogeneity in RGS9-2 detergent solubility along with its unique subcellular localization suggests that its mechanism of membrane anchoring and localization is complex and likely involves additional proteins beside R7BP. An important nuclear function for RGS9-2 seems unlikely.
Introduction
RGS (Regulators of G Protein Signaling) proteins regulate a wide range of GPCR pathways by virtue of their actions as GTPase accelerating proteins (GAPs) for heterotrimeric G proteins. The RGS family in mammals consists of more than 30 members defined by a conserved, catalytic RGS domain and distinguished by highly divergent N- and C-termini (Ross and Wilkie 2000). First identified as negative regulators of yeast pheromone signaling and C.elegans egg-laying behavior (Koelle and Horvitz 1996;Dohlman et al. 1996), RGS proteins have been found to regulate a number of signaling pathways in mammals, with the molecular mechanism most clearly established for the vertebrate phototransduction cascade (He et al. 1998).
In most cases the pathways regulated and the cellular and molecular mechanisms of regulation by specific RGS proteins remain unknown, but the sequence diversity and varied expression patterns of the RGS family members imply a wide range of functional specificity (Ross and Wilkie 2000;Gold et al. 1997). Assays with recombinant RGS fragments show that the catalytic activities of RGS domains are quite promiscuous for members of the Gαi/o and Gαq families with little activity on Gαs but individual differences in direct catalytic activity for specific Gα are insufficient to account for pathway specificity (Berman et al. 1996;Hooks et al. 2003).
The R7 family of RGS proteins includes multiple splice variants of RGS6, RGS7, RGS9, and RGS11 expressed primarily in the nervous system. All of these contain G-protein-gamma-like (GGL) domains that bind tightly to Gβ5, and DEP domains, which serve for at least some of them as binding sites for membrane anchors. The function of the photoreceptor-specific variant, RGS9-1, is well understood in a physiological context, and it provides an instructive example of mechanisms that can confer pathway specificity: (1) through cell-type specific co-expression with a specific GPCR pathway. RGS9-1 is expressed in rod and cone photoreceptors where it regulates their respective phototransduction cascades. (2) through subcellular co-localization. RGS9-1 is found exclusively in the outer segments of the photoreceptors where the phototransduction cascade takes place. (3) through interactions with specific anchoring or scaffolding proteins. RGS9-1 is anchored to photoreceptor outer segment membranes by its anchor R9AP, which stabilizes the complex and increases its catalytic activity for the phototransduction specific Gα transducin (Hu and Wensel 2002;Hu et al. 2003).
Two RGS9 isoforms are products of alternative RNA processing of the same Rgs9 gene. RGS9-2 is expressed in the central nervous system, primarily the dopamine receptor rich areas such as the caudate-putamen (dorsal striatum), nucleus accumbens (ventral striatum), and olfactory tubercle (Gold et al. 1997;Rahman et al. 1999). The neurons in these areas express multiple GPCRs, many with poorly understood molecular pathways that contribute to an intricate signaling network, complicating efforts to pinpoint the specific role of RGS9-2 (Wang et al. 2006). Consistent with a role in dopamine signaling, mice deficient in RGS9-2 are more sensitive to systemic administration of drugs of abuse such as cocaine and apomorphine (Rahman et al. 2003). Additionally, viral overexpression of RGS9-2 in rats without its known binding partners or introduction of purified recombinant RGS9 RGS domain through a patch pipette decreases the response to apomorphine and to a lesser extent to specific D2 dopamine receptor (Drd2) agonists (Cabrera-Vera et al. 2004). RGS9-2 mRNA and protein have been found by in situ hybridization and immunofluorescence staining in striatal medium spiny neurons, including the population expressing Drd2. RGS9-2 over-expressed in cultured cells without expression of a membrane anchor (R7BP or R9AP) is largely cytoplasmic, but co-expression of Drd2 causes it to be recruited to the plasma membrane (Rahman et al. 2003;Kovoor et al. 2005). These observations suggest that RGS9-2 may interact with or modulate Drd2 signaling networks but leave undiscovered the molecular mechanism and direct target of that regulation.
Understanding the molecular mechanisms of RGS9-2 function requires determining its subcellular environment governed by its interactions with its protein and lipid binding partners. Identical to RGS9-1 for its first 464 amino acids, RGS9-2 has a unique 25 kD C-terminus thought to increase its affinity for Gα much the same way as the γ subunit of PDE6 does for RGS9-1 (Figure 1A) (Martemyanov et al. 2003a;He et al. 2000). Like RGS9-1, RGS9-2 requires co-expression of a Gβ5 subunit (Chen et al. 2003) and a membrane anchor, the palmitoylated R7BP, for proper protein stability and localization (Martemyanov et al. 2003b;Anderson et al. 2007). Recombinant RGS9-2 expressed in cell culture with or without Gβ5, in the absence of any anchoring protein, has been reported to localize to the cell nucleus (Bouhamdan et al. 2004). Further, after densitometric fractionation of whole brain homogenates, RGS9-2 immunoreactivity is found in a crude microsome fraction that contains, among other structures, cell nuclei (Bouhamdan et al. 2004). Although the ramifications of nuclear localization of any RGS have been speculated upon, no potential function of RGS9-2 in the nucleus is known.
Figure 1.
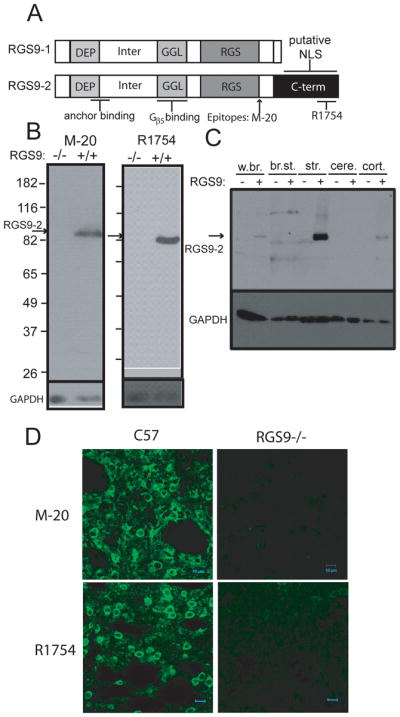
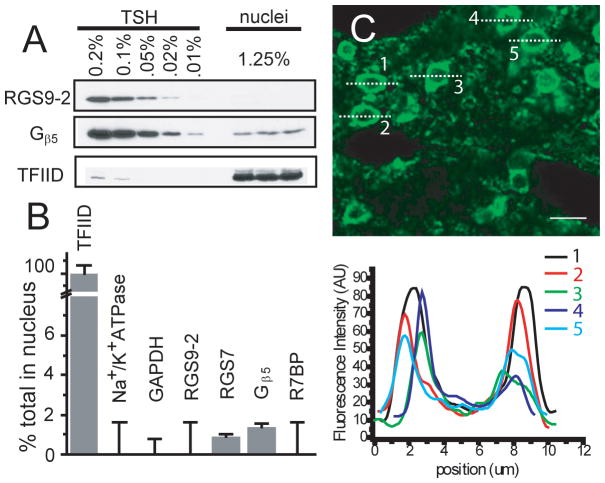
A. Domain structures of the RGS9-1 and RGS9-2 proteins and location of RGS9-2 specific antibody (M-20 and R1754) epitopes. DEP= disheveled, egl-10, plekstrin homology; Inter=area between the DEP and GGL domains; GGL= g gamma-like; RGS= regulator of G-protein signaling; C-term = RGS9-2 c-terminus; NLS = nuclear localization signal. B. Testing the specificity of anti-RGS9-2 antibodies by immunoblot. Both the M-20 and R1754 antibodies recognize a single band of approximately 82 kD in mouse striatal homogenate that is absent in the RGS9 −/− mouse. C. Immunoblot for RGS9-2 in a various brain tissue homogenates shows that RGS9-2 is present in the brain and highly enriched in the striatum. w.br. = whole brain; br.st. = brain stem; str. = striatum; cere. = cerebellum; cort. = cortex. D. Immunoflurescence staining of mouse striatal slices from C57/Bl6 and RGS9−/− using both RGS9-2 specific antibodies (M-20 and R1754). All signal is specific to RGS9-2 as it is absent in RGS9−/−.
We have analyzed the cellular and subcellular localization of the RGS9-2 protein using immunofluorescence staining with highly specific antibodies in brain slices as well as in cultured striatal neurons. We have coupled these studies with biochemical fractionation and quantitative immunoblotting in order to paint a qualitative and quantitative picture of the endogenous localization of the RGS9-2 protein with all of its binding partners.
Materials and Methods
Animals
All mice were housed and handled according to NIH guidelines as prescribed by Baylor College of Medicine’s Institutional Animal Care and Use Committee. Mice used for tissue samples were sacrificed by CO2 inhalation prior to dissection. Animals used were C57/Bl6 or RGS9−/− bred onto a C57/Bl6 background (>97.5% congenic) using a marker assisted congenic breeding protocol. The genetic background of these animals is different from those used in previous studies of the RGS9 knockout allele, which had a mixed background from 129/SvEv and C57/Bl6 strains.
Antibodies
The commercial goat anti-RGS9-2 (M20, sc-8142, Santa Cruz Biotechnology) was raised against a peptide the sequence of which corresponds to the region at the junction of the end of RGS domain and the start of the RGS9-2 specific C-terminal region. We have also developed a rabbit anti-RGS9-2 antibody (R1754) which was raised against a fusion protein with GST fused to the last 40 amino acids of mouse RGS9-2 (GST-RGS9-2c) by Bethyl Laboratories. The anti-GST component in the antisera was depleted by absorption with a column containing the lysates of bacteria (BL21) induced to express GST, and the anti-RGS9-2 component affinity purified on a GST-RGS9-2c column. The purified antibody recognized the exogenously expressed RGS9-2 protein specifically by western blot and immunocytochemistry. Anti-Gβ5 antibodies were raised in rabbit (R5214) or goat (G4718) using a conjugate of a previously described 15 amino acid peptide (Watson et al. 1996) corresponding to the N-terminus of Gβ5s and affinity purified on an immobilized peptide column. Anti-RGS7 antibody was raised and affinity purified using denatured recombinant His6-tagged bovine RGS7 (Morgans et al. 2007). The anti-R7BP antibody was raised in goat and affinity purified using a peptide corresponding to the N-terminus of R7BP CSAPNGRKKRPSRSTRSSI and used previously for antibody production(Martemyanov et al. 2005). Marker antibodies were anti-TFIID N12 (sc-204, Santa Cruz), anti-Na+/K+ATPase α-1 (05-369, Upstate Signaling), anti-GAPDH (MAB 6C5, Advanced Immunochemical), anti-synaptophysin (MAB5258, Chemicon), anti-PSD-95 (75-028, NeuroMab[UC Davis]), anti-spinophilin (AB-5669, Chemicon), and anti-DARPP-32 (611520, BD Transduction Lab).
Tissue preparation and fractionation
For all fractionation experiments whole mouse brain was removed and placed in ice cold PBS. 1 mm coronal slices were cut using a tissue block and striatal tissue was dissected out and placed directly on ice. All fractionation was carried out on ice and in the presence of a protease inhibitor cocktail (2 μg/ml aprotinin, 2 μg/ml chymostatin, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin A, 0.3 μg/ml trypsin inhibitor, 1.6 mg/ml Benzamide, 0.4 μg/ml e-64, and 0.4 mg/ml pefabloc).
Mouse striatal nuclei were isolated according to densitometric centrifugation (Rickwood et al. 1997). 8 striata were finely chopped in 5 ml nuclear homogenization buffer (0.25 M sucrose, 5 mM MgCl2, 10 mM Tris pH 7.4) and homogenized using a “pestle b” Dounce homogenizer. A small aliquot was removed as total striatal homogenate. The remaining homogenate was filtered through a 200 μm cell sieve to remove unbroken tissue and centrifuged at 600 × g for 10 minutes in a JA25.5 rotor. This pellet was resuspended in 2.5 ml homogenization buffer and re-centrifuged. This crude nuclear pellet was resuspended in 5 ml high sucrose nuclear buffer (2.2 M sucrose, 1 mM MgCl2, 10 mM Tris pH7.4), re-homogenized, and centrifuged at 70,000 × g for 80 minutes in two aliquots in a TLA 100.2 rotor. The cream colored pellet at the bottom of the tube consists of highly pure nuclei with no attached cellular structures. This was confirmed by comparing a fluorescence and light microscopy images of a Hoechst-stained aliquot of resuspended nuclei (not shown), and by immunoblotting with marker antibodies.
For detergent solubilization experiments, 2 striata were homogenized in 1 ml homogenization buffer (10 mM Tris pH7.4, 150 mM NaCl, 1 mM DTT) by pulling samples first through a series of syringe needles (18g, 22g, 26g) followed by sonication for 3 × 10 seconds using a Microson Ultrasonic Cell Disruptor. 200 μl was saved as starting material and the rest centrifuged at 88,000 × g for 15 minutes using a TLA 100.2 rotor in a Beckman Coulter Optima Ultracentrifuge to pellet membranes. The supernatant was saved as the soluble fraction and the pellet was resuspended in homogenization buffer with the appropriate detergent and the homogenization and centrifugation repeated for each step. The final “insoluble” pellet was dissolved in 1% SDS with protease inhibitors for electrophoresis.
Quantitative immunoblotting
Quantitative immunoblotting was performed on isolated nuclei and detergent solubilized homogenates for all proteins shown, using a standard curve of unfractionated starting material. All samples were transferred to supported nitrocellulose. Immunoreactivity was measured using either HRP-conjugated secondary antibodies and enhanced chemiluminescence or LI-COR IR-conjugated secondary antibodies and the Odyssey Infrared Imager. Measurements were either within the linear range of the standard curve or below the detection limit in which case they were plotted as an error bar at the value of the detection limit. Samples were run in triplicate and all data presented are the average and standard deviation of three measurements.
Cortico-striatal co-cultures
Striatal neurons were cultured by a procedure modified from that described by Malgaroli and Tsien (Malgaroli and Tsien 1992) for primary hippocampal neuronal cultures. Striatal regions of 0- to 3-day-old Sprague–Dawley rats were dissected out and dissociated mechanically. The cells were re-plated on coverslips and maintained in culture for 14–21 days on a Matrigel substrate (1:50 dilution, BD Biosciences) in minimal essential medium (MEM; Invitrogen) supplemented with B-27 (Invitrogen), 5-10% fetal calf serum (Sigma), 100 μg/ml bovine transferrin (Calbiochem), and 25 μg/ml insulin (Sigma). Proliferation of non-neuronal cells was prevented by the addition of 4 μM cytosine β-arabinofuranoside (Sigma) from the second day in culture onward.
Immunohistochemistry
Wildtype and RGS9 knockout mice were sacrificed by CO2 inhalation. The brains were removed and fixed in 4% paraformaldehyde/PBS, pH7.4 for 1 hour at 4°C. Such light fixation is necessary for the detection of endogenous RGS9-2 in the brain. Fixed brain blocks were cryoprotected with O.C.T medium (Sakura Finetechnical) and stored at -80°C. Coronal sections were cut at 20–30 μm using a cryo-microtome. Sections directly mounted on glass slides were stored at −80°C. Upon staining, sections were postfixed in methanol:acetone (1:1) for 10 minutes at room temperature and rehydrated by 3 PBS washes (5 minutes each). For single staining, an antigen retrieval procedure was used to enhance the signal. After 3 brief washes (2 minute each) in deionized water, the slides were boiled in 10 mM Na-citrate (pH6.0) for 10 minutes and cooled for 15-20 minutes at room temperature. Such a step is necessary for the detection of endogenous RGS9-2 immunoreactivity using R1754 antibody and allows the successful staining using M20 antibody at a lower concentration (see below). A tyramide signal amplification method was used for the detection of endogenous RGS9-2 immunoreactivity. Sections were permeabilized in 0.2% Triton/ PBS at room temperature for 15 minutes. After 3 PBS washes (10 minutes each) endogenous peroxidase activity was quenched by treatment of sections with 0.3% H2O2 (Sigma) in PBS for 1 hour at room temperature, followed by 3 PBS washes. The sections were then preblocked in PBS containing 1% blocking reagent (from Invitrogen tyramide amplication kit T-20922) and 0.2% Triton-100 for a period of from a few hours at room temperature or overnight at 4°C. Sections were then incubated with 0.5 μg/ml of R1754 or 1 μg/ml of M20 prepared in blocking buffer overnight at 4°C. After 3 PBS washes, sections were further stained with HRP-conjugated goat anti-rabbit IgG (1:100, for R1754, from Invitrogen Kit T-20922) or HRP-conjugated rabbit anti-goat IgG (1:100, Invitrogen R21459) prepared in 1% blocking reagent/PBS for 1–2 hours at room temperature. After 3 PBS washes, tyramide amplication reactions were performed by incubating the sections with 1:100 dilution of Alexa-488 conjugated tyramide in amplification buffer/0.0015% H2O2 (all from Invitrogen Kit T-20922) for 10 minutes at room temperature. Reactions were stopped by immersing the slices in large volumes of PBS. After 3 PBS wash, the sections were coverslipped with Gel/Mount (Biomeda, Cat# M01).
For sequential double staining, the antigen retrieval step was omitted as this step is not compatible with immunostaining procedure for the other antibodies. Only M-20 is suitable for staining and high concentration was required (4 μg/ml). For the secondary incubation step, a HRP-conjugated donkey-anti-goat IgG (Jackson Immunoresearch Laboratories 705-035-147) was used. After the tyramide amplification step, the sections were washed 3 times and then processed for staining with a marker antibody. Direct immunofluorescence was used for the detection of endogenous DARPP-32. Sections were incubated with DARP32 antibody (4 ug/ml) overnight at 4°C and with donkey anti-mouse Alexa 594 (1:200, Invitrogen A-21203) for 1–2 hours at room temperature. 3 PBS washes were included between the primary and secondary incubation steps. Sections were coverslipped with Gel/Mount.
A second tyramide amplification procedure was used to detect endogenous spinophilin. The HRP activity of the first TSA procedure was quenched by incubation with 3% H2O2/PBS for 30 minutes at room temperature. After 3 PBS washes, sections were incubated with spinophilin antibody(1 μg/ml) at 4°C overnight and HRP-conjugated donkey anti rabbit IgG (Immunoresearch Laboratories 711-035-152) for 1–2 hours at room temperature and processed for tyramide reaction as described above using Alexa594 conjugated tyramide (from Kit T-20925, Invitrogen) as substrate. 3 PBS washes were included between each step. Sections were coverslipped with Gel/Mount.
The specificity of the RGS9-2 antibodies was validated by the absence of staining in RGS9 knockout sections. Additional control experiments include incubating the sections without primary antibody or omitting one primary antibody to ensure that the secondary antibodies used reacted only with the appropriate primary antibody. Images were acquired with either an Olympus confocal microscope (Fluoview FV300) or a Zeiss confocal laser scan microscope LSM 510. For double-labeling experiments, dual images were obtained using sequential scanning to ensure no bleed through. Images from single representative optical sections are presented.
Fluorescence intensity profiles were determined by measuring fluorescence intensity along a single axis (as shown in figure 3C) using Image J software available from the NIH.
Figure 3.
Examination of endogenous RGS9-2 and associated proteins in cell nuclei of mouse striatum. A. Immunoblot used to determine relative protein levels in cell nuclei isolated by sucrose gradient versus a standard curve determined from mouse total striatal homogenate (TSH). Amounts shown are percentage of total with the nuclear amount normalized by the yield of nuclear marker TFIID. B. The relative amount of each protein found in the nuclear fraction as determined by immunoblotting normalized by the yield of the nuclear marker TFIID. C. Immunoflurescence staining of mouse striatal sections from C57/Bl6 using the R1754 antibody and antigen retrieval. Profiles of the fluorescence intensity through 5 representative neurons are plotted below. The vast majority of RGS9-2 staining appears around the cell periphery rather than the cell interior. Scale bar = 10 μm.
Immunocytochemistry
14 or 21 DIV cortico-striatal co-cultures grown on glass coverslips were fixed with 4% paraformaldehyde/4% sucrose in PBS for 1 hr at 4°C. After 3 PBS washes, the coverslips were permeabilized and preblocked with 10% normal donkey serum/0.2% Triton made in PBS. The coverslips were then incubated with M20 (4 μg/ml) together with anti-PSD95 (10 μg/ml), rabbit anti-Gβ5 (4 μg/ml), anti-spinophilin (1 μg/ml) or anti-synaptophysin (1 μg/ml) overnight at 4°C. After 3 PBS washes, the coverslips were incubated with a mixture of Alexa 488 conjugated donkey anti-goat IgG and Alexa 594 conjugated donkey anti-rabbit IgG for RGS9-2/Gβ5 co-staining and RGS9-2/spinophilin co-staining, or a mixture of Alexa 488 conjugated donkey anti-goat IgG and Alexa 594 conjugated donkey anti-mouse IgG for RGS9-2/synaptophysin, RGS9-2/DARRP-32 and RGS9-2/PSD95 co-staining. Dilutions of 1:200 were used for each individual secondary antibody (all from Invitrogen). After 3 PBS washes, the coverslips were mounted onto superfrost glass slides with Gel/Mount. Control experiments include staining by omitting both primary antibodies or omitting one primary antibody to ensure that the secondary antibodies used reacted only with the appropriate primary antibody.
Results
In order to make a confident assessment of RGS9-2 immunoreactivity for immunoblotting and tissue staining, a number of antibodies to a range of epitopes were tested. The two shown, M-20 and R1754, (Figure 1A) were selected due to their lack of cross-reactivity with RGS9−/− tissue by western blot (Figure 1B). As expected, RGS9-2 immunoreactivity is highly and specifically enriched in the striatum (Gold et al. 1997;Rahman et al. 1999) (Figure 1C). Consistent with the immunoblot results, immunohistochemical staining using both antibodies revealed strong and localized RGS9-2 immunoreactivity present in striatal sections of wild-type mice that is absent in sections from RGS9−/− mice (Fig 1D). Specific RGS9-2 immunoreactivity was found in both soma and neuropile. No obvious nuclear staining is visible.
In order to better characterize the specific cellular localization of RGS9-2 within mouse striatum, we performed immunofluorescence staining for RGS9-2 along with cell-type specific markers. Staining for RGS9-2 using our RGS9-2 C-terminus specific antibody R1754 required a harsh, denaturing heat-induced antigen retrieval technique, indicating this epitope is likely involved in tight protein-protein or protein-lipid interactions that render it inaccessible to antibody binding. The M-20 antibody was able to detect endogenous RGS9-2 immunoreactivity without the HIAR procedure at a higher concentration and was thus used in all co-labeling experiments with antibodies incompatible with HIAR. The HIAR procedure can also be used to enhance the signal when using the commercial M20 antibody (see methods).
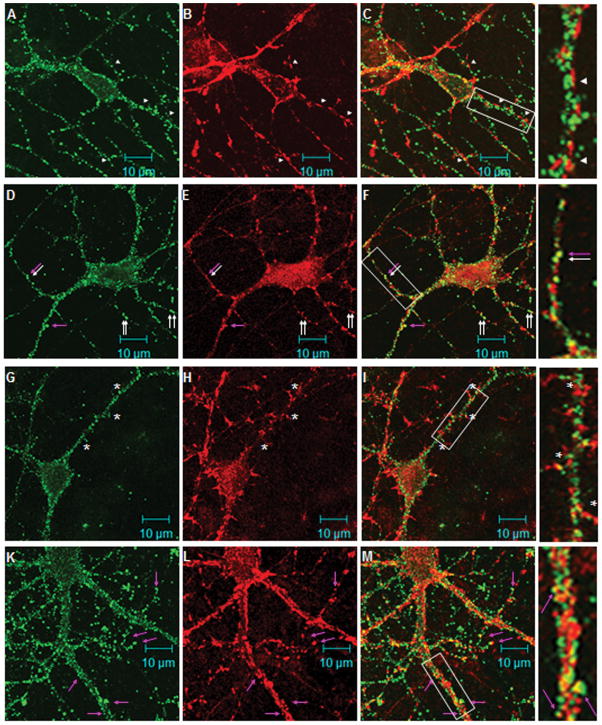
Co-labeling of mouse brain slices for RGS9-2 and DARPP-32, a marker for dopamine receptor expressing medium spiny neurons (MSNs) (Walaas and Greengard 1984;Ouimet et al. 1984), indicates that some MSNs exhibit RGS9-2 immunoreactivity at a higher level than others (Figure 2A–C). DARPP-32 immunoreactivity also appears to fill the cell soma more completely than the RGS9-2 immunoreactivity which is confined to the periphery of the soma. Co-labeling of mouse brain slices for RGS9-2 and spinophilin, an F-actin binding protein that is enriched in dendritic spines (Muly et al. 2004), yields overlapping signal throughout the striatum in the neuropil with no overlapping signal in the cell soma where spinophilin is not found (Figure 2E–G).
Figure 2.
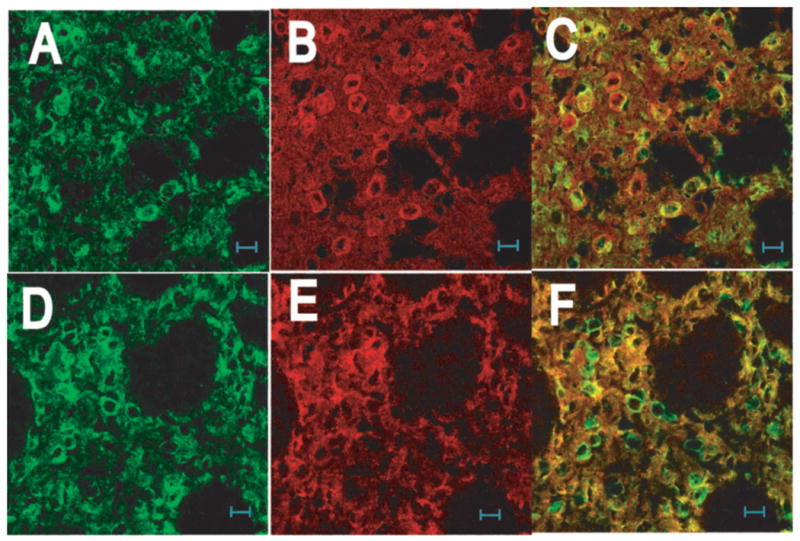
Cellular and subcellular distribution of RGS9-2 in striatum. Sections from wildtype mouse were sequentially stained with either the goat anti-RGS9-2 M20 and mouse anti-DARPP32 antibody (A –C) or the goat anti-RGS9-2 antibody and rabbit anti-spinophilin antibody (D –F). No heat-induced antigen retrieval treatment was used as such treatment abolished the staining for DARPP32. Images in the left (green), middle (red), and right columns are for RGS9-2, marker, and merges immunofluorescence, respectively.
To provide a better understanding of the subcellular localization of RGS9-2 and known binding partners we turned to cellular fractionation and immunoblotting of striatal tissue, which provides a more quantitative measure of protein quantities, and immunofluorescence staining of cultured striatal neurons, which allows better visualization of individual neuronal processes. We used fractionation and sucrose density centrifugation to obtain highly purified nuclei followed by quantitative immunoblotting for RGS9-2 and known interaction partners. Blotting for compartment specific markers was used to determine purity (< 2% contamination from cytoplasm and plasma membrane) and yield (25%) of the nuclear preparation. No RGS9-2 or R7BP and only trace amounts of Gβ5 and the R7 family member RGS7, comparable to the trace levels of membrane and cytoplasmic contamination, were found in nuclei (Figure 3A&B). Immunofluorescence staining of mouse brain sections confirms this observation with fluorescence intensity concentrated at the cell periphery in all cells examined (Figure 3C.) with little if any staining visible in the nuclei.
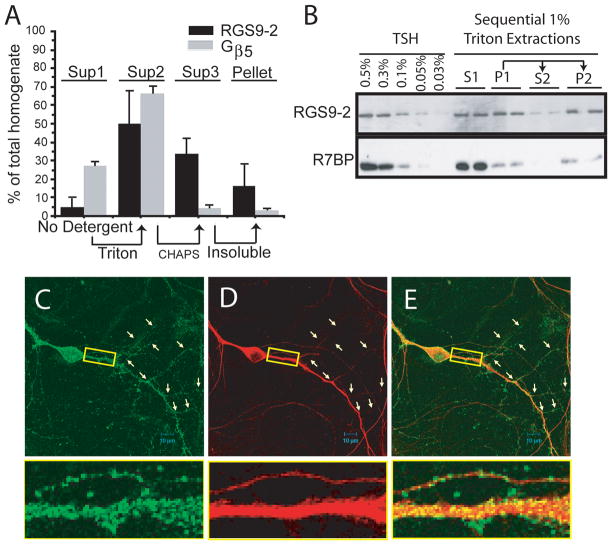
Using tissue extraction techniques and quantitative immunoblotting, we compared the relative solubilities of RGS9-2 and Gβ5 in the absence and presence of Triton X-100, a relatively mild, non-ionic detergent, or CHAPS, a harsher, zwitterionic detergent (Figure 4A). Very little RGS9-2 is soluble in the absence of detergent while about half is soluble in Triton, with most of the rest soluble in CHAPS. A small but significant fraction (~15% of the total) was insoluble in CHAPS. Gβ5, on the other hand, consists of a significant population that is soluble in the absence of detergent (~30%), with the remainder almost entirely soluble in Triton. Likely, this fraction of Gβ5 soluble in the absence of detergent corresponds to the immunoreactivity found throughout the cell body, where it may be associated with other R7 family RGS proteins such as RGS7. In order to determine whether the Triton X-100 solubility properties of RGS9-2 were due to its presence in two distinct populations or a low partition coefficient, we performed sequential detergent extractions in 1% Triton X-100 along with centrifugation. The pellet from the extraction was re-extracted in 1% Triton and centrifuged. The sequential extractions clearly show that RGS9-2 is present in two distinct populations and the Triton insoluble fraction co-fractionates with only a small fraction of the total Gβ5 and R7BP (Figure 4B). In additional experiments we found that a subpopulation of RGS9-2 is also insoluble in another non-ionic detergent, NP-40, that completely solubilizes RGS9-1 from retina (data not shown). RGS9-2 in complex with Gβ5 is an intrinsically soluble protein as revealed by heterologous expression; thus the insolubility is necessarily due to interactions with other insoluble cell components, e.g., cytoskeleton or membrane skeleton. We cannot rule out the possibility that detergent extraction removes one set of interactions that are replaced by aberrant interactions with insoluble components. However, even if this were the case the experimental results still point to distinct populations of RGS9-2, likely due to different sets of native interactions.
Figure 4.
RGS9-2 exhibits a distribution pattern distinct from that of its binding partners Gβ5 and R7BP. A. Detergent solubilization and fractionation of mouse striatal homogenate was followed by quantitative immunoblotting for RGS9-2 and Gβ5. The distinct solubilities of the two proteins correspond with their distinct immunofluorescence distributions. Arrows indicate that the pellets from the samples whose supernatant contents are shown at the bases of the arrows were subjected to a subsequent extraction to yield the supernatant samples at the tips of the arrows. The height of each bar indicates the amount soluble in the indicated detergent from the previous pellet. B. Western blots for RGS9-2 and R7BP performed on sequential Triton X-100 extractions of proteins from striatal homogenates. TSH indicates total striatal homogenate. (C–E) Rat striatal-cortical co-cultures at 14 DIV were double stained for RGS9-2 and Gβ5. Images in the left (green), middle column (red), and right columns are for RGS9-2, Gβ5, and merged immunofluorescence, respectively. The RGS9-2 protein assumed an even but discrete distribution along the dendrites including their secondary branches. The Gβ5 staining pattern on the dendrites was continuous with the intensity on the main shafts much stronger than that on the second order branches. Pairs of arrows are used to point out the positions of secondary dendritic branches. The inset is a 6.6x enlarged image of the boxed area to show RGS9-2 clusters decorating the dendrites with Gβ5 distributed homogeneously throughout the full volume of the section of dendrite.
Immunofluorescence staining of 14 DIV cortico-striatal co-cultures (Figure 4C–E) shows RGS9-2 immunoreactivity in discrete puncta along dendritic processes (Figure 4C). By contrast Gβ5 immunoreactivity is distributed homogeneously throughout the cytoplasm of both soma and dendritic processes (Figure 4D). While signals for Gβ5 immunoreactivity decrease towards the ends of secondary dendritic branches, RGS9-2 immunoreactivity remains constant throughout the entire length of the branches. Immunofluorescence staining in cultured striatal neurons shows that RGS9-2 and Gβ5 are found both in GABAergic medium spiny neurons based on co-staining with the marker GAD67 as well as other GAD67-nonreactive neurons (Supplementary Figure 1 A–F).
To better characterize these RGS9-2 immunoreactive puncta we co-labeled striatal-cortical co-cultures for RGS9-2 and a number of cell compartment-specific markers. The immunofluorescence pattern of synaptophysin, a marker for axon terminals, clearly does not overlap with that of RGS9-2. Synaptophysin-positive varicosities are found in direct opposition to RGS9-2-positive puncta, indicating that RGS9-2 is found entirely postsynaptically (Figure 5, A–C). Consistent with this, RGS9-2 staining frequently overlaps with staining for PSD-95, a marker for the excitatory postsynaptic density, and thus presumably of cortico-striatal synapses in the culture, (Fig. 5, D–F, arrows), but it is infrequent that this overlapping staining corresponds to clearly defined dendritic spines (Fig. 5, D–F, pink arrow). In 14 DIV striatal-cortical co-cultures labeling for spinophilin is found at dendritic filapodia/immature spines and is enriched especially at the tips. When the images of RGS9-2 immunoreactivity and spinophilin immunoreactivity are merged, the dendritic shafts decorated by RGS9-2 puncta are clearly distinct from the spines labeled by spinophilin antibody (Fig. 5G-L). We also examined staining in more mature 21 DIV cultured neurons to see if RGS9-2 moves to the tips of dendritic spines. Here RGS9-2 puncta do infrequently co-localize with immunoreactivity for spinophilin (Figure 5J–L, pink arrows) but the majority of RGS9-2 puncta are found away from the spineheads of cortico-striatal synapses and are rather found along the dendritic shafts and near the spine necks (Figure 5J–L). These results show that in cultured striatal neurons, under basal conditions, RGS9-2 is found near but not directly at synaptic sites likely corresponding to cortico-striatal synapses. Further, RGS9-2 immunoreactive puncta are often found directly opposed to staining for GAD65, a marker for GABA-ergic presynaptic terminals (Supplementary Figure 1 G–I). In this culture system those GABA-ergic axon terminals could be from the same neuron or from another neuron with a distant cell body. Using the high resolution of electron microscopy, synaptic release sites for inhibitory neurotransmitters have been characterized as symmetric and without an obvious post-synaptic density (Bolam J et al. 1985; Fujiyama et al. 2000). At this level of magnification, it is impossible to determine whether the post-synaptic RGS9-2 puncta are near to or immediately at the synaptic release site. The unique localization of RGS9-2 immunoreactivity at the base or neck of spines along dendritic shafts suggests that they are mainly localized at inhibitory postsynaptic sites or extrasynaptic sites near the excitatory synapses.
Figure 5.
RGS9-2 immunofluorescence is found postsynaptically along the dendritic shaft in cultured striatal neurons. Rat striatal-cortical co-cultures were double stained for RGS9-2 and synaptophysin (A–C, 14DIV), or PSD-95 (D–F, 14DIV), or spinophilin (G–I, 14DIV and J–L, 21DIV). Images in the left (green), middle (red), and right columns are for RGS9-2, marker, and merged immunofluorescence, respectively. In A–C, axons outlined by synaptophysin-positive varicosities intertwine with dendrites outlined by the RGS9-2 positive puncta. RGS9-2 immunostaining are found opposed the synaptophysin immunoreactive puncta (arrowheads). In D–F, the discrete staining pattern of RGS9-2 immunoreactivity and that of PSD95 overlaps to a great extent. White arrows indicate areas of co-localization. The pink arrows indicate RGS9-2/PSD95 puncta that exhibit a spine like structure. In G–I, RGS9-2 immunostaining does not share the same pattern with spinophilin. Note there are a few puncta (marked by asterisks) of RGS9-2 found at the base of the dendritic protrusions. In K-L, spinophilin antibody labels the mature form of spine in 21DIV co-cultures. Very few RGS9-2 positive puncta co-localize with spinophilin positive spines (pink arrows). The majority of RGS9-2 positive puncta remained positioned along the dendritic shaft. Boxed areas are a 2.67x enlargement.
Discussion
Striatal RGS9-2 is found primarily at plasma membrane and not nucleus
Our combination of specific tissue fractionation and quantitative Western blotting coupled with striatal immunofluorescence staining with multiple antibodies in both brain slices and neuronal cultures, provides conclusive evidence that, under basal conditions, virtually no RGS9-2 is found in the cell nucleus under any conditions tested; similar conclusions can be drawn, albeit based on fewer experiments, for RGS7 (Fig. 3B). Rather, RGS9-2 is primarily associated with the plasma membrane, consistent with a role in membrane-delimited G protein signaling. Further, the RGS9-2 C-terminal epitope for R1754, which is found in the putative nuclear localization sequence (Figure 1A) (Bouhamdan et al. 2004), is inaccessible to the antibody before a harsh denaturing treatment. This inaccessibility suggests that the NLS is also inaccessible to the nuclear localization machinery in vivo making it very unlikely that RGS9-2 could be transported into the nucleus. We cannot rule out the possibility, however, that under certain conditions RGS9-2 could dissociate from the plasma membrane (Anderson et al. 2009), find its C-terminus free, and be transported into the nucleus, but there is currently no compelling evidence for a nuclear role for endogenous RGS9-2. A previous report using a more crude nuclear preparation is consistent with the present results in that little RGS9-2 protein was found in the nuclear fraction (Song et al. 2006).
Striatal RGS9-2 is distributed in distinct subpopulations with differential resistance to extraction
Our tissue extraction data clearly show that endogenous RGS9-2 exists in at least two distinct populations, one soluble and the other insoluble in 1% Triton X-100. The Triton insoluble population of RGS9-2 co-fractionates with only a small percentage of striatal Gβ5 and R7BP implying that majority of Gβ5 and R7BP in the striatum are not likely associated with RGS9-2. The Triton insoluble RGS9-2 complex must be attached to the membrane skeleton or cytoskeleton through an interaction other than or in addition to binding palmitoylated R7BP and an interesting future direction is to investigate this interaction. Possibilities include RGS9-2 DEP domain interaction with Drd2 or another GPCR or through interactions with lipid microdomains that may exhibit greater Triton insolubility (London and Brown 2000). These potential interactions may be directly on RGS9-2 or with the RGS9-2 signaling complex through the small percentage of co-fractionating Gβ5 or R7BP.
There are a number of functionally important implications to the existence of two populations of RGS9-2. For instance, each population could be involved in the regulation of a separate signaling pathway diverging either at the level of GPCR interactions or further downstream in interactions with scaffolding proteins or effectors. Consistent with this idea is the observation that exogenously expressed RGS9-2 interacts with Drd2 but not Drd1 or M1 muscarinic acetylcholine receptors (Kovoor et al. 2005). Alternatively, RGS9-2 could exist in more active and less active states determined by its interactions with binding partners and the membrane. This idea is consistent with the observation that RGS9-1, which has the same DEP domain as RGS9-2, has lower affinity for its membrane anchor R9AP and reduced GAP activity when phosphorylated, and its phosphorylation state is rapidly changed by Ca2+ levels in the phototransduction cascade (Sokal et al. 2003; Hu et al. 2003). Further, differential RGS9-2 regulation has been proposed to play a role in transition between the normal Drd2 “downstate” and the hypersensitive Drd2 “upstate” (Seeman et al. 2006).
RGS9-2 is found along the dendritic shaft and near the necks of dendritic spines of striatal medium spiny neurons
It has previously been shown that RGS9-2 mRNA and protein are found in striatal medium spiny neurons including the population that expresses Drd2 (Rahman et al. 2003; Kovoor et al. 2005). We have employed immunofluorescence staining for endogeneous proteins in striatal slices and cultured striatal neurons to provide a more detailed picture of the subcellular localization patterns of RGS9-2. Our data from slices show that a high level of RGS9-2 protein is found primarily at the cell periphery consistent with membrane association. In cultured striatal neurons, RGS9-2 displays a particularly punctate pattern of postsynaptic staining along dendritic shafts and near spine necks that does not correlate entirely with any markers that we have tested. Rarely does this staining extend to the dendritic spine head and the post synaptic density, which are presumably sites of excitatory synapses. RGS9-2 immunoreactivity is found opposed to pre-synaptic terminals labeled by GAD65, a marker for GABAergic neurons, (Supplementary Figure 1 G–I). The distribution pattern of RGS9-2 immunoreactivity suggests that under the culture conditions we have used, RGS9-2 activity occurs postsynaptically along the dendritic shaft near where GABAergic inputs from other MSNs are received. It may also, to a lesser extent, regulate responses to GPCRs found at the neck of spines which modulate the response to glutamatergic excitatory input at the spine head.
Previously, RGS9-2 immunoreactivity has been reported at the postsynaptic density of excitatory synapses by postembedding immunogold electron microscopy, while extrasynaptic RGS9-2 immunoreactivity along dendritic shafts and spines was observed by preembedding immunogold electron microscopy of mouse striatal tissues (Anderson et al, 2007). Although the immunofluorescence technique we employed here does not share the same resolution as immunogold electron microscopy, our findings for the distribution pattern of RGS9-2 in cultured condition are in harmony with the distribution pattern of RGS9-2 in situ, and provide an overall view of the relative distribution throughout the cell membrane that suggests the levels at excitatory synapses are quite low compared to their levels elsewhere.
Disparities between observations with exogenous and endogenous proteins
Expression of signaling proteins in cell culture has been a frequently used tool for studying complex neuronal signaling pathways, because it provides high enough protein levels to facilitate imaging and biochemistry and allows the experimenter to establish proper controls for experiments that may be difficult in a more heterogeneous system such as the brain. However, because these experiments rarely involve all interacting partners for the studied protein and cannot possibly ensure that all interacting proteins are at endogenous relative levels, such experiments can only reveal potential functions and properties of the endogenous protein. The data presented here clearly show that the endogenous localization and membrane binding properties of one protein, RGS9-2, are quite different from those reported previously from well conducted, carefully controlled cell culture experiments with exogenously expressed protein. The observation that RGS9-2 exists in at least two distinct populations suggests a possible explanation for why interaction of the endogenous RGS9-2 signaling complex with Drd2, which it is proposed to regulate, has yet to be observed. It is clear that elucidating the molecular properties of similar endogenous proteins in complex neuronal signaling pathways remains an iterative process and of necessity researchers must employ as many approaches as possible.
Supplementary Material
Supplementary Figure 1. Immunofluorescence co-labeling for RGS9-2 or Gβ5 with markers for GABAergic neurons. A–C, costaining of RGS9-2 with GAD67, which mainly labels cell bodies of GABA-ergic neuons. D–F, costaining of G β5 with GAD67. G–I, co-staining of RGS9-2 with GAD65, which mainly labels presynaptic terminals of GABA-ergic axons. Boxed areas are enlarged.
Acknowledgments
This work was supported by NIH grants R01-DA15189 (TGW), R01-EY11900 (TGW), T32- DK007696 (JJM), and R01-NS055339 (GW)
Abbreviations
- RGS
regulator of G-protein signaling
- GAP
GTPase accelerating protein
- Drd2
D2 dopamine receptor
- DIV
days in vitro
- HIAR
heat-induced antigen retrieval
- HRP
horseradish peroxidase
- R7BP
R7 RGS protein binding protein
- R9AP
RGS9-1 binding protein
Reference List
- Anderson GR, Lujan R, Martemyanov KA. Changes in striatal signaling induce remodeling of RGS complexes containing Gbeta5 and R7BP subunits. Mol Cell Biol. 2009;29:3033–3044. doi: 10.1128/MCB.01449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Lujan R, Semenov A, Pravetoni M, Posokhova EN, Song JH, Uversky V, Chen CK, Wickman K, Martemyanov KA. Expression and localization of RGS9-2/G 5/R7BP complex in vivo is set by dynamic control of its constitutive degradation by cellular cysteine proteases. J Neurosci. 2007;27:14117–14127. doi: 10.1523/JNEUROSCI.3884-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- Bolam JP, Powell JF, Wu JY, Smith AD. Glutamate decarboxylase-immunoreactive structures in the rat striatum: a correlated light and electron microscopic study including a combination of Golgi impregnation with immunocytochemistry. J Comp Neurol. 1985;237:1–20. doi: 10.1002/cne.902370102. [DOI] [PubMed] [Google Scholar]
- Bouhamdan M, Michelhaugh SK, Calin-Jageman I, Ahern-Djamali S, Bannon MJ. Brain-specific RGS9-2 is localized to the nucleus via its unique proline-rich domain. Biochim Biophys Acta. 2004;1691:141–150. doi: 10.1016/j.bbamcr.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Cabrera-Vera TM, Hernandez S, Earls LR, Medkova M, Sundgren-Andersson AK, Surmeier DJ, Hamm HE. RGS9-2 modulates D2 dopamine receptor-mediated Ca2+ channel inhibition in rat striatal cholinergic interneurons. Proc Natl Acad Sci U S A. 2004;101:16339–16344. doi: 10.1073/pnas.0407416101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Eversole-Cire P, Zhang H, Mancino V, Chen YJ, He W, Wensel TG, Simon MI. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc Natl Acad Sci U S A. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG, Song J, Ma D, Courchesne WE, Thorner J. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein alpha subunit) Mol Cell Biol. 1996;16:5194–5209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama F, Fritschy JM, Stephenson FA, Bolam JP. Synaptic localization of GABAA receptor subunits in the striatum of the rat. J Comp Neurol. 2000;416:158–172. [PubMed] [Google Scholar]
- Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Cowan CW, Wensel TG. RGS9, a GTPase accelerator for phototransduction. Neuron. 1998;20:95–102. doi: 10.1016/s0896-6273(00)80437-7. [DOI] [PubMed] [Google Scholar]
- He W, Lu L, Zhang X, El Hodiri HM, Chen CK, Slep KC, Simon MI, Jamrich M, Wensel TG. Modules in the photoreceptor RGS9-1.Gbeta 5L GTPase-accelerating protein complex control effector coupling, GTPase acceleration, protein folding, and stability. J Biol Chem. 2000;275:37093–37100. doi: 10.1074/jbc.M006982200. [DOI] [PubMed] [Google Scholar]
- Hooks SB, Waldo GL, Corbitt J, Bodor ET, Krumins AM, Harden TK. RGS6, RGS7, RGS9, and RGS11 stimulate GTPase activity of Gi family G-proteins with differential selectivity and maximal activity. J Biol Chem. 2003;278:10087–10093. doi: 10.1074/jbc.M211382200. [DOI] [PubMed] [Google Scholar]
- Hu G, Wensel TG. R9AP, a membrane anchor for the photoreceptor GTPase accelerating protein, RGS9-1. Proc Natl Acad Sci U S A. 2002;99:9755–9760. doi: 10.1073/pnas.152094799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Zhang Z, Wensel TG. Activation of RGS9-1GTPase acceleration by its membrane anchor, R9AP. J Biol Chem. 2003;278:14550–14554. doi: 10.1074/jbc.M212046200. [DOI] [PubMed] [Google Scholar]
- Koelle MR, Horvitz HR. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- Kovoor A, Seyffarth P, Ebert J, Barghshoon S, Chen CK, Schwarz S, Axelrod JD, Cheyette BN, Simon MI, Lester HA, Schwarz J. D2 dopamine receptors colocalize regulator of G-protein signaling 9-2 (RGS9-2) via the RGS9 DEP domain, and RGS9 knock-out mice develop dyskinesias associated with dopamine pathways. J Neurosci. 2005;25:2157–2165. doi: 10.1523/JNEUROSCI.2840-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London E, Brown DA. Insolubility of lipids in triton X-100: physical origin and relationship to sphingolipid/cholesterol membrane domains (rafts) Biochim Biophys Acta. 2000;1508:182–195. doi: 10.1016/s0304-4157(00)00007-1. [DOI] [PubMed] [Google Scholar]
- Malgaroli A, Tsien RW. Glutamate-induced long-term potentiation of the frequency of miniature synaptic currents in cultured hippocampal neurons. Nature. 1992;357:134–139. doi: 10.1038/357134a0. [DOI] [PubMed] [Google Scholar]
- Martemyanov KA, Hopp JA, Arshavsky VY. Specificity of G protein-RGS protein recognition is regulated by affinity adapters. Neuron. 2003a;38:857–862. doi: 10.1016/s0896-6273(03)00320-9. [DOI] [PubMed] [Google Scholar]
- Martemyanov KA, Lishko PV, Calero N, Keresztes G, Sokolov M, Strissel KJ, Leskov IB, Hopp JA, Kolesnikov AV, Chen CK, Lem J, Heller S, Burns ME, Arshavsky VY. The DEP domain determines subcellular targeting of the GTPase activating protein RGS9 in vivo. J Neurosci. 2003b;23:10175–10181. doi: 10.1523/JNEUROSCI.23-32-10175.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martemyanov KA, Yoo PJ, Skiba NP, Arshavsky VY. R7BP, a novel neuronal protein interacting with RGS proteins of the R7 family. J Biol Chem. 2005;280:5133–5136. doi: 10.1074/jbc.C400596200. [DOI] [PubMed] [Google Scholar]
- Morgans CW, Wensel TG, Brown RL, Perez-Leon JA, Bearnot B, Duvoisin RM. Gbeta5-RGS complexes co-localize with mGluR6 in retinal ON-bipolar cells. Eur J Neurosci. 2007;26:2899–2905. doi: 10.1111/j.1460-9568.2007.05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muly EC, Smith Y, Allen P, Greengard P. Subcellular distribution of spinophilin immunolabeling in primate prefrontal cortex: localization to and within dendritic spines. J Comp Neurol. 2004;469:185–197. doi: 10.1002/cne.11001. [DOI] [PubMed] [Google Scholar]
- Ouimet CC, Miller PE, Hemmings HC, Jr, Walaas SI, Greengard P. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. III. Immunocytochemical localization. J Neurosci. 1984;4:111–124. doi: 10.1523/JNEUROSCI.04-01-00111.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman Z, Gold SJ, Potenza MN, Cowan CW, Ni YG, He W, Wensel TG, Nestler EJ. Cloning and characterization of RGS9-2: a striatal-enriched alternatively spliced product of the RGS9 gene. J Neurosci. 1999;19:2016–2026. doi: 10.1523/JNEUROSCI.19-06-02016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman Z, Schwarz J, Gold SJ, Zachariou V, Wein MN, Choi KH, Kovoor A, Chen CK, DiLeone RJ, Schwarz SC, Selley DE, Sim-Selley LJ, Barrot M, Luedtke RR, Self D, Neve RL, Lester HA, Simon MI, Nestler EJ. RGS9 modulates dopamine signaling in the basal ganglia. Neuron. 2003;38:941–952. doi: 10.1016/s0896-6273(03)00321-0. [DOI] [PubMed] [Google Scholar]
- Rickwood D, Messent A, Patel D. Isolation of nuclei and nuclear subfractions. In: Rickwood D, Graham JM, editors. Subcellular fractionation: a practical approach. IRL Press at Oxford University Press; New York: 1997. pp. 71–97. [Google Scholar]
- Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- Seeman P, Schwarz J, Chen JF, Szechtman H, Perreault M, McKnight GS, Roder JC, Quirion R, Boksa P, Srivastava LK, Yanai K, Weinshenker D, Sumiyoshi T. Psychosis pathways converge via D2high dopamine receptors. Synapse. 2006;60:319–346. doi: 10.1002/syn.20303. [DOI] [PubMed] [Google Scholar]
- Sokal I, Hu G, Liang Y, Mao M, Wensel TG, Palczewski K. Identification of protein kinase C isozymes responsible for the phosphorylation of photoreceptor-specific RGS9-1 at Ser475. J Biol Chem. 2003;278:8316–8325. doi: 10.1074/jbc.M211782200. [DOI] [PubMed] [Google Scholar]
- Song JH, Waataja JJ, Martemyanov KA. Subcellular targeting of RGS9-2 is controlled by multiple molecular determinants on its membrane anchor, R7BP. J Biol Chem. 2006;281:15361–15369. doi: 10.1074/jbc.M600749200. [DOI] [PubMed] [Google Scholar]
- Walaas SI, Greengard P. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. I. Regional and cellular distribution in the rat brain. J Neurosci. 1984;4:84–98. doi: 10.1523/JNEUROSCI.04-01-00084.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kai L, Day M, Ronesi J, Yin HH, Ding J, Tkatch T, Lovinger DM, Surmeier DJ. Dopaminergic control of corticostriatal long-term synaptic depression in medium spiny neurons is mediated by cholinergic interneurons. Neuron. 2006;50:443–452. doi: 10.1016/j.neuron.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Watson AJ, Aragay AM, Slepak VZ, Simon MI. A novel form of the G protein beta subunit Gbeta5 is specifically expressed in the vertebrate retina. J Biol Chem. 1996;271:28154–28160. doi: 10.1074/jbc.271.45.28154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Immunofluorescence co-labeling for RGS9-2 or Gβ5 with markers for GABAergic neurons. A–C, costaining of RGS9-2 with GAD67, which mainly labels cell bodies of GABA-ergic neuons. D–F, costaining of G β5 with GAD67. G–I, co-staining of RGS9-2 with GAD65, which mainly labels presynaptic terminals of GABA-ergic axons. Boxed areas are enlarged.