Background: Protein inhibitors of activated STAT (Pias) proteins can modulate the activity of transcription factors.
Results: Pias1 and Piasy enhance or repress pituitary homeobox 2 (PITX2) transcriptional activity.
Conclusion: Pias interactions regulate PITX2 transcriptional activity.
Significance: Pias proteins can differentially regulate PITX2 activity.
Keywords: Gene Regulation, Protein-Protein Interactions, Sumoylation, Transcription Coactivators, Transcription Regulation, BiFC, Lef-1, PIAS, PITX2, β-Catenin
Abstract
Protein inhibitors of activated STAT (Pias) proteins can act independent of sumoylation to modulate the activity of transcription factors and Pias proteins interacting with transcription factors can either activate or repress their activity. Pias proteins are expressed in many tissues and cells during development and we asked if Pias proteins regulated the pituitary homeobox 2 (PITX2) homeodomain protein, which modulates developmental gene expression. Piasy and Pias1 proteins are expressed during craniofacial/tooth development and directly interact and differentially regulate PITX2 transcriptional activity. Piasy and Pias1 are co-expressed in craniofacial tissues with PITX2. Yeast two-hybrid, co-immunoprecipitation and pulldown experiments demonstrate Piasy and Pias1 interactions with the PITX2 protein. Piasy interacts with the PITX2 C-terminal tail to attenuate its transcriptional activity. In contrast, Pias1 interacts with the PITX2 C-terminal tail to increase PITX2 transcriptional activity. The E3 ligase activity associated with the RING domain in Piasy is not required for the attenuation of PITX2 activity, however, the RING domain of Pias1 is required for enhanced PITX2 transcriptional activity. Bimolecular fluorescence complementation assays reveal PITX2 interactions with Piasy and Pias1 in the nucleus. Piasy represses the synergistic activation of PITX2 with interacting co-factors and Piasy represses Pias1 activation of PITX2 transcriptional activity. In contrast, Pias1 did not affect the synergistic interaction of PITX2 with transcriptional co-factors. Last, we demonstrate that Pias proteins form a complex with PITX2 and Lef-1, and PITX2 and β-catenin. Lef-1, β-catenin, and Pias interactions with PITX2 provide new molecular mechanisms for the regulation of PITX2 transcriptional activity and the activity of Pias proteins.
Introduction
Pias (protein inhibitor of activated STAT)3 proteins are a family of transcriptional regulators that possess SUMO E3 ligase activity (1–4). There are five members in the Pias superfamily, Pias1, Pias3, Piasxa, Piasxb, and Piasy, all of which possess E3 ligase activity linked to their conserved RING finger domains (5–9). However, Pias proteins differ in their regulation of a variety of transcription factors through distinct mechanisms. Pias proteins interacting with transcriptional factors can either activate or repress their transcriptional activity (10–12). Pias proteins may also influence the activity of targets via both sumoylation-dependent and sumoylation-independent mechanisms (6, 9, 13–16). These activities may be dependent upon the functional domains of the Pias proteins.
The members of the mammalian Pias family have several functional domains and motifs that are shared and an overall significant sequence identity (∼40%) between the family members. A SAP domain (scaffold-attachment factor A (SAF), acinus and Pias) is present in many chromatin-binding proteins. The SAP domain of Piasy and Pias1 can bind nonspecific A + T-rich DNA sequences (7). Therefore, the SAP domain may target these Pias proteins to chromatin. A PINIT amino acid motif found in both Piasy and Pias1 may be involved in nuclear retention (17). A conserved RING finger-like zinc-binding domain is observed in all Pias proteins and is required for the SUMO-E3-ligase activity (8). Pias1 contains a carboxyl (C)-terminal acidic domain (AD) and a serine and threonine-rich (S/T) region. The AD region also contains a SUMO1-interaction motif (18). However, Piasy contains only the AD domain without the associated SUMO1-interaction motif, the S/T domain. All of these domains are involved in protein-protein interactions and may confer different transcriptional activities to specific factors.
PITX2 (pituitary homobox 2), a paired-like homeobox transcription factor is the earliest transcription marker of tooth development and is also expressed in the brain, heart, pituitary, eye, and umbilicus (19–21). PITX2A, PITX2B, and PITX2C are the major PITX2 isoforms and differ in the amino terminus and all regulate transcription (22). PITX2A and PITX2C are widely expressed in vertebrate tissues and have similar activities. PITX2 is a homeodomain transcriptional activator and its activity can be modulated through protein-protein interactions and phosphorylation (23–27). The PITX2 C-terminal tail and homeodomain (HD) regions have been identified as sites for protein-protein interactions. However, exploring the molecular mechanisms and identifying new regulators and protein interactions of PITX2 are very important to better understand the fundamental roles of PITX2 during embryogenesis.
In a search to understand how factors regulate PITX2 transcriptional activity we employed yeast two-hybrid analysis to identify PITX2 interacting factors. We performed functional analyses to determine the effect of Pias proteins on PITX2 transcriptional activity. We demonstrate that Piasy and Pias1 physically interact with PITX2 to regulate PITX2 transcriptional activity. Piasy interacts with the PITX2 C-terminal tail and represses PITX2 transcriptional activity. Interestingly, Pias1 interacts with the PITX2 C-terminal tail and synergistically increases PITX2 transcriptional activity. Bimolecular fluorescence complementation assays reveal direct PITX2 interactions with Piasy and Pias1 in the nucleus of living cells. Piasy represses the activation of PITX2 transcriptional activity with known co-factors, whereas Pias1 does not affect these interactions. We demonstrate that PITX2 forms a multiple protein complex with Pias proteins and co-factors to regulate PITX2 transcriptional activity. This is the first report demonstrating independent binding of two Pias proteins to one transcription factor with each causing an opposite effect. Therefore, the tissue-specific expression of Pias proteins may control the activation or repression of PITX2.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screen
Several PITX2 constructs containing the full-length protein, homeodomain, and the C-terminal tail (C-HD) were cloned in the Gal4 DBD fusion vector (pGBKT7, BD-Clontech) as bait and transformed into yeast MATα strain AH109. A commercially available mouse E17 cDNA library in Gal4 AD fusion vector (pACT2, BD Biosciences-Clontech) and pre-transformed into yeast MATa strain Y187 (BD Biosciences-Clontech) was used to screen for interactors. pGBKT7 carries a gene for leucine (Leu), whereas pACT2 can encode tryptophan (Trp). The MATa reporter strain AH109 was used as a mating partner for the MATa Y187 strain. The AH109 strain contains the ADE2, HIS3, and lacZ markers, which are under the control of different GAL4 upstream activating sequences and TATA boxes. A large-scale mating was carried out according to the protocol provided with the Clontech MatchMaker3 yeast two-hybrid system between Y187 cells carrying the pre-transformed prey cDNA library and AH109 cells expressing the PITX2 C-HD bait. The mated yeast were plated onto 150-mm plates containing SD/-Leu/-Trp/-His and grown for 1 week. The resulting colonies were then replica plated onto plates containing SD/-Leu/-Trp/-His/-Ade/X-α-Gal and grown for another week. The 400 colonies showing the strongest blue color were selected visually. These 400 colonies were then grown on SD/-Leu/-Trp/-His/-Ade plates containing 10 mm 3-AT to select for the strongest interactors. The plasmid DNA from the mated yeast was obtained using standard protocols. This DNA containing both bait (Kan+) and prey (Amp+) plasmids was transformed into DH5α competent cells (Invitrogen) and grown on LB plates containing 100 μg/ml of ampicillin to select for prey vector. Once isolated, the vector was sequenced in both directions, checked for open reading frame maintenance, and the candidate identified through BLAST (NCBI) searches.
Individual colonies were taken from the mated yeast, grown in 2 ml of Leu-Trp media overnight, and transferred to 10 ml of Leu-Trp media to an A600 of 0.4–0.6. The yeast were collected by centrifugation and resuspended in 1 ml of buffer Z (BD Biosciences-Clontech). 100 μl of the resuspended yeast were lysed by freezing in liquid nitrogen (30–60 s) followed by thawing at 37 °C (30–60s) four times. The β-galactosidase activity was assayed by measuring chemiluminescence from the lysate (25 μl) in a 96-well plate luminometer (Dynex) using the Galacto-Light Plus kit (Tropix).
Expression and Reporter Constructs
Expression plasmids containing the cytomegalovirus (CMV) promoter linked to the PITX2 cDNA were constructed in pcDNA 3.1 MycHisC or pFLAG-CMV (Invitrogen) (24). The human LEF-1 promoter has been previously described and was PCR amplified and cloned into the luciferase vector (28). Mammalian expression plasmids for FLAG-Sumo-1, Sumo2, Ubc9, and HA-Pias1 and its Ring domain mutant were described (29, 30). The Piasy cDNA was cloned by PCR and inserted into the pcDNA3.1 MycHisC vector. The ΔPiasy clone was made using PCR primers that deleted the C-terminal region of PIASy. shRNA clones were constructed in the pLL3.7 vector containing the U6 promoter and the following short hairpin sequences targeting the Pias1 and Piasy transcripts. For Pias1 two shRNA sequences were used: sh1, forward, 5′-TGGATCATTCTAGAGCTTTATTCAAGAGATAAAGCTCTAGAATGATCCTTTTTTC-3′and reverse, 5′-TCGAGAAAAAAGGATCATTCTAGAGCTTTATCTCTTGAATAAAGCTCTAGAATGATCCA-3′; sh2, forward, 5′-TGTGACGAGATACAGTTTAATTCAAGAGATTAAACTGTATCTCGTCACTTTTTTC-3′ and reverse, 5′-TCGAGAAAAAAGTGACGAGATACAGTTTAATCTCTTGAATTAAACTGTATCTCGTCACA-3′. shPiasy was constructed using: forward, 5′-TGCTACTCTGTGGCTTTGTATTCAAGAGATACAAAGCCACAGAGTAGCTTTTTTC-3′ and reverse, 5′-TCGAGAAAAAAGCTACTCTGTGGCTTTGTATCTCTTGAATACAAAGCCACAGAGTAGCA-3′ primers. shRNA controls included scrambled sequences and shRNAs to other factors that had no adverse or off target effects on PIAS function. All constructs were confirmed by DNA sequencing. A simian virus 40 (SV-40) or CMV β-galactosidase reporter plasmid was cotransfected in all experiments as a control for transfection efficiency. All plasmids were double-banded CsCl purified.
Cell Culture, Transient Transfections, Luciferase, and β-Galactosidase Assays
CHO and LS-8 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 5 or 10% fetal bovine serum (FBS), penicillin/streptomycin, and transfected by electroporation. The cultures were fed 24 h prior to transfection, resuspended in PBS, and mixed with 2.5 μg of expression plasmids, 5 μg of reporter plasmid, and 0.25 μg of CMV or SV-40 β-galactosidase plasmid. Electroporation of CHO cells were performed at 360 V and 950 microfarads (Gene Pulser XL; Bio-Rad). LS-8 cells were transfected by electroporation as previously described (31). The transfected cells were incubated for 24 h in 60-mm culture dishes and fed 5 or 10% FBS and Dulbecco's modified Eagle's medium and then lysed and assayed for reporter activities, protein content by Bradford assay (Bio-Rad), Western blotting assay, and isolated total RNAs for real-time PCR assay. Luciferase was measured using reagents from Promega. β-Galactosidase was measured using the Galacto-Light Plus reagents (Tropix Inc.). All luciferase activities were normalized to β-galactosidase activity.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assays were performed as previously described using the ChIP assay kit (Upstate Biotechnology) with the following modifications (32, 33). LS-8 cells were fed for 24 h, harvested, and plated in 60-mm dishes. Cells were cross-linked with 1% formaldehyde for 10 min at 37 °C the next day. All PCR were done under an annealing temperature of 65 °C. The sense primer (5′-TCAGTCATCCCGAAGAGGAG-3′) and the antisense primer (5′-GAGGGACCAGGGCTCAAT-3′) were used to amplify the Lef-1 promoter containing the Pitx2 binding site. All PCR products were evaluated on a 2% agarose gel in 1× Tris borate/EDTA for appropriate size (193 bp) and confirmed by sequencing. As controls the primers were used in PCRs without chromatin; normal rabbit IgG was used, replacing the Pias1 or Piasy antibody to reveal nonspecific immunoprecipitation of the chromatin. Primers 4.7 kbp upstream (sense, 5′-GACACCACTTCCTGGGGTTA-3′; antisense, 5′-CGCAGTGTGCTGTCTTTAGC-3′) of the Pitx2 binding site were used as controls. Quantification of the Pias1/Piasy binding DNA fragment were performed using the same primers and condition as above. The same quantities of pulldown DNA were used in real-time PCR. Each reaction was done in triplicate and S.E. were calculated from 5 independent experiments.
Western Blot Assays
Expression of endogenous or transiently expressed PITX2 and Pias proteins were demonstrated using the PITX2 P2R10 antibody (21) and Pias antibodies (Santa Cruz Biotechnology). The Pias1 rabbit polyclonal antibody from Santa Cruz Biotechnology was raised against amino acids 471–645 mapping near the C terminus of Pias1 of human origin and recognizes the 78-kDa protein. The Piasy rabbit polyclonal antibody from Santa Cruz Biotechnology was raised against amino acids 61–130 mapping within an internal region of Piasy of mouse origin and recognizes the 57-kDa protein. Expression of transiently expressed and endogenous Lef-1 protein was demonstrated using the Lef-1 (C18A7) rabbit polyclonal antibody (Cell Signaling). Approximately 10 to 40 μg of transfected or nontransfected cell lysates were analyzed in Western blots. Following SDS-gel electrophoresis, the proteins were transferred to polyvinylidene difluoride (PVDF) filters (Millipore), immunoblotted, and detected using specific antibodies and enhanced chemiluminescence (ECL) reagents from GE HealthCare.
Expression and Purification of PIAS Proteins
Mammalian expression plasmids for Pias1, Piasy, and mutant constructs were previously described (29). The bacterial Pias plasmids were constructed and transformed into BL21 cells. Protein was isolated as described (34). Pias proteins were cleaved from the GST moiety using 80 units of PreScission Protease (Pharmacia Biotech) per ml of glutathione-Sepharose. Purified proteins used in the pulldown assays have been previously described (24). The cleaved proteins were analyzed on SDS-polyacrylamide gels by silver stain or Coomassie Blue stain and quantitated by the Bradford protein assay (Bio-Rad). All stained gels were directly quantitated using image analysis programs.
GST-PITX2 Pulldown Assays
Immobilized GST-PITX2 full-length and truncated fusion proteins were prepared as described above and suspended in binding buffer (20 mm HEPES, pH 7.5, 5% glycerol, 50 mm NaCl, 1 mm EDTA, 1 mm DTT, with 1% milk and 400 μg/ml of ethidium bromide). Bacterially overexpressed and purified Pias1, Piasy, β-catenin, or Lef-1 protein (75–400 ng) were added to 45 μg of immobilized GST-PITX2, GST-PITX2 fusion proteins, or GST, respectively, in a total volume of 150 μl, and incubated for 30 min at 4 °C. The beads were pelleted and washed three times with 300 μl of binding buffer. The bound proteins were eluted by boiling in SDS-sample buffer and separated on a 10% SDS-polyacrylamide gel. The experiment to analyze co-binding of Pias1, Piasy, β-catenin, and Lef-1 to PITX2 was performed by adding purified Pias1, Piasy, β-catenin, and Lef-1 to the GST-PITX2 construct. After incubation and extensive washing the beads were divided into 3 equal aliquots, run on an SDS gel, and probed with Pias1, Piasy, β-catenin, or Lef-1 antibodies. Thus, Pias1, β-catenin, and Lef-1 or Piasy were present in the binding reactions. Purified Pias1, Piasy, β-catenin, or Lef-1 proteins were analyzed in the Western blot. Following SDS-gel electrophoresis, the proteins were transferred to PVDF filters (Millipore), immunoblotted, and detected using the Pias1 (Santa Cruz), Piasy (Santa Cruz), β-catenin (Cell Signaling), or Lef-1 (Cell Signaling) antibodies and ECL reagents from GE HealthCare.
Immunoprecipitation Assay
Approximately 24 h after cell transfection with PITX2 and Pias1 or Piasy, CHO cells were rinsed with 1 ml of PBS, then incubated with 1 ml of ice-cold RIPA buffer for 15 min at 4 °C. Cells were harvested and disrupted by repeated aspiration through a 25-gauge needle attached to a 1-ml syringe. The lysates were then incubated on ice for 30 min. Cellular debris was pelleted by centrifugation at 10,000 × g for 10 min at 4 °C. An aliquot of lysate was saved for analysis as input control. Supernatant was transferred to a fresh 1.5-ml microfuge tube on ice and precleared using the mouse ExactaCruz F IP matrix (ExactaCruz F, Santa Cruz Biotechnology) for 30 min at 4 °C. Matrix was removed by brief centrifugation and the supernatant was transferred to a new tube. An IP antibody·IP matrix complex was prepared as per the manufacturer's instructions using primary anti-Pias1 or anti-Piasy antibody (Santa Cruz Biotechnology). The IP antibody·IP matrix complex was incubated with the pre-cleared cell lysate at 4 °C for 12 h. After incubation, the lysate was centrifuged to pellet the IP matrix. The matrix was washed two times with PBS and resuspended in 15 μl of ddH2O and 5 μl of 5× SDS loading dye. Samples were boiled for 5 min and resolved on a 10% polyacylamide gel. Western blotting was used with anti-PITX2 antibody and HRP-conjugated rabbit ExactaCruz C reagent to detect immunoprecipitated proteins.
In Vivo Sumoylation and Ubiquitination Assays
Wild type Nkx2.5, PITX2A, PITX2C, and mutant PITX2C were transfected into HeLa cells with FLAG-tagged SUMO-1 wild type (WT) or its mutant FLAG-Sumo-1-DGG. To determine whether Pias1 potentiated the sumoylation of PITX2 in vivo, the transfection assays were also done in the presence of Pias1 or its RING finger mutant expression vectors. The procedure for protein blotting and visualization was detailed previously (15, 16).
Immunofluorescent and Immunohistochemical Staining
LS-8 oral epithelial cells (35) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and penicillin/streptomycin seeded on 18 × 18-mm microscope glass coverslips in 60-mm dishes. Immunofluorescent double staining was performed as previously described (27). Fixed cells were washed with PBS for 2 × 5 min, incubated in 5% normal goat serum-PBS for 30 min at room temperature (RT), then incubated cells with PIAS1 primary antibodies at 4 °C overnight. After overnight incubation, cells were rinsed by washing in PBS for 3 × 5 min and cells were incubated with secondary antibodies, Alexa Fluor 555 or 488 goat anti-rabbit IgG and Alexa Fluor® 488 or 555 goat anti-mouse IgG (H+L) (Molecular Probes, Invitrogen), 30 min at room temperature or 37 °C. Cells were washed with PBS for 3 × 5 min. The cells were mounted with VECTASHIELD® Mounting Medium with DAPI (Vector Laboratories, Inc., Burlingame, CA) and images were acquired using an Olympus Fluoview FV1000 confocal microscope and FV10-ASW1.7 software.
Bimolecular Fluorescence Complementation Assay (BiFC)
BiFC assay was performed as previously described (36, 37). EYFP N-terminal fragment (amino acids 1–155) was inserted between the HindIII and NotI sites in the pFLAG-CMV-2 plasmid (Sigma). PITX2C cDNA was then inserted in the modified plasmid between the XbaI and BamHI sites to build the YN-PITX2C construct. The EYFP C-terminal fragment (amino acids 156–240, with stop code) was inserted between the XbaI and BamHI sites in the pFlAG-CMV-2 plasmid to build the FLAG-YC construct. Pias1 or Piasy cDNA was then inserted between HindIII and NotI sites in the FLAG-YC plasmid to build Pias1-YC and Piasy-YC constructs. The EYFP N-terminal fragment (amino acids 1–155, with stop code) was inserted between the XbaI and BamHI sites in the pFLAG-CMV-2 plasmid to build the FLAG-YN construct. The constructs are shown in Fig. 2. The complementation of the YN and YC fusion proteins produce a fluorescence complex. The constructs were transfected into CHO cells and fluorescence complementation in living cells were observed after 24 h using a Zeiss LSM510 confocal microscope equipped with a ×63 plan-apochromat oil objective and low noise fluorescence imaging capability.
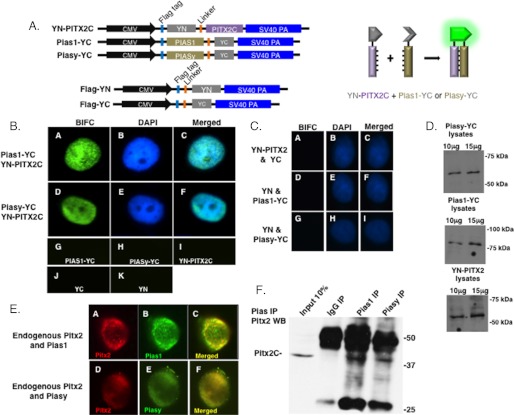
FIGURE 2.
Piasy and Pias1 co-localize with PITX2 in nuclear bodies. The BiFC assay was used to visualize protein interactions in living CHO cells. A, schematic of the constructs used in the BiFC assay. Piasy and Pias1 cDNA were cloned into the YC vector and PITX2C cDNA cloned into the YN vector. Controls included the YN and YC empty vectors. Schematic of the complementation that produces fluorescence YFP is also shown. B, the constructs were transfected into CHO cells. Fluorescence complementation in living cells was observed after 24 h using a Nikon Eclipse 80i microscope with low noise fluorescence imaging capability. C, additional controls for the BiFC experiments. D, Western blot showing expression of the constructs. E, immunofluorescence of endogenous Pitx2 and Pias proteins in Ls-8 cells. Pitx2 expression was detected with a Pitx2 antibody and visualized with a rodamine-conjugated secondary antibody. Pias1 was detected with a Pias1 antibody and visualized with a FITC-conjugated secondary antibody (A–C). Piasy was detected with a Piasy antibody and visualized with a FITC-conjugated secondary antibody (D–F). The images were merged to show co-localization. Fluorescence images were acquired using an Olympus Fluoview FV1000 confocal microscope and FV10-ASW1.7 software. F, endogenous immunoprecipitation of Pitx2·Pias1 and Pitx2·Piasy protein complexes from LS-8 cells. The complexes were IPed with Pias antibodies and the Pitx2 interaction shown by Western blot using a PITX2 antibody. Pitx2C was immunoprecipitated with Pias antibodies.
RT-PCR
Molar and incisor tooth germs were dissected from E16.5 mice with the use of a dissection microscope. Total RNAs were extracted from molar and incisor tooth germ using the miRNeasy Mini Kit from Qiagen (Valencia, CA), as per the manufacturer's instructions. RNA was eluted in 30 μl of nuclease-free water. The quantity of extracted RNA was evaluated spectrophotometrically and by gel analyses. To ensure the integrity of RNA, only samples with a 260/280 OD ratio of between 1.7 and 2.0 were used. Complementary DNA (cDNA) was created by adding 1 μg of RNA to 5× iScript select reaction and Oligo(dT) and reverse-transcribed using the iScriptTM Select cDNA Synthesis Kit (Bio-Rad) as per the manufacturer's instructions. The cDNA product was stored at −20 °C and for future use. The real-time PCR assay was performed in a 50-μl volume in a 96-well plate (iCycler iQTM) and iQ SYBR® Green Supermix (Bio-Rad) was used to detect amplification under the following amplification condition: pre-amplification cycle for 5 min at 95 °C, 40 amplification cycles for 30 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C. Pias1 primers were: sense, 5′-TGGCGGACAGTGCGGAACTA-3′ and antisense, 5′-AGACAAGTCCGCAGGCGTCA-3′; Piasy primers were: sense, 5′-ATGGCGGCAGAGCTGGTGGA-3′ and antisense, 5′-TCTTGGCATAGCGAGTCTCA-3′; Lef-1 primers were: sense, 5′-TCACTGTCAGGCGACACTTC-3′ and antisense, 5′-ATGAGGTCTTTTGGGCTCCT-3′, β-actin primers were: sense, 5′-GCCTTCCTTCTTGGGTATG-3′ and antisense, 5′-ACCACCAGACAGCACTGTG-3′.
RESULTS
PITX2 Physically Interacts with Piasy
The yeast two-hybrid assay identified Piasy protein interacting with PITX2 (data not shown). Co-immunoprecipitation and GST pulldown experiments were used to confirm the interaction between Pias proteins and PITX2A. Co-immunoprecipitation experiments detected an interaction of Piasy with PITX2A (Fig. 1B). CHO cells were co-transfected with PITX2A and Piasy and cell lysates were directly analyzed by Western blotting for PITX2A expression (Fig. 1B, lanes 2–5). The transfected PITX2A proteins migrate slightly slower due to a myc/His tag compared with the purified protein in lane 1. Immunoprecipitation (IP) experiments utilized the Piasy antibody to pulldown the PITX2A·Piasy complex and the PITX2 antibody (Western blot) to probe for PITX2A protein on the Western blot. Because CHO cells endogenously express Piasy, endogenous Piasy immunoprecipitated transfected PITX2A (Fig. 1B, lane 6). The PITX2A·Piasy complex formation was observed when both PITX2A and Piasy were co-transfected (Fig. 1B, lane 7). In Piasy-transfected cell lysates, the Piasy·PITX2A complex was not immunoprecipitated because CHO cells do not endogenously express PITX2A (Fig. 1B, lane 8).
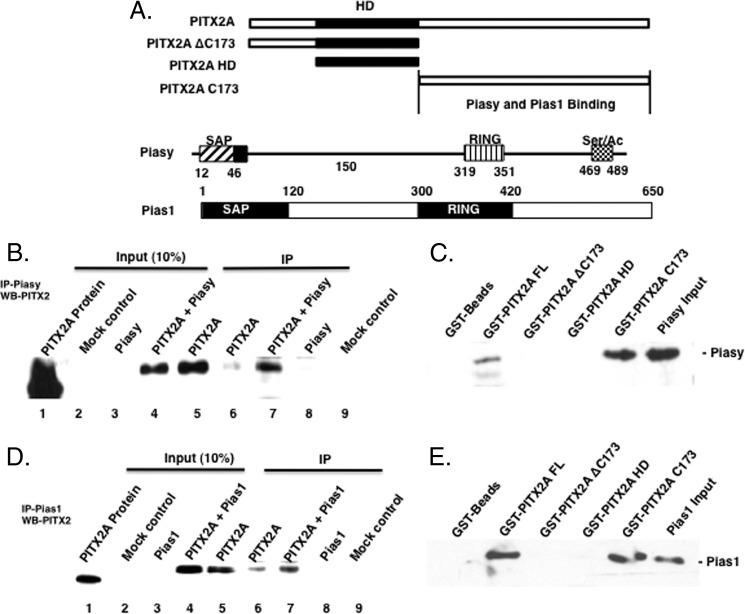
FIGURE 1.
Piasy and Pias1 interact with PITX2. A, schematic of PITX2A deletion proteins used to map the interaction with Piasy and Pias1; a schematic of the Pias proteins is shown. B, Piasy and/or PITX2A expression plasmids (5 μg) were transfected into CHO cells (2–60 mm dishes) and incubated for 24 h. Cells were harvested, lysates prepared, and the Piasy-PITX2A protein complex was immunoprecipitated using the Piasy antibody. The immunoprecipitated complexes were resolved on a 10% polyacrylamide gel and transferred to a PVDF filter, and Western blotting was done using a PITX2 antibody. Input controls are shown in lanes 2–5 and the IP PITX2 shown in lanes 6–9. C, bacteria expressed and purified Piasy (200 ng) was incubated with GST-PITX2A constructs (10 μg) as previously described (33). The bound Piasy protein was detected by Piasy antibody on Western blots. Piasy bound to the PITX2 full-length protein, and the PITX2 C-terminal tail (PITX2 C173). The C-terminal deletion (PITX2 ΔC173) and the PITX2 HD did not bind Piasy. D, Pias1 and/or PITX2A expression plasmids (5 μg) were transfected into CHO cells and the Pias1·PITX2A protein complex was immunoprecipitated using the Pias1 antibody. The immunoprecipitated complexes were resolved on a 10% polyacrylamide gel and transferred to a PVDF filter, and Western blotting was done using a PITX2 antibody. Input controls are shown in lanes 2–5 and the IP PITX2 shown in lanes 6–9. E, bacteria expressed and purified Pias1 (200 ng) was incubated with GST-PITX2A constructs (10 μg). Pias1 bound to the full-length PITX2A and the PITX2 C173 C-terminal tail but not to the PITX2-HD (homeodomain) or PITX2 ΔC173 protein.
To determine the region of PITX2 that interacts with Piasy, GST-PITX2, and GST-PITX2 truncated proteins were immobilized on Sepharose beads and incubated with purified Piasy expressed in bacteria (Fig. 1, A and C). Piasy bound to PITX2A full-length protein (FL), but not to GST beads (Fig. 1C). Piasy did not bind to PITX2 ΔC-173 (C-terminal deletion) or the PITX2 HD (homeodomain only) but did bind PITX2 Cys-173 (C terminus) (Fig. 1C). Thus, Piasy interacts with the PITX2 C-terminal tail a known protein interaction region (24).
PITX2 Physically Interacts with Pias1
Pias1 was co-immunoprecipitated with PITX2A in transfected CHO cells (Fig. 1D). IP experiments utilized the Pias1 antibody to immunopreciptiate the PITX2A·Pias1 complex and the PITX2 antibody to probe for PITX2A protein on the Western blot. The Pias1 antibody immunoprecipitated the PITX2A·Pias1 complex in PITX2A-transfected lysates (Fig. 1D, lane 6). The PITX2A·Pias1 complex was observed when both PITX2A and Pias1 were co-transfected (Fig. 1D, lane 7). In Pias1-transfected cell lysates, the PITX2·Pias1 complex was not immunoprecipitated because CHO cells do not endogenously express PITX2 (Fig. 1D, lane 8). Mock (empty vector) transfected lysates served as a control. The input controls demonstrate expression of PITX2A (Fig. 1D, lanes 4 and 5). PITX2A GST pulldown experiments with bacteria-purified Pias1 protein demonstrate Pias1 binding to the PITX2A C-terminal tail (Fig. 1E). Interestingly, the PITX2A C-terminal tail interacts with both Piasy and Pias1 proteins. We next asked if these interactions regulated PITX2 nuclear/cell localization as it has been shown that Pias proteins can sequester interacting factors to regions of the nucleus (7, 38–40).
Piasy and Pias1 Co-localize with PITX2 in the Nucleus
Pias proteins can redistribute interacting factors to nuclear bodies or the nuclear periphery (3, 4). All mammalian Pias proteins concentrate in nuclear bodies, which can trigger recruitment of Pias-associated transcriptional regulators to these structures or the nuclear periphery (3, 4). To determine the cellular localization of the PITX2·Pias complexes in living cells we used the BiFC assay. This technique has been successfully used to identify protein interactions in cells (36, 37). PITX2 was cloned with the YN moiety next to its N terminus and Piasy and Pias1 with the YC moiety adjacent to their C termini (Fig. 2A). A linker separates the YN and YC peptides from the PITX2 and Pias proteins. If YN-PITX2 and Pias-YC proteins interact in the living cells the YN and YC moieties form a fluorescence protein complex. The YN-PITX2·Pias1-YC complex is evenly distributed in small nuclear bodies (Fig. 2B). In contrast, the Piasy-YC·YN-PITX2 complex was localized to the nuclear periphery and in larger nuclear bodies (Fig. 2B). As controls we demonstrate that Pias1-YC, Piasy-YC, YN-PITX2, YN, and YC proteins do not fluoresce on their own (Fig. 2B). Additional controls include YN-PITX2 and only the YC DNA, YN, and the Pias-YC constructs (Fig. 2C). The expression of YN-PITX2, Pias1-YC, and Piasy-YC are shown by Western blot (Fig. 2D). The advantage of this system is detecting protein interactions in living cells. These results were also observed using specific antibodies to PITX2 and Pias endogenous proteins in immunocytochemistry experiments, demonstrating nuclear co-localization in LS-8 cells (Fig. 2E). These data reveal nuclear interactions between PITX2 and Pias1 and Piasy, however, we further demonstrate protein interactions in LS-8 cells by immunoprecipitation of endogenous proteins. The endogenous interaction was identified between Pitx2 and Pias1 or Piasy (Fig. 2F). We next asked if Pias interactions resulted in the sumoylation of PITX2.
PITX2 Is Not Sumoylated
Analyses of the PITX2C protein revealed three potential sumoylation sites located at residues Lys-5, Lys-77, and Lys-149 (only two in the PITX2A isoform protein due to a different N terminus) (Fig. 3A). PITX2A and PITX2C isoforms have similar transcriptional activities and expression patterns (22). PITX2 sumoylation was determined using a previously reported assay for Nkx2.5 sumoylation (30). Nkx2.5 served as a positive control and we detected sumoylated Nkx2.5 as previously reported (30) (Fig. 3B). PITX2A was subjected to the sumoylation assay in the presence of SUMO-1, Pias proteins, and Ubc9, in HeLa cells and a mutant Sumo-1 protein (Sumo-1-Δ-GG) was used as a control (Fig. 3B). PITX2A was not sumoylated in these assays (Fig. 3B). PITX2C was not sumoylated and mutation of each PITX2C lysine (Lys) residue (Lys-5, Lys-77, and Lys-149) to an arginine (Arg) residue did not affect sumoylation (Fig. 3B). Purified PITX2 proteins were also tested for sumoylation in cell extracts and we did not detect sumoylated PITX2 (data not shown). Thus, PITX2 is not sumoylated and the sumoylation of PITX2 was not required for its interaction with Pias proteins. We next asked if each Pias protein regulated PITX2 transcriptional activity.
FIGURE 3.
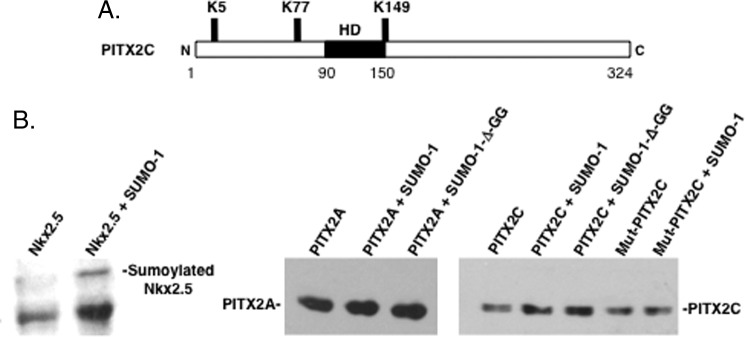
PITX2 is not sumoylated. A, schematic of the conserved potential sumoylation sites in PITX2C. The homeodomain is noted (HD) and the lysine residues associated with sumoylation are indicated. B, as a control we demonstrate that Nkx2.5 was modified by SUMO-1 in vivo. The Western blot was executed on cell lysates from HeLa cells transfected with Nkx2.5-encoding vector in the presence or absence of SUMO-1 as indicated and blotted with anti-Nkx2.5 antibody. The retarded migratory band revealed with anti-Nkx2.5 antibody is sumoylated Nkx2.5 (30). Transfected PITX2A, PITX2C, and mutant PITX2C proteins were subjected to the same assay with SUMO-1 or the SUMO-1-D-GG mutant. PITX2 proteins were detected by Western blot using a PITX2 antibody and ECL reagents from GE Healthcare.
Piasy Represses PITX2 Transcriptional Activity
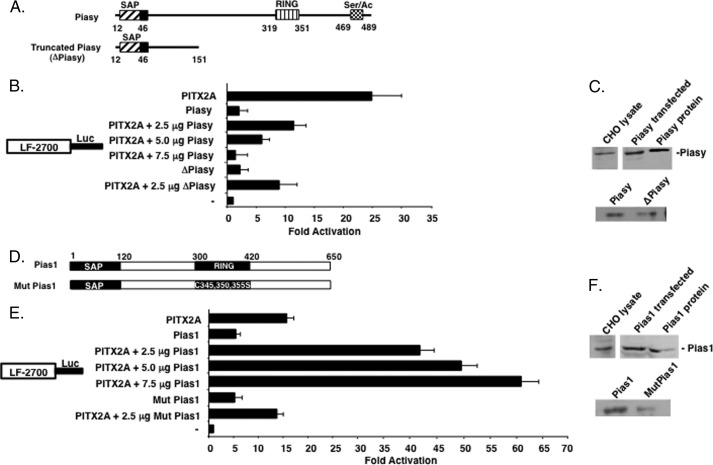
Because Piasy and Pias1 interacted with PITX2 we asked if they played a role in regulating PITX2 transcriptional activity. Two Piasy constructs were used to study the role of Piasy in regulating PITX2 transcriptional activity (Fig. 4A). We have previously shown that PITX2 activates the LEF-1 promoter (33). PITX2 activates the LEF-1 promoter at ∼24-fold compared with empty vector control (Fig. 4B). Co-transfection of full-length Piasy and PITX2A resulted in repression of PITX2A activation of the LEF-1 promoter (Fig. 4B). PITX2 was transfected at 2.5 μg and Piasy DNA was titrated from 2.5 to 7.5 μg to reveal a dose-response of Piasy repression of PITX2 transcriptional activation (Fig. 4B). Piasy and truncated Piasy (ΔPiasy) minimally activate the LEF-1 promoter. Co-transfection of equal amounts of PITX2A with ΔPiasy (2.5 μg each) repressed PITX2A activation from 24- to ∼8-fold (Fig. 4B). Thus, Piasy interacts with PITX2 to repress its transcriptional activity requiring only the N-terminal SAP domain of Piasy (Fig. 4A). Furthermore, this repression of PITX2 activation does not require the RING domain. Western blot of PIASy endogenous and transfected lysates are shown (Fig. 4C).
FIGURE 4.
Piasy represses PITX2A and Pias1 activates PITX2 transcriptional activation of the LEF-1 promoter. A, schematic of the Piasy constructs used in the transfection experiments. The SAP, RING, and C-terminal activation domain are indicated with the specific residue numbers shown. B, PITX2A and Piasy or ΔPiasy constructs (2.5 μg) were co-transfected in CHO cells with the 2.7-kb LEF-1 luciferase promoter (5 μg). To control for transfection efficiency, all transfections included the SV-40 β-galactosidase reporter (0.5 μg). Cells were incubated for 24 h, and then assayed for luciferase and β-galactosidase activities. The activities are shown as mean fold-activation compared with the LEF-1 promoter plasmid without PITX2 or Piasy expression and normalized to β-galactosidase activity (± S.E. from five independent experiments). C, expression of endogenous and transfected Piasy shown by Western blot using Piasy antibody (purified Piasy was used as a control). D, schematic of the Pias1 constructs used in the transfection experiments. E, CHO cells were transfected with the LEF-1 luciferase reporter gene as in panel B and normalized to β-galactosidase activity (± S.E. from five independent experiments). F, expression of endogenous and transfected Pias1 shown by Western blot using Pias1 antibody (purified Pias1 was used as a control).
Pias1 Enhances PITX2 Transcriptional Activity
We next asked if Pias1-regulated PITX2 transcriptional activity. CHO cells were co-transfected with PITX2 and/or Pias1 or Mut Pias1 (Fig. 4D). In these sets of experiments, PITX2A activated the LEF-1 promoter by ∼15-fold (Fig. 4E). Co-transfection of PITX2A and titrated Pias1 DNA (2.5 to 7.5 μg) resulted in a dose-responsive synergistic activation of the LEF-1 promoter (Fig. 4E). However, co-transfection of PITX2A with Mut Pias1 did not synergistically activate the LEF-1 promoter, suggesting that the E3 ligase activity (RING domain) of Pias1 was required for synergistic transcriptional activation (Fig. 4E). However, this mutation did not inhibit the interaction with PITX2 (data not shown). A Western blot of endogenous and transfected Pias1 is shown (Fig. 4F). Because Piasy and Pias1 are co-expressed with PITX2 in the oral epithelium and both interact with different regions of the PITX2 protein we asked if the two Pias proteins would act together to regulate the transcriptional activity of PITX2.
Piasy Represses Pias1 Activation of PITX2
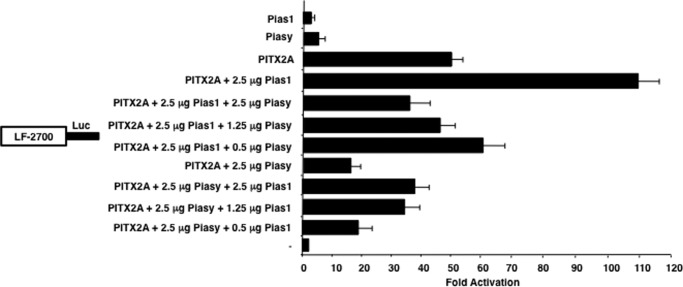
Because both Piasy and Pias1 are co-expressed in tissues and cells we asked if they work together to regulate PITX2 transcriptional activity. In these sets of experiments CHO cells that co-transfected Pias1 and PITX2 resulted in a 105-fold activation of the LEF-1 promoter (Fig. 5). However, co-transfection of equal amounts of Pias1 and Piasy (2.5 μg) with PITX2 resulted in an attenuation of the Pias1 synergistic effect on PITX2 transcriptional activity (Fig. 5). Titration of Piasy (2.5 to 0.5 μg) with 2.5 μg of PITX2 and Pias1 demonstrated attenuation of the Pias1 synergistic effect even at the lower amount of Piasy DNA (0.5 μg) (Fig. 5). Pias1 (2.5 to 0.5 μg) was titrated with equal amounts of PITX2 and Piasy DNA (2.5 μg) resulting in an increased repressive effect of Piasy (with lower amounts of Pias1) on PITX2 transcriptional activity (Fig. 5). The Piasy repressive effect on PITX2 transcriptional activity is greater than the Pias1 synergistic activation of PITX2 transcriptional activity. We next asked if activities of the Pias proteins with PITX2 were cell dependent.
FIGURE 5.
Piasy antagonizes the Pias1 synergistic activation of PITX2. CHO cells were transfected with the LEF-1 2700 luciferase reporter gene (5 μg). The cells were co-transfected with the PITX2A and/or the Pias1 and Piasy expression plasmids, or the CMV plasmid without PITX2, Pias1, or Piasy (2.5 μg). Piasy DNA was titrated (0.5 to 2.5 μg) with constant amounts of PITX2 and Pias1 DNA (2.5 μg) or Pias1 DNA was titrated with constant amounts of PITX2 and Piasy DNA. The cells were incubated for 24 h and then assayed for luciferase and β-galactosidase activities. The activities are shown as mean activation (n-fold) compared with the LEF-1 promoter plasmid without PITX2, Pias1, or Piasy expression and normalized to β-galactosidase activity (mean ± S.E. from 5 independent experiments).
Piasy Represses and Pias1 Activates PITX2 in Oral Epithelial Cells
CHO cells do not endogenously express Pitx2 and promoter activities in this cell line are independent of the response to endogenous activation. However, the LS-8 oral epithelial cell line endogenously expresses Pitx2, β-catenin, and Lef-1 isoforms (31, 33, 35). We compare the transcriptional activities of PITX2 in these two cell lines to understand a potential role for cell-specific endogenous factors in regulating PITX2. Furthermore, both cells endogenously express Piasy and Pias1 (Figs. 2, E and F, and 6).
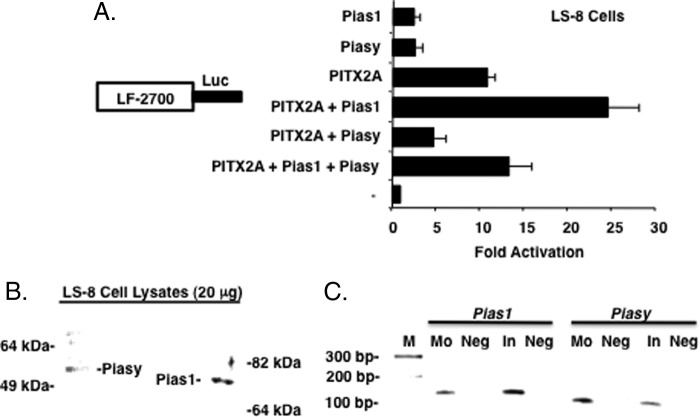
FIGURE 6.
Piasy and Pias1 regulation of PITX2 is not cell specific. A, LS-8 oral epithelial cells (endogenously express Pitx2, Piasy, Pias1, and Lef-1) were transfected with the LEF-1 luciferase reporter gene (5 μg) as previously described. The activities are shown as mean activation (n-fold) compared with the LEF-1 promoter plasmid without PITX2, Pias1, or Piasy expression and normalized to β-galactosidase activity (± S.E. from 5 independent experiments). B, endogenous expression of Pias proteins in LS-8 oral epithelial cells. C, RT-PCR of Pias transcripts in E16.5 tooth epithelium, molars (Mo) and incisors (In), all PCR products were sequenced to confirm their identity.
LS-8 cell transfections demonstrate that Piasy repressed, whereas Pias1 increased PITX2 transcriptional activation of the LEF-1 promoter, respectively (Fig. 6A). Therefore, the individual activities of the Pias proteins with PITX2 are not cell specific. Furthermore, co-transfection of Piasy with Pias1 and PITX2 yielded repression of the PITX2 and Pias1 synergistic activation as seen in CHO cells (Fig. 6A).
Pitx2 is highly expressed during craniofacial development in the oral and dental epithelium and we asked if both Pias proteins were expressed in these tissues. Pias1 and Piasy proteins are endogenously expressed in the LS-8 oral epithelial cell line (Fig. 6B) and their transcripts were detected in E16.5 mouse incisor and molar tooth germs (Fig. 6C). Altogether our data suggest that co-expression of PITX2 and Pias proteins could regulate PITX2 transcriptional activity dependent on the relative expression levels of the Pias proteins during development. We further speculate that these activities could be due to other endogenous factors interacting with PITX2. Therefore, we next asked if Piasy and Pias1 would modulate the interaction and transcriptional activation of known PITX2 co-factors.
Piasy Represses the Synergistic Activation of PITX2 with Co-factors
We have previously shown that PITX2 can interact with LEF-1 and β-catenin to synergistically activate the LEF-1 promoter (28, 33). To understand if the Piasy interaction and repression of PITX2 was dependent on these PITX2 co-factors, Piasy was co-transfected with PITX2, Lef-1, and/or β-catenin. Lef-1 FL (full-length isoform) and β-catenin each synergistically activated the LEF-1 promoter in concert with PITX2, whereas Piasy repressed PITX2 activation (Fig. 7A). Piasy further repressed the synergistic activation of PITX2 by either Lef-1 FL or β-catenin (Fig. 7A). Thus, Piasy repression of PITX2 appears dominate to the activation of PITX2 by Lef-1 or β-catenin.
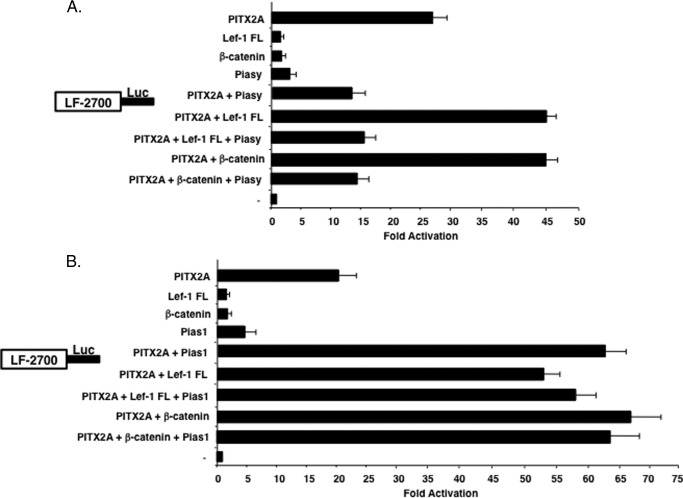
FIGURE 7.
Piasy represses the synergistic activation of PITX2 with other co-factors. A, CHO cells were co-transfected with PITX2A, Lef-1 FL, β-catenin, and Piasy or combinations of factors with the LEF-1 luciferase promoter. Transfections were performed as described in the legend to Fig. 4. Piasy repressed PITX2 transcriptional activity in the presence of PITX2 co-factors. The activities are shown as mean fold-activation compared with the LEF-1 promoter plasmid without PITX2A, Lef-1, β-catenin, or Piasy expression and normalized to β-galactosidase activity (± S.E. from 5 independent experiments). B, Pias1 was transfected with co-factors as in panel A without Piasy. Pias1 did not enhance or repress PITX2 transcriptional activity in the presence of PITX2 co-factors.
Pias1 Stimulation of PITX2 Activity Is Independent of Co-factors
To determine whether the synergistic activation of Pias1 with PITX2 was affected by PITX2 interacting factors, Pias1 was co-transfected with PITX2 and Lef-1 and/or β-catenin. Individually Lef-1 FL, β-catenin, and Pias1 each synergistically activate the LEF-1 promoter in combination with PITX2 (Fig. 7B). Co-expression of Pias1 with PITX2 and Lef-1 or PITX2 and β-catenin did not disrupt or increase the synergism between PITX2 and Lef-1 or β-catenin (Fig. 7B). The increase in PITX2 transcriptional activity with Pias1 appears to be independent of other factors and mutually exclusive of those factors.
Endogenous Piasy and Pias1 Interact with Pitx2 on Chromatin
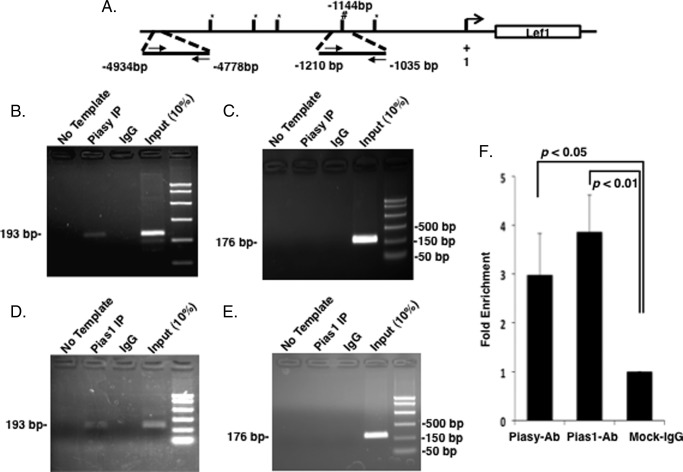
We have previously shown by ChIP that endogenous Pitx2 binds to the Lef-1 promoter (33). Pias proteins do not bind DNA alone and we demonstrate using Pias antibodies that Piasy and Pias1 bind to the Lef-1 promoter containing Pitx2 binding elements. A schematic of the Lef-1 promoter with Pitx2 binding sites and primers used to amplify the IPed chromatin is shown in Fig. 8A. ChIP assays using the Piasy antibody IP Piasy binding to the Pitx2-bound element and the IgG control did not IP the chromatin (Fig. 8B). Furthermore, controls demonstrate that Piasy does not bind to the Lef-1 promoter without a Pitx2 binding element (Fig. 8C). Pias1 binds to the Pitx2-bound element and IgG control does not IP the chromatin (Fig. 8D). Controls also reveal that Pias1 does not bind to DNA without Pitx2 binding to DNA (Fig. 8E). Pias proteins interact with Pitx2 on chromatin. Quantitative PCR of the ChIP products reveals a 3-fold enrichment of Piasy binding and a 4-fold enrichment of Pias1 binding to chromatin (Fig. 8F).
FIGURE 8.
Endogenous Pias proteins bind to the Lef-1 promoter. A, schematic representation and location of the Pitx2 binding site in the Lef-1 promoter. # indicates the Pitx2 binding element (TAATCC). B, ChIP of endogenous Piasy binding to the Pitx2 element within the Lef-1 promoter in LS-8 cells. Rabbit antisera used as a control IP and Piasy antisera from Santa Cruz were used to IP Piasy binding to the chromatin. We have previously shown Pitx2 binding to this element (33). The input chromatin is shown as a positive control for the ChIP. C, control ChIP using the Piasy antisera and primers to a 4.7-kb upstream region of the Lef-1 promoter. This chromatin does not contain a Pitx2 binding site and was not IPed using Piasy antisera, the primers did amplify the input chromatin. D, ChIP of endogenous Pias1 binding to the Pitx2 element within the Lef-1 promoter in LS-8 cells. Rabbit antisera was used as a control IP and Pias1 antisera from Santa Cruz was used to IP Pias1 binding to the chromatin. The input chromatin is shown as a positive control for the ChIP. E, control ChIP using the Pitx2 antisera and primers to an upstream region of the Lef-1 promoter. This chromatin does not contain a Pitx2 binding site and was not IPed using Pias1 antisera, the primers did amplify the input chromatin. F, ChIP DNA fragment concentrations were evaluated by real-time PCR to calculate the fold-enrichment of Pias1 and Piasy antibody IP group, respectively. Both IP enrichments were normalized to the IgG group.
Knockdown of Endogenous Pias Proteins Relieves the Effect on Pitx2 Transcriptional Activity
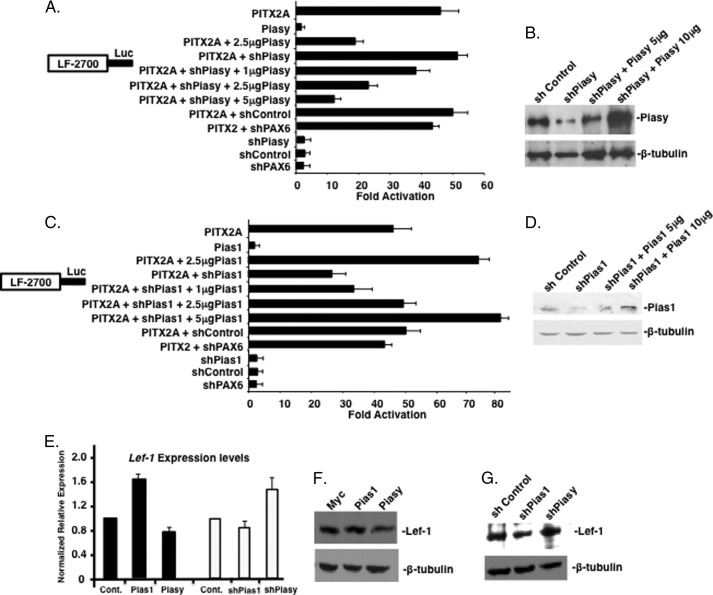
Knockdown of endogenous Piasy (shPiasy) increased PITX2 activation of the LEF-1 promoter compared with shControl and shPAX6, a transcription factor control (Fig. 9A). To demonstrate specificity of shPiasy, Piasy plasmid was titrated against a constant amount of shPiasy plasmid (2.5 μg). Increasing amounts of Piasy plasmid overcame the shPiasy inhibition of Piasy expression (Fig. 9A). Knockdown of endogenous Pias1 (shPias1) reduced activation of the LEF-1 promoter by transfected PITX2A by ∼30%, p = 0.02 (Fig. 9C). Titration of increasing amounts of Pias1 plasmid overcame the inhibition of Pias1 expression by shPias1 (Fig. 9C). shControl and shPAX6 had no effect on PITX2 activation of the LEF-1 promoter.
FIGURE 9.
Knockdown of endogenous Pias expression relieves the Pias protein modulation of Pitx2 transcriptional activity. A, CHO cell transfections were performed as described in the legend to Fig. 4, however, shPiasy was used to inhibit endogenous Piasy expression. Piasy plasmid DNA was increased from 1 to 5 μg against a constant amount of shPiasy. shControl (scrambled sequence) and shPAX6 were used as controls for shRNA effects. B, Western blot of endogenous Piasy expression with and without shPiasy, and titration of Piasy expression plasmid to rescue shPiasy knockdown, β-tubulin was used as a loading control. C, shPias1 was used to inhibit endogenous Pias1 expression in CHO cells (transfection were performed as in panel A). Pias1 plasmid DNA was increased from 1–5 μg against a constant amount of shPias1. shControl (scrambled sequence) and shPAX6 were used as controls for shRNA effects. D, Western blot of endogenous Pias1 expression with and without shPias1, and titration of Pias1 expression plasmid to rescue shPias1 knockdown, β-tubulin was used as a loading control. E, overexpression of Pias1 and Piasy expression constructs and shPias1 and shPiasy affects Lef-1 mRNA expression in LS-8 cells. Quantification by real-time PCR of Lef-1 in LS-8 cells transfected with Pias1 and Piasy or shPias1 and shPiasy for 48 h. The data are means of three independent experiments with all measurements made in triplicate. F, Western blot of endogenous Lef-1 protein in LS-8 cells after transfection of Pias1 or Piasy expression constructs. G, Western blot of endogenous Lef-1 protein in LS-8 cells after transfection of shPias1 or shPiasy constructs. β-Tubulin was used as a loading control in both E and F.
Western blots demonstrate knockdown of the Pias proteins by their shRNA inhibitors, and rescue by Pias expression plasmids (Fig. 9, B and D). Endogenous Lef-1 expression levels were assayed by real-time PCR in the presence of increased Pias expression and decreased Pias expression by shRNAs. Pias1 overexpression increased Lef-1 transcripts compared with control vector (Fig. 9E). Conversely Piasy overexpression decreased Lef-1 transcript levels (Fig. 9E). These data correlate with increased and decreased endogenous Pitx2 transcriptional activities, respectively. Knockdown of endogenous Pias1 decreased Lef-1 transcript levels and Piasy knockdown increased Lef-1 transcripts (Fig. 9E). Lef-1 protein levels are also similarly affected (Fig. 9, F and G). Pias proteins endogenously regulate Pitx2 activation of endogenous Lef-1 expression. We have previously shown that Pitx2 activates endogenous Lef-1 expression (28, 33).
Pias Proteins Do Not Inhibit the Interactions of Lef-1 and β-Catenin with PITX2
To further understand how these PITX2 co-factors interact with PITX2 we performed GST-PITX2 pulldown experiments using purified Pias, Lef-1, and β-catenin proteins. Piasy, β-catenin, and Lef-1 all interact with the PITX2 C-terminal tail (28, 33). We asked if these proteins bound to PITX2 in a complex. We show that Piasy, β-catenin, and Lef-1 all bind PITX2 independently and that Piasy and β-catenin bind PITX2; and Lef-1 and Piasy bind PITX2 (Fig. 10B). A schematic of the binding interactions is depicted in Fig. 10A. Piasy binding to PITX2 does not inhibit the binding of these co-factors, however, the repressive activity of Piasy negates the synergistic activity of the co-factors with PITX2. The GST fusion proteins used in the assay are shown in Fig. 10C.
FIGURE 10.
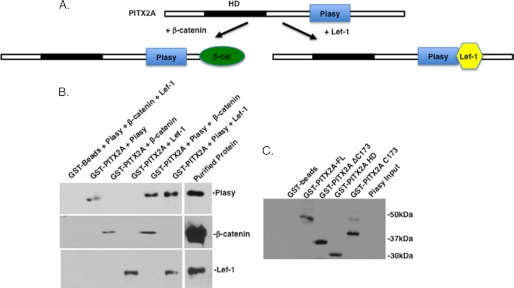
Piasy binds to PITX2 in the presence of Lef-1 or β-catenin. A, schematic of the co-factor interactions with PITX2 in the presence of Piasy. B, GST-PITX2 pulldown assay with bacterial expressed Piasy, β-catenin, or Lef-1. Proteins were incubated with immobilized GST-PITX2 or GST-beads, the beads were washed extensively and the bound proteins visualized using specific antibodies to each protein by Western blot and the ECL method (GE Healthcare).
Pias1 and β-catenin as well as Pias1 and Lef-1 also bind PITX2 (Fig. 11B). A schematic of the protein interactions is shown in Fig. 11A. The clearest interpretation of these results demonstrates that Piasy or Pias1 binding to PITX2 does not inhibit other co-factors from binding to PITX2. The GST fusion proteins used in the assays are shown in Fig. 11C. It has been shown that Lef-1 can directly interact with Piasy to regulate Lef-1 transcriptional activity (7). Thus, Lef-1 may also interact with Piasy bound to PITX2. Thus, Pias proteins interact with PITX2 in the presence of other interacting factors. Interestingly, Piasy represses PITX2 transcriptional activity when Lef-1 and β-catenin directly interact with PITX2. However, Pias1 does not increase the synergistic activation of PITX2 in the presence of Lef-1 or β-catenin.
FIGURE 11.
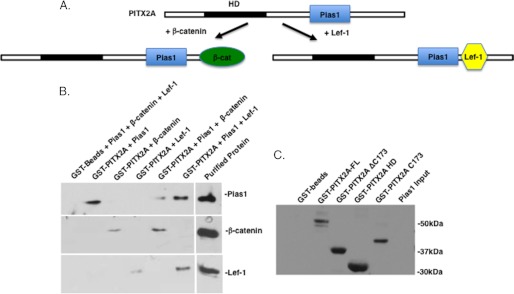
Pias1 binds to PITX2 in the presence of Lef-1 or β-catenin. A, schematic of the co-factor interactions with PITX2 in the presence of Pias1. B, GST-PITX2 pulldown assay with bacterial expressed Pias1, β-catenin, or Lef-1. Proteins were incubated with immobilized GST-PITX2 or GST-beads, the beads were washed extensively and the bound proteins visualized using specific antibodies to each protein by Western blot and the ECL method (GE Healthcare).
DISCUSSION
PITX2 is an important homeodomain transcription factor required for early embryonic development of several organs and tissues (19–21, 41–43). PITX2 is a transcriptional activator, however, depending on co-factor interactions its activity can be enhanced or attenuated. The PITX2 homeodomain is a 60-residue highly structured motif that binds specific DNA elements (34). The homeodomain can also interact with other factors to regulate the transcriptional activity of PITX2. MEF2A, FoxJ1, HMGN2, and β-catenin all directly bind to the PITX2 homeodomain to repress or enhance PITX2 transcriptional activity (27, 33, 44, 45). The PITX2 C-terminal tail is 173 residues and is relatively unstructured and contains a 39-residue OAR domain that can interact with other factors (24). Lef-1, β-catenin, and Pit-1 transcription factors interact with the PITX2 C-terminal tail as well as PITX2 dimerization occurring through the C-terminal tail (24, 33, 34). Protein interactions play a major role in the activity of transcription factors and their cell-specific activities and PITX2 transcriptional activity is modulated through its interactions with a variety of cell/tissue-specific factors. PIASy and PIAS1 proteins reveal a new mechanism for the regulation of PITX2 interacting co-factors.
Piasy and Pias1 Interact with PITX2 in the Nucleus
The nuclear periphery is associated with inner nuclear membrane proteins that can regulate transcription and Pias proteins can sequester co-factors to discrete regions of the nucleus. Piasy represses Lef-1 transcriptional activity by sumoylation-independent sequestration of Lef-1 in promyelocytic leukemia nuclear bodies (7). Piasy also represses the transcriptional activation of Nurr1 and Ets-1 by targeting to the nuclear matrix, independent of sumoylation (38, 39). Piasy-mediated repression of C/EBPδ is sumoylation independent and sequesters C/EBPδ to the nuclear periphery (40). It appears that sequestration of Pias·transcription complexes to the nuclear periphery acts to repress the transcriptional activity of these factors.
The BiFC assay revealed that the Pias1·PITX2 complex was distributed to nuclear bodies throughout the nucleus. However, the Piasy·PITX2 complex was distributed in larger bodies toward the nuclear periphery. The endogenous immunofluorescence assays revealed similar nuclear distribution. These data could reflect the differences in the activation or repression of PITX2, where Piasy represses PITX2 in complexes at the nuclear periphery, whereas Pias1 enhances PITX2 activity in the nuclear bodies as has been previously reported for other factors.
PIAS Proteins Modulate PITX2 Transcriptional Activity
The significance of Piasy and Pias1 as PITX2 interacting partners is underscored by the Lef-1 connection, as both Pitx2 and Lef-1 are co-expressed in the oral/dental epithelium during craniofacial development (33). The fact that Piasy also interacts with Lef-1 via its N terminus (SAP domain) and suppresses its activity by sequestration to nuclear bodies suggests a similar mechanism of suppression of PITX2 activity (7). In contrast to Lef-1, the SAP domain is not required for the interaction and sumoylation of Smad4 (29). The SAP domain is highly conserved among all members of the Pias family. The SAP domain and ∼104 residues C-terminal to the SAP domain contain sufficient activity to repress PITX2A transcriptional activity. Piasy can also repress the activity of the androgen receptor and a SUMO-E3-ligase-defective mutant of Piasy repressed the activity of the androgen receptor (46). Thus, similar to the androgen receptor, PITX2 activity was also repressed by an E3 ligase-deficient Piasy protein. The Piasy SAP domain represses PITX2 transcriptional activity indicating that the SAP domain acts as a transcriptional repressor independent of sumoylation.
Pias1 synergism with PITX2 required the E3 ligase activity of the RING domain. However, the physical interaction with PITX2 did not require the E3 ligase activity as a Pias1 RING domain mutant bound PITX2 (data not shown). Although Pias proteins are mainly known as transcriptional repressors, they can also positively regulate transcription factor activity (3). These activities may be regulated through direct sumoylation of the transcription factor or independent of sumoylation through unknown mechanisms. PITX2 is not sumoylated and sumoylation was not required for the Pias proteins to regulate PITX2 transcriptional activity.
Piasy but Not Pias1 Modulate PITX2 Transcriptional Regulation by Co-factors
How these Pias proteins regulate transcription factor activity associated with other factors is not known. We have attempted to show that Pias proteins form complexes with other proteins to regulate transcription. Pias1 had no effect on the synergistic activation of the LEF-1 promoter by PITX2 and Lef-1 or β-catenin. However, Piasy repressed the synergistic activation of PITX2 by both Lef-1 and β-catenin, suggesting that Piasy has a dominant repressive effect on PITX2 activity. Both PIAS proteins interact with the PITX2 C-terminal tail with these factors to differentially modulate the transcriptional mechanisms of PITX2. The PITX2 C-terminal tail is unstructured and may freely interact with a variety of factors independently (24). These results suggest that Piasy may bind more efficiently to PITX2 than Pias1 or the repressive activity of Piasy is more effective. Because Pias proteins are ubiquitously expressed, these data suggest that Pias proteins may mediate the transcriptional responses between interacting factors.
Differential Regulation of PITX2 by Pias Proteins, a New Molecular Mechanism
We have shown a new mechanism where a transcription factor can be differentially regulated by Pias proteins interacting with a similar region of the protein (Fig. 12). Recent studies have shown that Pias proteins can have multiple effects on androgen receptor (AR) activity. AR-dependent transcription can be repressed by Pias1 with exogenous Sumo1 and Pias RING domain expression or enhanced in the absence of sumoylation (39). Pias3 inhibits AR transactivation in some cells but activates it in other cell types. Sumo3 can have a negative or positive effect on AR depending on the type of prostate cancer cells (47). In addition, Pias3 activates, whereas Piasy represses the transcriptional activity of SMAD3 (48). Taken together these data demonstrate that different Pias proteins can interact with the same factor to repress or activate the transcriptional potential of that factor.
FIGURE 12.
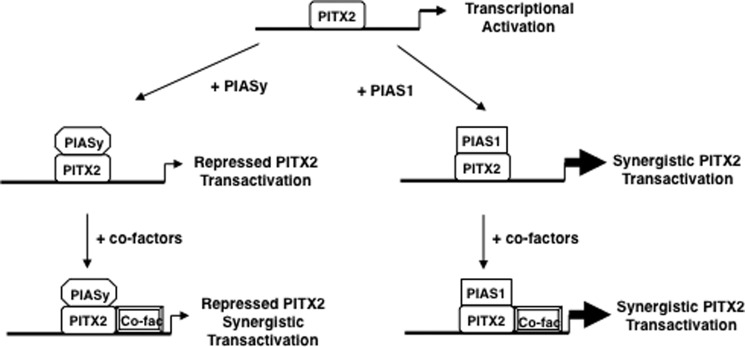
Model for Pias protein activities with PITX2. PITX2 activates gene expression and addition of Piasy represses PITX2 transcriptional activation and Piasy represses the synergism between PITX2 and co-factors. PITX2 and Pias1 synergistically activate gene expression. Pias1 does not affect the synergism between PITX2 and co-factors.
In summary, the repression of PITX2 by Piasy and activation of PITX2 by Pias1 are not cell specific and may be a general mechanism for the regulation of PITX2. There may be other factors participating in this regulation as both Pias proteins sequester PITX2 to subnuclear compartments, however, the RING domain is required for Pias1-enhanced PITX2 transcriptional activation, whereas the RING domain is not required for Piasy repression of PITX2 activity. The Pias interactions also differentially affect the activity of PITX2 interacting cofactors, which could play an important part in early developmental gene expression. Further work will investigate the functional interactions between PITX2, Pias proteins, and specific Pias cofactors as potential mechanisms to regulate PITX2 transcriptional activity.
Acknowledgments
We thank Drs. Tord A. Hjalt (University of Lund, Lund, Sweden), Grace Gill (Harvard University), and Gary Hayward (Johns Hopkins University) for reagents and technical advice and members of the Amendt laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants EY011930 and EY010608 from the NEI, Grants DE 016511 (to A. F. R.) and DE 13941 (to B. A. A.) from the NIDCR, and Robert A. Welch Foundation Grant G-0010 (to W. H. K.).
- Pias
- protein inhibitors of activated STAT
- PITX2
- pituitary homeobox 2
- HD
- homeodomain
- AD
- acidic domain
- BiFC
- bimolecular fluorescence complementation
- IP
- immunoprecipitation
- AR
- androgen receptor
- SAP
- scaffold-attachment factor A.
REFERENCES
- 1. Johnson E. S., Gupta A. A. (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106, 735–744 [DOI] [PubMed] [Google Scholar]
- 2. Jackson P. (2001) A new RING for SUMO. Wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes. Dev. 15, 3053–3058 [DOI] [PubMed] [Google Scholar]
- 3. Shuai K., Liu B. (2005) Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 5, 593–605 [DOI] [PubMed] [Google Scholar]
- 4. Schmidt D., Müller S. (2003) PIAS/SUMO. New partners in transcriptional regulation. Cell. Mol. Life Sci. 60, 2561–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chung C. D., Liao J., Liu B., Rao X., Jay P., Berta P., Shuai K. (1997) Specific inhibition of Stat3 signal transduction by PIAS3. Science 278, 1803–1805 [DOI] [PubMed] [Google Scholar]
- 6. Liu B., Liao J., Rao X., Kushner S. A., Chung C. D., Chang D. D., Shuai K. (1998) Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. U.S.A. 95, 10626–10631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sachdev S., Bruhn L., Sieber H., Pichler A., Melchior F., Grosschedl R. (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15, 3088–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kotaja N., Karvonen U., Jänne O. A., Palvimo J. J. (2002) PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 22, 5222–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wong K. A., Kim R., Christofk H., Gao J., Lawson G., Wu H. (2004) Protein inhibitor of activated STAT Y (PIASy) and a splice variant lacking exon 6 enhance sumoylation but are not essential for embryogenesis and adult life. Mol. Cell. Biol. 24, 5577–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu B., Yang R., Wong K. A., Getman C., Stein N., Teitell M. A., Cheng G., Wu H., Shuai K. (2005) Negative regulation of NF-κB signaling by PIAS1. Mol. Cell. Biol. 25, 1113–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oh H. J., Kido T., Lau Y. F. (2007) PIAS1 interacts with and represses SOX9 transactivation activity. Mol. Reprod. Dev. 74, 1446–1455 [DOI] [PubMed] [Google Scholar]
- 12. Ilmarinen T., Kangas H., Kytömaa T., Eskelin P., Saharinen J., Seeler J. S., Tanhuanpää K., Chan F. Y., Slattery R. M., Alakurtti K., Palvimo J. J., Ulmanen I. (2008) Functional interaction of AIRE with PIAS1 in transcriptional regulation. Mol. Immunol. 45, 1847–1862 [DOI] [PubMed] [Google Scholar]
- 13. Imoto S., Sugiyama K., Yamamoto T., Matsuda T. (2004) The RING domain of PIASy is involved in the suppression of bone morphogenetic protein-signaling pathway. Biochem. Biophys. Res. Commun. 319, 275–282 [DOI] [PubMed] [Google Scholar]
- 14. Gross M., Yang R., Top I., Gasper C., Shuai K. (2004) PIASy-mediated repression of the androgen receptor is independent of sumoylation. Oncogene 23, 3059–3066 [DOI] [PubMed] [Google Scholar]
- 15. Wang J., Feng X. H., Schwartz R. J., (2004) SUMO-1 modification activated GATA4-dependent cardiogenic gene activity. J. Biol. Chem. 279, 49091–49098 [DOI] [PubMed] [Google Scholar]
- 16. Wang J., Li A., Wang Z., Feng X., Olson E.N., Schwartz R. (2007) Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol. Cell. Biol. 27, 622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duval D., Duval G., Kedinger C., Poch O., Boeuf H. (2003) The “PINIT” motif, of a newly identified conserved domain of the PIAS protein family, is essential for nuclear retention of PIAS3L. FEBS Lett. 554, 111–118 [DOI] [PubMed] [Google Scholar]
- 18. Minty A., Dumont X., Kaghad M., Caput D. (2000) Covalent modification of p73α by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 275, 36316–36323 [DOI] [PubMed] [Google Scholar]
- 19. Semina E. V., Reiter R., Leysens N. J., Alward W. L., Small K. W., Datson N. A., Siegel-Bartelt J., Bierke-Nelson D., Bitoun P., Zabel B. U., Carey J. C., Murray J. C. (1996) Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat. Genet. 14, 392–399 [DOI] [PubMed] [Google Scholar]
- 20. Mucchielli M.-L., Mitsiadis T. A., Raffo S., Brunet J.-F., Proust J.-P., Goridis C. (1997) Mouse Otlx2/RIEG expression in the odontogenic epithelium precedes tooth initiation and requires mesenchyme-derived signals for its maintenance. Dev. Biol. 189, 275–284 [DOI] [PubMed] [Google Scholar]
- 21. Hjalt T. A., Semina E. V., Amendt B. A., Murray J. C. (2000) The Pitx2 protein in mouse development. Dev. Dyn. 218, 195–200 [DOI] [PubMed] [Google Scholar]
- 22. Cox C. J., Espinoza H. M., McWilliams B., Chappell K., Morton L., Hjalt T. A., Semina E. V., Amendt B. A. (2002) Differential regulation of gene expression by PITX2 isoforms. J. Biol. Chem. 277, 25001–25010 [DOI] [PubMed] [Google Scholar]
- 23. Espinoza H. M., Ganga M., Vadlamudi U., Martin D. M., Brooks B.P., Semina E. V., Murray J. C., Amendt B. A. (2005) Protein kinase C phosphorylation modulates N- and C-terminal regulatory activities of the PITX2 homeodomain protein. Biochemistry 44, 3942–3954 [DOI] [PubMed] [Google Scholar]
- 24. Amendt B. A., Sutherland L. B., Russo A. F. (1999) Multifunctional role of the Pitx2 homeodomain protein C-terminal tail. Mol. Cell. Biol. 19, 7001–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ganga M., Espinoza H. M., Cox C. J., Morton L., Hjalt T. A., Lee Y., Amendt B. A. (2003) PITX2 isoform-specific regulation of atrial natriuretic factor expression. Synergism and repression with Nkx2.5. J. Biol. Chem. 278, 22437–22445 [DOI] [PubMed] [Google Scholar]
- 26. Berry F. B., Lines M. A., Oas J. M., Footz T., Underhill D. A., Gage P. J., Walter M. A. (2006) Functional interactions between FOXC1 and PITX2 underlie the sensitivity to FOXC1 gene dose in Axenfeld-Rieger syndrome and anterior segment dysgenesis. Hum. Mol. Genet. 15, 905–919 [DOI] [PubMed] [Google Scholar]
- 27. Amen M., Espinoza H. M., Cox C., Liang X., Wang J., Link T. M., Brennan R. G., Martin J. F., Amendt B. A. (2008) Chromatin-associated HMG-17 is a major regulator of homeodomain transcription factor activity modulated by Wnt/β-catenin signaling. Nucleic Acids Res. 36, 462–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vadlamudi U., Espinoza H. M., Ganga M., Martin D. M., Liu X., Engelhardt J. F., Amendt B. A. (2005) PITX2, β-catenin and LEF-1 interact to synergistically regulate the LEF-1 promoter. J. Cell. Sci. 118, 1129–1137 [DOI] [PubMed] [Google Scholar]
- 29. Liang M., Melchior F., Feng X. H., Lin X. (2004) Regulation of Smad4 sumoylation and transforming growth factor-β signaling by protein inhibitor of activated STAT1. J. Biol. Chem. 279, 22857–22865 [DOI] [PubMed] [Google Scholar]
- 30. Wang J., Zhang H., Iyer D., Feng X.-H., Schwartz R. J. (2008) Regulation of cardiac specific nkx2.5 gene activity by small ubiquitin-like modifier. J. Biol. Chem. 283, 23235–23243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Green P. D., Hjalt T. A., Kirk D. E., Sutherland L. B., Thomas B. L., Sharpe P. T., Snead M. L., Murray J. C., Russo A. F., Amendt B. A. (2001) Antagonistic regulation of Dlx2 expression by PITX2 and Msx2. Implications for tooth development. Gene Expr. 9, 265–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Diamond E., Amen M., Hu Q., Espinoza H. M., Amendt B. A. (2006) Functional interactions between Dlx2 and lymphoid enhancer factor regulate Msx2. Nucleic Acids Res. 34, 5951–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amen M., Liu X., Vadlamudi U., Elizondo G., Diamond E., Engelhardt J. F., Amendt B. A. (2007) PITX2 and β-catenin interactions regulate Lef-1 isoform expression. Mol. Cell. Biol. 27, 7560–7573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amendt B. A., Sutherland L. B., Semina E. V., Russo A. F. (1998) The molecular basis of Rieger syndrome. Analysis of Pitx2 homeodomain protein activities. J. Biol. Chem. 273, 20066–20072 [DOI] [PubMed] [Google Scholar]
- 35. Chen L. S., Couwenhoven R. I., Hsu D., Luo W., Snead M. L. (1992) Maintenance of amelogenin gene expression by transformed epithelial cells of mouse enamel organ. Arch. Oral Biol. 37, 771–778 [DOI] [PubMed] [Google Scholar]
- 36. Hu C.-D., Chinenov Y., Kerppola T. K. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 [DOI] [PubMed] [Google Scholar]
- 37. Kerppola T. K. (2006) Visualization of molecular interactions by fluorescence complementation. Nature Rev. 7, 449–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Galleguillos D., Vecchiola A., Fuentealba J. A., Ojeda V., Alvarez K., Gómez A., Andrés M. E. (2004) PIASγ represses the transcriptional activation induced by the nuclear receptor Nurr1. J. Biol. Chem. 279, 2005–2011 [DOI] [PubMed] [Google Scholar]
- 39. Nishida T., Terashima M., Fukami K. (2006) PIASy-mediated repression of the Ets-1 is independent of its sumoylation. Biochem. Biophys. Res. Commun. 345, 1536–1546 [DOI] [PubMed] [Google Scholar]
- 40. Zhou S., Si J., Liu T., DeWille J. W. (2008) PIASy represses CCAAT/enhancer-binding protein δ (C/EBPδ) transcriptional activity by sequestering C/EBPδ to the nuclear periphery. J. Biol. Chem. 283, 20137–20148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin C. R., Kioussi C., O'Connell S., Briata P., Szeto D., Liu F., Izpisúa-Belmonte J. C., Rosenfeld M. G. (1999) Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 401, 279–282 [DOI] [PubMed] [Google Scholar]
- 42. Lu M. F., Pressman C., Dyer R., Johnson R. L., Martin J. F. (1999) Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature 401, 276–278 [DOI] [PubMed] [Google Scholar]
- 43. Gage P. J., Suh H., Camper S. A. (1999) The bicoid-related Pitx gene family in development. Mammalian Genome 10, 197–200 [DOI] [PubMed] [Google Scholar]
- 44. Toro R., Saadi I., Kuburas A., Nemer M., Russo A. F. (2004) Cell-specific activation of the atrial natriuretic factor promoter by PITX2 and MEF2A. J. Biol. Chem. 279, 52087–52094 [DOI] [PubMed] [Google Scholar]
- 45. Venugopalan S. R., Amen M. A., Wang J., Wong L., Cavender A. C., D'Souza R. N., Akerlund M., Brody S. L., Hjalt T. A., Amendt B. A. (2008) Novel expression and transcriptional regulation of FoxJ1 during oro-facial morphogenesis. Hum. Mol. Genet. 17, 3643–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gross M., Liu B., Tan J., French F. S., Carey M., Shuai K. (2001) Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene 20, 3880–3887 [DOI] [PubMed] [Google Scholar]
- 47. Zheng Z., Cai C., Omwancha J., Chen S.-Y., Baslan T., Shemshedini L. (2006) SUMO-3 enhances androgen receptor transcriptional activity through a sumoylation-independent mechanism in prostate cancer cells. J. Biol. Chem. 281, 4002–4012 [DOI] [PubMed] [Google Scholar]
- 48. Long J., Wang G., Matsuura I., He D., Liu F. (2004) Activation of Smad transcriptional activity by protein inhibitor of activated STAT3 (PIAS3). Proc. Natl. Acad. Sci. U.S.A. 101, 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]