Background: Information about the structure-function relationship and activation mechanism of 7TM receptors is needed.
Results: Single mutations in CCR5 induce biased signaling with increased activation through Gαi but decreased β-arrestin recruitment.
Conclusion: The TM6/7 interface controls the G protein-dependent and -independent activity state of CCR5.
Significance: Knowledge about specific 7TM receptor regions targeted by pathway-selective (biased) ligands is vital for future drug design.
Keywords: 7-Helix Receptor, Chemokines, G Protein-coupled Receptors (GPCR), G Proteins, Pharmacology, Receptor Structure-Function, Signal Transduction, Biased Signaling
Abstract
The equilibrium state of CCR5 is manipulated here toward either activation or inactivation by introduction of single amino acid substitutions in the transmembrane domains (TMs) 6 and 7. Insertion of a steric hindrance mutation in the center of TM7 (G286F in position VII:09/7.42) resulted in biased signaling. Thus, β-arrestin recruitment was eliminated, whereas constitutive activity was observed in Gαi-mediated signaling. Furthermore, the CCR5 antagonist aplaviroc was converted to a full agonist (a so-called efficacy switch). Computational modeling revealed that the position of the 7TM receptor-conserved Trp in TM6 (Trp-248 in position VI:13/6.48, part of the CWXP motif) was influenced by the G286F mutation, causing Trp-248 to change orientation away from TM7. The essential role of Trp-248 in CCR5 activation was supported by complete inactivity of W248A-CCR5 despite maintaining chemokine binding. Furthermore, replacing Trp-248 with a smaller aromatic amino acid (Tyr/Phe) impaired the β-arrestin recruitment, yet with maintained G protein activity (biased signaling); also, here aplaviroc switched to a full agonist. Thus, the altered positioning of Trp-248, induced by G286F, led to a constraint of G protein active, but β-arrestin inactive and thus biased, CCR5 conformation. These results provide important information on the molecular interplay and impact of TM6 and TM7 for CCR5 activity, which may be extrapolated to other chemokine receptors and possibly to other 7TM receptors.
Introduction
Chemokine receptors are critical mediators of leukocyte trafficking and thereby regulation and development of the immune system. They belong to class A of the seven transmembrane spanning (7TM)2 receptors, which are the largest family of membrane proteins in the human genome and are activated by highly diverse ligands (1). In accordance with the large variety of endogenous agonists, 7TM receptors are involved in regulating most aspects of normal physiology and pathophysiology. Consequently, they have enormous potential as drug targets, and studying their activation process is of great importance. Despite the chemical diversity among endogenous ligands, it is generally believed that all 7TM receptors share the overall same activation mechanism (2). Recently, crystal structures of fully activated β2-adrenergic receptors (receptors in complex with both agonist and G protein) were published (3, 4). Compared with the crystal structures of inactive receptors (5–11), the most pronounced structural changes are at the cytosolic face. Here, transmembrane helix 5 (TM5) and TM6 move outward away from the center of the receptor, whereas TM3 and TM7 move slightly inward (3, 9, 10). Three-dimensional structures of inactive chemokine receptors have also been published; that is, crystal structures of CXCR4 in complex with a peptide and a small-molecule compound as the first in 2010 (11) followed by a structure of CXCR1 determined by nuclear magnetic resonance (NMR) spectroscopy in 2012 (12).
Even though we have insights into the overall movements during receptor activation, the details of the structural events have not been clearly established. Importantly, the difference in receptor structure depending on which effector molecule is bound (e.g. G protein versus β-arrestin) has gained a lot of attention recently. Ligands that bind to a receptor and can elicit different responses in different pathways (termed “biased signaling” or “functional selectivity”) have been recognized for their therapeutic potential (13). Thus, selective targeting of signaling events that contribute to disease while preserving other functions would reduce the negative effects of some therapeutics. Ever since the chemokine receptor CCR5 was discovered for its pivotal role in HIV entry into host cells (14, 15), several CCR5-targeting compounds have been developed as anti-HIV treatment. Among these, biased ligands have been described; one of the first was an N-terminal-modified form of the endogenous ligand RANTES (or CCL5), AOP-RANTES (16). AOP-RANTES was shown to cause CCR5 internalization in peripheral blood mononuclear cells (17) but was incapable of inducing chemotaxis of monocytes (16). Another N-terminal-modified form of CCL5 (5P14-RANTES) induced significant amounts of CCR5 internalization in activated CD4+ T cells but no G protein-mediated signaling, assessed in HeLa cells (18). Both ligands effectively inhibited HIV entry, very likely because of their ability to sequester CCR5 inside the cell. Meanwhile, the lack of G protein-mediated signaling could decrease the risk of unwanted effects (for review, see Ref. 19).
Biased signaling has also been described in several other 7TM receptors, e.g. CXCR7 (20), the herpesvirus-encoded CXC-chemokine receptor ECRF3 (21), the β2-adrenergic receptor (22), and the nicotinic receptor GPR109A (23).
Here we describe how manipulation of the interface between TM6 and TM7 in CCR5 can be used to determine receptor areas important for G protein signaling relative to β-arrestin recruitment. Whereas most of the single amino acid mutations shift the equilibrium toward a more active state of CCR5 in G protein-coupled signaling (i.e. increase in basal activity and “efficacy switch” of an antagonist to agonist), β-arrestin binding is impaired. Computational modeling of CCR5 showed that a likely mechanism could be an interaction between two residues; one in TM7 (Gly-286, VII:09/7.42,3 which was mutated to a Phe) and one in TM6 (Trp-248, VI:13/6.48). Together these results show that the interface of TM6 and TM7 is very important for the activation state of CCR5 and that compounds targeting this area very likely could have biased properties. The results also show that small local alterations can lead to large overall changes, i.e. biased and constitutive signaling, without alteration in ligand binding.
EXPERIMENTAL PROCEDURES
Materials
The human chemokines CCL3 and CCL5 were purchased from Peprotech. The human CCR5 WT cDNA was cloned from a spleen-derived cDNA library. The small molecule CCR5 antagonists Merck, SCH-C, TAK-779, and aplaviroc were kindly provided by Gary Bridger (AnorMED, Langley, British Columbia, Canada). Iodinated CCL3 was purchased from PerkinElmer Life Sciences. The promiscuous chimeric G protein GαΔ6qi4myr (Gqi4myr), which converts Gαi-related signaling into a Gαq readout (24, 25), was kindly provided by Evi Kostenis (University of Bonn, Bonn, Germany).
Molecular Biology
FLAG-tagged receptor cDNA was cloned into expression vectors pcDNA3.1(+) (Invitrogen) (phosphatidylinositol (PI)-turnover, cAMP, and competition binding) and pCMV-ProLinkTM1 (DiscoveRx, Birmingham, UK) (β-arrestin recruitment). Mutations were constructed by PCR using the QuikChangeTM site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. All mutations were verified by restriction endonuclease mapping and subsequent DNA sequence analysis.
Transfections and Tissue Culture
COS-7 cells were grown at 10% CO2 and 37 °C in Dulbecco's modified Eagle's medium 1885 supplemented with 10% fetal bovine serum, 2 mm glutamine, 180 units/ml penicillin, and 45 μg/ml streptomycin. PathHunter U2OS β-Arrestin 2 Parental cell line (DiscoveRx) were grown at 5% CO2 and 37 °C in Ham's F-12 medium with glutamine supplemented with 10% fetal bovine serum, 180 units/ml penicillin, 45 μg/ml streptomycin, and 0.25 mg/ml hygromycin B (Invitrogen). Transfection of COS-7 cells for PI-turnover and competition binding was performed using the calcium phosphate precipitation method with chloroquine addition as previously described (26, 27) or by using LipofectamineTM 2000 (Invitrogen) as described by the manufacturer for cAMP determination and ELISA. U2OS cells were transfected using FuGENE® 6 Transfection reagent (Roche Applied Science).
PI-turnover Assay
COS-7 cells were co-transfected with receptor cDNA and Gqi4myr, which converts the Gαi signal into a Gαq signal, making it possible to measure the chemokine receptor activation as PI-turnover (24, 25). One day after transfection, the cells were seeded in 24-well plates (1.5 × 105 cells/well) and incubated with 2 μCi of myo-[3H]inositol in 0.3 ml of growth medium for 24 h. Cells were washed twice with Hanks' buffered salt solution supplemented with CaCl2 and MgCl2 and afterward incubated for 15 min in 0.3 ml of buffer supplemented with 10 mm LiCl before ligand addition followed by 90 min of incubation. When used, the antagonists were added 10 min before the agonist. The generated [3H]inositol phosphates were purified on AG 1X8 anion exchange resin. Determinations were made in duplicate.
cAMP Assay
COS-7 cells (3.5 × 104 cells/well) were seeded in 96-well plates 1 day before transfection with receptor DNA. Two days after transfection the cells were washed twice with Hepes buffered saline (HBS) buffer and incubated with HBS and 1 mm 3-isobutyl-1-methylxanthine for 30 min at 37 °C. Forskolin (Sigma) was added in various concentrations, and the cells were incubated for 30 min at 37 °C. The HitHunterTM cAMP XS+ assay (DiscoveRx) was carried out according to the manufacturer's instructions. Determinations were made in triplicate.
β-Arrestin Recruitment Assay
PathHunter U2OS β-arrestin cells were seeded in 96-well plates (2.0 × 104 cells per well). Overnight transient transfection with PK1-tagged receptor DNA was started the following day and stopped a day before the assay. PathHunterTM β-arrestin GPCR assay (DiscoveRx) were carried out according to the manufacturer's instructions.
125I-CCL3 Competition Binding Assay
COS-7 cells were seeded in wells 1 day after transfection with receptor DNA with the number of cells seeded per well aimed at obtaining 5–10% specific binding of the added radioactive ligand (3–15 × 105 cells/well for the different CCR5 constructs). Two days after transfection, cells were assayed by competition binding for 3 h at 4 °C using 20–70 pm 125I-CCL3 as well as unlabeled ligand in 50 mm Hepes buffer, pH 7.4, supplemented with 1 mm CaCl2, 5 mm MgCl2, and 0.5% (w/v) bovine serum albumin (BSA). After incubation, cells were washed twice in ice-cold binding buffer supplemented with 0.5 m NaCl. Nonspecific binding was determined as the binding in the presence of 0.1 μm unlabeled CCL3. Determinations were made in duplicate.
Cell Surface Expression Measurement (ELISA)
COS-7 cells were transfected with FLAG-tagged (M1) receptor DNA in 96-well plates (3.5 × 104 cells/well). Two days after transfection, cells were washed 3 times in Tris-buffered saline (TBS), fixed in 3.7% formaldehyde for 15 min at room temperature, washed, and incubated in blocking solution (TBS supplemented with 2% BSA) for 30 min. Cells were kept at room temperature for subsequent steps. Cells were incubated for 2 h with anti-FLAG (M1) antibody (Sigma) at 2 μg/ml in TBS with 1 mm CaCl2 and 1% BSA. After 3 washes with TBS/CaCl2/BSA, the cells were incubated with goat anti-mouse horseradish peroxidase-conjugated antibody (Abcam, Cambridge, UK) at 1:1000 dilution. After extensive washing, the immunoreactivity was revealed by the addition of TMB Plus substrate (Kem-En-Tec, Taastrup, Denmark), and the reaction was stopped with 0.2 m H2SO4. Absorbance was measured at 450 nm on a Wallac VICTOR2 platereader (PerkinElmer Life Sciences).
Calculations
IC50, EC50, and Kd/Ki values were determined by nonlinear regression, and Bmax values were calculated using the GraphPad Prism 4.0 software (GraphPad, San Diego, CA).
CCR5 Comparative Models and Docking of Aplaviroc
A pair-wise sequence alignment and the construction of a comparative homology model of the human CCR5 receptor was produced in the Internal Coordinate Mechanics software package (Molsoft LLC, La Jolla, CA) using the human CXCR4 (11) (PDB entry 3ODU) receptor as the structural template. In brief, manual adjustment of the alignment was necessary to ensure proper alignment of the loop regions and eliminate gaps in the TM regions. During the model construction, a disulfide bridge between Cys-101 (III:01/3.25) and Cys-197 (in ECL2) together with a disulfide bridge between Cys-20 (in the N terminus) and Cys-269 (in ECL3) was applied as a structural constraint. The developed CCR5 models were subjected to full-atom structure relaxation using the ROSETTA membrane force field (28) in Rosetta 3.2.1 (29). A total of 100 models were generated, and a set of 10 representative low energy CCR5 receptor models was selected as a receptor ensemble for the subsequent docking of aplaviroc. Full flexible ligand docking was performed using the biased probability Monte Carlo docking routine in ICM under softened van der Waals conditions using 4D grids represented by six grid potentials of 0.5 Å spacing, including three van der Waals grid potentials for a carbon probe, large atom probe, or hydrogen probe, a hydrogen bonding grid potential, an electrostatic grid potential, and a hydrophobic grid potential ICM (30, 31). The docking grids were defined to encompass a binding pocket described by all corresponding receptor residues within 4.5 Å of the IT1t ligand (11) in the CXCR4 template structure when superimposed onto the stack of generated CCR5 models. The final docking grid was extended ∼10 Å toward TM5 to allow aplaviroc to interact with pocket formed between TM3, -4, -5, and -6. Individual best scored docking poses were subsequently optimized using a combined Monte Carlo and minimization procedure (using the MMFF94 force field), keeping ligand and surrounding protein residues (in an 8 Å radius from the starting position) flexible. All backbone coordinates were held fixed. Two rounds of optimization were performed: an initial refinement under a softened van der Waals potential and a second refinement with the full van der Waals potential. A final stack of 50 conformations was generated that was scored and manually analyzed to identify the complexes between aplaviroc and CCR5.
Conformational Sampling and Statistics of Side Chain Rotamer States
In brief, the G286F-CCR5 receptor variant was constructed from the initial CCR5 WT model using the residue substitution function in the Rosetta 3.2.1 (29). CCR5 WT as well as G286F-CCR5 was subjected to full-atom structure relaxation using the ROSETTA membrane force field (28) in Rosetta 3.2.1 (29) to optimize the structures and repack the side chain packing. A total of 1000 models were generated of both CCR5 WT and G286F-CCR5. Statistics on side chain rotamer states for both receptors were analyzed using customized scripts and function in the CCP4 software package (32).
RESULTS
Regulation of Basal Receptor Activity by a Space-filling Substitution in the Center of TM7
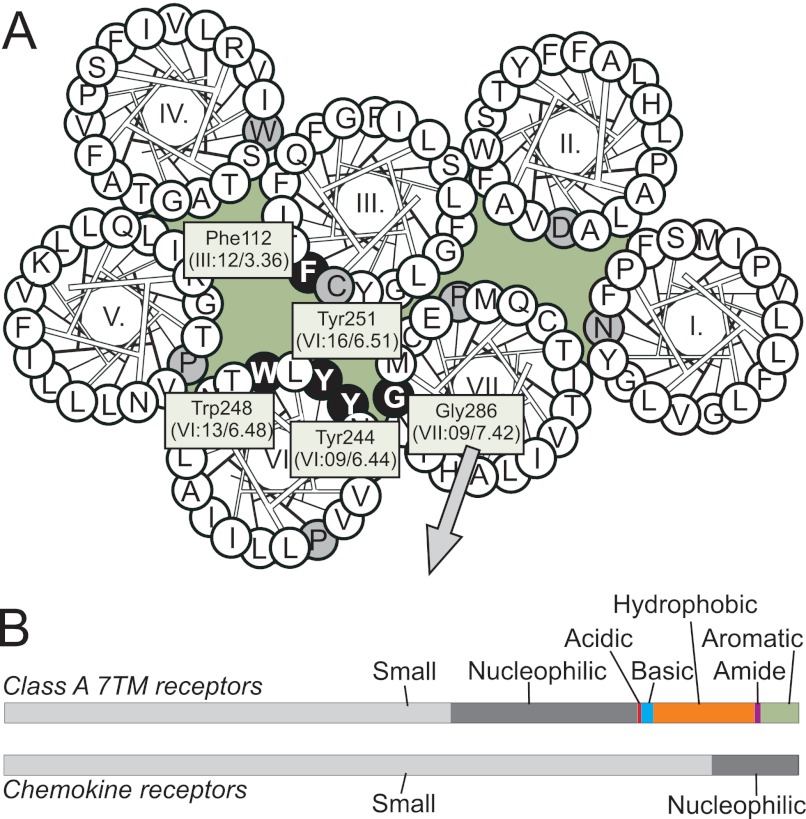
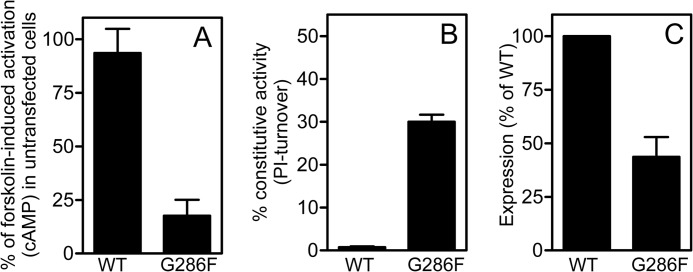
Approximately half of the amino acids at position VII:09/7.42 in class A 7TM receptors are small, whereas this number is much higher among chemokine receptors (89%) (Fig. 1). In CCR5, a Gly (Gly-286) occupies position VII:09. To explore the role of this small amino acid, Gly-286 was mutated to a Phe, G286F (i.e. introduction of a steric hindrance). The mutant was tested in competition binding with 125I-CCL3 and in G protein-mediated signaling assays; that is, inhibition of forskolin-induced cAMP accumulation and PI-turnover (phosphatidylinositol) measurements, where co-transfection with the chimeric G protein Gqi4myr ensures transmission of a Gαi-coupled receptor activity into a Gαq readout (24, 25). The level of cAMP accumulation in G286F-CCR5 induced by forskolin was decreased to 20% that of the WT level, indicating constitutive activity via Gαi. In contrast, no constitutive activity was observed in CCR5 WT (Fig. 2A). Moreover, G286F displayed a 30-fold increase in basal activity in PI-turnover (Fig. 2B and Table 1). As in cAMP accumulation, virtually no agonist-independent (i.e. constitutive) PI-turnover was observed for CCR5 WT. Importantly, the enhanced basal signaling was not a consequence of enhanced receptor surface expression, as the expression of G286F, determined by ELISA against an N-terminal M1 tag, was ∼50% decreased compared with WT (Fig. 2C). The potency of CCL3 was basically unaltered, and the potency of CCL5 was decreased 7-fold compared with WT (Table 1). Despite unaltered CCL3 potency, the affinity determined by homologous competition binding experiments was increased 3.5-fold compared with WT (Table 2). Thus, the presence of a large aromatic side chain in VII:09/7.42 in CCR5 induces constitutive signaling via Gαi.
FIGURE 1.
Distribution of amino acids in position VII:09/7.42. A, shown is a helical wheel diagram of CCR5 as viewed from the extracellular side with the residues examined indicated with white letters in black circles and the most conserved residue in each helix indicated in gray. B, shown is distribution of amino acids in position VII:09/7.42 (corresponding to Gly-286 in CCR5) in non-chemokine class A 7TM receptors (upper panel) and chemokine receptors (lower panel). Small amino acids (aa) are Ala and Gly; nucleophilic aa are Cys, Ser, and Thr; acidic aa are Asp and Glu; basic aa are His, Lys, and Arg; hydrophobic aa are Val, Leu, Ile, Met, and Pro; amide aa are Asn and Gln; aromatic aa are Trp, Phe, and Tyr.
FIGURE 2.
Activation of G286F in G protein-coupled signaling. Ligand-independent activity upon introduction of Phe in position 286 in cAMP accumulation after forskolin stimulation (A) and in PI-turnover (B) in COS-7 cells is shown. The data are normalized to the level obtained in untransfected cells (A) or Emax of CCL3 activation on CCR5 WT and G286F (B). The surface expression of the receptors was estimated in ELISA in COS-7 cells using an N-terminal FLAG tag, and the data are normalized to CCR5 WT (C).
TABLE 1.
Functional analysis of agonists on CCR5 WT and mutants.
PI-turnover was measured in COS-7 cells co-transfected with receptor and chimeric G protein Gqi4myr (which converts Gαi signals into Gαq) in response to the endogenous chemokines CCL3 and CCL5. The expression level was measured in ELISA by an anti-FLAG antibody (M1). The number of independent experiments is shown in parentheses (n), and -fold EC50 indicates the difference between the potency of the mutant and CCR5 WT (EC50 (mutant)/EC50 (WT)). Constitutive activity is calculated as (basal level of activity/Emax) × 100. NA, no activity.
| Residue | Expression level | Constitutive activity | CCL3 |
CCL5 |
||||
|---|---|---|---|---|---|---|---|---|
| Log EC50 ± S.E. | -Fold EC50 | (n) | Log EC50 ± S.E. | -Fold EC50 | (n) | |||
| % of WT ± S.E. | % Basal/Emax ×100 | |||||||
| CCR5 WT | 100 ± 0.00 | 1.6 ± 1.8 | −8.1 ± 0.07 | 1.0 | (40) | −9.1 ± 0.07 | 1.0 | (36) |
| F112L | 87 ± 5.3 | 2.9 ± 1.8 | −8.0 ± 0.11 | 1.2 | (5) | −9.2 ± 0.18 | 0.86 | (3) |
| Y244A | 5.3 ± 0.69 | NA | NA | (3) | NA | (3) | ||
| W248A | 0.66 ± 0.55 | NA | NA | (3) | NA | (3) | ||
| W248F | 21 ± 4.4 | 1.6 ± 4.1 | −7.9 ± 0.10 | 1.4 | (7) | −9.2 ± 0.10 | 0.73 | (4) |
| W248Y | 19 ± 3.4 | 4.9 ± 3.8 | −8.1 ± 0.11 | 0.91 | (8) | −9.6 ± 0.12 | 0.34 | (5) |
| Y251A | 20 ± 7.4 | 0.08 ± 3.6 | −7.7 ± 0.19 | 2.6 | (4) | −8.3 ± 0.24 | 6.7 | (4) |
| G286F | 44 ± 9.3 | 31 ± 2.0 | −7.8 ± 0.06 | 2.0 | (4) | −8.3 ± 0.19 | 7.1 | (3) |
TABLE 2.
Affinity of CCL3 and four small-molecule antagonists on CCR5 WT and mutants.
The data were obtained from competition binding with 125I-labeled CCL3 as radioligand on transiently transfected COS-7 cells. Values in parentheses represent number of experiments (n), and -fold Kd/Ki is a measure of the differences between the affinities on mutant receptors compared to CCR5 WT (Kd/Ki (mutant)/Kd/Ki (WT)). Bmax values are calculated from the homologous binding curves and indicated for each receptor. ND, no displacement.
| Residue | Bmax ± S.E. | CCL3 |
Merck |
SCH C |
TAK-779 |
Aplaviroc |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Log Kd ± S.E. | -Fold Kd | (n) | Log Ki ± S.E. | -Fold Ki | (n) | Log Ki ± S.E. | -Fold Ki | (n) | Log Ki ± S.E. | -Fold Ki | (n) | log Ki ± S.E. | -Fold Ki | (n) | ||
| % of WT | ||||||||||||||||
| CCR5 WT | 100 ± 0.0 | −8.4 ± 0.06 | 1.0 | (19) | −7.0 ± 0.05 | 1.0 | (15) | −7.4 ± 0.05 | 1.0 | (15) | −7.6 ± 0.05 | 1.0 | (15) | −7.3 ± 0.03 | 1.0 | (15) |
| F112L | 76 ± 25 | −8.1 ± 0.11 | 1.9 | (4) | −7.0 ± 0.02 | 1.0 | (3) | −7.3 ± 0.02 | 1.0 | (3) | −7.5 ± 0.01 | 1.2 | (3) | −7.5 ± 0.01 | 0.6 | (3) |
| Y244A | 14 ± 10 | −6.9 ± 0.20 | 29 | (7) | ND | (3) | ND | (3) | ND | (3) | ND | (3) | ||||
| W248A | 6.0 ± 1.4 | −9.2 ± 0.15 | 0.16 | (4) | −7.1 ± 0.02 | 0.77 | (3) | −7.2 ± 0.04 | 1.5 | (3) | −7.8 ± 0.01 | 0.70 | (3) | −7.1 ± 0.03 | 1.4 | (3) |
| W248F | 29 ± 7.9 | −8.5 ± 0.16 | 0.83 | (4) | −7.1 ± 0.03 | 0.89 | (3) | −7.0 ± 0.03 | 2.1 | (3) | −7.7 ± 0.05 | 0.93 | (3) | −6.9 ± 0.04 | 2.5 | (3) |
| W248Y | 52 ± 0.90 | −9.0 ± 0.06 | 0.25 | (4) | −7.2 ± 0.16 | 0.66 | (3) | −7.1 ± 0.06 | 1.8 | (3) | −7.6 ± 0.00 | 0.99 | (3) | −7.0 ± 0.03 | 1.7 | (3) |
| Y251A | 28 ± 8.6 | −7.3 ± 0.23 | 11 | (8) | ND | (3) | −7.7 ± 0.06 | 0.45 | (3) | −8.3 ± 0.14 | 0.21 | (3) | −7.4 ± 0.19 | 0.68 | (3) | |
| G286F | 52 ± 15 | −8.9 ± 0.22 | 0.28 | (3) | −7.1 ± 0.04 | 0.74 | (3) | −7.0 ± 0.10 | 2.2 | (3) | −7.1 ± 0.03 | 3.5 | (3) | −7.0 ± 0.05 | 2.0 | (3) |
The Constitutively Active G286F Promotes Efficacy Switch of Aplaviroc
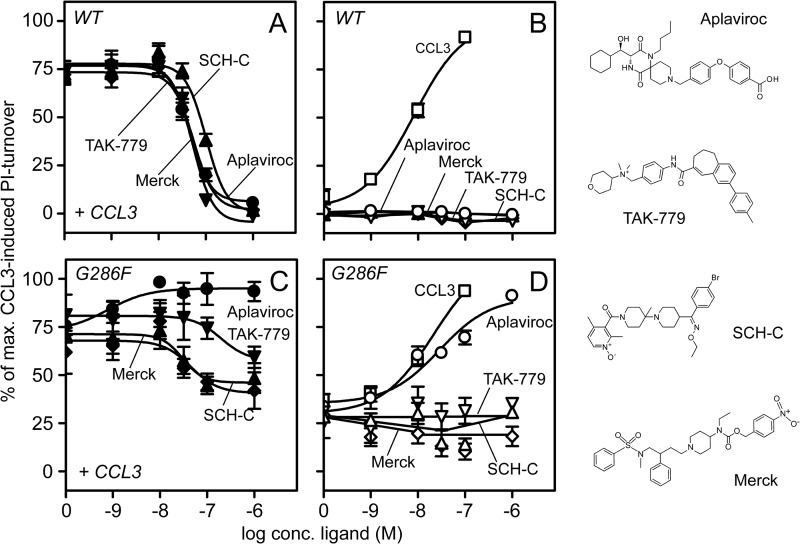
Due to the agonist-prone nature of G286F, we analyzed its influence on binding and action of four small-molecule antagonists: SCH-C, TAK-779, aplaviroc, and a ligand structurally similar to compounds previously described by Merck (33) (Fig. 3). All four have previously been reported to inhibit CCR5 activation and HIV cell-entry via CCR5 with nanomolar potencies (33–37). Consistently, they all completely inhibited CCL3-induced PI-turnover in CCR5 WT with EC50 values between 41 and 104 nm and displayed no intrinsic activities (Fig. 3, A and B, and Table 3). The potencies were matched by similar affinities as determined in heterologous competition binding experiments against 125I-CCL3 (Table 2). In contrast to the complete inhibition in CCR5 WT, the CCL3 activity was only partially inhibited in G286F by SCH-C, TAK-779, and the Merck-derived compound, which acted with EC50 values in the same range as on WT (SCH-C) or slightly increased (Merck and TAK-779) (Fig. 3C, Table 3). Aplaviroc, however, did not antagonize CCL3 at all, but instead potentiated it with a potency drastically increased compared with its antagonistic counterpart on CCR5 WT (∼30-fold increase, Table 3 and Fig. 3C). The affinities of the antagonists did not differ more than 3.5-fold from WT, and intriguingly, despite its radically different efficacy profile, the affinity of aplaviroc was basically unaltered (Table 2). The partial inhibition by SCH-C, TAK-779, and the Merck-derived compound as well as the CCL3-potentiation by aplaviroc prompted us to determine their effects alone. Interestingly, none of the antagonists acted as inverse agonists on G286F (Fig. 3D). Instead, aplaviroc activated the receptor with the same efficacy as CCL3, i.e. it was converted from a full antagonist on WT to a full agonist on this mutant (Fig. 3D). The potency of aplaviroc when acting solo was increased ∼3-fold compared with its (antagonistic) potency on WT (Table 3).
FIGURE 3.
Effect of small-molecule antagonists on G286F-CCR5 in PI-turnover. Four small-molecule antagonists (Merck (♦/♢), SCH C (▴/▵), TAK-779 (▾/▿), and aplaviroc (●/○)) were tested in PI-turnover. They were tested in combination with 10 nm CCL3 (black symbols) or alone (white) on CCR5 WT (A and B, respectively) and on G286F (C and D, respectively). CCL3 activation is shown (□) (B and D). The curves are normalized to Emax of CCL3 on each receptor. The chemical structures of the antagonists are shown to the right.
TABLE 3.
Functional analysis of antagonists on CCR5 WT and mutants
PI-turnover was measured in COS-7 cells co-transfected with receptor and chimeric G protein Gqi4myr (which converts Gαi signals into Gαq). The actions of the four antagonists on CCL3-induced activity are shown. The number of experiments is shown in parentheses, and -fold IC50 indicates the difference between the potency of antagonist on the mutant and on CCR5 WT (IC50 (mutant)/IC50 (WT)). Full inhibition is indicated in magenta, and partial inhibition is in orange (Merck and SCH-C). Mutations where aplaviroc induces activation are indicated in green, and -fold EC50 is compared to IC50 on WT (EC50 (mutant)/IC50 (WT)). NA, no activity. NT, not tested.

Molecular Modeling of CCR5 WT and G286F-CCR5
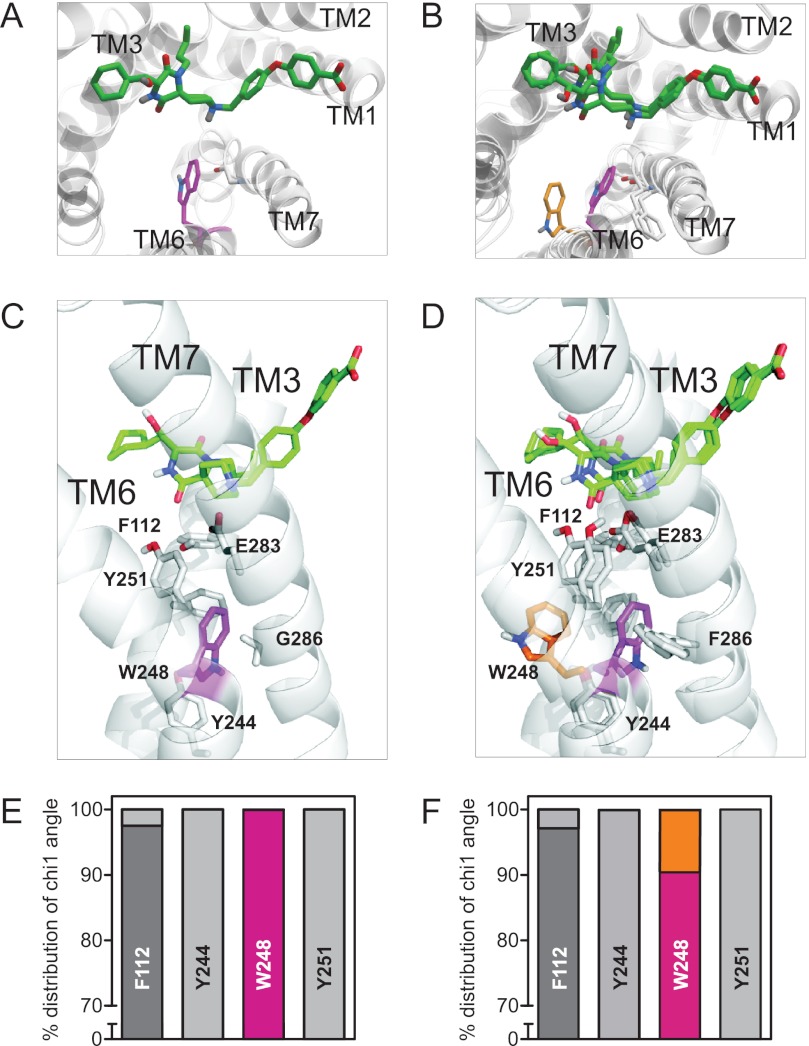
To further explore the mechanism behind the increased basal activity and efficacy switch on G286F-CCR5, docking of aplaviroc in CCR5 WT was performed (Fig. 4, A and C), and Phe-286 was subsequently introduced (Fig. 4, B and D). The docking of aplaviroc suggested three different poses all in contact with residues shown to be important based on previous mutational mapping (38). One was selected, but the other poses were equally likely. We focused on the dynamics of aromatic amino acids in the vicinity of position 286: three in TM6 (Tyr-244 (VI:09/6.44), Trp-248 (VI:13/6.48), and Tyr-251 (VI:16/6.51)) and one in TM3 (Phe-112 (III:12/3.36)). To create an overview of potential discrepancies in the positions of these amino acids between WT and G286F, the χ1 angle was computed in the 1000 models from the Rosetta protocol, and the numerical value was categorized into either of two preferred rotamer states, g+ or trans (the performed normalization was not more than 35 degrees in any case). Fig. 4C illustrates the position of the side chains of the five amino acids in CCR5 WT, with the corresponding distribution of the χ1 angles in this model illustrated in Fig. 4E. In CCR5 WT only the side chain of Phe-112 varied slightly between the models. In G286F, the distribution of the χ1 angles of Tyr-244, Tyr-251, and Phe-112 did not differ from CCR5 WT. However, the rotamer position of Trp-248 was changed in 10% of the models (Fig. 4, D and F). Thus, in CCR5 WT and 90% of the models of G286F, the side chain of Trp-248 was oriented toward TM7, corresponding to g+ (shown in magenta). However, in the 10% models of the G286F model, where the side chain of Trp-248 was rotated to trans (shown in orange), the residue pointed toward TM5.
FIGURE 4.
Computational modeling of CCR5 WT and G286F. In silico models of CCR5 WT (A and C) and G286F (B and D) viewed from the extracellular space (top row) or from the side (middle row) in complex with aplaviroc (green) are shown. The relevant amino acids and the chemokine receptor-conserved Glu-283 (VII:06/7.39) are shown in sticks, and Trp-248 is highlighted in magenta (χ1 angle: g+) and orange (χ1 angle: trans). E and F, the percentage distribution of χ1 angles of aromatic amino acid in the vicinity of position VII:09/7.42 (G/F286) in the WT (E) and G286F (F) computational models is shown. The χ1 angles are color-coded as follows; g+ are shown in dark gray in Phe-112, Tyr-244, and Tyr-251, whereas the light gray symbolizes trans. In the case of Trp-248, g+ is represented in magenta, whereas trans is shown in orange.
Trp-248 in the CWXP Motif in TM6 Is Important for CCR5 Signaling
The computational modeling of G286F-CCR5 suggested that the side chain of Trp-248 changes to an alternative rotamer state in 10% of the models (Fig. 4). During the history of 7TM receptor activation much focus has been put on this conserved position (39–41). Therefore, Trp-248 was mutated to Ala, which resulted in complete elimination of chemokine-induced activity (Fig. 5, A and B, and Table 1). However, CCL3 binding was still maintained (Table 2) despite a low cell surface expression (Table 1).
FIGURE 5.
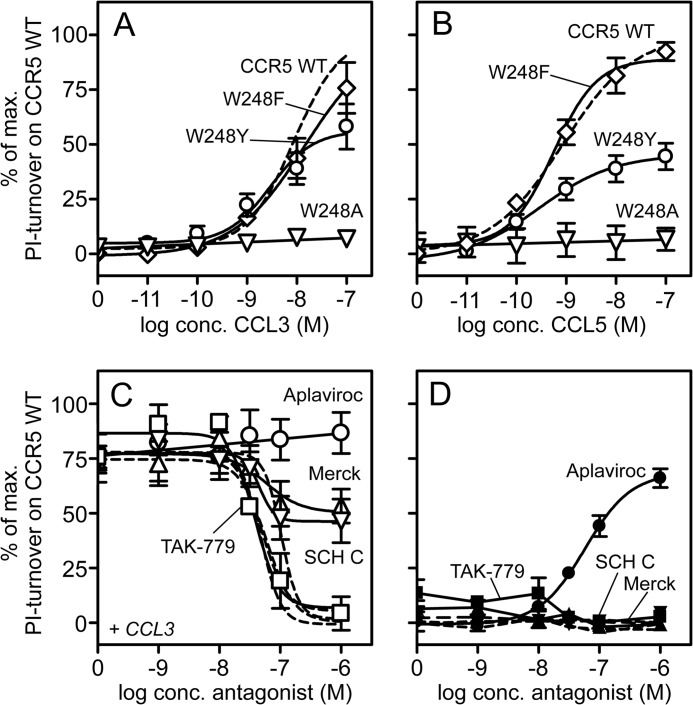
Effect of the elimination or alteration of the aromatic side chain of Trp-248 in PI-turnover. Shown are CCL3- (A) and CCL5-induced (B) PI turnover in COS-7 cells for CCR5 WT (dashed lines), W248A (▿), W248F (♢), and W248Y (○). The four small molecule antagonists, Merck (▾/▿) and SCH-C (▴/▵), TAK-779 (□/■), and aplaviroc (O/●) were tested in the presence (white symbols, C) and absence (black, D) of 10 nm CCL3 (for simplicity, only results for W248Y are shown). For comparison, the similar action of these on CCR5 WT (from Fig. 3A) is indicated as dashed lines in C and D.
To further determine the role of the particular characteristics of Trp as opposed to those of aromatic amino acids in general, Trp-248 was substituted with Phe and Tyr (W248(F/Y)). This resulted in WT-like efficacy and potency of CCL3-induced PI-turnover and no constitutive activity, whereas the potency of CCL5 was increased up to 3.5-fold (Table 1 and Fig. 5, A and B). Furthermore, the affinity of CCL3 measured in homologous competition binding was increased up to 4-fold as compared with WT (Table 2). Thus, substitution to a non-aromatic residue (Ala) resulted in maintained chemokine binding but completely blocked activation. In contrast, substitution with other aromatic residues (Tyr and Phe) resulted in WT-like properties both with respect of agonist binding and activation (Table 2 and Fig. 5, A and B).
Among the four antagonists, TAK-779 completely inhibited CCL3-induced activity on W248(F/Y) with WT-like potencies, whereas both the Merck-derived compound and SCH-C only inhibited to ∼50% (Fig. 5C). Moreover, they displayed up to ∼20-fold decreased potencies as compared with WT (Table 3). Intriguingly, aplaviroc was unable to inhibit CCL3-induced activation (Fig. 5C), and furthermore, in the absence of CCL3 it induced activation to 70% that of the CCL3-induced PI-turnover (Fig. 5D and Table 3). No intrinsic activity was observed for the Merck compound and SCH-C despite the partial inhibition. Thus, aplaviroc displayed the same efficacy switch from antagonist to agonist on W248(F/Y) as seen on G286F with an agonistic potency equal to the antagonistic potency at WT (Table 3). Despite huge changes in efficacies, the affinities of these four compounds did not differ more than 2.5-fold from WT (Table 2).
Impact of Aromatic Residues Above and Below Trp-248
In addition to Trp-248, TM6 in CCR5 contains two other aromatic amino acids in the proximity of Gly-286, namely Tyr-244 and Tyr-251 located below and above Trp-248, respectively (Fig. 1). Both residues have been implicated in 7TM receptor activation previously (39). Although the computational modeling did not show side chain alterations of these in the G286F mutation (Fig. 4), they were both substituted with Ala. CCL3 was able to bind both mutant receptors, albeit with reduced affinity (11–29-fold) (Table 2). With regard to the signaling, Y251A resulted in a 2–5-fold decrease in potency of chemokine-induced PI-turnover, whereas Y244A completely abolished this (Table 1). The small molecule antagonists acted as full antagonists on Y251A-CCR5, with potencies similar to WT (TAK-779 and the Merck-derived compound) or slightly increased (SCH-C and aplaviroc, Table 3).
Biased Signaling; Impaired β-Arrestin Recruitment Despite Constitutive Gαi Activity and Agonist-prone Nature
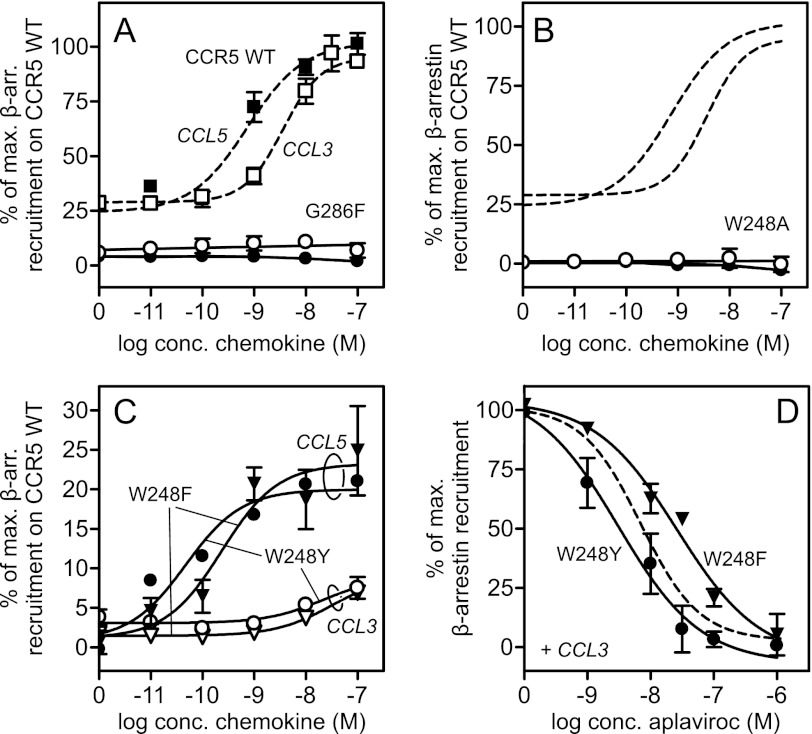
Lately much attention has been drawn to the signaling pathways beyond the ones coupled to G proteins, e.g. recruitment and signaling via β-arrestins. We, therefore, investigated whether the signaling mediated by β-arrestin was similar to the G protein-coupled pathway. Fig. 6A shows that CCR5 WT recruited β-arrestin even in absence of chemokines, with a basal level of 30% of Emax. The potency of CCL3 and CCL5 (EC50 of 8.0 and 2.2 nm, respectively) was in the same range as the one obtained in PI-turnover (Table 1). Surprisingly, despite the enhanced signaling through Gαi, G286F-CCR5 resulted in a complete loss of the basal β-arrestin recruitment. Furthermore, it was completely unable to be activated by the chemokines (Fig. 6A). In accordance with the lack of Gαi activity in W248A, no β-arrestin recruitment was observed here (Fig. 6B). Ala substitution of the other two aromatic amino acids in TM6 (Y251A and Y244A) also completely eliminated chemokine-induced β-arrestin recruitment (data not shown) despite maintained Gαi activity for Y251A. Furthermore, the efficacy of W248F and W248Y was highly impaired with regard to β-arrestin recruitment, as an 80% (CCL5-mediated) to 90% (CCL3) decrease was observed compared with CCR5 WT (Fig. 6C). The potency of CCL3 was decreased ∼4-fold, whereas the potency of CCL5 actually was increased 10-fold, the same tendency as seen in PI-turnover (Fig. 6). However, unlike the efficacy switch of aplaviroc observed for W248(F/Y) in the G protein-coupled pathway (Fig. 5, C and D), no aplaviroc-induced β-arrestin recruitment was observed. Instead, when added together with CCL3, aplaviroc acted as an antagonist with a potency similar to that on WT (Fig. 6D). No aplaviroc-induced β-arrestin recruitment was observed on G286F either (data not shown). Thus, all of the mutations constructed in TM6 and TM7 either completely eliminated or heavily impaired β-arrestin recruitment.
FIGURE 6.
β-Arrestin recruitment in CCR5 WT and mutations. The receptors were tested in enzyme fragment complementation-based β-arrestin recruitment in U2OS cells. CCL3 (white symbols)- and CCL5 (black symbols)-induced β-arrestin recruitment in G286F (A), W248A (B), and W248(F/Y) (C) with corresponding CCR5 WT dose-response curves (dashed lines) are shown (A–C). D, aplaviroc inhibition of CCL3-induced β-arrestin recruitment on W248F (▾) and Y (●) is shown. The curves are normalized to maximum CCL3-induced β-arrestin recruitment on each receptor.
DISCUSSION
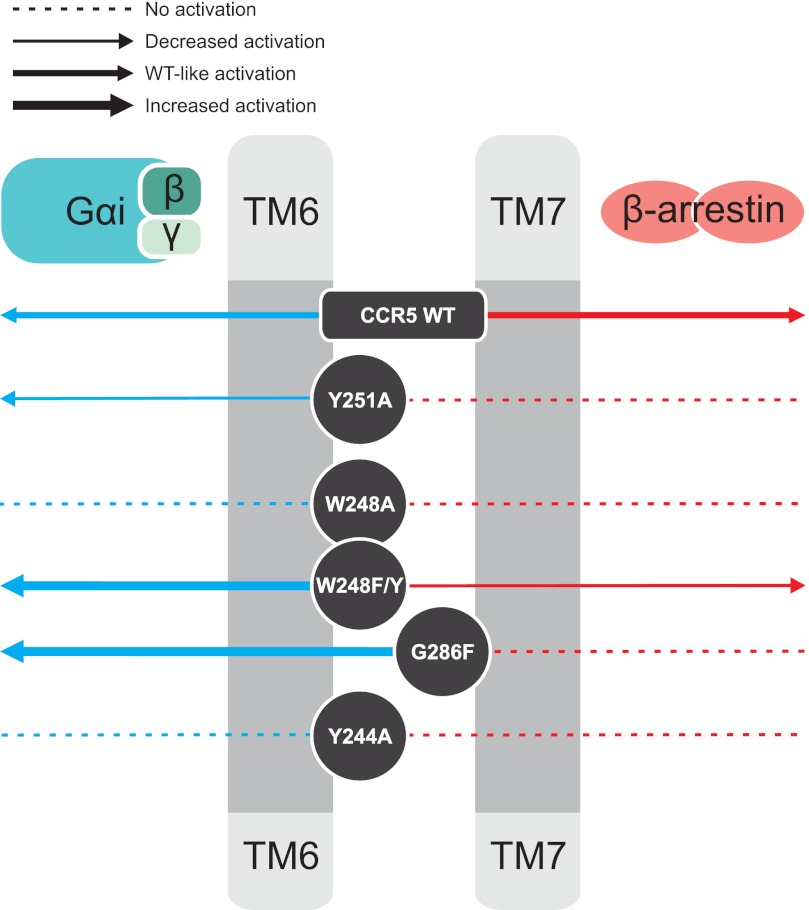
By single point mutagenesis we show here that the interplay between TM6 and TM7 is critical for CCR5 activation, in particular for the balance between G protein-dependent and -independent pathways (summarized in Fig. 7).
FIGURE 7.
Impact of the mutations for G protein-coupling and β-arrestin recruitment. Illustration of the activation pattern for CCR5 WT and mutants is shown. The approximate position of the residues is indicated in TM6 and TM7. The thickness of the arrows represents the degree of activation (dashed line, no activation; thin arrow, ≥5-fold decrease in agonist potency compared with WT; medium arrow, WT-like activation; thick arrow, increase in basal activity/agonist potency/efficacy switch). Blue arrows signify G protein-coupled signaling, whereas red arrows indicate β-arrestin recruitment.
Manipulation with the TM6/7 Interface Induces Biased Agonism
Lately, the concept of biased signaling has gained a lot of attention because of its potential in drug development (42). A drug that selectively targets receptor endocytosis without affecting signaling or the opposite would benefit from the lack of side effects determined by the altered pharmacodynamics. For example, it has been shown that the GPR109A agonist, MK-0354, selectively signals via Gαi, which mediates beneficial anti-lipolytic effects in vivo but does not affect the β-arrestin pathway and thus does not cause the unwanted side effect of flushing that normally accompanies GPR109A agonists (43, 44). Since the discovery of CCR5 and CXCR4 as co-receptors for HIV entry (14, 15), much focus has been drawn toward development of receptor antagonists (45, 46). Yet, agonists are also able to inhibit HIV cell entry by blocking the interaction between gp120 and CCR5/CXCR4, and by inducing receptor internalization they thereby obliterate the gate for virus cell entry. A biased drug with agonism toward β-arrestin recruitment and not G protein signaling would be superior as functional antagonist for HIV cell entry. Such a biased drug would cause receptor internalization, thereby preventing virus interaction with CCR5 and a lack of potential side effects after unwanted cellular signaling. Importantly, these properties were recently presented in ESN-196, a novel small-molecule CCR5-targeting ligand that induced CCR5 sequestration without promoting chemotaxis (47). Theoretically, targeting human components involved in the HIV infection cycle instead of the viral proteins would decrease the risk of resistance (48, 49). Currently, the CCR5 antagonist maraviroc is the only entry inhibitor targeting a chemokine receptor on the market. Despite its human target, some HIV clones have developed resistance by altered CCR5 recognition in the presence of maraviroc (48, 50). It is conceivable that a biased ligand inducing receptor internalization and thereby decreasing the availability of receptors on the cell surface would decrease viral resistance. Individuals homozygous for a deletion in CCR5 (Δ32 CCR5) do not express CCR5 on cell surfaces and are highly resistant to HIV, proving that this will be an effective treatment strategy (51–54).
To develop biased ligands it is vital to know which areas of the receptors are involved in activation of, for example, β-arrestin but not G protein or the opposite. Our results clearly show a bias of G286F-CCR5 toward G protein-coupled signaling as opposed to β-arrestin recruitment. The same bias was observed for Y251A, whereas W248A and Y244A were overall silent. The activation of W248(F/Y)-CCR5 induced by CCL3 was highly biased toward G protein signaling. On the other hand, the CCL5 potency was increased compared with WT in β-arrestin recruitment. Thus, tampering with the TM6/7 interface had a huge impact on β-arrestin recruitment. The biased activity was seen with both endogenous ligands, CCL3 and CCL5, despite different binding patterns (55). In accordance with our results, recently published structural data from an NMR study of the β2-adrenergic receptor (56) and from crystal structures of the avian β1-adrenergic receptor in complex with a biased as well as unbiased ligands (57) showed that β-arrestin biased ligands are more prone to interact with residues in especially TM7 than non-biased ligands. It is important to stress that a crystal structure of a 7TM receptor in complex with β-arrestin has yet to be published, and therefore, we do not know the conformation of a receptor in such complex. However, our results combined with structural data like those obtained from the β1- and β2-adrenergic receptors (56, 57) suggest that TM6 and -7 play a pivotal role both in the G protein signaling and β-arrestin recruitment.
G286F Affects the Rotameric State of Trp-248
It was already pointed out by Holst et al. in 2004 (58) that a small side chain (Ala, Gly, or Ser) is found in 73% of all 7TM receptors in position VII:09/7.42 (59), and mutational analysis in members of the ghrelin receptor family revealed that a large aromatic Phe or Tyr naturally occurring at this position was associated with high constitutive activity (58). In 2010, the same group showed that insertion of a Val instead of an Ala in the bombesin receptor BB3 increased the level of basal activity (60). In line with these results, we here show that mutation of this residue (Gly to Phe, G286F) in CCR5, which is otherwise not constitutively active through Gαi, raised the basal activity through Gαi to 30% that of maximum CCL3 activity. In 10% of the models obtained by the computational simulation protocol, the side chain of Trp-248 had an alternative rotamer state in G286F compared with CCR5 WT (trans versus g+). Confirming the importance of this residue, a complete elimination of chemokine activity was observed upon mutation to Ala despite maintained high affinity chemokine binding. In several different 7TM receptors it has been shown that elimination of this conserved aromatic side chain (84% aromatic, of which 64% is a Trp) has a negative impact on the G protein activation (e.g. the 5-HT4 and the β2-adrenergic receptor (40, 41) among others). Before crystal structures of active 7TM receptors were published, Schwartz and co-workers (2, 39) speculated that the side chain of this Trp serves as a rotamer switch, which is important for the overall activation mechanism of the receptor. In fact, when the χ1 angle was trans, the Trp was proposed to be in the active conformation, which correlates well with the increase in activity of G286F in G protein signaling. However, despite major helical movements of TM6, none of the agonist-bound crystal structures confirmed this (3, 4, 61–64). Even though we have seen structures of what, by definition are truly activated receptors (a ternary complex of an agonist, the receptor itself, and a G protein or equivalent), these structures do not provide an overview of the dynamics in receptor activation. We observed a change in the side chain of Trp-248 in 10% of the cases, i.e. a relatively low fraction that could explain why no side chain rotations is seen in the two crystal structures of truly active receptors (3, 4). Another possibility could be that the conformational change of the conserved Trp differs between receptors. Indeed, one-third of the chemokine receptors carry a Gln in VI:13/6.48, and accordingly, a different mechanism may be in play here. Alteration of TM kink properties or displacement of potential water molecules could also contribute. However, in CCR5 we clearly see a linkage between the G286F mutation and a rotation of the side chain of Trp-248, which could contribute to the altered conformational equilibrium in Gαi-mediated signaling.
Identification of an Efficacy Switch Region for a Small Molecule CCR5 Antagonist
In this study four different well known small molecule CCR5 antagonists were included. Changing a single amino acid (G286F or W248(F/Y)) completely reversed the efficacy of one (aplaviroc) from full antagonist to full agonist in Gαi signaling. There have been several reports on single amino acid mutations resulting in an efficacy switch, i.e. conversion of an antagonist to an agonist or vice versa, in 7TM receptors. For example, Holst et al. (65) showed that space-generating mutations in the top of TM3 in the ghrelin receptor led to a switch from inverse agonism to agonism in Gαq coupling, presumably by allowing the ligand to bind more superficially. Similar switches have been seen by a steric hindrance mutation in the center of TM6 in the dopamine D1 receptor (66) and in TM3 and -6 in the bradykinin B2 receptor (67). All of the above-mentioned examples of increased ligand efficacy also caused a rise in the basal G protein activity. Accordingly, G286F-CCR5 displayed high constitutive activity in our experiments. This could indicate that the mechanism behind the efficacy switches is a skewed equilibrium toward the activated G protein-coupled receptor state(s). Hence, the probability of the compound binding to a receptor in the active conformation is higher, and the compound will in this case stabilize the active conformation instead of the inactive, assuming that the affinity of the compound is the same for both conformations. Indeed, introduction of Phe-286 in the aplaviroc-CCR5 WT complex did not affect the position of aplaviroc in the in silico screening (Fig. 4), indicating that it binds to the same site. This is also supported by the unaltered affinity (Table 2). Thus, presumably aplaviroc does not induce an alternate receptor conformation but rather stabilizes the existing structure.
The four included antagonists interact differently with CCR5. Thus, mutagenesis as well as docking studies of Merck compounds have indicated a binding pocket near the top of TM2, -3, -6, and -7 (33). Similar binding modes have been proposed for TAK-779 and SCH-C (68, 69). We have recently shown in a chimeric CCR2/CCR5 receptor approach that whereas TAK-779 and SCH-C solely interact with residues in the transmembrane domain, aplaviroc is also dependent on the extracellular loop 2 (37), in accordance with a previously suggested binding mode (38). Thus, among these four, aplaviroc is the only one that depends upon residues in the extracellular regions, which could be the reason why it alone acquires agonistic properties in the present study.
The present study indicates that the interface between TM6 and -7 in CCR5 is highly important for the activation state both when coupling to G protein and β-arrestin. It is generally acknowledged that 7TM receptors share a common activation mechanism, and thus, it is possible that parallels could be drawn between the results presented here for CCR5 and other chemokine receptors, even to other 7TM receptors.
Acknowledgments
We thank Thue W. Schwartz, Joshua M. Farber, and Alexander Hovard Sparre-Ulrich for fruitful discussions of the manuscript. We also thank Randi Thøgersen, Inger Smith Simonsen, and Olav Larsen for excellent technical assistance.
This work was supported by The Danish Council for Independent Research Medical Sciences, the Aase and Einar Danielsen Foundation, and the Novo Nordisk Foundation.
- 7TM
- 7-transmembrane helix
- TM
- transmembrane domain
- PI
- phosphatidylinositol.
REFERENCES
- 1. Sharman J. L., Mpamhanga C. P., Spedding M., Germain P., Staels B., Dacquet C., Laudet V., Harmar A. J., NC-IUPHAR (2011) IUPHAR-DB. New receptors and tools for easy searching and visualization of pharmacological data. Nucleic Acids Res. 39, D534–D538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nygaard R., Frimurer T. M., Holst B., Rosenkilde M. M., Schwartz T. W. (2009) Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 30, 249–259 [DOI] [PubMed] [Google Scholar]
- 3. Rasmussen S. G., DeVree B. T., Zou Y., Kruse A. C., Chung K. Y., Kobilka T. S., Thian F. S., Chae P. S., Pardon E., Calinski D., Mathiesen J. M., Shah S. T., Lyons J. A., Caffrey M., Gellman S. H., Steyaert J., Skiniotis G., Weis W. I., Sunahara R. K., Kobilka B. K. (2011) Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rasmussen S. G., Choi H.-J., Fung J. J., Pardon E., Casarosa P., Chae P. S., Devree B. T., Rosenbaum D. M., Thian F. S., Kobilka T. S., Schnapp A., Konetzki I., Sunahara R. K., Gellman S. H., Pautsch A., Steyaert J., Weis W. I., Kobilka B. K. (2011) Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Crystal structure of rhodopsin. A G protein-coupled receptor. Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 6. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H.-J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rasmussen S. G., Choi H.-J., Rosenbaum D. M., Kobilka T. S., Thian F. S., Edwards P. C., Burghammer M., Ratnala V. R., Sanishvili R., Fischetti R. F., Schertler G. F., Weis W. I., Kobilka B. K. (2007) Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature 450, 383–387 [DOI] [PubMed] [Google Scholar]
- 8. Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Structure of a β1-adrenergic G-protein-coupled receptor. Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jaakola V.-P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jardón-Valadez E., Ulloa-Aguirre A., Piñeiro A. (2008) Modeling and molecular dynamics simulation of the human gonadotropin-releasing hormone receptor in a lipid bilayer. J. Phys. Chem. B 112, 10704–10713 [DOI] [PubMed] [Google Scholar]
- 11. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., Stevens R. C. (2010) Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park S. H., Das B. B., Casagrande F., Tian Y., Nothnagel H. J., Chu M., Kiefer H., Maier K., De Angelis A. A., Marassi F. M., Opella S. J. (2012) Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature 491, 779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Urban J. D., Clarke W. P., von Zastrow M., Nichols D. E., Kobilka B., Weinstein H., Javitch J. A., Roth B. L., Christopoulos A., Sexton P. M., Miller K. J., Spedding M., Mailman R. B. (2007) Functional selectivity and classical concepts of quantitative pharmacology. J. Pharmacol. Exp. Ther. 320, 1–13 [DOI] [PubMed] [Google Scholar]
- 14. Deng H., Liu R., Ellmeier W., Choe S., Unutmaz D., Burkhart M., Di Marzio P., Marmon S., Sutton R. E., Hill C. M., Davis C. B., Peiper S. C., Schall T. J., Littman D. R., Landau N. R. (1996) Identification of a major co-receptor for primary isolates of HIV-1. Nature 381, 661–666 [DOI] [PubMed] [Google Scholar]
- 15. Alkhatib G., Combadiere C., Broder C. C., Feng Y., Kennedy P. E., Murphy P. M., Berger E. A. (1996) CC CKR5. a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272, 1955–1958 [DOI] [PubMed] [Google Scholar]
- 16. Simmons G., Clapham P. R., Picard L., Offord R. E., Rosenkilde M. M., Schwartz T. W., Buser R., Wells T. N., Proudfoot A. E. (1997) Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science 276, 276–279 [DOI] [PubMed] [Google Scholar]
- 17. Mack M., Luckow B., Nelson P. J., Cihak J., Simmons G., Clapham P. R., Signoret N., Marsh M., Stangassinger M., Borlat F., Wells T. N., Schlöndorff D., Proudfoot A. E. (1998) Aminooxypentane-RANTES induces CCR5 internalization but inhibits recycling. A novel inhibitory mechanism of HIV infectivity. J. Exp. Med. 187, 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gaertner H., Cerini F., Escola J.-M., Kuenzi G., Melotti A., Offord R., Rossitto-Borlat I., Nedellec R., Salkowitz J., Gorochov G., Mosier D., Hartley O. (2008) Highly potent, fully recombinant anti-HIV chemokines. Reengineering a low-cost microbicide. Proc. Natl. Acad. Sci. U.S.A. 105, 17706–17711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O'Hayre M., Salanga C. L., Handel T. M., Hamel D. J. (2010) Emerging concepts and approaches for chemokine-receptor drug discovery. Expert Opin. Drug Discov. 5, 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rajagopal S., Kim J., Ahn S., Craig S., Lam C. M., Gerard N. P., Gerard C., Lefkowitz R. J. (2010) β-arrestin, but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proc. Natl. Acad. Sci. U.S.A. 107, 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rosenkilde M. M., McLean K. A., Holst P. J., Schwartz T. W. (2004) The CXC chemokine receptor encoded by herpesvirus saimiri, ECRF3, shows ligand-regulated signaling through Gi, Gq, and G12/13 proteins but constitutive signaling only through Gi and G12/13 proteins. J. Biol. Chem. 279, 32524–32533 [DOI] [PubMed] [Google Scholar]
- 22. Wisler J. W., DeWire S. M., Whalen E. J., Violin J. D., Drake M. T., Ahn S., Shenoy S. K., Lefkowitz R. J. (2007) A unique mechanism of β-blocker action. Carvedilol stimulates β-arrestin signaling. Proc. Natl. Acad. Sci. U.S.A. 104, 16657–16662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Walters R. W., Shukla A. K., Kovacs J. J., Violin J. D., DeWire S. M., Lam C. M., Chen J. R., Muehlbauer M. J., Whalen E. J., Lefkowitz R. J. (2009) β-Arrestin1 mediates nicotinic acid-induced flushing, but not its antilipolytic effect, in mice. J. Clin. Invest. 119, 1312–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heydorn A., Ward R. J., Jorgensen R., Rosenkilde M. M., Frimurer T. M., Milligan G., Kostenis E. (2004) Identification of a novel site within G protein α subunits important for specificity of receptor-G protein interaction. Mol. Pharmacol. 66, 250–259 [DOI] [PubMed] [Google Scholar]
- 25. Kostenis E., Zeng F. Y., Wess J. (1998) Functional characterization of a series of mutant G protein αq subunits displaying promiscuous receptor coupling properties. J. Biol. Chem. 273, 17886–17892 [DOI] [PubMed] [Google Scholar]
- 26. Graham F. L., van der Eb A. J. (1973) A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52, 456–467 [DOI] [PubMed] [Google Scholar]
- 27. Kissow H., Hartmann B., Holst J. J., Viby N.-E., Hansen L. S., Rosenkilde M. M., Hare K. J., Poulsen S. S. (2012) Glucagon-like peptide-1 (GLP-1) receptor agonism or DPP-4 inhibition does not accelerate neoplasia in carcinogen-treated mice. Regul. Pept. 179, 91–100 [DOI] [PubMed] [Google Scholar]
- 28. Barth P., Schonbrun J., Baker D. (2007) Toward high-resolution prediction and design of transmembrane helical protein structures. Proc. Natl. Acad. Sci. U.S.A. 104, 15682–15687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leaver-Fay A., Tyka M., Lewis S. M., Lange O. F., Thompson J., Jacak R., Kaufman K., Renfrew P. D., Smith C. A., Sheffler W., Davis I. W., Cooper S., Treuille A., Mandell D. J., Richter F., Ban Y.-E., Fleishman S. J., Corn J. E., Kim D. E., Lyskov S., Berrondo M., Mentzer S., Popović Z., Havranek J. J., Karanicolas J., Das R., Meiler J., Kortemme T., Gray J. J., Kuhlman B., Baker D., Bradley P. (2011) ROSETTA3. An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Totrov M., Abagyan R. (2008) Flexible ligand docking to multiple receptor conformations. A practical alternative. Curr. Opin. Struct. Biol. 18, 178–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bottegoni G., Kufareva I., Totrov M., Abagyan R. (2009) Four-dimensional docking. A fast and accurate account of discrete receptor flexibility in ligand docking. J. Med. Chem. 52, 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Castonguay L. A., Weng Y., Adolfsen W., Di Salvo J., Kilburn R., Caldwell C. G., Daugherty B. L., Finke P. E., Hale J. J., Lynch C. L., Mills S. G., MacCoss M., Springer M. S., DeMartino J. A. (2003) Binding of 2-aryl-4-(piperidin-1-yl)butanamines and 1,3,4-trisubstituted pyrrolidines to human CCR5. a molecular modeling-guided mutagenesis study of the binding pocket. Biochemistry 42, 1544–1550 [DOI] [PubMed] [Google Scholar]
- 34. Shiraishi M., Aramaki Y., Seto M., Imoto H., Nishikawa Y., Kanzaki N., Okamoto M., Sawada H., Nishimura O., Baba M., Fujino M. (2000) Discovery of novel, potent, and selective small-molecule CCR5 antagonists as anti-HIV-1 agents. Synthesis and biological evaluation of anilide derivatives with a quaternary ammonium moiety. J. Med. Chem. 43, 2049–2063 [DOI] [PubMed] [Google Scholar]
- 35. Strizki J. M., Xu S., Wagner N. E., Wojcik L., Liu J., Hou Y., Endres M., Palani A., Shapiro S., Clader J. W., Greenlee W. J., Tagat J. R., McCombie S., Cox K., Fawzi A. B., Chou C. C., Pugliese-Sivo C., Davies L., Moreno M. E., Ho D. D., Trkola A., Stoddart C. A., Moore J. P., Reyes G. R., Baroudy B. M. (2001) SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 98, 12718–12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maeda K., Nakata H., Koh Y., Miyakawa T., Ogata H., Takaoka Y., Shibayama S., Sagawa K., Fukushima D., Moravek J., Koyanagi Y., Mitsuya H. (2004) Spirodiketopiperazine-based CCR5 inhibitor which preserves CC-chemokine/CCR5 interactions and exerts potent activity against R5 human immunodeficiency virus type 1 in vitro. J. Virol. 78, 8654–8662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thiele S., Steen A., Jensen P. C., Mokrosinski J., Frimurer T. M., Rosenkilde M. M. (2011) Allosteric and orthosteric sites in CC chemokine receptor (CCR5), a chimeric receptor approach. J. Biol. Chem. 286, 37543–37554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maeda K., Das D., Ogata-Aoki H., Nakata H., Miyakawa T., Tojo Y., Norman R., Takaoka Y., Ding J., Arnold G. F., Arnold E., Mitsuya H. (2006) Structural and molecular interactions of CCR5 inhibitors with CCR5. J. Biol. Chem. 281, 12688–12698 [DOI] [PubMed] [Google Scholar]
- 39. Schwartz T. W., Frimurer T. M., Holst B., Rosenkilde M. M., Elling C. E. (2006) Molecular mechanism of 7TM receptor activation. A global toggle switch model. Annu. Rev. Pharmacol. Toxicol. 46, 481–519 [DOI] [PubMed] [Google Scholar]
- 40. Pellissier L. P., Sallander J., Campillo M., Gaven F., Queffeulou E., Pillot M., Dumuis A., Claeysen S., Bockaert J., Pardo L. (2009) Conformational toggle switches implicated in basal constitutive and agonist-induced activated states of 5-hydroxytryptamine-4 receptors. Mol. Pharmacol. 75, 982–990 [DOI] [PubMed] [Google Scholar]
- 41. Holst B., Nygaard R., Valentin-Hansen L., Bach A., Engelstoft M. S., Petersen P. S., Frimurer T. M., Schwartz T. W. (2010) A conserved aromatic lock for the tryptophan rotameric switch in TM-VI of seven-transmembrane receptors. J. Biol. Chem. 285, 3973–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rajagopal S., Rajagopal K., Lefkowitz R. J. (2010) Teaching old receptors new tricks. biasing seven-transmembrane receptors. Nat. Rev. Drug Discov. 9, 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Richman J. G., Kanemitsu-Parks M., Gaidarov I., Cameron J. S., Griffin P., Zheng H., Guerra N. C., Cham L., Maciejewski-Lenoir D., Behan D. P., Boatman D., Chen R., Skinner P., Ornelas P., Waters M. G., Wright S. D., Semple G., Connolly D. T. (2007) Nicotinic acid receptor agonists differentially activate downstream effectors. J. Biol. Chem. 282, 18028–18036 [DOI] [PubMed] [Google Scholar]
- 44. Semple G., Skinner P. J., Gharbaoui T., Shin Y.-J., Jung J.-K., Cherrier M. C., Webb P. J., Tamura S. Y., Boatman P. D., Sage C. R., Schrader T. O., Chen R., Colletti S. L., Tata J. R., Waters M. G., Cheng K., Taggart A. K., Cai T.-Q., Carballo-Jane E., Behan D. P., Connolly D. T., Richman J. G. (2008) 3-(1H-tetrazol-5-yl)-1,4,5,6-tetrahydro-cyclopentapyrazole (MK-0354). A partial agonist of the nicotinic acid receptor, G-protein coupled receptor 109a, with antilipolytic but no vasodilatory activity in mice. J. Med. Chem. 51, 5101–5108 [DOI] [PubMed] [Google Scholar]
- 45. Singh I. P., Chauthe S. K. (2011) Small molecule HIV entry inhibitors. Part I. Chemokine receptor antagonists. 2004 - 2010. Expert Opin. Ther. Pat. 21, 227–269 [DOI] [PubMed] [Google Scholar]
- 46. Chen W., Zhan P., De Clercq E., Liu X. (2012) Recent progress in small molecule CCR5 antagonists as potential HIV-1 entry inhibitors. Curr. Pharm. Des. 18, 100–112 [DOI] [PubMed] [Google Scholar]
- 47. Ferain T., Hoveyda H., Ooms F., Schols D., Bernard J., Fraser G. (2011) Agonist-induced internalization of CC chemokine receptor 5 as a mechanism to inhibit HIV replication. J. Pharmacol. Exp. Ther. 337, 655–662 [DOI] [PubMed] [Google Scholar]
- 48. Tilton J. C., Wilen C. B., Didigu C. A., Sinha R., Harrison J. E., Agrawal-Gamse C., Henning E. A., Bushman F. D., Martin J. N., Deeks S. G., Doms R. W. (2010) A maraviroc-resistant HIV-1 with narrow cross-resistance to other CCR5 antagonists depends on both N-terminal and extracellular loop domains of drug-bound CCR5. J. Virol. 84, 10863–10876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Steen A., Schwartz T. W., Rosenkilde M. M. (2009) Targeting CXCR4 in HIV cell-entry inhibition. Mini. Rev. Med. Chem. 9, 1605–1621 [DOI] [PubMed] [Google Scholar]
- 50. Westby M., Smith-Burchnell C., Mori J., Lewis M., Mosley M., Stockdale M., Dorr P., Ciaramella G., Perros M. (2007) Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 81, 2359–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ditzel H. J., Rosenkilde M. M., Garred P., Wang M., Koefoed K., Pedersen C., Burton D. R., Schwartz T. W. (1998) The CCR5 receptor acts as an alloantigen in CCR5Δ32 homozygous individuals. Identification of chemokineand HIV-1-blocking human antibodies. Proc. Natl. Acad. Sci. U.S.A. 95, 5241–5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dean M., Carrington M., Winkler C., Huttley G. A., Smith M. W., Allikmets R., Goedert J. J., Buchbinder S. P., Vittinghoff E., Gomperts E., Donfield S., Vlahov D., Kaslow R., Saah A., Rinaldo C., Detels R., O'Brien S. J. (1996) Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273, 1856–1862 [DOI] [PubMed] [Google Scholar]
- 53. Samson M., Libert F., Doranz B. J., Rucker J., Liesnard C., Farber C. M., Saragosti S., Lapoumeroulie C., Cognaux J., Forceille C., Muyldermans G., Verhofstede C., Burtonboy G., Georges M., Imai T., Rana S., Yi Y., Smyth R. J., Collman R. G., Doms R. W., Vassart G., Parmentier M. (1996) Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382, 722–725 [DOI] [PubMed] [Google Scholar]
- 54. Liu R., Paxton W. A., Choe S., Ceradini D., Martin S. R., Horuk R., MacDonald M. E., Stuhlmann H., Koup R. A., Landau N. R. (1996) Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply exposed individuals to HIV-1 infection. Cell 86, 367–377 [DOI] [PubMed] [Google Scholar]
- 55. Blanpain C., Doranz B. J., Bondue A., Govaerts C., De Leener A., Vassart G., Doms R. W., Proudfoot A., Parmentier M. (2003) The core domain of chemokines binds CCR5 extracellular domains, while their amino terminus interacts with the transmembrane helix bundle. J. Biol. Chem. 278, 5179–5187 [DOI] [PubMed] [Google Scholar]
- 56. Liu J. J., Horst R., Katritch V., Stevens R. C., Wüthrich K. (2012) Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science 335, 1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Warne T., Edwards P. C., Leslie A. G., Tate C. G. (2012) Crystal structures of a stabilized β1-adrenoceptor bound to the biased agonists bucindolol and carvedilol. Structure 20, 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Holst B., Holliday N. D., Bach A., Elling C. E., Cox H. M., Schwartz T. W. (2004) Common structural basis for constitutive activity of the ghrelin receptor family. J. Biol. Chem. 279, 53806–53817 [DOI] [PubMed] [Google Scholar]
- 59. Mirzadegan T., Benkö G., Filipek S., Palczewski K. (2003) Sequence analyses of G-protein-coupled receptors. Similarities to rhodopsin. Biochemistry 42, 2759–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gbahou F., Holst B., Schwartz T. W. (2010) Molecular basis for agonism in the BB3 receptor. An epitope located on the interface of transmembrane-III, -VI, and -VII. J. Pharmacol. Exp. Ther. 333, 51–59 [DOI] [PubMed] [Google Scholar]
- 61. Standfuss J., Edwards P. C., D'Antona A., Fransen M., Xie G., Oprian D. D., Schertler G. F. (2011) The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature 471, 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xu F., Wu H., Katritch V., Han G. W., Jacobson K. A., Gao Z.-G., Cherezov V., Stevens R. C. (2011) Structure of an agonist-bound human A2A adenosine receptor. Science 332, 322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rosenbaum D. M., Zhang C., Lyons J. A., Holl R., Aragao D., Arlow D. H., Rasmussen S. G., Choi H.-J., Devree B. T., Sunahara R. K., Chae P. S., Gellman S. H., Dror R. O., Shaw D. E., Weis W. I., Caffrey M., Gmeiner P., Kobilka B. K. (2011) Structure and function of an irreversible agonist-β2 adrenoceptor complex. Nature 469, 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lebon G., Warne T., Edwards P. C., Bennett K., Langmead C. J., Leslie A. G., Tate C. G. (2011) Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature 474, 521–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Holst B., Mokrosinski J., Lang M., Brandt E., Nygaard R., Frimurer T. M., Beck-Sickinger A. G., Schwartz T. W. (2007) Identification of an efficacy switch region in the ghrelin receptor responsible for interchange between agonism and inverse agonism. J. Biol. Chem. 282, 15799–15811 [DOI] [PubMed] [Google Scholar]
- 66. Cho W., Taylor L. P., Akil H. (1996) Mutagenesis of residues adjacent to transmembrane prolines alters D1 dopamine receptor binding and signal transduction. Mol. Pharmacol. 50, 1338–1345 [PubMed] [Google Scholar]
- 67. Marie J., Koch C., Pruneau D., Paquet J. L., Groblewski T., Larguier R., Lombard C., Deslauriers B., Maigret B., Bonnafous J. C. (1999) Constitutive activation of the human bradykinin B2 receptor induced by mutations in transmembrane helices III and VI. Mol. Pharmacol. 55, 92–101 [DOI] [PubMed] [Google Scholar]
- 68. Dragic T., Trkola A., Thompson D. A., Cormier E. G., Kajumo F. A., Maxwell E., Lin S. W., Ying W., Smith S. O., Sakmar T. P., Moore J. P. (2000) A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. U.S.A. 97, 5639–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tsamis F., Gavrilov S., Kajumo F., Seibert C., Kuhmann S., Ketas T., Trkola A., Palani A., Clader J. W., Tagat J. R. (2003) Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J. Virol. 77, 5201–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Baldwin J. M. (1993) The probable arrangement of the helices in G protein-coupled receptors. EMBO J. 12, 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schwartz T. W. (1994) Locating ligand-binding sites in 7TM receptors by protein engineering. Curr. Opin. Biotechnol. 5, 434–444 [DOI] [PubMed] [Google Scholar]
- 72. Ballesteros J. A., Weinstein H. (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 [Google Scholar]