Background: The relationship between connective tissue growth factor (CCN2) and hypoxia-inducible factor (HIF)-1 in hypoxic nucleus pulposus is unknown.
Results: HIF-1α suppresses CCN2 expression, whereas CCN2 represses basal HIF-1α levels and transcriptional activity.
Conclusion: In nucleus pulposus, HIF-1α and CCN2 form a negative regulatory circuit.
Significance: Tight control of CCN2 and HIF-1 activities may play a role in disc homeostasis.
Keywords: Cartilage Biology, Chondrocytes, Extracellular Matrix, Hypoxia, Hypoxia-inducible Factor (HIF), CCN2, CTGF, Intervertebral Disc, Nucleus Pulposus
Abstract
The objective of the study was to investigate if hypoxia-inducible factor (HIF)-1α and connective tissue growth factor (CCN2) form a regulatory network in hypoxic nucleus pulposus (NP) cells. A decrease in CCN2 expression and proximal promoter activity was observed in NP cells after hypoxic culture. Analysis of both human and mouse CCN2 promoters using the JASPAR core database revealed the presence of putative hypoxia response elements. Transfection experiments showed that both promoter activities and CCN2 expression decreases in hypoxia in a HIF-1α-dependent fashion. Interestingly, deletion analysis and mutation of the hypoxia responsive elements individually or in combination resulted in no change in promoter activity in response to hypoxia or in response to HIF-1α, suggesting an indirect mode of regulation. Notably, silencing of endogenous CCN2 increased HIF-1α levels and its target gene expression, suggesting a role for CCN2 in controlling basal HIF-1α levels. On the other hand, treatment of cells with rCCN2 resulted in a decrease in the ability of HIF-1α transactivating domain to recruit co-activators and diminished target gene expression. Last, knockdown of CCN2 in NP cells results in a significant decrease in GAG synthesis and expression of AGGRECAN and COLLAGEN II. Immunohistochemical staining of intervertebral discs of Ccn2 null embryos shows a decrease in aggrecan. These findings reveal a negative feedback loop between CCN2 and HIF-1α in NP cells and demonstrate a role for CCN2 in maintaining matrix homeostasis in this tissue.
Introduction
The intervertebral disc is a unique tissue that separates adjoining vertebrae and helps to accommodate biomechanical forces on the spine. Cartilagenous end plates form its superior and inferior boundaries, whereas between these end plates a ligamentous annulus fibrosus circumferentially surrounds the proteoglycan-rich nucleus pulposus. Nutrients and oxygen reach the nucleus pulposus through diffusion from blood vessels that infiltrate only the outer annulus and end plate cartilage, resulting in the characteristically hypoxic microenvironment (1–3).
Nucleus pulposus cells have adapted to survive in this hypoxic niche through robust expression of hypoxia inducible factor (HIF-1α),4 a transcription factor responsive to local oxygen tension (4). HIF is a basic helix-loop-helix transcription factor comprising a constitutively expressed β subunit and an α subunit that undergoes degradation by both oxygen-dependent and -independent pathways (5, 6). When stabilized, the α subunit dimerizes with the β subunit and binds to hypoxia response elements (HREs) in the promoter of target genes. In the disc, HIF-1α is critical for maintenance of nucleus pulposus cell survival, glycolytic metabolism, and functional activities including proteoglycan matrix synthesis (7–11).
Connective tissue growth factor (CCN2/CTGF) is a member of the CCN family of proteins that plays an important role in chondrogenesis and angiogenesis, where it regulates cell proliferation and migration, as well as extracellular matrix production (12–18). Signaling through integrins and other cell surface receptors, CCN proteins function to modulate growth factor activity in response to a plethora of stimuli, including oxygen tension and mechanical force (16, 17, 19–24). Mice null for Ccn2 die immediately after birth due to severe skeletal and cartilage dysplasia, indicating its importance in development (13). In a pathological context, hypoxia induces CCN2 expression in a number of cell types, including fibroblasts and lung and pancreatic adenocarcinoma, where it is associated with enhanced fibrosis and tumor growth (22, 25–28). On the other hand, in a tubular epithelial cell line, CCN2 expression is shown to decrease with hypoxia, underlying the cell type specificity of CCN2 regulation and function in response to hypoxia (30). In contrast, little information is available on the role of CCN2 in regulating HIF-1α expression (24, 31).
In the context of the disc, CCN2 has been shown to stimulate extracellular matrix production by nucleus pulposus cells (32). However, little is known about the regulation of CCN in the nucleus pulposus under normal and pathological conditions. CCN2 may play a role in the development of the notochord in zebrafish, the embryonic anlagen of the intervertebral disc (33), but a similar effect on the notochord is not seen in mice lacking CCN2 (13). We have observed increasing levels of CCN2 in the degenerate nucleus pulposus and have identified TGF-β as an important regulator of its expression. However, it is yet to be determined if CCN2 expression is controlled by hypoxia in the disc (23). Resolution of this question is important because CCN2 is a major target and regulator of hypoxic pathways in many pathological conditions. Thus, the objective of this investigation is to determine whether CCN2 expression in nucleus pulposus cells is regulated by hypoxia and HIF-1α and if CCN2, in turn, regulates HIF-1α. Results of the study show that in nucleus pulposus cells hypoxia decreases CCN2 expression through HIF-1α and that CCN2 suppresses HIF-1α activity.
MATERIALS AND METHODS
Plasmids and Reagents
Human CCN2 promoter plasmids were obtained from Dr. George Yang, Stanford University, and the mouse Ccn2 promoter plasmids were as described previously (34). Expression plasmids were provided by Dr. Celeste Simon, University of Pennsylvania (pCA-HIF-2α with a triple mutation (P405A/P530A/N851A)) and Dr. Eric Huang, National Cancer Research Institute, Bethesda (pARNT). siHIF-1α (PBS/pU6-HIF1α) (number 21103) developed by Dr. Connie Cepko, HA-HIF1αP402A/P564A-pcDNA3 (number 18955) by William Kaelin, HRE-Luc (number 26731) by Navdeep Chandeland, and psPAX2 (number 12260) and pMD2.G (number 12259) by Didier Trono were obtained from Addgene. Lentiviral shHIF-1α that co-expresses YFP was a gift Dr. Andree Yeremian, University of Lleida, Spain, and pre-validated pLKO.1Sh-CCN2 (TRC#0000061951) was purchased from Sigma. For transactivation studies of HIF-1, the binary Gal4 reporter plasmids (N+C TAD, amino acids 530–826; N-TAD, amino acids 530–778; and C-TAD, amino acids 740–826) were provided by Dr. Nianli Sang. Backbone plasmid pM (Clontech) contains no transactivation domain (TAD) but expresses the Gal4DBD. pFR-Luc (Stratagene) reporter contains the yeast Gal4-binding site upstream of a minimal promoter and the firefly luciferase gene. pRL-TK (Promega) containing the Renilla reniformis luciferase gene was used as an internal transfection control. The amount of transfected plasmid, the pre-transfection period after seeding, and the post-transfection period before harvesting have been previously optimized for rat nucleus pulposus cells using pSVβ-galactosidase plasmid (Promega) (7).
Isolation of Nucleus Pulposus Cells and Treatments of Cells
Rat nucleus pulposus cells were isolated using a method reported earlier by Risbud et al. (7). Nucleus pulposus cells were maintained in Dulbecco's modified Eagle's medium (DMEM) and 10% fetal bovine serum (FBS) supplemented with antibiotics. Cells were cultured in a Hypoxia Work Station (Invivo2 300, Ruskinn, UK) with a mixture of 1% O2, 5% CO2, and 94% N2 for 8 to 24 h. In some experiments, cells were treated with CCN2 (100 ng/ml, Prospec, Israel), TGF-β1 (10 ng/ml, Peptrotech, NJ), or 1 mm dimethyl oxalylglycine (DMOG).
Real Time RT-PCR Analysis
Total RNA was extracted from nucleus pulposus cells using RNAeasy mini columns (Qiagen). Before elution from the column, RNA was treated with RNase-free DNase I (Qiagen). The purified, DNA-free RNA was converted to cDNA using Superscript III Reverse Transcriptase (Invitrogen). Template cDNA and gene-specific primers were added to the SYBR Green master mixture (Applied Biosystems) and mRNA expression was quantified using the Step One Plus Real-time PCR System (Applied Biosystems). Hprt1 and β-ACTIN were used to normalize gene expression. Melting curves were analyzed to verify the specificity of the RT-PCR and the absence of primer dimer formation. Each sample was analyzed in duplicate and included a template-free control. All primers used were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
Western Blotting
Cells were placed on ice immediately following treatment and washed with ice-cold Hanks' balanced salt solution. All wash buffers and final re-suspension buffer included 1× protease inhibitor mixture (Roche Applied Science), NaF (5 mm), and Na3VO4 (200 μm). Total cell proteins were resolved on 8–12% SDS-polyacrylamide gels and transferred by electroblotting to polyvinylidene difluoride membranes (Bio-Rad). The membranes were blocked with 5% nonfat dry milk in TBST (50 mm Tris, pH 7.6, 150 mm NaCl, 0.1% Tween 20) and incubated overnight at 4 °C in 3% nonfat dry milk in TBST with the anti-CCN2 (1:900, Santa Cruz), anti-HIF-1α (1:1000, R&D Systems), or anti-β-tubulin antibody (1:2000, Developmental Studies Hybridoma Bank). Immunolabeling was detected using the ECL reagent (Amersham Biosciences).
Immunofluorescence Microscopy
Cells were plated in flat bottom 96-well plates (4 × 103/well) and cultured in hypoxia for 4–24 h. After incubation, cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS for 10 min, blocked with phosphate buffered saline containing 5% FBS, and incubated with antibodies against CCN2 (1:200) (Abcam) at 4 °C overnight. As a negative control, cells were reacted with isotype IgG under similar conditions. After washing, the cells were incubated with Alexa Fluor-488-conjugated anti-mouse secondary antibody (Invitrogen), at a dilution of 1:50 for 45 min at room temperature. Cells were imaged using a laser scanning confocal microscope (Olympus Fluoview, Japan).
Transfections and Dual Luciferase Assay
Cells were transferred to 24-well plates at a density of 4 × 104 cells/well 1 day before transfection. To measure the effect of hypoxia, cells were transfected with 500 ng of CCN2 reporter plasmids and 500 ng of pRL-TK plasmid. To investigate the effect of HIF-α on CCN2 promoter activity, cells were co-transfected with 100–300 ng of CA-HIF-1α, CA-HIF-2α, siHIF-1α, shHIF-2α or the respective backbone vector along with 400 ng of CCN2 reporter and 300 ng of pRL-TK plasmid. Lipofectamine 2000 (Invitrogen) was used as the transfection reagent; for each transfection, plasmids were premixed with transfection reagent. The next day, the cells were harvested and a Dual-LuciferaseTM reporter assay system (Promega) was used for sequential measurements of firefly and Renilla luciferase activities. Quantification of luciferase activities and calculation of relative ratios were carried out using a luminometer (TD-20/20, Turner Designs, CA). At least three independent transfections were performed, and all analyses were carried out in triplicate.
Site-directed Mutagenesis
Site-directed mutagenesis of the human and mouse CCN2 promoters was performed according to the manufacturer's protocol using the In-Fusion HD Cloning Kit (Clontech). Primers used for human CCN2 promoter mutants with mutated HRE sites underlined are: HRE MT1 F, 5′-AGAGAACAAAGCAACGTGTGACTCAGGATGCAGTCTCCTG-3′; HRE MT2 F, 5′-GAAGGGTAGGGCCTTGCAGTGCGAGTCTCACCTAAGGTGG-3′; HRE MT3 F, 5′-CAGGGAAGTCATGTGTAGGAGCCATATTCCCATTTCTGTT-3′. Primers used for mouse CCN2 promoter mutants with the mutated HRE site underlined were: HRE MT1 F, 5′-TGGCTTTGAGTCGAATTCCACCAGTATGTTTCCCTGAGAC-3′; HRE MT2 F, 5′-CACACACACACAAAAGCAAAGAGAGACAGAGAGAGAGAGA-3′. Mutations were verified by sequencing using the Applied Biosystems 3730 DNA Sequencer.
Lentiviral Particle Production and Viral Transduction
HEK 293T cells were seeded in 10-cm plates (1.3 × 106 cells/plate) in DMEM with 10% heat-inactivated FBS 2 days before transfection. Cells were transfected with 2.5 μg of pLKO.1-Ctr, shHIF-1α, or shCCN2 plasmids along with 1.875 μg of psPAX2 and 0.625 μg of pMD2.G. After 16 h, transfection media was removed and replaced with DMEM with 5% heat-inactivated FBS and penicillin-streptomycin. Lentiviral particles were harvested at 48 and 60 h post-transfection. Nucleus pulposus cells were plated in DMEM with 5% heat-inactivated FBS 1 day before transduction. Cells in 10-cm plates were transduced with 5 ml of conditioned media containing viral particles along with 6 μg/ml of Polybrene. After 24 h, conditioned media was removed and replaced with DMEM, 5% heat-inactivated FBS. Cells were harvested for protein extraction 5 days after viral transduction.
Dimethylmethylene Blue Assay
The proteoglycan content of conditioned media was measured as sulfated glycosaminoglycan by colorimetric assay with 1,9-dimethylmethylene blue (Blyscan, Biolcolor Ltd., Carrickfergus, UK) with chondroitin-4-sulfate as a standard following the manufacturer's instructions. GAGs were precipitated from conditioned media and stained with dimethylmethylene blue and staining was quantified by measuring absorbance at 656 nm. Results were calculated as GAG (μg)/total DNA (ng) and expressed relative to the value obtained for untreated controls.
Immunohistochemisty of Disc Tissue
Paraffin-embedded sections of wild type and mutant mice were boiled for 15 min in citrate buffer to reverse cross-links and unmask epitopes. Sections were blocked with 5% goat serum for 1 h and incubated with anti-CCN2 (CCN2) (Abcam, Cambridge MA) or anti-aggrecan (Millipore, Danvers, MA) at a dilution of 1:250 overnight at 4 °C. The following day sections were incubated with secondary antibody (Alexa Fluor 555 or 488 goat anti-rabbit, Invitrogen) for 1 h at room temperature. Sections were counterstained with DAPI (Vectashield, Vector Laboratories, Burlingame, CA). FAST staining was performed as described by Leung et al. (35).
Statistical Analysis
All measurements were performed in triplicate, data are presented as mean ± S.E. Differences between groups were analyzed by the Student's t test; *, p < 0.05.
RESULTS
Hypoxia Decreases CCN2 Expression in Nucleus Pulposus Cells
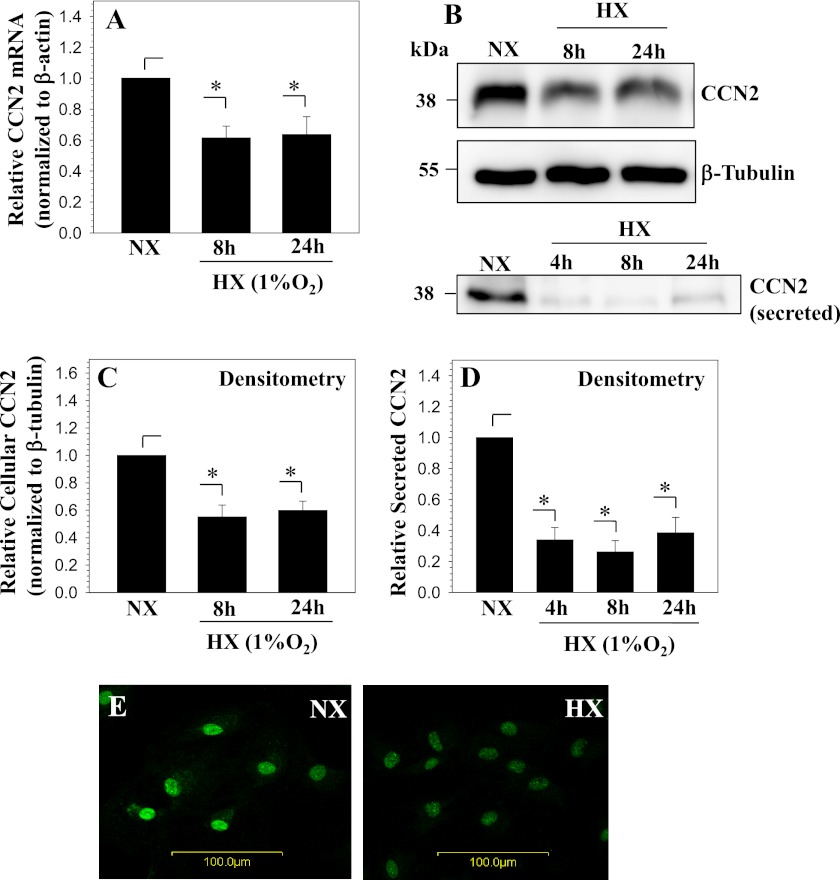
Nucleus pulposus cells were cultured in hypoxia (1% O2) and expression of Ccn2 was analyzed using real time RT-PCR. Fig. 1A shows that hypoxia results in a significant decrease in Ccn2 mRNA expression in nucleus pulposus cells. Western blot analysis of cellular lysates and secreted protein in conditioned media shows that Ccn2 levels are decreased by hypoxia as well (Fig. 1B). Densitometric analysis of Western blot experiments confirmed a significant decrease in both cellular and secreted CCN2 in hypoxia (Fig. 1, C and D). Immunofluorescence microscopy of cells cultured in hypoxia for 24 h confirmed that there is a decrease in CCN2 levels compared with normoxic controls (Fig. 1E).
FIGURE 1.
Hypoxia decreases CCN2 expression in nucleus pulposus cells. A, real time RT-PCR analysis of CCN2 expression by nucleus pulposus cells cultured in hypoxia (HX) for 8 and 24 h. CCN2 expression decreases with hypoxia. B, Western blot analysis shows a decrease in both cellular and secreted CCN2 in nucleus pulposus cells cultured in hypoxia up to 24 h. C and D, densitometric analysis of three independent experiments described in B confirms a significant decline in levels of cellular (C) and secreted (D) CCN2 in hypoxia. E, immunofluorescent analysis of CCN2 expression in nucleus pulposus cells cultured in hypoxia for 24 h, CCN2 staining decreases in hypoxia. Magnification ×20. Values shown are mean ± S.E. from three independent experiments, *, p < 0.05. NX, normoxia.
CCN2 Promoter Contains Conserved HREs
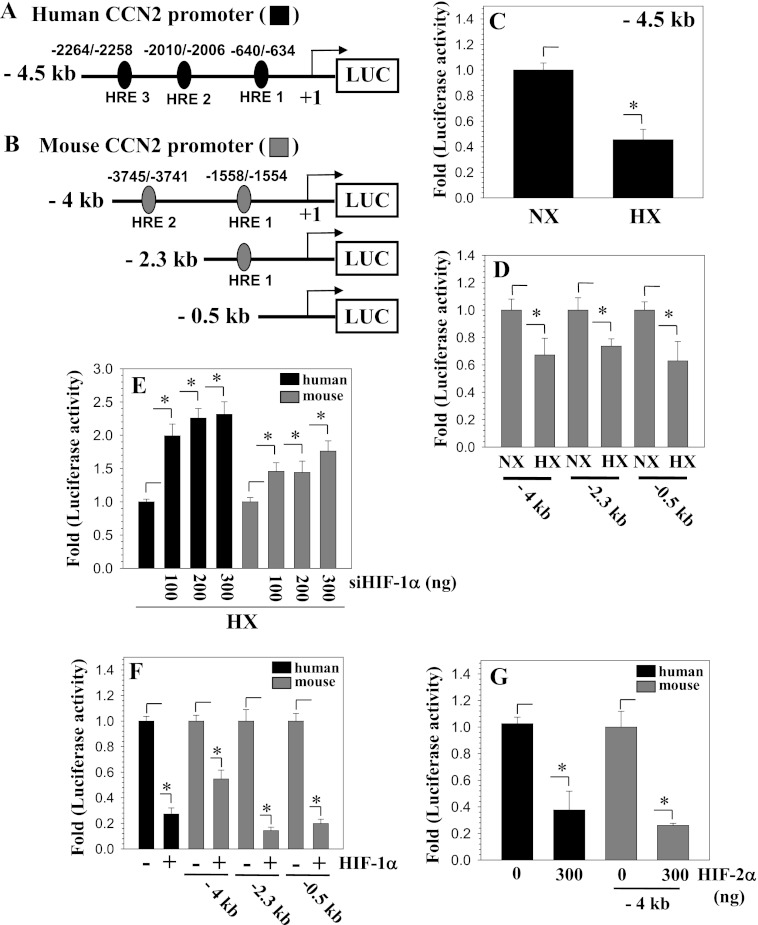
To investigate the mechanism of transcriptional regulation of CCN2 by hypoxia, we examined the response of both human and mouse CCN2 promoters to 1% O2. First, using the Evolutionarily Conserved Region Browser and UCSC genome browser, we observed that the first 5 kb of the CCN2 promoter is highly conserved between several vertebrate species including human, mouse, and rat (Fig. 2A). Next, we scanned the first 5 kb of the human CCN2 promoter for the presence of HREs using the JASPAR core database (36). Previous studies have experimentally validated two functional HREs within the first 5 kb of the mouse Ccn2 promoter; thus, the profile score threshold used was first optimized by analyzing the mouse promoter sequence in JASPAR as a positive control (22). At an 85% profile score threshold, the JASPAR database returned three putative HRE sites at −640/−634, −2010/−2006, and −2264/−2258 bp on the human CCN2 promoter, whose specific sequences are shown in bold in Fig. 2B. The HREs in the mouse Ccn2 promoter, however, are not conserved in location in the human promoter and vice versa, as verified by the Multiz alignment (Fig. 2B).
FIGURE 2.
Analysis of human CCN2 promoter for conserved HREs. A, evolutionarily conserved regions of the CCN2 gene between several vertebrate species, including chimpanzee, bovine, dog, rat, and mouse, are shown using the Evolutionarily Conserved Region (ECR) genome browser. ECR threshold is set at 80%. CCN2 promoter: red, 5′ UTR; yellow, exons; blue, introns; peach, transposons; and simple repeats, green. B, Multiz alignment from UCSC genome database of the three HREs located by the JASPAR database in the human CCN2 promoter (HRE1, HRE2, and HRE3). Non-conserved bases are underlined. *, gaps in alignment; −, no alignment.
Hypoxia and HIF Regulation of CCN2 Promoter Activity
We then used plasmids containing either the human or mouse CCN2 promoter driving luciferase expression to measure the effect of hypoxia on promoter activity in nucleus pulopsus cells (Fig. 3, A and B). Fig. 3, C and D, shows that hypoxia significantly decreases the activity of the human and mouse CCN2 promoter fragments. Serial deletion analysis of the mouse Ccn2 promoter showed that activity of smaller promoter fragments including a 0.5-kb reporter that lacked HREs is suppressed by hypoxia (Fig. 3D). To test the hypothesis that HIFs were involved in mediating the hypoxia-induced decrease in promoter activity we performed loss- and gain-of-function experiments by co-transfecting cells with siHIF-α plasmid. Both human and mouse promoters show an increase in promoter activity in the presence of siHIF-1α (Fig. 3E). Similarly, overexpression of HIF-1α or HIF-2α strongly suppresses the activity of both human and mouse CCN2 promoters (Fig. 3, F and G).
FIGURE 3.
Hypoxia and HIF regulation of CCN2 promoter activity. A and B, schematic of human CCN2 promoter construct with putative HRE at −640/−634, −2010/−2006, and −2264/−2258 bp (A) and of mouse Ccn2 promoter constructs with HRE at −3745/−3741 and −1558/−1554 bp (B). C and D, NP cells transfected with human or mouse Ccn2 promoter constructs and activity were measured following a 24-h culture in hypoxia (black and gray bars, respectively). Activity of both human (C) and mouse (D) CCN2 reporters was decreased in hypoxia. E and F, nucleus pulposus cells were co-transfected with siHIF-1α (E) or shHIF-2α (F) and CCN2 reporter activities were measured in hypoxia. Both human and mouse CCN2 promoter activities increased with siHIF-1α (E). Overexpression of HIF-1α (F) or HIF-2α (G) suppressed activities of human and mouse CCN2 promoter. Values shown are mean ± S.E. from three independent experiments, *, p < 0.05. HX, hypoxia; NX, normoxia.
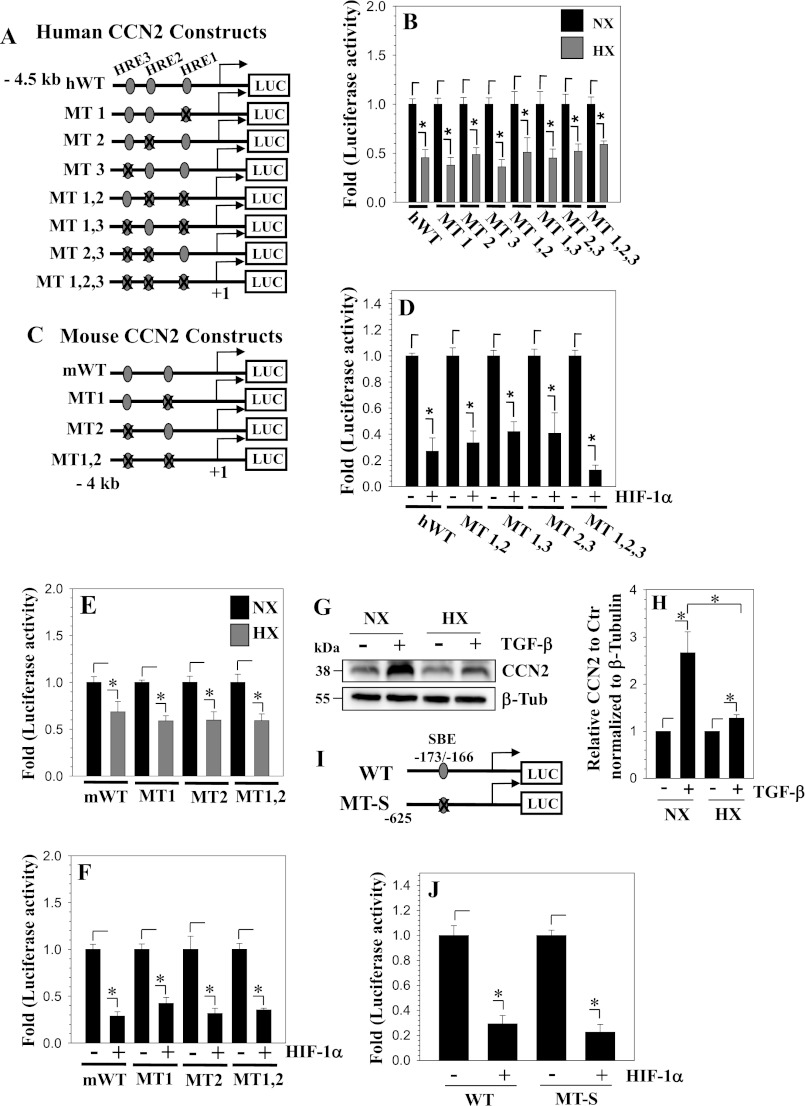
HIF-1 Suppresses CCN2 Promoter Activity Independent of HREs
To further examine if the HREs are necessary for promoter regulation by hypoxia, we performed site-directed mutagenesis to mutate HREs individually or in combination. Fig. 4A shows a schematic of the wild type and mutant human CCN2 promoter constructs. Cells were transfected with these constructs and luciferase activity was measured following a 24-h culture in hypoxia (Fig. 4B) or co-transfection with HIF-1α (Fig. 4C). Results show that neither individual nor any combination of HRE mutations cause a difference in response to hypoxia or HIF-1α overexpression compared with the wild type promoter (Fig. 4, B and C). We also mutated mouse Ccn2 promoter constructs (Fig. 4D) and similarly observed hypoxic suppression of the wild type and mutant reporters (Fig. 4E). Additionally, overexpression of HIF-1α caused a decrease in promoter activity despite the mutation of one or both of the HREs, indicating HRE independent regulation of the promoter activity by HIF-1α (Fig. 4F). To delineate the possible mechanisms of this indirect regulation of CCN2 expression by hypoxia and HIF-1, we examined if hypoxia decreases the responsiveness of nucleus pulposus cells to TGF-β, an autocrine factor that maintains the CCN2 expression levels in these cells (23). Our results clearly show that hypoxia diminishes the effect of TGF-β on CCN2 expression by NP cells (Fig. 4, G and H). We then measured the activity of a wild type or CCN2 reporter containing a mutation in the Smad binding element (Fig. 4I) following co-transfection with HIF-1α. It is noteworthy that the activity of both the reporters was strongly suppressed by HIF-1α (Fig. 4J).
FIGURE 4.
HIF-1 suppresses CCN2 promoter activity independent of HREs. A, schematic of human CCN2 promoter constructs with putative HREs located at −640/−632, −2010/−2006, and −2264/−2258 bp. HREs were mutated individually (MT1, MT2, and MT3) or in combination (MT1,2; MT1,3; and MT1,2,3). B and C, human CCN2 reporters were transfected into NP cells and cultured under normoxia (black) or hypoxia (gray). Individual HRE mutations or mutations in combination did not change the response of the CCN2 promoter to hypoxia (B) or overexpression of HIF-1α (C) when compared with wild type reporter. D, schematic of the mouse Ccn2 promoter constructs used (WT, wild type; MT1, HRE 1 mutant; MT2, HRE 2 mutant; MT1,2, HRE 1 and 2 double mutant). E and F, cells were transfected with wild type or mutant promoter constructs and cultured under normoxia (black) or hypoxia (gray). Both hypoxia (E) and HIF-1α overexpression (F) decreased activity of WT as well as all the HRE mutant constructs. G and H, nucleus pulposus cells were treated with TGF-β (10 ng/ml) for 24 h in either normoxia or hypoxia and CCN2 expression was measured by Western blot. H, densitometric analysis shows that in hypoxia, a much smaller increase in CCN2 levels by TGF-β was seen compared with normoxia. I, schematic of the 625-bp Smad3 wild type (WT) and mutant (MT-S) human CCN2 promoter constructs used in transfection. J, activity of Smad3 WT and MT-S construct was measured in nucleus pulposus cells following co-transfection with HIF-1α. HIF-1 suppressed activity of both the reporters. Values shown are mean ± S.E. from three independent experiments, *, p < 0.05. HX, hypoxia; NX, normoxia.
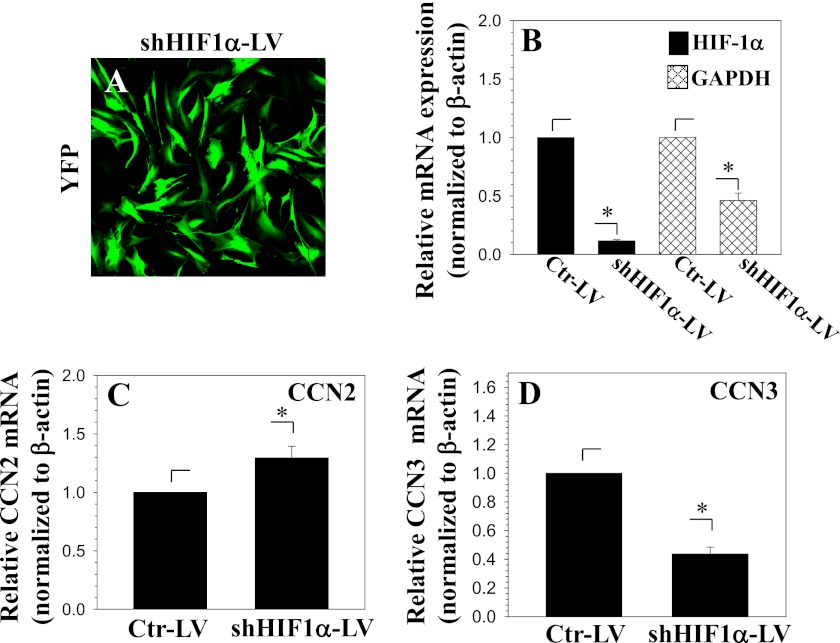
HIF-1α Is a Negative Regulator of CCN2 Expression in Nucleus Pulposus Cells
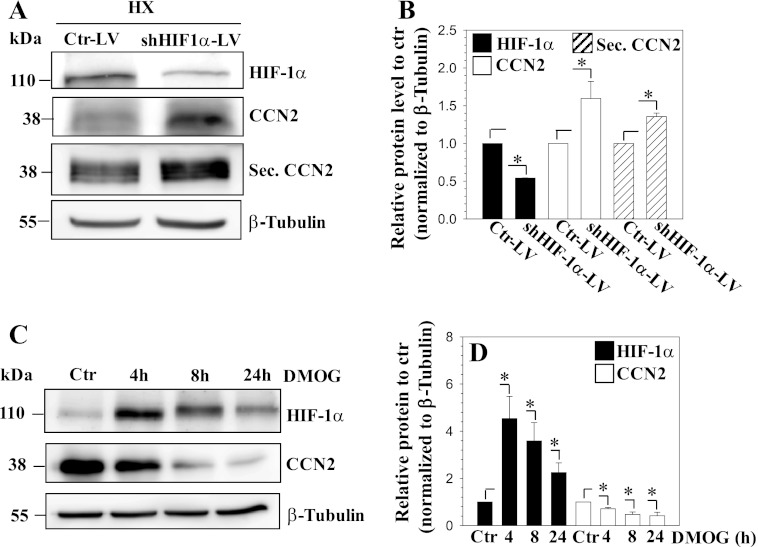
To confirm the involvement of HIF-1α in hypoxic regulation of CCN2 expression, we performed HIF-1α loss-of-function studies. First, HIF-1α was stably knocked down in nucleus pulposus cells using lentiviral transduction of shHIF-1α. Fig. 5A shows the robust expression of YFP in lentivirus-infected cells, indicating high transduction efficiency. As expected, the mRNA expression of HIF-1α and its target gene, GAPDH, is significantly decreased following shHIF-1α expression (Fig. 5B). Correspondingly, CCN2 mRNA expression is significantly increased in HIF-1α-silenced cells (Fig. 5C). Interestingly, we found that CCN3 mRNA expression responded in a reciprocal manner to CCN2 (Fig. 5D). Furthermore, suppression of HIF-1α protein levels results in a significant increase in both cellular and secreted CCN2 (Fig. 6, A and B). To complement the loss-of-function experiments, we perform gain-of-function studies by treating cells with the prolyl hydroxylase inhibitor DMOG. As expected, this treatment causes an accumulation of HIF-1α; there is a simultaneous decline in CCN2 expression. Densitometric analysis of three independent experiments confirms significant induction of HIF-1α and concomitant decrease in CCN2 protein levels at 4, 8, and 24 h (Fig. 6D).
FIGURE 5.
Suppression of HIF-1α increases CCN2 expression in nucleus pulposus cells. A, YFP expression by nucleus pulposus cells infected with a lentivirus co-expressing shHIF-1α and YFP shows high transduction efficiency. B–D, nucleus pulposus cells transduced with shHIF-1α show significantly decreased mRNA expression of HIF-1α (black bars) and a corresponding HIF-1α target gene, GAPDH (hatched bars) (B). Correspondingly, CCN2 expression is significantly increased (C), whereas CCN3 expression is decreased (D). Values shown are mean ± S.E. from three independent experiments, *, p < 0.05.
FIGURE 6.
Knockdown of HIF-1α in nucleus pulposus cells increases CCN2 protein levels. A and B, Western blot analysis of nucleus pulposus cells transduced with control-shRNA (Ctr-LV) or shHIF-1α (shHIF-1α-LV) expressing lentivirus. Cellular and secreted CCN2 levels were increased in HIF-1α-silenced cells (A). Densitometric analysis of three independent experiments shows that transduction of cells with shHIF-1α significantly decreases HIF-1α (black bars), whereas increasing cellular CCN2 (open bars) and secreted CCN2 (diagonally striped bars) levels (B). C, Western blot of cellular lysate of cells treated with DMOG for 4, 8, and 24 h. HIF-1α shows accumulation at 4, 8, and 24 h, whereas CCN2 expression is correspondingly decreased. D, densitometric analysis of three independent experiments shows that DMOG treatment increases HIF-1α protein levels (black bars), whereas decreasing CCN2 protein expression (open bars) at all time points. Values shown are mean ± S.E. from three independent experiments, *, p < 0.05.
CCN2 Antagonizes HIF-1 in Nucleus Pulposus Cells
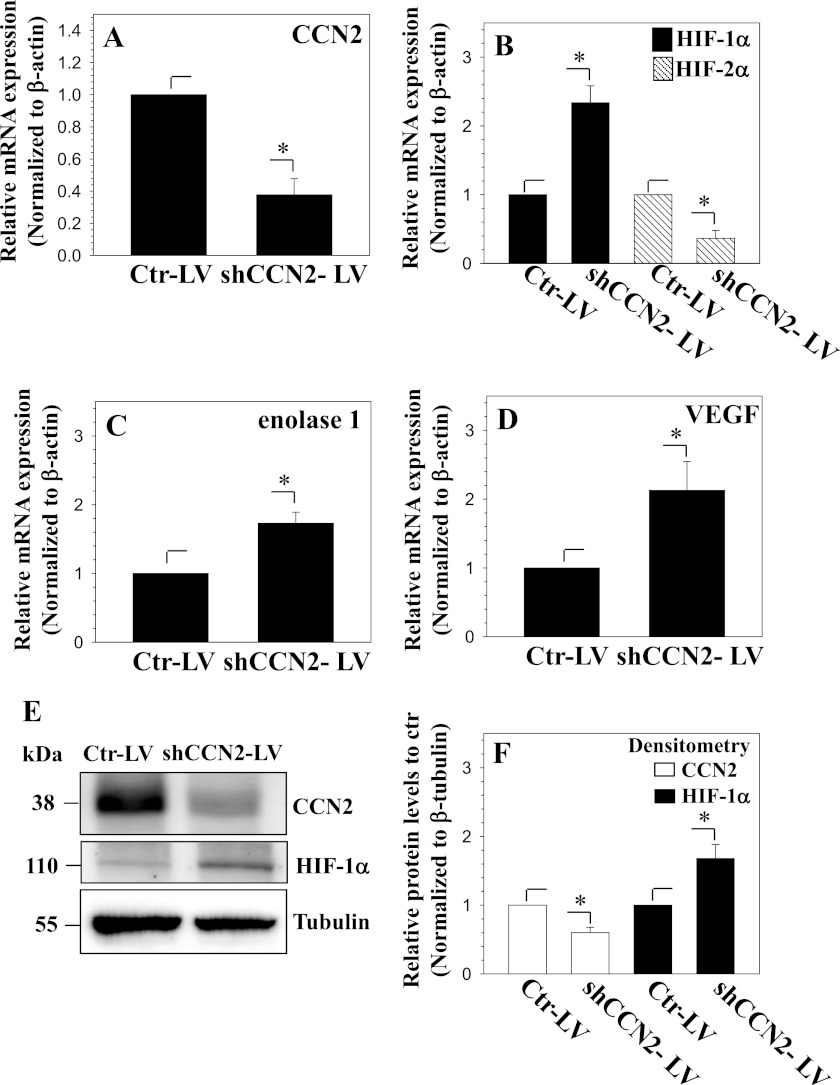
Next, we sought to determine the effect of CCN2 on the regulation of HIF-1α expression and activity. To suppress endogenous CCN2, cells were transduced with a lentiviral construct expressing shRNA against CCN2 (shCCN2-LV) or shRNA control (Ctr-LV). Real time RT-PCR analysis shows that CCN2 mRNA is decreased with shCCN2-LV transduction (Fig. 7A), whereas mRNA expression of HIF-1α and its target genes, ENOLASE 1 and VEGF, increased (Fig. 7, B–D). In contrast to HIF-1α, HIF-2α mRNA levels declined in CCN2-silenced cells (Fig. 7B). Western blot analysis was then used to follow changes at the protein level. Results show that CCN2 levels are significantly decreased in shCCN2-LV-transduced cells (Fig. 7, E and F). Moreover, in agreement to changes in mRNA expression, HIF-1α protein levels increased in CCN2-silenced cells (Fig. 7, E and F).
FIGURE 7.
Knockdown of CCN2 in nucleus pulposus cells increases HIF-1α. A–C, real time RT-PCR analysis of cells transduced with control (Ctr-LV) or shCCN2 (shCCN2-LV) expressing lentivirus shows that CCN2 expression is decreased (A). Correspondingly, whereas HIF-1α expression is increased, that of HIF-2α is decreased (B). Expression of HIF-1 target genes, ENOLASE 1 (C) and VEGF expression (D), is increased in CCN2-silenced cells. E, Western blot analysis of nucleus pulposus cells transduced with control shRNA (Ctr-LV) or shCCN2 lentivirus (shCCN2-LV). HIF-1α protein increases with knockdown of CCN2. F, densitometric analysis of three independent experiments shows that transduction of cells with shCCN2 significantly decreases CCN2 expression (open bars), whereas increasing HIF-1α (black bars). Values shown are mean ± S.E. from three independent experiments, *, p < 0.05.
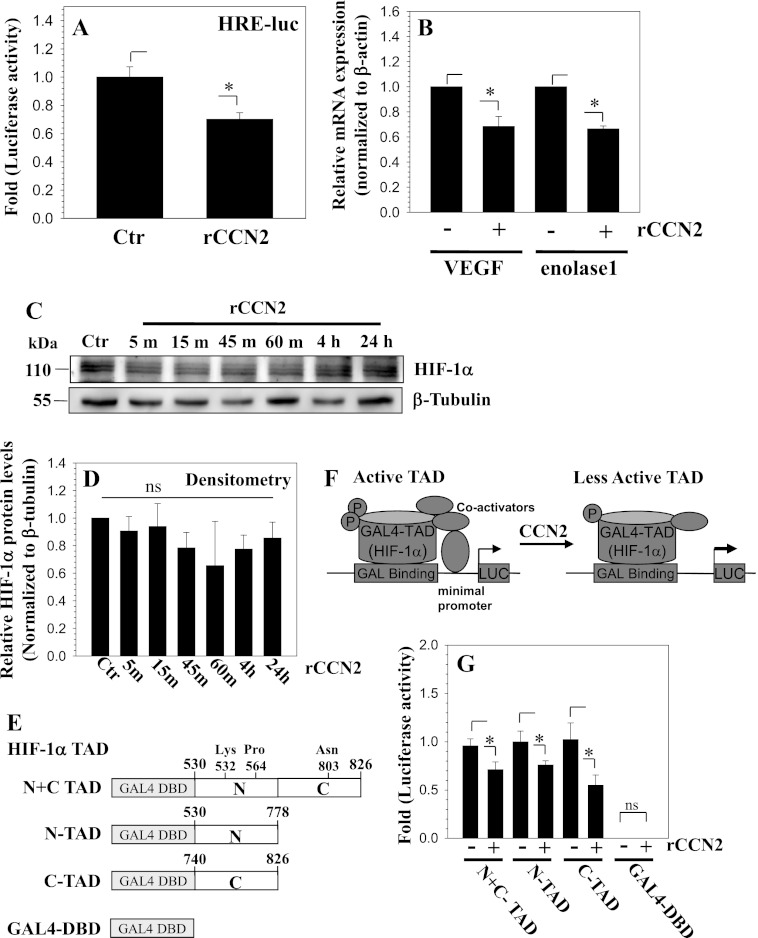
Next, we treated cells with recombinant CCN2 to examine if endogenous HIF-1α activity and expression is affected in a corresponding manner. Treatment with recombinant CCN2 for 24 h results in a significant decrease in HRE reporter activity (Fig. 8A). We then examined the expression of HIF-1α target genes, VEGF and ENOLASE 1, in response to CCN2 treatment and found that the expression of both genes is significantly decreased (Fig. 8B). Interestingly, Western blot and corresponding densitometric analysis showed that HIF-1α protein expression is not significantly affected by CCN2 treatment (Fig. 8, C and D). Next, we examined the mechanism by which CCN2 regulates HIF-1α activity. For this purpose, we tested if CCN2 treatment affects interactions between the transactivation domain of HIF-1α and its co-activators. Reporter constructs expressing both the N- and C-terminal TAD (Fig. 8E) were transfected into cells and their activity was measured following CCN2 treatment. Fig. 8, F and G, shows that the addition of CCN2 caused a significant decrease in the activity of all HIF-1 α-TAD reporters, indicating a decline in transcriptional activity. As expected the empty Gal4-DBD showed almost undetectable activity that remained unaffected by CCN2 treatment.
FIGURE 8.
CCN2 treatment decreases the activity of HIF-1α in nucleus pulposus cells. A, cells were transfected with a HIF-1 responsive luciferase reporter (HRE-luc) and activity measured following treatment with 100 ng/ml of rCCN2. CCN2 treatment significantly decreases HRE reporter activity. B, real time RT-PCR analysis of VEGF and ENOLASE 1 mRNA expression in cells treated with rCCN2. Both VEGF and ENOLASE 1 expression decreases with rCCN2 treatment. C, Western blot of HIF-1α in cells treated with rCCN2 for 5 min to 24 h. HIF-1α levels were largely unaffected by the treatment. D, densitometric analysis of three independent experiments in C shows that rCCN2 treatment does not significantly affect HIF-1α levels. E, schematic of TAD constructs used for transactivation studies of HIF-1α. F, schematic summarizing the role of CCN2 in controlling HIF-1α-TAD activity. G, cells were transfected with a Gal4 binary reporter system consisting of Gal4-HIF1α-TAD (HIF-1α N+C TAD, amino acids 530–826; N-TAD, amino acids 530–778; and C-TAD, amino acids 740–826) and pFR-Luc reporter and luciferase activity measured following rCCN2 treatment. Gal4DBD (empty vector pM) was used to measure background pFR-Luc activity. rCCN2 decreased the reporter activity indicating a decreased ability of all HIF-1α-TAD fusion proteins to recruit transcriptional co-activators. Background activity of Gal4DBD was almost undetectable and remained unaffected by CCN2 treatment. Values shown are mean ± S.E. from three independent experiments, *, p < 0.05.
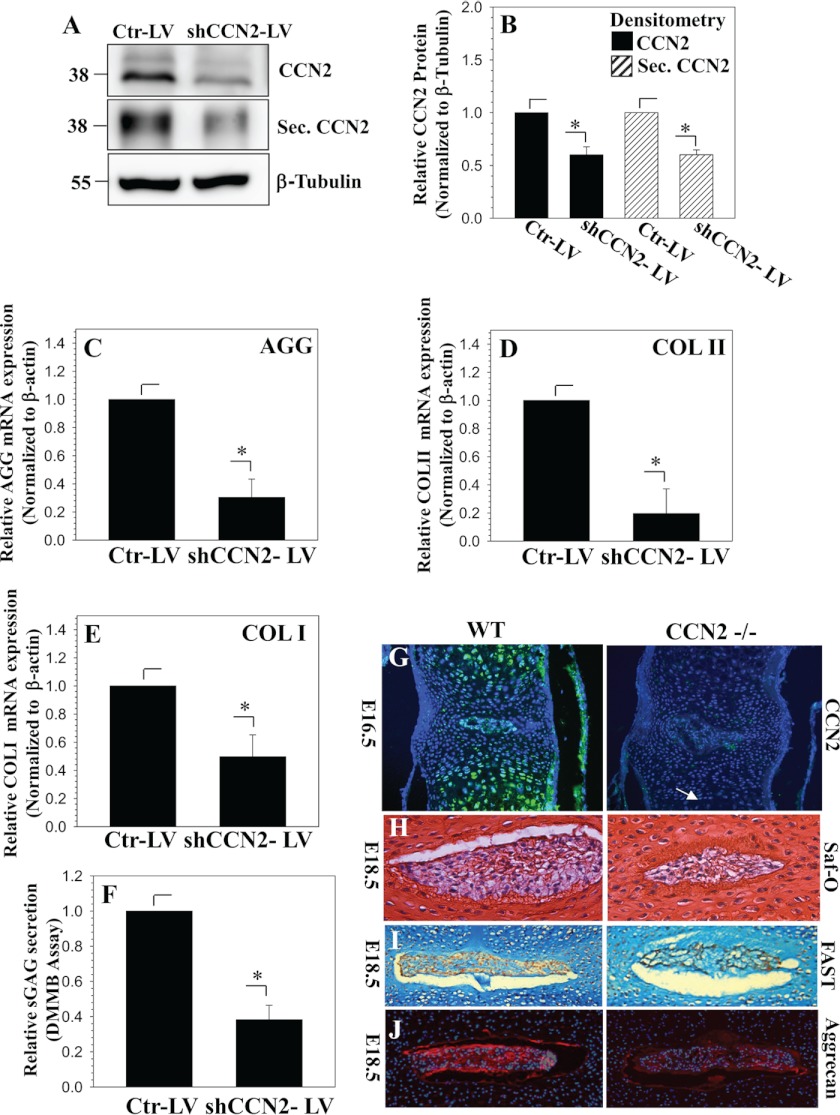
Finally, we measured the role of CCN2 in controlling the expression of AGGRECAN and COLLAGEN II, two important extracellular matrix genes for nucleus pulposus cells. Fig. 9, A and B, confirms that the lentiviral shCCN2 efficiently surpasses the level of cellular and secreted CCN2. Results show that the suppression of endogenous CCN2 results in a significant decrease in AGGRECAN (Fig. 9C) and COLLAGEN II (Fig. 9D) and COLLAGEN I (Fig. 9E) mRNA expression. Moreover, the level of sulfated glycosaminoglycans is significantly declined in CCN2-suppressed cells (Fig. 9F).
FIGURE 9.
CCN2 controls expression of extracellular matrix genes and accumulation of the proteoglycan matrix in nucleus pulposus cells. A, Western blot analysis of nucleus pulposus cells transduced with control shRNA (Ctr-LV) or shCCN2 (shCCN2-LV) lentivirus. B, densitometric analysis shows that cellular and secreted CCN2 is significantly decreased in cells transduced with shCCN2-LV (CCN2, black; secreted CCN2, diagonally striped). C and D, real time RT-PCR analysis shows that suppression of CCN2 results in a decrease of AGGRECAN (C), COLLAGEN II (D), and COLLAGEN I (E) expression. F, dimethylmethylene blue (DMMB) assay shows that knockdown of CCN2 results in decreased accumulation of sulfated glycosaminoglycan. Values shown are mean ± S.E. from three independent experiments, *, p < 0.05. G, immunofluorescence analysis of CCN2 expression in vertebral bodies from E16.5 WT (left) and CCN2−/− littermates (right). In the WT mouse, CCN2 is abundant in the hypertrophic cartilage of the vertebral body and in the nucleus pulposus. Hypertrophy of chondrocytes in the vertebral bodies appears to be delayed in mutants (arrow), but there are no obvious differences in the nucleus pulposus. H, sections through the spines of E18.5 WT (left) and mutant (right) littermates, stained with safranin-o (Saf-O). The relative level of safranin-o-stained material is decreased in mutants in the nucleus pulposus. I, images of the nucleus pulposus at E18.5 stained using the FAST protocol. J, immunofluorescence staining for aggrecan demonstrates decreased expression in the nucleus pulposus of CCN2 mutants.
CCN2 Promotes Extracellular Matrix Production by Nucleus Pulposus Cells
To support these in vitro silencing studies, we measured expression levels of proteoglycans in general and aggrecan in particular in discs of Ccn2 null mice. Fig. 9G shows that Ccn2 is expressed in the nucleus pulposus of the developing intervertebral disc. At E16.5, the nucleus pulposus is present in both WT and Ccn2 null littermates, and there are no obvious differences. However, differences were apparent in E18.5 by safranin-o (Fig. 9H) and FAST staining (Fig. 9I). FAST staining is a multiple dye staining procedure that includes fast green, Alcian blue, safranin-o, and tartrazine, and thus detects collagens and proteoglycans. As shown in Fig. 9, H and I, there is a relative paucity of safranin-o-stained proteoglycan in the nucleus pulposus of the mutant. Finally, we examined aggrecan levels by immunofluorescence (Fig. 9J). Consistent with the FAST staining analysis, lower levels of aggrecan are seen in the nucleus pulposis in Ccn2−/− mutants. Other details of the phenotypic analysis will be reported elsewhere, but the above results support the hypothesis that CCN2 is a regulator of aggrecan expression in the nucleus pulposus.
DISCUSSION
The experiments described in this investigation show that hypoxia regulates CCN2 expression through HIF-1α in nucleus pulposus cells. We demonstrate that although hypoxia decreases CCN2 expression and promoter activity in a HIF-1α-dependent manner, direct binding of HIF-1 to HREs in the proximal CCN2 promoter is not required. We also observed that CCN2 regulates HIF-1α expression and activity, which may play a role in tissue homeostasis in both normal and degenerate discs.
Initially, we focused on the regulation of CCN2 by hypoxia. We and others have shown that hypoxia is an overriding aspect of disc biology that regulates the expression of important cell survival and matrix genes in this tissue (7–10). Results of this study show that hypoxia decreased CCN2 transcription and protein levels in nucleus pulposus cells. Interestingly, in HCS-2/8 chondrosarcoma cells that are used to model chondrocyte behavior, hypoxia stabilizes CCN2 expression (37). Similarly, in several other cell types hypoxia was also shown to increase CCN2 expression (12, 27, 38); which in turn mediated the downstream effects of pathological hypoxia. Evidence to support the notion that CCN2 expression must be tightly regulated in tissues is apparent from transgenic mice that overexpress CCN2. Mice with two copies of Col1a2-Ccn2, which drives expression in fibroblasts, developed accelerated systemic multiorgan fibrosis compared with mice with only one copy of the transgene, which showed a much later onset of fibrosis and longer lifespan (39). Likewise, increased fibrosis was observed in mice with liver-specific CCN2 overexpression, but only upon tissue injury. This observation suggests that perhaps induced inflammatory mediators could interact with CCN2 to promote increased fibrosis or that the inflammatory microenvironment could change the response of cells to CCN2 (40). This is relevant because CCN2 levels are elevated during disc degeneration, a condition characterized by tissue inflammation (23). Thus, it is reasonable to assume that whereas CCN2 is expressed in the developing nucleus pulposus and plays a role in maintenance of normal levels of aggrecan in this hypoxic niche, tight control of CCN2 expression is necessary to prevent excessive accumulation and to regulate its biological function(18, 23, 32, 34).
We next examined the mechanism by which hypoxia suppresses CCN2 expression in nucleus pulposus cells. For this purpose, we chose to examine 4.0-kb proximal promoters of mouse and human CCN2. Importantly, studies of transgenic mice in which expression of nuclear-localized lacZ is driven by the 4-kb CCN2 proximal promoter have shown that this region is sufficient to drive high expression in the nucleus pulposus (34). We found that HIF-1α overexpression decreased CCN2 promoter activity, whereas siHIF-1α relieved hypoxic suppression of promoter activity. Further support for the role of HIF-1α in controlling CCN2 expression was forthcoming from loss- and gain-of-function experiments, which clearly showed a reciprocal relationship between HIF-1α and CCN2 levels in nucleus pulposus cells. These results were in accordance with previous studies showing the involvement of HIF-1α in hypoxic regulation of CCN2 in other cell types (12, 22, 27, 30, 42). Noteworthy, deletion and mutation analysis showed that the binding of HIF-1α to HREs contained within the studied promoter was not necessary for hypoxic repression of the activity.
The observation that CCN2 expression was less responsive to TGF-β treatment in hypoxia raised the possibility that hypoxia and HIF-1 may modulate canonical Smad signaling that is critical in maintenance of CCN2 expression in nucleus pulposus cells (23). Surprisingly, suppressive effects of hypoxia/HIF-1 did not involve interfering with the binding of Smad3 to the CCN2 promoter. This clearly suggests that in nucleus pulposus cells, regulation of CCN2 by HIF-1α is complex and may occur indirectly through modulation of binding of other regulatory factor(s) beside Smad3. This mode of regulation is similar to what we have previously observed in the context of HIF-1α regulation of Hsp70 and GlcAT-I promoters in nucleus pulposus cells (6, 11). Related to this discussion, one possibility is that hypoxia may exert its effects on CCN2 through the modulation of Fli1 or Ets-1, members of the Ets family of transcription factors. These factors function as molecular switches regulating a subset of target genes in response to physiological stimuli including hypoxia (43–45). Nakerakanti et al. (46) showed that Fli1 and Ets1 had differential effects on CCN2 expression in fibroblasts through differential occupancy of the Ets binding site, located at −114 bp in the proximal CCN2 promoter. Occupancy of this site by Fli1 had a suppressive effect on CCN2 gene expression, whereas occupancy by Ets-1 had a positive effect (46). It will be of interest to examine if, in nucleus pulposus cells, hypoxia controls the levels and binding ability of these factors to the CCN2 promoter in a HIF-1α-dependent fashion. This possibility is currently being examined.
We then investigated whether HIF-1α expression and activity is in turn controlled by CCN2. We observed an increase in HIF-1α and target gene expression with CCN2 knockdown. Correspondingly, decreased HRE reporter activity and decreased HIF-1α target gene expression following treatment of cells with rCCN2 suggested that CCN2 may be involved in the maintenance of basal HIF-1α levels. Although these results are in contradiction to studies that reported a CCN2-mediated increase in HIF-1α levels in stromal fibroblasts and chondrosarcoma cells (24, 47), they are in agreement with a previous report of human lung adenocarcinoma (31). These investigators showed that lung adenocarcinomas with high levels of CCN2 exhibited lower HIF-1α levels and that in cultured cells, CCN2 decreased HIF-1α levels by enhancing its degradation through acetylation of Lys-532 and subsequent association with pVHL. Noteworthy, in nucleus pulposus cells, unlike previous studies, CCN2 treatment decreased the activity of HIF-1α-TAD reporters, suggesting that in addition to maintenance of basal levels it may affect the activity of the HIF-1α transcriptional complex. Furthermore, our study shows that in contrast to HIF-1α, CCN2 is a positive regulator of HIF-2α expression. This is relevant, as we have previously reported that in nucleus pulposus, HIF-2α controls the expression of cited2, a p300 interacting protein (29). By competing with HIF-α for p300 binding, cited2 suppresses transcriptional activity of HIF (29). Thus, it is possible that through induction of the HIF-2α-cited2 circuit, CCN2 may decrease HIF-1α activity and target gene expression. The details of the mechanism by which CCN2 controls HIF-1α levels/activity in the nucleus pulposus is not yet known and is a subject of future investigation.
Last, our study provided some insight into the role of endogenous CCN2 in nucleus pulposus cells. Both in vitro loss-of-function studies using postnatal nucleus pulposus cells and analysis of Ccn2 null mouse embryos showed a decrease in proteoglycan levels and a specific decrease in aggrecan accumulation, although the effect on proteoglycan matrix levels in the embryonic discs seemed modest. However, as matrix accumulation and turnover in a healthy disc is a slow process (41), it is possible that the cumulative effect of a small but continuous decrease in matrix production over time will have a significant impact on the structure and function of postnatal discs. Because Ccn2 global null is perinatally lethal, a nucleus pulposus-specific deletion of Ccn2 will be required to further investigate this possibility in postnatal tissue. Taken together, our results confirm that basal CCN2 levels in nucleus pulposus cells play a role in maintenance of proteoglycan-rich extracellular matrix and tissue homeostasis. However, as both CCN2 and HIF-1α have anabolic function in nucleus pulposus cells (10, 23, 32), it is plausible that their effect on matrix production is likely to be independent of each other. Importantly, this negative feedback loop may serve to tightly control both CCN2 and HIF-1α levels in the healthy disc. In summary, we show for the first time that in the nucleus pulposus, hypoxia regulates CCN2 in a HIF-1α-dependent manner, whereas CCN2 controls HIF-1α levels and activity. Further studies are needed to determine how HIF-1α regulation of CCN2 and vice versa could impact the progression of degenerative disc disease.
This work was supported, in whole or in part, by National Institutes of Health Grants AR050087 and AR055655 (to M. R.) and AR052686 (to K. M. L.).
- HIF-1α
- hypoxia inducible factor-1α
- CCN2
- connective tissue growth factor
- HRE
- hypoxia response element
- DMOG
- dimethyl oxalylglycine
- TAD
- transactivation domain.
REFERENCES
- 1. Bartels E. M., Fairbank J. C., Winlove C. P., Urban J. P. (1998) Oxygen and lactate concentrations measured in vivo in the intervertebral discs of patients with scoliosis and back pain. Spine (Phila Pa 1976) 23, 1–7 [DOI] [PubMed] [Google Scholar]
- 2. Hassler O. (1969) The human intervertebral disc. A micro-angiographical study on its vascular supply at various ages. Acta Orthop. Scand. 40, 765–772 [DOI] [PubMed] [Google Scholar]
- 3. Rudert M., Tillmann B. (1993) Lymph and blood supply of the human intervertebral disc. Cadaver study of correlations to discitis. Acta Orthop. Scand. 64, 37–40 [DOI] [PubMed] [Google Scholar]
- 4. Rajpurohit R., Risbud M. V., Ducheyne P., Vresilovic E. J., Shapiro I. M. (2002) Phenotypic characteristics of the nucleus pulposus. Expression of hypoxia inducing factor-1, glucose transporter-1 and MMP-2. Cell Tissue Res. 308, 401–407 [DOI] [PubMed] [Google Scholar]
- 5. Fujita N., Chiba K., Shapiro I. M., Risbud M. V. (2012) HIF-1α and HIF-2α degradation is differentially regulated in nucleus pulposus cells of the intervertebral disc. J. Bone Miner. Res. 27, 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gogate S. S., Fujita N., Skubutyte R., Shapiro I. M., Risbud M. V. (2012) Tonicity enhancer binding protein (TonEBP) and hypoxia-inducible factor (HIF) coordinate heat shock protein 70 (Hsp70) expression in hypoxic nucleus pulposus cells. Role of Hsp70 in HIF-1α degradation. J. Bone Miner. Res. 27, 1106–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Risbud M. V., Guttapalli A., Stokes D. G., Hawkins D., Danielson K. G., Schaer T. P., Albert T. J., Shapiro I. M. (2006) Nucleus pulposus cells express HIF-1α under normoxic culture conditions. A metabolic adaptation to the intervertebral disc microenvironment. J. Cell. Biochem. 98, 152–159 [DOI] [PubMed] [Google Scholar]
- 8. Zeng Y., Danielson K. G., Albert T. J., Shapiro I. M., Risbud M. V. (2007) HIF-1α is a regulator of Galectin-3 expression in the intervertebral disc. J. Bone Miner. Res. 22, 1851–1861 [DOI] [PubMed] [Google Scholar]
- 9. Skubutyte R., Markova D., Freeman T. A., Anderson D. G., Dion A. S., Williams C. J., Shapiro I. M., Risbud M. V. (2010) Hypoxia-inducible factor regulation of ANK expression in nucleus pulposus cells. Possible implications in controlling dystrophic mineralization in the intervertebral disc. Arthritis Rheum. 62, 2707–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Agrawal A., Guttapalli A., Narayan S., Albert T. J., Shapiro I. M., Risbud M. V. (2007) Normoxic stabilization of HIF-1α drives glycolytic metabolism and regulates aggrecan gene expression in nucleus pulposus cells of the rat intervertebral disk. Am. J. Physiol. Cell Physiol. 293, C621–631 [DOI] [PubMed] [Google Scholar]
- 11. Gogate S. S., Nasser R., Shapiro I. M., Risbud M. V. (2011) Hypoxic regulation of β-1,3-glucuronyltransferase 1 expression in nucleus pulposus cells of the rat intervertebral disc. Role of hypoxia-inducible factor proteins. Arthritis Rheum. 63, 1950–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shimo T., Kubota S., Kondo S., Nakanishi T., Sasaki A., Mese H., Matsumura T., Takigawa M. (2001) Connective tissue growth factor as a major angiogenic agent that is induced by hypoxia in a human breast cancer cell line. Cancer Lett. 174, 57–64 [DOI] [PubMed] [Google Scholar]
- 13. Ivkovic S., Yoon B. S., Popoff S. N., Safadi F. F., Libuda D. E., Stephenson R. C., Daluiski A., Lyons K. M. (2003) Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development 130, 2779–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Braig S., Wallner S., Junglas B., Fuchshofer R., Bosserhoff A. (2011) CTGF is overexpressed in malignant melanoma and promotes cell invasion and migration. Br. J. Cancer. 105, 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hall-Glenn F., De Young R. A., Huang B. L., van Handel B., Hofmann J. J., Chen T. T., Choi A., Ong J. R., Benya P. D., Mikkola H., Iruela-Arispe M. L., Lyons K. M. (2012) CCN2/connective tissue growth factor is essential for pericyte adhesion and endothelial basement membrane formation during angiogenesis. PLoS One 7, e30562-e30562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoshijima M., Hattori T., Inoue M., Araki D., Hanagata H., Miyauchi A., Takigawa M. (2006) CT domain of CCN2/CTGF directly interacts with fibronectin and enhances cell adhesion of chondrocytes through integrin α5β1. FEBS Lett. 580, 1376–1382 [DOI] [PubMed] [Google Scholar]
- 17. Inoki I., Shiomi T., Hashimoto G., Enomoto H., Nakamura H., Makino K., Ikeda E., Takata S., Kobayashi K., Okada Y. (2002) Connective tissue growth factor binds vascular endothelial growth factor (VEGF) and inhibits VEGF-induced angiogenesis. FASEB J. 16, 219–221 [DOI] [PubMed] [Google Scholar]
- 18. Aoyama E., Hattori T., Hoshijima M., Araki D., Nishida T., Kubota S., Takigawa M. (2009) N-terminal domains of CCN family 2/connective tissue growth factor bind to aggrecan. Biochem. J. 420, 413–420 [DOI] [PubMed] [Google Scholar]
- 19. Gao R., Brigstock D. R. (2004) Connective tissue growth factor (CCN2) induces adhesion of rat activated hepatic stellate cells by binding of its C-terminal domain to integrin αvβ3 and heparan sulfate proteoglycan. J. Biol. Chem. 279, 8848–8855 [DOI] [PubMed] [Google Scholar]
- 20. Nishida T., Kawaki H., Baxter R. M., Deyoung R. A., Takigawa M., Lyons K. M. (2007) CCN2 (connective tissue growth factor) is essential for extracellular matrix production and integrin signaling in chondrocytes. J. Cell. Commun. Signal. 1, 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abreu J. G., Ketpura N. I., Reversade B., De Robertis E. M. (2002) Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-β. Nat. Cell Biol. 4, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Higgins D. F., Biju M. P., Akai Y., Wutz A., Johnson R. S., Haase V. H. (2004) Hypoxic induction of Ctgf is directly mediated by Hif-1. Am. J. Physiol. Renal Physiol. 287, F1223–1232 [DOI] [PubMed] [Google Scholar]
- 23. Tran C. M., Markova D., Smith H. E., Susarla B., Ponnappan R. K., Anderson D. G., Symes A., Shapiro I. M., Risbud M. V. (2010) Regulation of CCN2/CTGF expression in the nucleus pulposus of the intervertebral disc. Role of Smad and AP1 signaling. Arthritis Rheum. 62, 1983–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nishida T., Kondo S., Maeda A., Kubota S., Lyons K. M., Takigawa M. (2009) CCN family 2/connective tissue growth factor (CCN2/CTGF) regulates the expression of Vegf through Hif-1α expression in a chondrocytic cell line, HCS-2/8, under hypoxic condition. Bone 44, 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ponticos M., Holmes A. M., Shi-wen X., Leoni P., Khan K., Rajkumar V. S., Hoyles R. K., Bou-Gharios G., Black C. M., Denton C. P., Abraham D. J., Leask A., Lindahl G. E. (2009) Pivotal role of connective tissue growth factor in lung fibrosis. MAPK-dependent transcriptional activation of type I collagen. Arthritis Rheum. 60, 2142–2155 [DOI] [PubMed] [Google Scholar]
- 26. Lee C. H., Shah B., Moioli E. K., Mao J. J. (2010) CTGF directs fibroblast differentiation from human mesenchymal stem/stromal cells and defines connective tissue healing in a rodent injury model. J. Clin. Invest. 120, 3340–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hong K. H., Yoo S. A., Kang S. S, Choi J. J., Kim W. U., Cho C. S. (2006) Hypoxia induces expression of connective tissue growth factor in scleroderma skin fibroblasts. Clin. Exp. Immunol. 146, 362–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bennewith K. L., Huang X., Ham C. M., Graves E. E., Erler J. T., Kambham N., Feazell J., Yang G. P., Koong A., Giaccia A. J. (2009) The role of tumor cell-derived connective tissue growth factor (CTGF/CCN2) in pancreatic tumor growth. Cancer Res. 69, 775–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Agrawal A., Gajghate S., Smith H., Anderson D. G., Albert T. J., Shapiro I. M., Risbud M. V. (2008) Cited2 modulates hypoxia-inducible factor-dependent expression of vascular endothelial growth factor in nucleus pulposus cells of the rat intervertebral disc. Arthritis Rheum. 58, 3798–3808 [DOI] [PubMed] [Google Scholar]
- 30. Kroening S., Neubauer E., Wullich B., Aten J., Goppelt-Struebe M. (2010) Characterization of connective tissue growth factor expression in primary cultures of human tubular epithelial cells. Modulation by hypoxia. Am. J. Physiol. Renal Physiol. 298, F796–806 [DOI] [PubMed] [Google Scholar]
- 31. Chang C. C, Lin M. T., Lin B. R., Jeng Y. M., Chen S. T., Chu C. Y., Chen R.J., Chang K. J., Yang P. C., Kuo M. L. (2006) Effect of connective tissue growth factor on hypoxia-inducible factor 1α degradation and tumor angiogenesis. J. Natl. Cancer Inst. 98, 984–995 [DOI] [PubMed] [Google Scholar]
- 32. Erwin W.M., Ashman K., O'Donnel P., Inman R. D. (2006) Nucleus pulposus notochord cells secrete connective tissue growth factor and up-regulate proteoglycan expression by intervertebral disc chondrocytes. Arthritis Rheum. 54, 3859–3867 [DOI] [PubMed] [Google Scholar]
- 33. Chiou M. J., Chao T. T., Wu J. L., Kuo C. M., Chen J. Y. (2006) The physiological role of CTGF/CCN2 in zebrafish notochond development and biological analysis of the proximal promoter region. Biochem. Biophys. Res. Commun. 349, 750–758 [DOI] [PubMed] [Google Scholar]
- 34. Huang B. L., Brugger S. M., Lyons K. M. (2010) Stage-specific control of connective tissue growth factor (CTGF/CCN2) expression in chondrocytes by Sox9 and β-catenin. J. Biol. Chem. 285, 27702–27712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leung V. Y., Chan W. C., Hung S. C., Cheung K. M., Chan D. (2009) Matrix remodeling during intervertebral disc growth and degeneration detected by multichromatic FAST staining. J. Histochem. Cytochem. 57, 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sandelin A., Alkema W., Engström P., Wasserman W. W., Lenhard B. (2004) JASPAR. An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 32, D91–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kondo S., Kubota S., Mukudai Y., Nishida T., Yoshihama Y., Shirota T., Shintani S., Takigawa M. (2011) Binding of glyceraldehyde-3-phosphate dehydrogenase to the cis-acting element of structure-anchored repression in ccn2 mRNA. Biochem. Biophys. Res. Commun. 405, 382–387 [DOI] [PubMed] [Google Scholar]
- 38. Gupta S., Clarkson M. R., Duggan J., Brady H. R. (2000) Connective tissue growth factor. Potential role in glomerulosclerosis and tubulointerstitial fibrosis. Kidney Int. 58, 1389–1399 [DOI] [PubMed] [Google Scholar]
- 39. Sonnylal S., Shi-Wen X., Leoni P., Naff K., Van Pelt C. S., Nakamura H., Leask A., Abraham D., Bou-Gharios G., de Crombrugghe B. (2010) Selective expression of connective tissue growth factor in fibroblasts in vivo promotes systemic tissue fibrosis. Arthritis Rheum. 62, 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tong Z., Chen R., Alt D. S., Kemper S., Perbal B., Brigstock D. R. (2009) Susceptibility to liver fibrosis in mice expressing a connective tissue growth factor transgene in hepatocytes. Hepatology 50, 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sivan S. S., Tsitron E., Wachtel E., Roughley P. J., Sakkee N., van der Ham F., DeGroot J., Roberts S., Maroudas A. (2006) Aggrecan turnover in human intervertebral disc as determined by the racemization of aspartic acid. J. Biol. Chem. 281, 13009–13014 [DOI] [PubMed] [Google Scholar]
- 42. Rimon E., Chen B., Shanks A. L., Nelson D. M., Sadovsky Y. (2008) Hypoxia in human trophoblasts stimulates the expression and secretion of connective tissue growth factor. Endocrinology 149, 2952–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oikawa M., Abe M., Kurosawa H., Hida W., Shirato K., Sato Y. (2001) Hypoxia induces transcription factor ETS-1 via the activity of hypoxia-inducible factor-1. Biochem. Biophys. Res. Commun. 289, 39–43 [DOI] [PubMed] [Google Scholar]
- 44. Aryee D. N., Niedan S., Kauer M., Schwentner R., Bennani-Baiti I. M., Ban J., Muehlbacher K., Kreppel M., Walker R. L., Meltzer P., Poremba C., Kofler R., Kovar H. (2010) Hypoxia modulates EWS-FLI1 transcriptional signature and enhances the malignant properties of Ewing's sarcoma cells in vitro. Cancer Res. 70, 4015–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Klappacher G. W., Lunyak V. V., Sykes D. B., Sawka-Verhelle D., Sage J., Brard G., Ngo S. D., Gangadharan D., Jacks T., Kamps M. P., Rose D. W., Rosenfeld M. G., Glass C. K. (2002) An induced Ets repressor complex regulates growth arrest during terminal macrophage differentiation. Cell 109, 169–180 [DOI] [PubMed] [Google Scholar]
- 46. Nakerakanti S. S., Kapanadze B., Yamasaki M., Markiewicz M., Trojanowska M. (2006) Fli1 and Ets1 have distinct roles in connective tissue growth factor/CCN2 gene regulation and induction of the profibrotic gene program. J. Biol. Chem. 281, 25259–25269 [DOI] [PubMed] [Google Scholar]
- 47. Capparelli C., Whitaker-Menezes D., Guido C., Balliet R., Pestell T. G., Howell A., Sneddon S., Pestell R. G., Martinez-Outschoorn U., Lisanti M. P., Sotgia F. (2012) CTGF drives autophagy, glycolysis and senescence in cancer-associated fibroblasts via HIF1 activation, metabolically promoting tumor growth. Cell Cycle 11, 2272–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]