Background: Regulation of virus entry by host lipids is poorly understood.
Results: Imaging of low pH-mediated fusion between single vesicular stomatitis pseudoviruses and lipid bilayers revealed a striking reliance on anionic lipids.
Conclusion: The dependence of fusion on late endosome-resident anionic lipids suggests a new means for regulating the virus entry sites.
Significance: Reliance on specific lipids for fusion may shed light on future antiviral strategies.
Keywords: Fluorescence, Membrane Bilayer, Membrane Fusion, Microscopic Imaging, Virus Entry, Fusion Pore, Hemifusion, Single Virus Imaging
Abstract
Viral glycoproteins mediate fusion between viral and cellular membranes upon binding to cognate receptors and/or experiencing low pH. Although activation of viral glycoproteins is thought to be necessary and sufficient for fusion, accumulating evidence suggests that additional cellular factors, including lipids, can modulate the fusion process. Understanding the role of lipids in virus entry via endocytosis is impeded by poor accessibility and the highly diverse nature of endosomes. Here we imaged fusion of single retroviral particles pseudotyped with the vesicular stomatitis virus (VSV) G protein with dextran-supported lipid bilayers. Incorporation of diffusible fluorescent labels into the viral membrane and the viral interior enabled detection of the lipid mixing (hemifusion) and content transfer (full fusion) steps of VSV G-mediated fusion at low pH. Although single virus fusion with supported bilayers made of zwitterionic lipids could not be detected, inclusion of anionic lipids, phosphatidylserine, and bis(monoacylglycero)phosphate (BMP), greatly enhanced the efficiency of hemifusion and permitted full fusion. Importantly, lipid mixing always preceded the opening of a fusion pore, demonstrating that VSV G-mediated fusion proceeds through a long-lived hemifusion intermediate. Kinetic analysis of lipid and content transfer showed that the lags between lipid and content mixing defining the lifetime of a hemifusion intermediate were significantly shorter for BMP-containing compared with PS-containing bilayers. The strong fusion-enhancing effect of BMP, a late endosome-resident lipid, is consistent with the model that VSV initiates fusion in early endosomes but releases its core into the cytosol after reaching late endosomal compartments.
Introduction
Entry of enveloped viruses into a host cell is a complex, multistep process mediated by viral glycoproteins, which are activated upon encountering cellular receptor(s) and/or low pH (1, 2). Generally, the entry of enveloped viruses proceeds through attachment to a host cell, binding to cognate receptor(s), and either fusion of the viral membrane with the cell plasma membrane or virus internalization and low pH-dependent fusion with endosomes (3). Recent evidence suggests that viruses that do not rely on low pH can also enter through endocytic pathways (4–7). Viral envelope glycoproteins are thought to mediate energetically unfavorable membrane fusion events by releasing the energy stored in their native structure upon refolding into a thermodynamically favorable conformation (8–11). Despite the highly diverse structures of viral fusion glycoprotein, they are categorized into three classes on the basis of their structural and biochemical properties (2, 12).
VSV2 G protein is a class III viral fusion protein that is well characterized both structurally and functionally (13–15). Interestingly, pH-induced conformational changes in VSV G are reversible, as the protein assumes the native conformation upon returning to neutral pH in the absence of a target membrane (16–18). This unusual feature of VSV G indicates that the difference between free energies of the initial “metastable” and final “stable” conformations is not large and that, therefore, little energy can be released by this protein to drive membrane fusion (19). These considerations warrant further studies of VSV G-mediated fusion to gain critical insights into the universal principles by which viral proteins promote membrane merger.
Despite extensive efforts, the host cell receptor(s) for VSV entry has not been unambiguously identified. Phosphatidylserine (PS) has been shown to promote VSV fusion and entry into cells (20–23). However, this lipid is unlikely to be the primary VSV receptor because VSV binding and infection do not correlate with the level of PS in the plasma membrane, and blocking PS with annexin V does not inhibit VSV infection (24). Moreover, the fact that VSV can merge with lipid vesicles made of phosphatidylcholine at low pH (21, 25) indicates the lack of reliance on a proteinaceous receptor or a specific lipid. On the other hand, lipid composition is known to modulate the kinetics and efficiency of viral fusion (26) and could, therefore, play a role in the regulation of G protein-mediated fusion. Recent studies highlight the importance of an endosome-specific anionic lipid, bis(monoacylglycero)phosphate (BMP), also referred to as lysobisphosphatidic acid (27), in promoting fusion of unrelated viruses entering cells via endocytosis (28–31). For instance, dengue virus fusion with liposomes and with cell membranes has been shown to depend on anionic lipids, including BMP (30), which is one of the major lipid species in late endosomes and multivesicular bodies (27, 32). BMP is thought to facilitate cellular entry of different pathogens by regulating back-fusion between intralumenal vesicles and the limiting membrane of an endosome (33, 34). Interestingly, a BMP-dependent back-fusion process has also been implicated in the VSV nucleocapsid release into the cytosol following virus fusion with intralumenal vesicles (28, 29). However, this two-step VSV entry model is not universally accepted because this virus appears to quickly fuse with early endosomes prior to entry into multivesicular bodies (35, 36).
Single virus trafficking in host cells is an increasingly popular tool to study the viral entry process (37). However, real-time detection of fusion events that culminate in the viral content release is technically challenging and, therefore, less common (4, 38–40). The main advantage of these approaches is the ability to study events at a single virus level, which allows distinguishing specific subpopulations of particles or characteristics hidden in bulk assay data.
Although single virus imaging in live cells provides important insights into the entry process, the complexity of vesicular trafficking and the heterogeneity of endosomal compartments impede mechanistic studies of viral fusion. Dissecting the viral entry processes in a controlled environment can shed light on the mechanism of fusion and on the host factors required for completion of this reaction. A powerful approach to address these questions is to reconstitute viral fusion in model systems such as liposomes and supported lipid bilayers (SLB). Recent advances in imaging of membrane fusion using supported lipid bilayers or tethered single liposomes have brought new mechanistic insights into the membrane merger mediated by cellular fusion proteins (41–43) and viral glycoproteins (44–46). Rapid pH-dependent hemifusion/fusion of single influenza and Sindbis viruses with SLB was observed by monitoring redistribution of a lipophilic dye incorporated into the viral membrane (hemifusion) (46) or by visualizing the transfer of both viral membrane and content markers (full fusion) (44, 45).
Here, we report the direct visualization of VSV G-pseudotyped particle (VSVpp) fusion with dextran-supported lipid bilayers. We took advantage of the murine leukemia virus (MLV) pseudotyping system, which allows incorporation of a releasable fluorescent protein marker into the virus (5) and of a membrane dye into the viral envelope. This labeling strategy permitted the detection of both lipid mixing (hemifusion) and content release (full fusion) steps of the VSVpp fusion process. Imaging of single VSVpp revealed that both hemifusion and fusion with supported bilayers were markedly promoted by anionic lipids, POPS, or BMP. Although BMP-containing bilayers supported faster conversion of hemifusion to full fusion compared with POPS bilayers, similar probabilities of fusion and identical effective sizes of nascent fusion pores were consistent with the lack of requirement for a specific lipid for productive VSV entry. The strong fusion-stimulating effect of BMP indicates, however, that this endosome-resident lipid may modulate the outcome of low pH-induced VSV fusion with intracellular compartments.
EXPERIMENTAL PROCEDURES
Virus Production
Fluorescently labeled pseudoviruses were produced in HEK293T/17 cells using PolyFect transfection reagent (Qiagen, Valencia, CA). Cells grown on 10-cm dishes were transfected with 2 μg MLV-Gag-Pol, 1 μg MLV-Gag-mKO, 3 μg pMLV-LTR-LacZ, and 3 μg of pMDG-VSV-G. Twenty-four hours post-transfection, cells were labeled with 10 μm 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine (DiD) for 4 h in a CO2 incubator at 37 °C. Cells were washed, covered with 6 ml of fresh phenol red-free growth medium, and incubated for an additional 24 h. Virus-containing medium was collected 48 h post-transfection, passed through a 0.45 μm filter, aliquoted, and stored at −80 °C. The infectious titer was determined by a β-galactosidase assay in TZM-bl cells (47). Vectors expressing MLV-Gag-pol and MLV-LTR LacZ (48) were provided by Dr. Walther Mothes (Yale University). The pMDG-VSV-G expression vector was provided by Dr. John Young (Salk Institute). The construction of the MLV-Gag-mKo expression vector was described in Ref. 5.
Preparation of Small Unilamellar Vesicles
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), BMP (S,R isomer) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(carboxyfluorescein) (CF-PE) were purchased from Avanti Polar Lipids (Alabaster, AL). Cholesterol (Chol) was from Sigma. Lipids were mixed in chloroform and dried to a thin film in a round-bottom glass flask under an argon stream. Traces of solvent were removed by placing the flask in a vacuum chamber for at least 3 h. The lipid film was hydrated in PBS buffer (Cellgro, Mediatech, Inc., Manassas, VA) to make a final lipid concentration of 1 mm. The lipid suspension was subjected to several freeze/thaw cycles and sonicated with a probe sonicator (VWR, Radnor, PA) on ice to avoid sample heating. The obtained lipid suspension was cleared by centrifugation at 10,000 × g for 5 min.
Coverslip Cleaning and Functionalization with Dextran
Glass coverslips (no. 1.5, 22 × 22 mm, Corning, NY) were thoroughly cleaned using the following steps: 1) immersion into hot 2% Hellmanex (Hellma Analytics, Müllheim, Germany) for 30 min, 2) sonication in Hellmanex for 15 min while warm, 3) rinsing extensively with Milli-Q water, 4) immersion into Piranha solution (3:1 concentrated sulfuric acid/hydrogen peroxide 30% without stabilizer) for 30 min, 5) rinsing extensively with Milli-Q water, 6) sonication in Milli-Q water for 10 min, and 7) rinsing extensively with Milli-Q water. The cleaned coverslips were dried in an oven for 1 h at ∼100 °C.
Dextran functionalization was carried out as described previously (45). Briefly, cleaned coverslips were immersed in 0.2% (3-glycidyloxypropyl)trimethoxysilane (Sigma) in isopropanol for 5 min under gentle agitation. After rinsing with pure isopropanol, the coverslips were cured for 1 h at ∼80 °C. A 30% (w/v) dextran (Mr 5 × 105, Sigma-Fluka) solution was prepared using Milli-Q water, and air bubbles were removed in a vacuum chamber. The coverslips were carefully covered with dextran solution (∼1 ml) and incubated in a humidified chamber for ∼24 h. Excess dextran was rinsed off by dipping repeatedly in water and leaving the coverslips in water for 2 days. The coverslips were dried in an oven for 1 h at 80 °C and stored in a vacuum desiccator.
Microfluidic Chamber Design and Fluorescence Microscopy
The microfluidic polydimethylsiloxane (PDMS) chamber was fabricated using both standard photolithography and soft lithography. To prepare the soft mold for the microfluidic channels, a negative photoresist (MicoChem, SU-8) was spin-coated and patterned on a 4-inch silicon wafer. The patterned wafers were silanized using hexamethyldisilazane so that the cured PDMS could be easily peeled off from the soft mold. PDMS mixed at a 10:1 ratio (w/w) of polymer to curing agent was then poured onto the silanized wafer and cured at 60 °C for about 24 h. The cured device was then peeled off from the wafer. The PDMS pieces were then rinsed thoroughly with deionized water and set to dry at 60 °C in an oven.
The 25 × 25-mm PDMS block had two 10-mm grooves that were 1 mm wide and 75 μm deep. Small holes were punched at each end of the grooves, and polytetrafluoroethylene tubing (microbore 0.03-inch (0.76-mm) diameter) was inserted into the holes (Fig. 1, A and B). The PDMS block was adhered to top of a dextran-functionalized coverslip, thus creating two flow channels between the PDMS block and the coverslip. Tight adhesion of the polymer to the glass surface was sufficient to seal the channels. This approach was similar to a method described previously for SLB formation in microfluidic cells (49).
FIGURE 1.
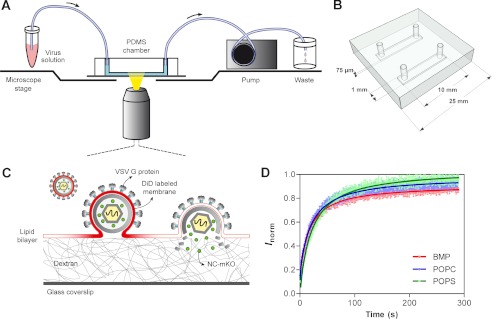
Schematic representation of single virus fusion experiments. A, schematic drawing of the microfluidic chamber mounted on a microscope stage. B, sketch of the microfluidic PDMS block (top view) that was attached to the glass coverslip to form two channels where the SLB were formed. C, single VSVpp fusion triggered by low pH detected by double fluorescent labeling with Gag-mKO (green) and DiD (red). VSV G-mediated lipid mixing is manifested in radial diffusion of DiD into the SLB. After pore formation, the fluorescent content marker is released into the dextran layer underneath the bilayer and disappears because of diffusion. D, FRAP assay to measure the fluidity of supported lipid bilayers. The recovery of CF-PE fluorescence after photobleaching of a defined circular region (w = 8.02 μm) was followed over time. Plotted values are means with error bars from 95% confidence intervals (n = 5). Recovery data were fitted with equation 1. The resulting mean ± S.E. diffusion coefficients for CF-PE were 0.946 ± 0.022 μm2/s in POPC, 0.904 ± 0.026 μm2/s in POPS, and 1.16 ± 0.035 μm2/s in BMP bilayers.
All imaging experiments were carried out at room temperature using a confocal laser scanning microscope Zeiss LSM 780 and oil immersion Plan-Neofluar ×40/1.3 numerical aperture objective. The laser lines used to excite fluorophores were 488 (CF-PE), 561 (mKO), and 633 nm (DiD). The PDMS block with coverslip was affixed to a microscope stage and connected to a peristaltic pump (Fig. 1A). Solution flow during the imaging experiments was adjusted to ∼48 μl/min.
Supported Lipid Bilayer Formation
SLB were formed by the vesicle fusion method (50). Small unilamellar vesicle suspensions with 0.1 mm of total lipids in PBS were injected into both microfluidic channels. After incubation for 30 min, the excess of liposomes was removed by flowing PBS for 5 min. The compositions used for the SLB were POPC:Chol:CF-PE, 84.5:15:0.5 mol %; POPC:Chol:POPS:CF-PE, 64.5:15:20:0.5 mol %; and POPC:Chol:BMP:CF-PE, 64.5:15:20:0.5 mol %. These mixtures are designated throughout the text as POPC, POPS, and BMP, respectively. Small amounts of the fluorescent lipid CF-PE were included for three reasons: to assess the uniformity of SLBs, to test whether lipids would diffuse freely in SLBs by performing FRAP experiments (see below), and to determine the exact time of the pH drop upon perfusion with acidic buffers (similar to Ref. 45).
FRAP Assay
SLB obtained by spontaneous rupture/fusion of small unilamellar vesicles over the hydrated dextran layer of the glass coverslip were usually uniform, as observed by the fluorescence of CF-PE incorporated in the lipids mixtures. Next, SLB were tested for fluidity using FRAP (51). Briefly, a circular region of interest (radius 8 μm) was photobleached using a brief pulse of the 488-nm line of an argon laser set at 100% power. The fluorescence recovery was monitored by low-intensity laser excitation for at least 5 min. The fluorescence of a distant circular region of interest (ROI) served as a reference signal.
Intensity profiles for the bleached and reference ROIs were analyzed using ImageJ software. The fluorescence recovery trace was double-normalized to take into account a possible inadvertent bleaching effect upon image acquisition
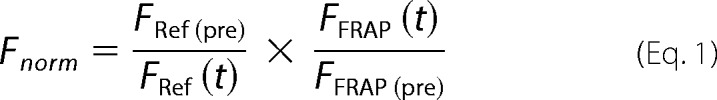 |
where FRef is the mean fluorescence intensity of the reference ROI, FFRAP is the intensity of the bleached ROI, t is the time after photobleaching, and pre stands for the mean intensity before photobleaching.
The normalized traces of the bleached ROI were fitted using a diffusion model for a circular spot (52)
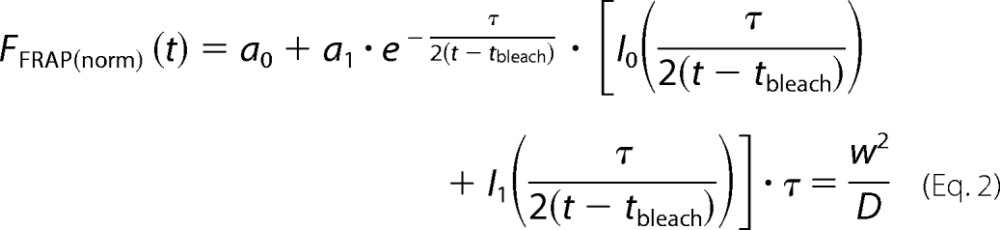 |
where tbleach is the time of bleaching, τ is the characteristic diffusion time, w is the radius of the ROI, D is the diffusion coefficient, and In(x) are modified Bessel functions. Nonlinear fit of the data was performed in GraphPad Prism version 5.
The results of the FRAP assay for each type of lipid mixture used are shown in Fig. 1D, and the corresponding diffusion coefficients are in the figure legend. Diffusion coefficients around 1 μm2/s, corresponding to free lipid diffusion in bilayers, were deemed acceptable for virus fusion experiments. Systematic verification of the bilayer fluidity is important because a fraction of bilayers did not exhibit fluorescence recovery after photobleaching (see also Ref. 49).
Imaging of VSVpp Fusion
All experiments were performed at room temperature. For each imaging experiment, a sample of 5 μl of the virus stock solution was diluted in 100 μl of PBS and vortexed briefly. Approximately 90 μl of viral suspension was continuously flowing into a channel. Viruses adhered to the bilayer reasonably well, permitting subsequent washing with BPS for 1–2 min to remove unbound viruses. Next, image acquisition was started, and perfusion with an MES (pH 5.5) buffer (144 mm NaCl, 2 mm CaCl2, 30 mm MES) was initiated. This acidic buffer quenched the CF-PE fluorescence, marking the exact time of the pH change (similar to an approach introduced in Ref. 45). Images were acquired with 2-s intervals for 11 min. At the end of the acquisition, the flow channel was cleaned by flowing ∼1 ml of 70% ethanol at a higher flow rate before removing the chamber from the microscope.
Image Analysis
Hemifusion and fusion events were initially identified by visual inspection of image sequences. Next, double-labeled (DiD and mKO) viral particles were detected, and their mean fluorescence intensities over time were measured using Speckle TrackerJ, a recent freely available ImageJ plug-in (53). The program identified fluorescent particles and tracked them until the fluorescence intensity/particle size decreased below a defined threshold. DiD-only labeled particles (likely dye aggregates) were excluded from the analysis. Virus hemifusion was defined as DiD redistribution into SLBs without the loss of mKO, whereas full fusion was defined as release of DiD followed by loss of the content marker. The mean fluorescence intensity profiles for each marker (CF-PE, DiD, and mKO) were obtained using small circular regions encompassing the particles. Analysis of these profiles yielded the following key events used for kinetic analysis. The pH drop was assessed by decay of the CF-PE signal; the onset of lipid mixing was defined as the onset of DiD dequenching or, when dequenching was weak, by the onset of fluorescence decay because of DiD dilution; the onset of content release was detected by mKO fluorescence decrease; and completion of content release was defined as the time point at which the mKO signal reached the background level. The non-parametric Mann-Whitney rank sum test was used to evaluate the differences among the kinetic parameters for bilayers of different composition.
RESULTS
Direct Visualization of Single VSVpp Hemifusion and Fusion Events
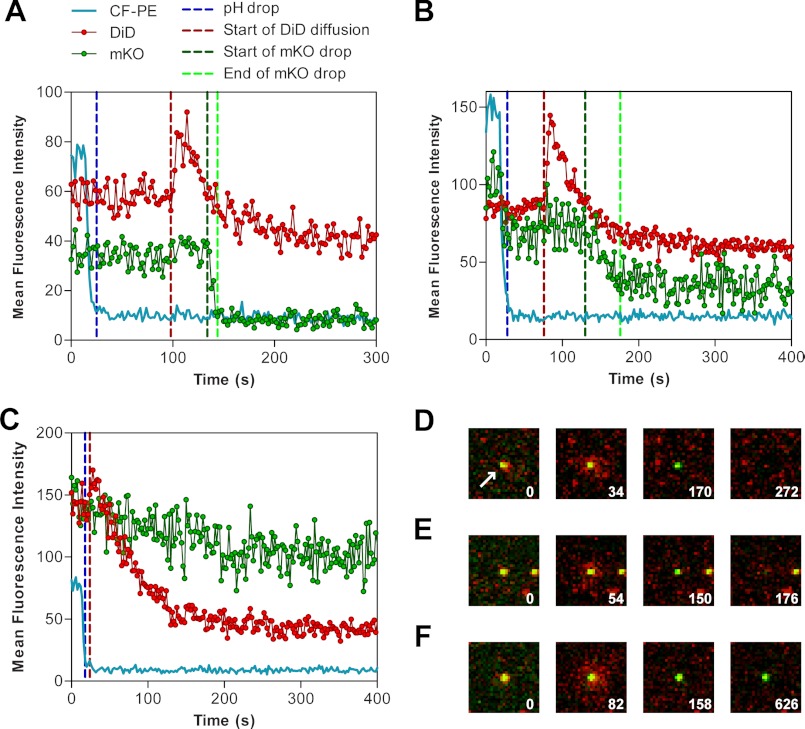
The microfluidic PDMS chamber allowed for an easy solution change and for a quick formation of supported lipid bilayers, as described under “Experimental Procedures” (Fig. 1, A and B). After testing the SLB fluidity by FRAP (Fig. 1D), double-labeled VSV pseudoviruses were drawn into the chamber and allowed to attach to the bilayer for 5 min. VSVpp readily adhered to bilayers and remained relatively immobile throughout the experiments despite the solution changes. The same volume of diluted viral stock was passed through a flow channel in each experiment, resulting in adhesion of 60–100 particles/image field. VSVpp were colabeled with the membrane dye, DiD, and the viral content marker, the MLV-Gag polyprotein tagged with mKO (monomeric Kusabira Orange, Ref. 54), as described in Ref. 5. The Gag-mKO marker is cleaved upon MLV maturation, producing the nucleocapsid-mKO (NC-mKO) fragment, which is not associated with the viral core and is, therefore, readily released upon viral fusion (4, 5, 55). For low pH-triggered fusion, mKO is a better marker than GFP because its resistance to quenching by low pH (pKa ≤ 5 (54)) minimizes the possibility that the loss of mKO signal is due to acidification of the viral interior. This strategy allowed the detection of DiD transfer into SLB as a marker for hemifusion as well as detection of the viral content release as a marker for full fusion (Fig. 1C).
VSVpp fusion with SLB was initiated by perfusion with an acidic (pH 5.5) buffer. The exact time of acidification was determined on the basis of quenching of CF-PE fluorescence, similarly to the approach described in Ref. 45. Shortly after acidification, radial diffusion of DiD into a bilayer, away from the viral particles, was observed for a fraction of the virions (Fig. 2). Typical fluorescence intensity profiles for fusion events are shown in Fig. 2, A and B, and Fig. 3. The DiD signal usually increased because of fluorescence dequenching and then decayed upon spreading into the supported bilayer. These lipid mixing events reflecting virus-SLB hemifusion were not observed at neutral pH. Following the lipid mixing events, a fraction of particles lost their content marker, NC-mKO. Because the mKO signal from neighboring particles that had no change in DiD fluorescence remained stable, the loss of the mKO signal following the hemifusion step signified the viral content release through a fusion pore (Fig. 2, D–E, and supplemental movie 1). The existence of a hydrated dextran layer supporting the membrane provided sufficient space to accommodate the viral content marker and to permit its diffusion away from a particle. The four sequential events (acidification of the flow channel, onset of lipid mixing, as well as the onset and time of completion of content release) are marked by vertical lines in Fig. 2, A and B. These four parameters were used to compare the kinetics of VSVpp fusion with SLB of a different lipid composition (see examples in Fig. 3).
FIGURE 2.
Imaging single VSV pseudovirus fusion. A, example of fluorescence intensity profiles for a single VSVpp-SLB fusion event (POPS). The drop of CF-PE signal (blue) marks the acidification of the image area to pH 5.5. Key events used for kinetic analysis are marked by vertical lines representing the time of acidification (blue), onset of lipid mixing (red), onset of content release (green), and end of content release (light green). B, intensity profiles for a single fusion event with BMP bilayers exhibiting a slow mKO release. The slight reduction in the mKO signal upon acidification reflects a spectral bleed-through from the CF-PE signal which was fully quenched by low pH. C, intensity profiles for a virus undergoing hemifusion (decay of the DiD signal without loss of the mKO signal). D and E, images of full VSVpp fusion events with POPS (D) and BMP (E) bilayers. The double-labeled pseudoviruses (yellow) released DiD (red) into the SLB by radial diffusion, followed by disappearance of green puncta because of release of NC-mKO. The first images in each panel are taken before acidification, the second and third images show the dequenching and spread of DiD while the mKO signal remains steady, and the last images were taken after completion of fusion (mKO release). F, example of a hemifusion event with BMP bilayer that did not culminate in full fusion. Here the mKO signal remained steady throughout the experiment. White numbers (D–F) indicate time after acidification in seconds. The image fields are 12 × 12 μm. The white arrow is pointed at the central particle of interest that will undergo fusion.
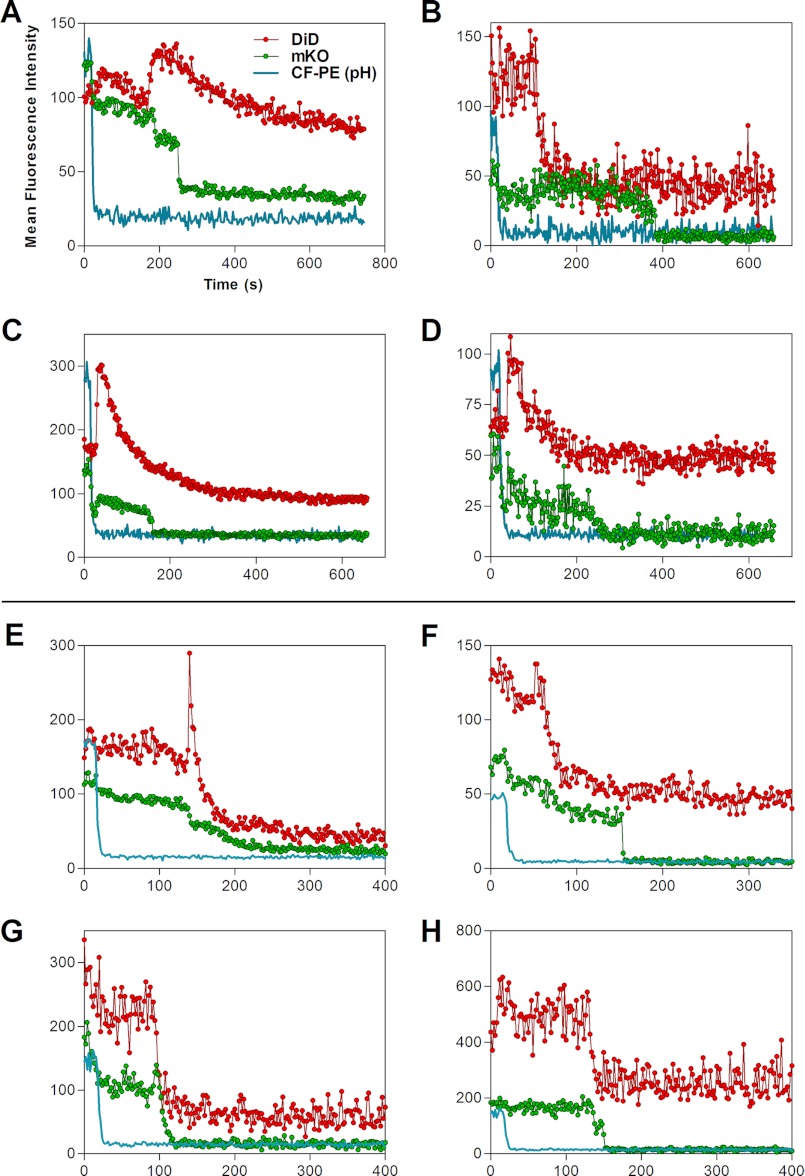
FIGURE 3.
Mean fluorescence intensity profiles of representative single virus fusion events with POPS (A–D) and BMP (E–H) bilayers. MFIs of arbitrary circular regions encompassing individual particles are plotted. Red traces show the DiD signal, green traces show the mKO, and blue traces show the CF-PE signal (the pH indicator). Note that quenching of the CF-PE fluorescence upon acidification was often associated with a concomitant reduction in the mKO signal because of a spectral bleed-through. These initial changes in the mKO signal were ignored.
Anionic Lipids Are Essential for VSV G-mediated Hemifusion and Fusion
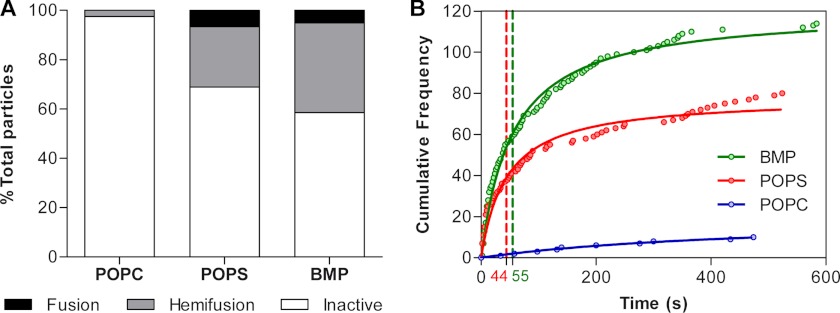
The total number of double-labeled VSVpp attached to POPS and BMP membranes was comparable, but a larger number of pseudoviruses adhered to POPC-based membranes lacking anionic lipids (see legend for Fig. 4A). However, despite the larger number of adhered particles, POPC bilayers did not support complete VSVpp fusion, and only 2.5% of double-labeled particles exhibited lipid mixing (hemifusion) with these bilayers (Fig. 4A). In sharp contrast, 31.1% and 41.5% of VSV pseudoviruses released DiD into POPS and BMP bilayers, respectively (Fig. 4A). BMP more potently facilitated lipid mixing than an equal amount of POPS. Under our experimental conditions, double-labeled particles released their lipid markers much more frequently than their contents. Complete fusion was detected for 6.6% of particles following their hemifusion with POPS bilayers and for 5.1% particles adhered to BMP bilayers. In other words, a significant fraction of viruses underwent hemifusion but did not form a fusion pore large enough to allow NC-mKO escape into the dextran layer underneath SLB (Figs. 1C and 2, C and F, and supplemental movie 2). Hemifusion always preceded full fusion. The fluorescence signal from double-labeled particles that did not exhibit lipid or content mixing was stable over the course of the experiment, demonstrating that inadvertent photobleaching of fluorophores was negligible (supplemental Fig. 1). In conclusion, 20 mol % of POPS or BMP greatly enhanced the efficiency of VSVpp hemifusion and, importantly, enabled complete fusion compared with the basal lipid mixture lacking anionic lipids (Fig. 4A).
FIGURE 4.
Analyses of the extent and kinetics of VSV pseudovirus fusion. A, the relative frequencies of fusion outcomes (no fusion, hemifusion, and full fusion) are shown for fusion of double-labeled VSVpp with SLBs of different lipid compositions. Hemifusion is defined as dequenching/loss of the DiD signal without the content release, whereas fusion is defined as the loss of both membrane and content markers (DiD and mKO, respectively). The total number of membrane-bound particles immediately after the pH drop was 398 for POPC, 257 for POPS, and 275 for BMP bilayers using four independent image fields for each lipid composition. B, distribution of the time elapsed between the pH drop (CF-PE fluorescence quenching) and the onset of lipid mixing for double-labeled particles that underwent hemifusion or fusion. The difference in the lag times before the lipid mixing for POPS and BMP was not significant (p > 0.93), but the differences between the hemifusion kinetics for these bilayers and POPC-based membranes were significant (p < 0.006 for POPS and p < 0.023 for BMP). The hemifusion half-time (t50) was determined by fitting the data with equation N = (A·t)/(t50 + t), where A is the total number of events and t is the time. The green and red dashed vertical lines with corresponding numbers at the x axis correspond to the t50 values for BMP and POPS bilayers, respectively.
BMP Shortens the Lag Time between Lipid Mixing and Fusion Pore Formation
As indicated above, lipid mixing events were rare for POPC bilayers lacking anionic lipids. This is in contrast with POPS or BMP bilayers supporting a much greater number of lipid mixing events. To further assess the role of anionic lipids in VSVpp fusion, we analyzed the distribution of delay times from the pH drop to the onset of lipid mixing (Fig. 4B). After acidification, VSVpp hemifusion with POPS and BMP bilayers occurred with the half-times of 44 s and 55 s, respectively (Fig. 4B). The difference in lag times to the onset of lipid mixing with BMP and POPS bilayers was not statistically significant (p > 0.93). These results suggest that although anionic lipids promote VSVpp hemifusion, this process does not require a specific lipid. It thus appears that the enhanced VSVpp fusion with bilayers containing anionic lipids is due to an electrostatic effect.
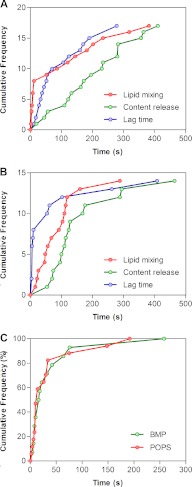
The kinetics of VSVpp fusion were analyzed in more detail using the following parameters illustrated in Figs. 2 and 5: 1) time to the onset of lipid mixing/hemifusion, 2) time to the onset of content release, 3) lag time between lipid mixing and content release for a given particle, and 4) the time from the onset to completion of the NC-mKO release as a measure of the relative size of a fusion pore. All full fusion events were preceded by lipid mixing (Figs. 2 and 3), demonstrating that VSVpp fusion proceeded through a relatively long-lived hemifusion intermediate. This analysis revealed that the distribution of the times from acidification to content release was slightly faster for BMP bilayers than for POPS bilayers, but the difference was not statically significant (Fig. 5, A and B). Importantly, the lag time between lipid mixing and content release was significantly shorter for BMP when compared with POPS bilayers (Fig. 5, A and B). These results indicate that BMP can accelerate the conversion of a hemifusion intermediate into a functional fusion pore. Although POPS supports the efficient formation of fusion pores, this process is slower than with BMP bilayers.
FIGURE 5.
Kinetics of distinct steps of single VSV pseudovirus fusion. A and B, distributions of the delay times from acidification to the onset of lipid mixing (hemifusion, red circles) to the onset of content release (green circles) as well as the lag times between hemifusion and fusion for a given particle (blue circles) for bilayers containing POPS (A) and BMP (B). Only the lag time between lipid and content transfer was significantly different between POPS and BMP (p < 0.045). C, distributions of the duration of mKO release (i.e. time from the onset of mKO signal decrease to reaching the background fluorescence level) that reflect the relative diameter of fusion pores. The total number of full fusion events was 17 for the POPS mixture (red circles) and 14 for the BMP mixture (green circles). No fusion events were observed for the POPC-based mixture lacking the anionic lipids.
We next asked whether the effective sizes of VSV G-mediated fusion pores are lipid-dependent. By analyzing the mKO release profiles for individual events on the basis of the fluorescence signal decay (Fig. 3), we found that the time required for complete release of NC-mKO varied. In many cases the mKO signal dropped virtually instantaneously (e.g. Figs. 2A and 3, A, C, and F), but there were also particles exhibiting a slow decrease of the signal (e.g. Figs. 2B and 3, D and E). Thus, VSV G formed fusion pores of different initial sizes. To assess whether lipids can alter the time between the onset and the end of the mKO signal decrease, we compared the VSVpp fusion with POPS and BMP bilayers and found that the distributions of this parameter were identical (Fig. 5C). These findings argue against a specific effect of anionic lipids on the apparent size of a fusion pore.
Another prominent feature of the VSV G-mediated fusion was the stepwise release of the mKO marker (Fig. 3, A and H). The two-step release of the viral content likely reflected the pore flickering. The transient halting of content release is likely due to pore closure or shrinkage, whereas resumption of release reflects subsequent pore dilation (56, 57). Thus, fusion pores formed by VSV G in supported bilayers exhibited dynamic features similar to those formed by other viral proteins (38, 56, 58, 59).
DISCUSSION
In this study, we successfully reconstituted VSV G-mediated membrane fusion in a minimal model system that enabled time-resolved imaging of single lipid mixing and content release events. The ability to mediated fusion with lipid bilayers at low pH implies that VSV G does not strictly require a proteinaceous receptor to induce fusion. On the other hand, VSVpp fusion was markedly augmented by anionic lipids, POPS, or BMP. Only bilayers containing POPS or BMP supported complete VSVpp fusion, whereas a POPC-based mixture supported only limited lipid mixing activity. Our results are in full agreement with the previous report that BMP somewhat selectively facilitates VSV G-mediated lipid mixing with liposomes (31). However, in that study, the beneficial effect of BMP was observed in the background of 50% POPS, thus complicating the evaluation of relative roles of these lipids in VSVpp fusion. We found that, although BMP supported more efficient lipid mixing and faster conversion from hemifusion to full fusion than POPS, these differences were modest. In addition, the average time required to complete the release of viral content, which is inversely proportional to the size of a fusion pore (60), was identical for BMP and POPS bilayers. These findings argue against POPS as the specific receptor for VSV and highlight the potential direct role of BMP in entry of this virus.
The BMP content in late endosomes was reported to be as high as 15% of total phospholipids, with the POPC:BMP molar ratio ∼3:1 (32) being close to the lipid composition of the SLB used in our study. By contrast, late endosomal membranes contain only 5% of PS (32), which appears to be even lower that the PS content of the plasma membrane (61, 62). Furthermore, because VSV infection is not inhibited by the PS-binding protein annexin V (22), it is unlikely that the incoming VSV encounters PS at the cell surface or during intracellular trafficking. On the other hand, recent evidence suggests a role for BMP in VSV entry from late endosomes (28). The observation that lipid mixing between VSV and endosomes is not attenuated by BMP depletion with specific antibodies (28) suggests that, similarly to dengue virus (30), VSV may undergo hemifusion with early endosomes. It is conceivable that complete fusion can be delayed until VSV enters into late compartments enriched in BMP (28). However, given the evidence that VSV enters from early endosomes (36, 63), further studies are required to test this model.
The relatively low probability of full fusion between VSVpp and SLBs containing BMP or POPS may be due to incorporation of fewer G proteins compared with bona fide VSV (64, 65). In addition, fusion with liposomes appears to occur at the base of a bullet-shaped VSV particle (65), suggesting that the architecture of the virus and perhaps the interactions between G and M proteins regulate the fusion activity. Another explanation for the poor fusion efficiency is that low pH and anionic lipids may not be the only factors required for VSV fusion. Future studies of single VSV fusion with lipid bilayers should distinguish between these possibilities.
Regardless of the lipid composition of SLBs tested in this study, hemifusion always preceded or full fusion. For some particles, the lag time from lipid mixing to the onset of content release reached several minutes. These results agree with previous data on influenza (45), HIV-1 (55) and avian sarcoma and leucosis virus (66) fusion and show that hemifusion is a true and long-lived intermediate stage of VSV G-mediated fusion. Note, however, that for 10% of single influenza virus fusion events with SLB, viral content transfer preceded lipid mixing (45). No explanation for this rather unusual phenotype has been offered by the authors. In addition, our experiments detected transient closing/shrinkage of fusion pores formed by VSV G, a phenomenon known as pore flickering (38, 56, 58, 59).
In conclusion, the SLB model offers a number of important advantages for mechanistic studies of viral fusion at the single particle level. Also, pseudotyping the MLV core with VSV G or other viral glycoproteins provides a universal platform for future mechanistic studies of single virus fusion. The virus labeling and imaging strategies introduced in this study enable detailed studies of the role of lipids at distinct intermediate steps of VSVpp fusion: hemifusion and formation of a small fusion pore. Future studies should help delineate the effects of other anionic and neutral lipids, as well as lipid domains, in regulation of membrane fusion mediated by VSV G and other viral glycoproteins. It would also be interesting to assess the pH dependence of VSVpp-SLB hemifusion and fusion. Of particular relevance are the efficiency and kinetics of viral fusion at a higher pH, around 6.0, typical of early endosomes (67, 68). Because the pH threshold of VSV fusion is only somewhat higher than 6.0 (65, 69, 70), fusion experiments carried out at this suboptimal pH can accentuate the stimulating effect of various lipids and cofactors compared with the optimal pH of 5.5 employed in this study.
Acknowledgments
We thank Dr. Erdem Karatekin (Yale University) for expert advice on techniques for supported bilayer formation and Matthew Smith (Lehigh University) for helpful hints in using the Speckle TrackerJ. We also thank José Rino (Instituto de Medicina Molecular) and Tanay Desai (Emory University) for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI053668 (to G. B. M.). This work was also supported by Fundação para a Ciência e Tecnologia, Ministério da Educação e Ciência (FCT-MEC, Portugal) Project Grant PTDC/QUI-BIQ/104787/2008 (to N. C. S.) and by Fellowship SFRH/BD/42205/2007 (to P. M. M.).

This article contains supplemental Fig. 1 and Movies 1 and 2.
- VSV
- vesicular stomatitis virus
- PS
- phosphatidylserine
- BMP
- bis(monooleoylglycero)phosphate
- SLB
- supported lipid bilayer(s)
- VSVpp
- vesicular stomatitis virus G protein-pseudotyped particle(s)
- DiD
- 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- CF-PE
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(carboxyfluorescein)
- Chol
- cholesterol
- PDMS
- polydimethylsiloxane
- mKO
- CoralHue® monomeric Kusabira Orange fluorescent protein
- FRAP
- fluorescence recovery after photobleaching
- ROI
- region of interest
- NC
- nucleocapsid
- MLV
- murine leukemia virus.
REFERENCES
- 1. Cohen F. S., Melikyan G. B. (2004) The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J. Membr. Biol. 199, 1–14 [DOI] [PubMed] [Google Scholar]
- 2. Harrison S. C. (2008) Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marsh M., Helenius A. (2006) Virus entry. Open sesame. Cell 124, 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miyauchi K., Kim Y., Latinovic O., Morozov V., Melikyan G. B. (2009) HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137, 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de la Vega M., Marin M., Kondo N., Miyauchi K., Kim Y., Epand R. F., Epand R. M., Melikyan G. B. (2011) Inhibition of HIV-1 endocytosis allows lipid mixing at the plasma membrane, but not complete fusion. Retrovirology 8, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akhtar J., Shukla D. (2009) Viral entry mechanisms. Cellular and viral mediators of herpes simplex virus entry. FEBS J. 276, 7228–7236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kolokoltsov A. A., Deniger D., Fleming E. H., Roberts N. J., Jr., Karpilow J. M., Davey R. A. (2007) Small interfering RNA profiling reveals key role of clathrin-mediated endocytosis and early endosome formation for infection by respiratory syncytial virus. J. Virol. 81, 7786–7800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carr C. M., Kim P. S. (1993) A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73, 823–832 [DOI] [PubMed] [Google Scholar]
- 9. Earp L. J., Delos S. E., Park H. E., White J. M. (2005) The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285, 25–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kielian M., Rey F. A. (2006) Virus membrane-fusion proteins. More than one way to make a hairpin. Nat. Rev. Microbiol. 4, 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Melikyan G. B. (2008) Common principles and intermediates of viral protein-mediated fusion. The HIV-1 paradigm. Retrovirology 5, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. White J. M., Delos S. E., Brecher M., Schornberg K. (2008) Structures and mechanisms of viral membrane fusion proteins. Multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43, 189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Albertini A. A., Baquero E., Ferlin A., Gaudin Y. (2012) Molecular and cellular aspects of rhabdovirus entry. Viruses 4, 117–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Backovic M., Jardetzky T. S. (2009) Class III viral membrane fusion proteins. Curr. Opin. Struct. Biol. 19, 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roche S., Albertini A. A., Lepault J., Bressanelli S., Gaudin Y. (2008) Structures of vesicular stomatitis virus glycoprotein. Membrane fusion revisited. Cell. Mol. Life Sci. 65, 1716–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blumenthal R., Bali-Puri A., Walter A., Covell D., Eidelman O. (1987) pH-dependent fusion of vesicular stomatitis virus with Vero cells. Measurement by dequenching of octadecyl rhodamine fluorescence. J. Biol. Chem. 262, 13614–13619 [PubMed] [Google Scholar]
- 17. Puri A., Winick J., Lowy R. J., Covell D., Eidelman O., Walter A., Blumenthal R. (1988) Activation of vesicular stomatitis virus fusion with cells by pretreatment at low pH. J. Biol. Chem. 263, 4749–4753 [PubMed] [Google Scholar]
- 18. Gaudin Y., Tuffereau C., Segretain D., Knossow M., Flamand A. (1991) Reversible conformational changes and fusion activity of rabies virus glycoprotein. J. Virol. 65, 4853–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaudin Y. (2000) Reversibility in fusion protein conformational changes. The intriguing case of rhabdovirus-induced membrane fusion. Subcell. Biochem. 34, 379–408 [DOI] [PubMed] [Google Scholar]
- 20. Carneiro F. A., Bianconi M. L., Weissmüller G., Stauffer F., Da Poian A. T. (2002) Membrane recognition by vesicular stomatitis virus involves enthalpy-driven protein-lipid interactions. J. Virol. 76, 3756–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eidelman O., Schlegel R., Tralka T. S., Blumenthal R. (1984) pH-dependent fusion induced by vesicular stomatitis virus glycoprotein reconstituted into phospholipid vesicles. J. Biol. Chem. 259, 4622–4628 [PubMed] [Google Scholar]
- 22. Schlegel R., Tralka T. S., Willingham M. C., Pastan I. (1983) Inhibition of VSV binding and infectivity by phosphatidylserine. Is phosphatidylserine a VSV-binding site? Cell 32, 639–646 [DOI] [PubMed] [Google Scholar]
- 23. Schlegel R., Wade M. (1983) Neutralized vesicular stomatitis virus binds to host cells by a different “receptor.” Biochem. Biophys. Res. Commun. 114, 774–778 [DOI] [PubMed] [Google Scholar]
- 24. Coil D. A., Miller A. D. (2004) Phosphatidylserine is not the cell surface receptor for vesicular stomatitis virus. J. Virol. 78, 10920–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Puri A., Grimaldi S., Blumenthal R. (1992) Role of viral envelope sialic acid in membrane fusion mediated by the vesicular stomatitis virus envelope glycoprotein. Biochemistry 31, 10108–10113 [DOI] [PubMed] [Google Scholar]
- 26. Waheed A. A., Freed E. O. (2010) The role of lipids in retrovirus replication. Viruses 2, 1146–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kobayashi T., Stang E., Fang K. S., de Moerloose P., Parton R. G., Gruenberg J. (1998) A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature 392, 193–197 [DOI] [PubMed] [Google Scholar]
- 28. Le Blanc I., Luyet P. P., Pons V., Ferguson C., Emans N., Petiot A., Mayran N., Demaurex N., Fauré J., Sadoul R., Parton R. G., Gruenberg J. (2005) Endosome-to-cytosol transport of viral nucleocapsids. Nat. Cell Biol. 7, 653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luyet P. P., Falguières T., Pons V., Pattnaik A. K., Gruenberg J. (2008) The ESCRT-I subunit TSG101 controls endosome-to-cytosol release of viral RNA. Traffic 9, 2279–2290 [DOI] [PubMed] [Google Scholar]
- 30. Zaitseva E., Yang S. T., Melikov K., Pourmal S., Chernomordik L. V. (2010) Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog. 6, e1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roth S. L., Whittaker G. R. (2011) Promotion of vesicular stomatitis virus fusion by the endosome-specific phospholipid bis(monoacylglycero)phosphate (BMP). FEBS Lett. 585, 865–869 [DOI] [PubMed] [Google Scholar]
- 32. Kobayashi T., Beuchat M. H., Chevallier J., Makino A., Mayran N., Escola J. M., Lebrand C., Cosson P., Kobayashi T., Gruenberg J. (2002) Separation and characterization of late endosomal membrane domains. J. Biol. Chem. 277, 32157–32164 [DOI] [PubMed] [Google Scholar]
- 33. Abrami L., Lindsay M., Parton R. G., Leppla S. H., van der Goot F. G. (2004) Membrane insertion of anthrax protective antigen and cytoplasmic delivery of lethal factor occur at different stages of the endocytic pathway. J. Cell Biol. 166, 645–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van der Goot F. G., Gruenberg J. (2006) Intra-endosomal membrane traffic. Trends Cell Biol. 16, 514–521 [DOI] [PubMed] [Google Scholar]
- 35. Sieczkarski S. B., Whittaker G. R. (2003) Differential requirements of Rab5 and Rab7 for endocytosis of influenza and other enveloped viruses. Traffic 4, 333–343 [DOI] [PubMed] [Google Scholar]
- 36. Johannsdottir H. K., Mancini R., Kartenbeck J., Amato L., Helenius A. (2009) Host cell factors and functions involved in vesicular stomatitis virus entry. J. Virol. 83, 440–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brandenburg B., Zhuang X. (2007) Virus trafficking. Learning from single-virus tracking. Nat. Rev. Microbiol. 5, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Melikyan G. B., Barnard R. J., Abrahamyan L. G., Mothes W., Young J. A. (2005) Imaging individual retroviral fusion events. From hemifusion to pore formation and growth. Proc. Natl. Acad. Sci. U.S.A. 102, 8728–8733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koch P., Lampe M., Godinez W. J., Müller B., Rohr K., Kräusslich H. G., Lehmann M. J. (2009) Visualizing fusion of pseudotyped HIV-1 particles in real time by live cell microscopy. Retrovirology 6, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Padilla-Parra S., Matos P. M., Kondo N., Marin M., Santos N. C., Melikyan G. B. (2012) Quantitative imaging of endosome acidification and single retrovirus fusion with distinct pools of early endosomes. Proc. Natl. Acad. Sci. U.S.A. 109, 17627–17632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karatekin E., Di Giovanni J., Iborra C., Coleman J., O'Shaughnessy B., Seagar M., Rothman J. E. (2010) A fast, single-vesicle fusion assay mimics physiological SNARE requirements. Proc. Natl. Acad. Sci. U.S.A. 107, 3517–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fix M., Melia T. J., Jaiswal J. K., Rappoport J. Z., You D., Söllner T. H., Rothman J. E., Simon S. M. (2004) Imaging single membrane fusion events mediated by SNARE proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 7311–7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bowen M. E., Weninger K., Brunger A. T., Chu S. (2004) Single molecule observation of liposome-bilayer fusion thermally induced by soluble N-ethyl maleimide sensitive-factor attachment protein receptors (SNAREs). Biophys. J. 87, 3569–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Costello D. A., Lee D. W., Drewes J., Vasquez K. A., Kisler K., Wiesner U., Pollack L., Whittaker G. R., Daniel S. (2012) Influenza virus-membrane fusion triggered by proton uncaging for single particle studies of fusion kinetics. Anal. Chem. 84, 8480–8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Floyd D. L., Ragains J. R., Skehel J. J., Harrison S. C., van Oijen A. M. (2008) Single-particle kinetics of influenza virus membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 105, 15382–15387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wessels L., Elting M. W., Scimeca D., Weninger K. (2007) Rapid membrane fusion of individual virus particles with supported lipid bilayers. Biophys. J. 93, 526–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kimpton J., Emerman M. (1992) Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66, 2232–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sherer N. M., Lehmann M. J., Jimenez-Soto L. F., Ingmundson A., Horner S. M., Cicchetti G., Allen P. G., Pypaert M., Cunningham J. M., Mothes W. (2003) Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4, 785–801 [DOI] [PubMed] [Google Scholar]
- 49. Karatekin E., Rothman J. E. (2012) Fusion of single proteoliposomes with planar, cushioned bilayers in microfluidic flow cells. Nat. Protoc. 7, 903–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brian A. A., McConnell H. M. (1984) Allogeneic stimulation of cytotoxic T cells by supported planar membranes. Proc. Natl. Acad. Sci. U.S.A. 81, 6159–6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kenworthy A. K. (2007) Fluorescence recovery after photobleaching studies of lipid rafts. Methods Mol. Biol. 398, 179–192 [DOI] [PubMed] [Google Scholar]
- 52. Soumpasis D. M. (1983) Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys. J. 41, 95–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smith M. B., Karatekin E., Gohlke A., Mizuno H., Watanabe N., Vavylonis D. (2011) Interactive, computer-assisted tracking of speckle trajectories in fluorescence microscopy. Application to actin polymerization and membrane fusion. Biophys. J. 101, 1794–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Karasawa S., Araki T., Nagai T., Mizuno H., Miyawaki A. (2004) Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem. J. 381, 307–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Markosyan R. M., Cohen F. S., Melikyan G. B. (2005) Time-resolved imaging of HIV-1 Env-mediated lipid and content mixing between a single virion and cell membrane. Mol. Biol. Cell 16, 5502–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Melikyan G. B., Niles W. D., Peeples M. E., Cohen F. S. (1993) Influenza hemagglutinin-mediated fusion pores connecting cells to planar membranes. Flickering to final expansion. J. Gen. Physiol. 102, 1131–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Spruce A. E., Iwata A., Almers W. (1991) The first milliseconds of the pore formed by a fusogenic viral envelope protein during membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 88, 3623–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Spruce A. E., Iwata A., White J. M., Almers W. (1989) Patch clamp studies of single cell-fusion events mediated by a viral fusion protein. Nature 342, 555–558 [DOI] [PubMed] [Google Scholar]
- 59. Markosyan R. M., Cohen F. S., Melikyan G. B. (2000) The lipid-anchored ectodomain of influenza virus hemagglutinin (GPI-HA) is capable of inducing nonenlarging fusion pores. Mol. Biol. Cell 11, 1143–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Padilla-Parra S., Marin M., Kondo N., Melikyan G. B. (2012) Synchronized retrovirus fusion in cells expressing alternative receptor isoforms releases the viral core into distinct sub-cellular compartments. PLoS Pathog. 8, e1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Leventis P. A., Grinstein S. (2010) The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39, 407–427 [DOI] [PubMed] [Google Scholar]
- 62. Leidl K., Liebisch G., Richter D., Schmitz G. (2008) Mass spectrometric analysis of lipid species of human circulating blood cells. Biochim. Biophys. Acta 1781, 655–664 [DOI] [PubMed] [Google Scholar]
- 63. Mire C. E., White J. M., Whitt M. A. (2010) A spatio-temporal analysis of matrix protein and nucleocapsid trafficking during vesicular stomatitis virus uncoating. PLoS Pathog. 6, e1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ge P., Tsao J., Schein S., Green T. J., Luo M., Zhou Z. H. (2010) Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science 327, 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Libersou S., Albertini A. A., Ouldali M., Maury V., Maheu C., Raux H., de Haas F., Roche S., Gaudin Y., Lepault J. (2010) Distinct structural rearrangements of the VSV glycoprotein drive membrane fusion. J. Cell Biol. 191, 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jha N. K., Latinovic O., Martin E., Novitskiy G., Marin M., Miyauchi K., Naughton J., Young J. A., Melikyan G. B. (2011) Imaging single retrovirus entry through alternative receptor isoforms and intermediates of virus-endosome fusion. PLoS Pathog. 7, e1001260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sieczkarski S. B., Whittaker G. R. (2002) Dissecting virus entry via endocytosis. J. Gen. Virol. 83, 1535–1545 [DOI] [PubMed] [Google Scholar]
- 68. Mercer J., Schelhaas M., Helenius A. (2010) Virus entry by endocytosis. Annu. Rev. Biochem. 79, 803–833 [DOI] [PubMed] [Google Scholar]
- 69. Fredericksen B. L., Whitt M. A. (1996) Mutations at two conserved acidic amino acids in the glycoprotein of vesicular stomatitis virus affect pH-dependent conformational changes and reduce the pH threshold for membrane fusion. Virology 217, 49–57 [DOI] [PubMed] [Google Scholar]
- 70. Shokralla S., He Y., Wanas E., Ghosh H. P. (1998) Mutations in a carboxy-terminal region of vesicular stomatitis virus glycoprotein G that affect membrane fusion activity. Virology 242, 39–50 [DOI] [PubMed] [Google Scholar]