Abstract
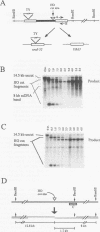
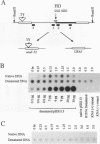
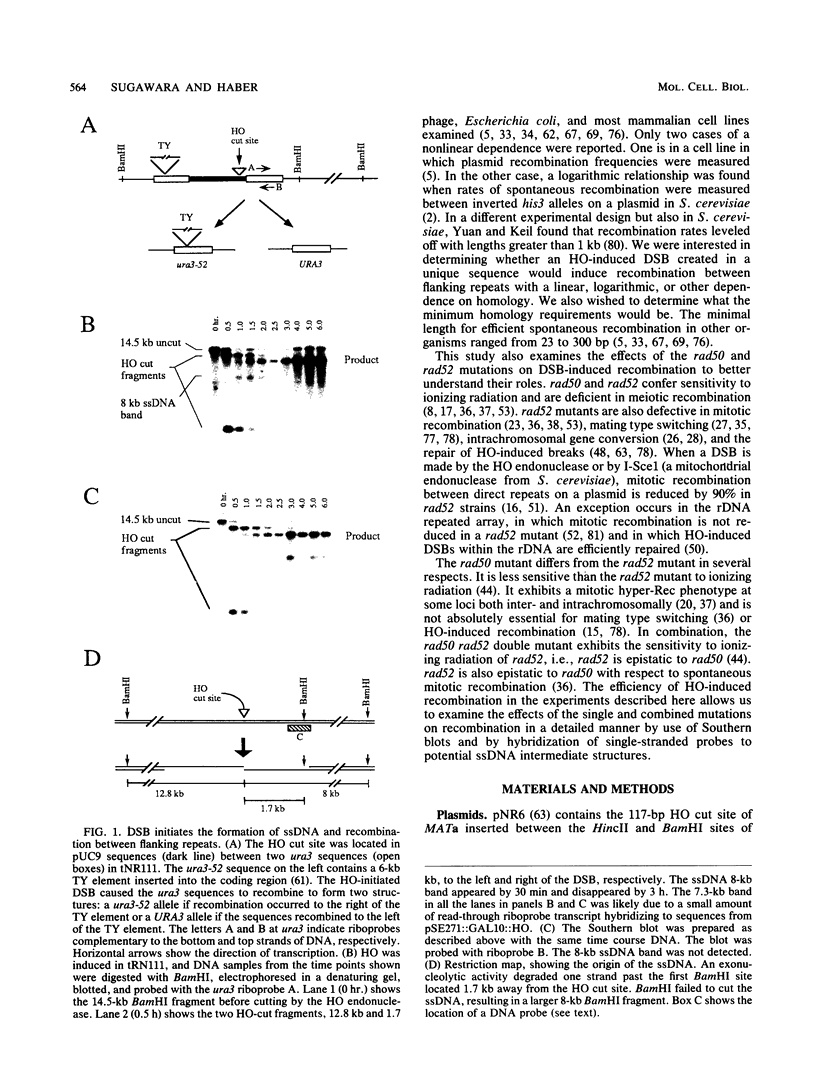
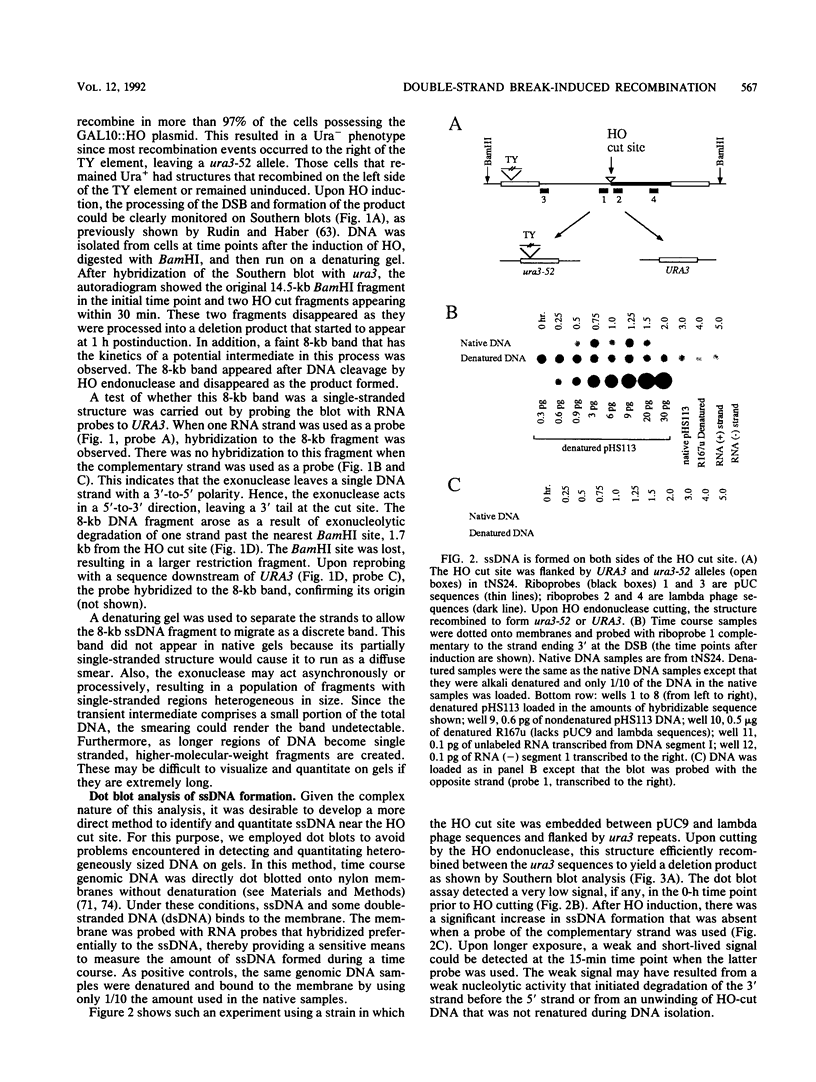
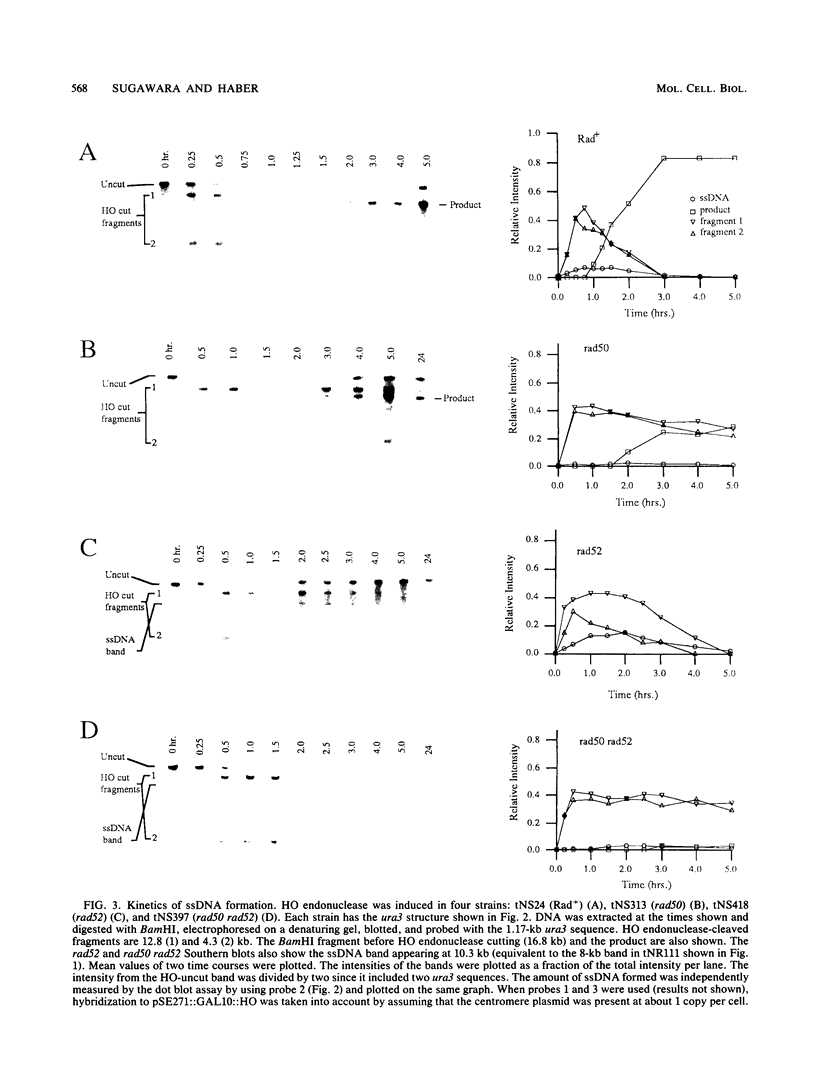
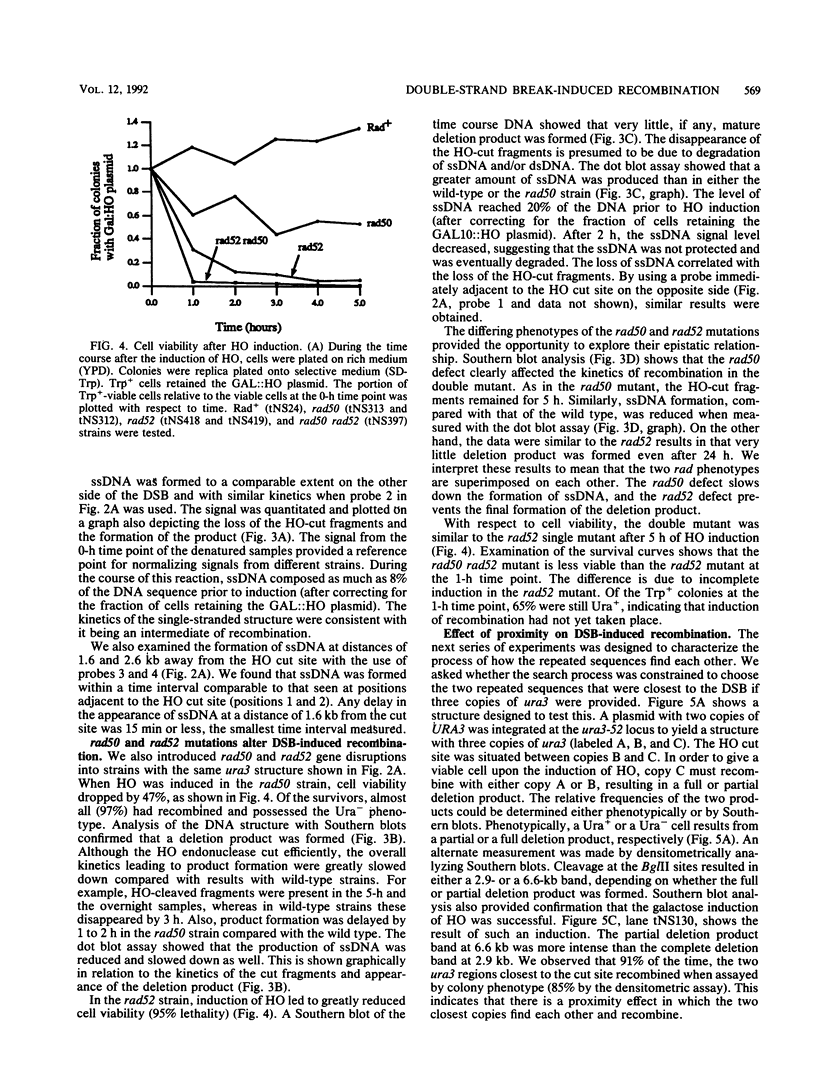
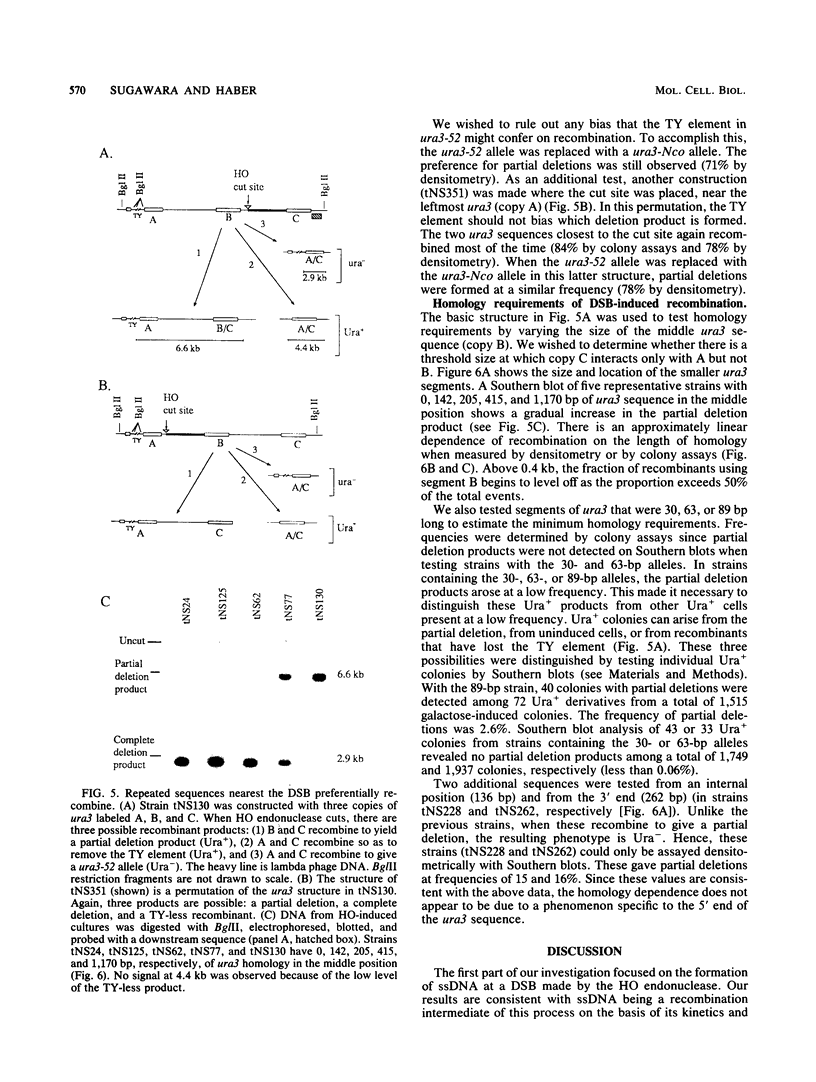
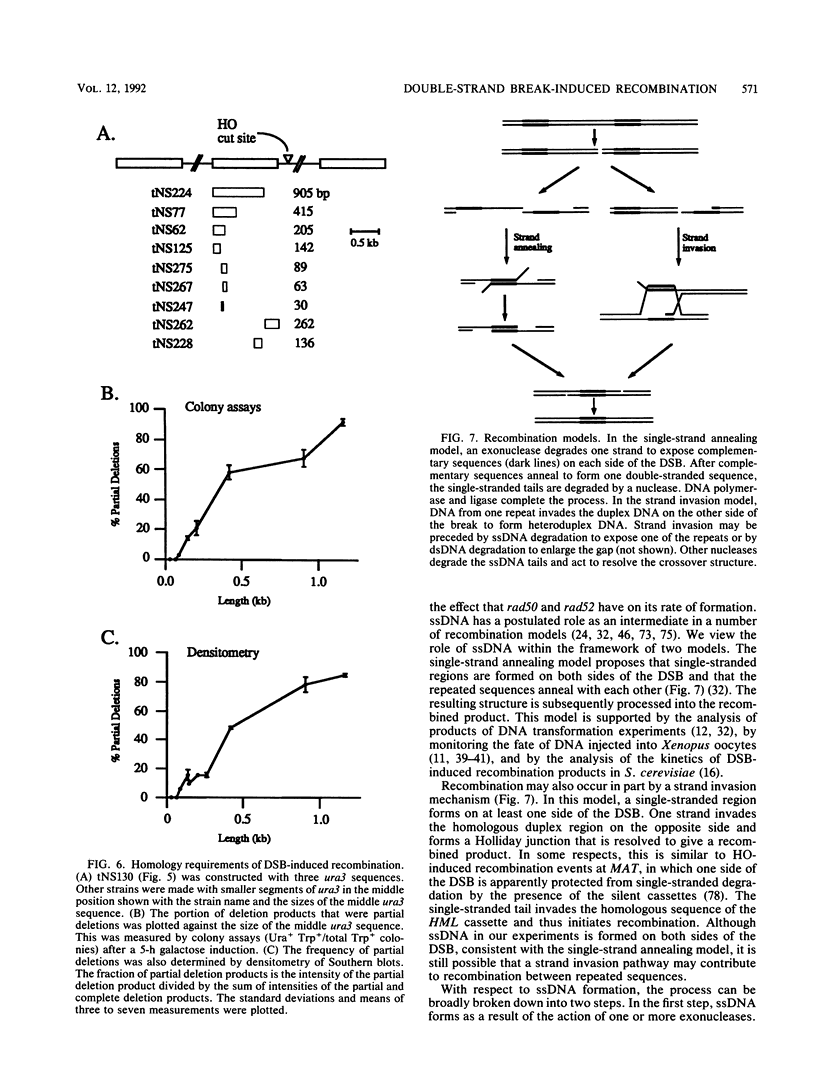
In the yeast Saccharomyces cerevisiae, a double-strand chromosome break created by the HO endonuclease is frequently repaired in mitotically growing cells by recombination between flanking homologous regions, producing a deletion. We showed that single-stranded regions were formed on both sides of the double-strand break prior to the formation of the product. The kinetics of the single-stranded DNA were monitored in strains with the recombination-deficient mutations rad52 and rad50 as well as in the wild-type strain. In rad50 mutants, single-stranded DNA was generated at a slower rate than in the wild type, whereas rad52 mutants generated single-stranded DNA at a faster rate. Product formation was largely blocked in the rad52 mutant. In the rad50 rad52 double mutant, the effects were superimposed in that the exonucleolytic activity was slowed but product formation was blocked. rad50 appears to act before or at the same stage as rad52. We constructed strains containing two ura3 segments on one side of the HO cut site and one ura3 region on the other side to characterize how flanking repeats find each other. Deletions formed preterentially between the homologous regions closest to the double-strand break. By varying the size of the middle ura3 segment, we determined that recombination initiated by a double-strand break requires a minimum homologous length between 63 and 89 bp. In these competition experiments, the frequency of recombination was dependent on the length of homology in an approximately linear manner.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adzuma K., Ogawa T., Ogawa H. Primary structure of the RAD52 gene in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2735–2744. doi: 10.1128/mcb.4.12.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn B. Y., Dornfeld K. J., Fagrelius T. J., Livingston D. M. Effect of limited homology on gene conversion in a Saccharomyces cerevisiae plasmid recombination system. Mol Cell Biol. 1988 Jun;8(6):2442–2448. doi: 10.1128/mcb.8.6.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E., Cao L., Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987 Aug;116(4):541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E., Padmore R., Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990 May 4;61(3):419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Ayares D., Chekuri L., Song K. Y., Kucherlapati R. Sequence homology requirements for intermolecular recombination in mammalian cells. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5199–5203. doi: 10.1073/pnas.83.14.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M., DasGupta C., Radding C. M. Synapsis and the formation of paranemic joints by E. coli RecA protein. Cell. 1983 Oct;34(3):931–939. doi: 10.1016/0092-8674(83)90550-0. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Borts R. H., Lichten M., Haber J. E. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics. 1986 Jul;113(3):551–567. doi: 10.1093/genetics/113.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M., Mortimer R. K. Repair of double-strand breaks in a temperature conditional radiation-sensitive mutant of Saccharomyces cerevisiae. Mutat Res. 1982 Jan;103(1):19–24. doi: 10.1016/0165-7992(82)90080-x. [DOI] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990 Jun 15;61(6):1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S., Seidman M. M. Intramolecular recombination between transfected repeated sequences in mammalian cells is nonconservative. Mol Cell Biol. 1986 Jul;6(7):2520–2526. doi: 10.1128/mcb.6.7.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Game J. C., Sitney K. C., Cook V. E., Mortimer R. K. Use of a ring chromosome and pulsed-field gels to study interhomolog recombination, double-strand DNA breaks and sister-chromatid exchange in yeast. Genetics. 1989 Dec;123(4):695–713. doi: 10.1093/genetics/123.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda D. K., Radding C. M. By searching processively RecA protein pairs DNA molecules that share a limited stretch of homology. Cell. 1983 Sep;34(2):647–654. doi: 10.1016/0092-8674(83)90397-5. [DOI] [PubMed] [Google Scholar]
- Gottlieb S., Wagstaff J., Esposito R. E. Evidence for two pathways of meiotic intrachromosomal recombination in yeast. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7072–7076. doi: 10.1073/pnas.86.18.7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., Thorburn P. C. Healing of broken linear dicentric chromosomes in yeast. Genetics. 1984 Feb;106(2):207–226. doi: 10.1093/genetics/106.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A., Bloom K. Acquisition and processing of a conditional dicentric chromosome in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Mar;9(3):1368–1370. doi: 10.1128/mcb.9.3.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra M. F., Naughton T., Malone R. E. Properties of spontaneous mitotic recombination occurring in the presence of the rad52-1 mutation of Saccharomyces cerevisiae. Genet Res. 1986 Aug;48(1):9–17. doi: 10.1017/s0016672300024599. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. A., Fink G. R. Gene conversion between duplicated genetic elements in yeast. Nature. 1981 Jul 23;292(5821):306–311. doi: 10.1038/292306a0. [DOI] [PubMed] [Google Scholar]
- Klar A. J., Strathern J. N., Abraham J. A. Involvement of double-strand chromosomal breaks for mating-type switching in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1984;49:77–88. doi: 10.1101/sqb.1984.049.01.011. [DOI] [PubMed] [Google Scholar]
- Klein H. L. Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics. 1988 Oct;120(2):367–377. doi: 10.1093/genetics/120.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin A. L., Klar A. J., Stahl F. W. Double-strand breaks can initiate meiotic recombination in S. cerevisiae. Cell. 1986 Aug 29;46(5):733–740. doi: 10.1016/0092-8674(86)90349-1. [DOI] [PubMed] [Google Scholar]
- Kostriken R., Heffron F. The product of the HO gene is a nuclease: purification and characterization of the enzyme. Cold Spring Harb Symp Quant Biol. 1984;49:89–96. doi: 10.1101/sqb.1984.049.01.012. [DOI] [PubMed] [Google Scholar]
- Kostriken R., Strathern J. N., Klar A. J., Hicks J. B., Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983 Nov;35(1):167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay R. M., Letsou A., Stachelek J. L. Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics. 1987 Jan;115(1):161–167. doi: 10.1093/genetics/115.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay R. M., Stachelek J. L., Letsou A. Homologous recombination between repeated chromosomal sequences in mouse cells. Cold Spring Harb Symp Quant Biol. 1984;49:183–189. doi: 10.1101/sqb.1984.049.01.021. [DOI] [PubMed] [Google Scholar]
- Malone R. E., Esposito R. E. Recombinationless meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Oct;1(10):891–901. doi: 10.1128/mcb.1.10.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone R. E., Esposito R. E. The RAD52 gene is required for homothallic interconversion of mating types and spontaneous mitotic recombination in yeast. Proc Natl Acad Sci U S A. 1980 Jan;77(1):503–507. doi: 10.1073/pnas.77.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone R. E., Montelone B. A., Edwards C., Carney K., Hoekstra M. F. A reexamination of the role of the RAD52 gene in spontaneous mitotic recombination. Curr Genet. 1988 Sep;14(3):211–223. doi: 10.1007/BF00376741. [DOI] [PubMed] [Google Scholar]
- Maryon E., Carroll D. Characterization of recombination intermediates from DNA injected into Xenopus laevis oocytes: evidence for a nonconservative mechanism of homologous recombination. Mol Cell Biol. 1991 Jun;11(6):3278–3287. doi: 10.1128/mcb.11.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryon E., Carroll D. Degradation of linear DNA by a strand-specific exonuclease activity in Xenopus laevis oocytes. Mol Cell Biol. 1989 Nov;9(11):4862–4871. doi: 10.1128/mcb.9.11.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryon E., Carroll D. Involvement of single-stranded tails in homologous recombination of DNA injected into Xenopus laevis oocyte nuclei. Mol Cell Biol. 1991 Jun;11(6):3268–3277. doi: 10.1128/mcb.11.6.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- McGill C., Shafer B., Strathern J. Coconversion of flanking sequences with homothallic switching. Cell. 1989 May 5;57(3):459–467. doi: 10.1016/0092-8674(89)90921-5. [DOI] [PubMed] [Google Scholar]
- McKee R. H., Lawrence C. W. Genetic analysis of gamma-ray mutagenesis in yeast. III. Double-mutant strains. Mutat Res. 1980 Mar;70(1):37–48. doi: 10.1016/0027-5107(80)90056-1. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff J. A., Chen E. Y., Heffron F. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7831–7835. doi: 10.1073/pnas.83.20.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff J. A., Singer J. D., Hoekstra M. F., Heffron F. Double-strand breaks stimulate alternative mechanisms of recombination repair. J Mol Biol. 1989 Jun 5;207(3):527–541. doi: 10.1016/0022-2836(89)90462-2. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozenberger B. A., Roeder G. S. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol Cell Biol. 1991 Mar;11(3):1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Prakash L., Burke W., Montelone B. A. Effects of the RAD52 Gene on Recombination in SACCHAROMYCES CEREVISIAE. Genetics. 1980 Jan;94(1):31–50. doi: 10.1093/genetics/94.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh D., Hughes S. H., Shafer B. K., Strathern J. N. Analysis of the HO-cleaved MAT DNA intermediate generated during the mating type switch in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1989 Dec;220(1):33–42. [PubMed] [Google Scholar]
- Ray A., Machin N., Stahl F. W. A DNA double chain break stimulates triparental recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6225–6229. doi: 10.1073/pnas.86.16.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Siddiqi I., Kolodkin A. L., Stahl F. W. Intra-chromosomal gene conversion induced by a DNA double-strand break in Saccharomyces cerevisiae. J Mol Biol. 1988 May 20;201(2):247–260. doi: 10.1016/0022-2836(88)90136-2. [DOI] [PubMed] [Google Scholar]
- Resnick M. A., Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976 Jan 16;143(2):119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- Resnick M. A. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976 Jun;59(1):97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Rose M., Grisafi P., Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- Rose M., Winston F. Identification of a Ty insertion within the coding sequence of the S. cerevisiae URA3 gene. Mol Gen Genet. 1984;193(3):557–560. doi: 10.1007/BF00382100. [DOI] [PubMed] [Google Scholar]
- Rubnitz J., Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984 Nov;4(11):2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin N., Haber J. E. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol Cell Biol. 1988 Sep;8(9):3918–3928. doi: 10.1128/mcb.8.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin N., Sugarman E., Haber J. E. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989 Jul;122(3):519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P., Huang H. V. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986 Mar;112(3):441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. S., Gold L., Gauss P., Doherty D. H. Determination of the amount of homology required for recombination in bacteriophage T4. Cell. 1982 Nov;31(1):25–33. doi: 10.1016/0092-8674(82)90401-9. [DOI] [PubMed] [Google Scholar]
- Strathern J. N., Klar A. J., Hicks J. B., Abraham J. A., Ivy J. M., Nasmyth K. A., McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982 Nov;31(1):183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Schultes N. P., Szostak J. W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989 Mar 2;338(6210):87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Szostak J. W. Extensive 3'-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991 Mar 22;64(6):1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Viret J. F., Alonso J. C. Generation of linear multigenome-length plasmid molecules in Bacillus subtilis. Nucleic Acids Res. 1987 Aug 25;15(16):6349–6367. doi: 10.1093/nar/15.16.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Vernaleone F., Wilson J. H. Topological requirements for homologous recombination among DNA molecules transfected into mammalian cells. Mol Cell Biol. 1985 Aug;5(8):2080–2089. doi: 10.1128/mcb.5.8.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt V. M., Ingles C. J., Urdea M. S., Rutter W. J. Homology requirements for recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4768–4772. doi: 10.1073/pnas.82.14.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiffenbach B., Haber J. E. Homothallic mating type switching generates lethal chromosome breaks in rad52 strains of Saccharomyces cerevisiae. Mol Cell Biol. 1981 Jun;1(6):522–534. doi: 10.1128/mcb.1.6.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. I., Haber J. E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990 Mar;9(3):663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L. W., Keil R. L. Distance-independence of mitotic intrachromosomal recombination in Saccharomyces cerevisiae. Genetics. 1990 Feb;124(2):263–273. doi: 10.1093/genetics/124.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]