Background: Prostaglandin E2 (PGE2) induces tumor growth and angiogenesis.
Results: PGE2-induced ERK activation in endothelial cells and angiogenesis are driven by syndecan-4-dependent PKCα activation.
Conclusion: The syndecan-4/PKCα/ERK pathway is important for PGE2-induced angiogenesis in vitro and in vivo.
Significance: Sdc4/PKCα activation is a novel finding in PGE2 signaling and may represent a pharmacological target.
Keywords: Angiogenesis, Cell Signaling, Prostaglandins, Protein Kinase C (PKC), Proteoglycan, Syndecan 4
Abstract
Prostaglandin E2 (PGE2) is regarded as the main mediator of inflammatory symptoms. In addition, it also plays an important role in tumor growth and angiogenesis. In this study, we examined the mechanism of PGE2-induced angiogenic response. We show that in the absence of proteoglycan syndecan-4 (Sdc4), PGE2-induced ERK activation is decreased significantly, as is endothelial cell migration and cord formation in a two-dimensional Matrigel assay. In vivo, PGE2-induced angiogenesis is reduced dramatically in Sdc4−/− mice. The mechanism was traced to Sdc4-dependent activation of protein kinase Cα (PKCα). Transduction of an Sdc4 S183E mutant (a cytoplasmic domain mutation that blocks Sdc4-dependent PKCα activation) into Sdc4−/− endothelial cells was not able to rescue the loss of PGE2-induced ERK activation, whereas a transduction with full-length Sdc4 resulted in full rescue. Furthermore, PGE2-induced angiogenesis was also reduced in PKCα−/− mice. Taken together, these results demonstrate that PGE2-induced activation of angiogenesis is mediated via syndecan-4-dependent activation of PKCα.
Introduction
Prostaglandins (PGs)2 and thromboxane A2 (TXA2), termed together prostanoids, are a class of lipid mediators generated by cyclooxygenase enzymes and terminal synthase enzymes starting from arachidonic acid. Among prostanoids, prostaglandin E2 (PGE2) production has been linked with acute inflammatory symptoms (redness, swelling, fever, and pain). However a large body of evidence indicates that uncontrolled PGE2 synthesis plays a major role in sustaining chronic inflammation (1) and promoting angiogenesis and tumor growth (2).
PGE2 exerts its effects via four related G-protein coupled receptors, termed E-prostanoid receptors (EP1–4). EP receptors couple to a striking variety of signaling pathways. EP2 and EP4 couple to Gs, which increases intracellular cAMP and activates protein kinase A. EP1 activates Gq, which initiates the Phospholipase C/IP3 pathway. EP3 can induce Gi activation, which leads to inhibition of cAMP synthesis. Furthermore, the complexity of PGE2-induced signaling is increased by the existence of cross-talk between EPs and tyrosine kinase receptors such FGFR1 (3) or EGFR (4). Among PGE2 receptors, EP4 is particularly important in the vasculature. A specific endothelial deletion of EP4 decreases reperfusion after ischemia and increases infarct size in the brain (5). EP4 has also been identified as the major EP receptor mediating effects of PGE2-induced tumor angiogenesis and lymphangiogenesis (6).
In this study, we investigated the role of syndecan-4 (Sdc4), a transmembrane proteoglycan that belongs to the four-member syndecan family (Sdc1–4) in PGE2 signaling. Sdc4 can act as a FGFR1 coreceptor and modulate its signaling (7), or it can also signal in an independent manner (8). Sdc4 signaling can lead to either activation of ERK and cell proliferation (8, 9) or of Rac1, leading to cell migration (10). Both are dependent on activation of PKCα, an event that requires dephosphorylation of a Ser-183 site in its cytoplasmic domain (11).
Sdc4 null mice are viable but show a number of defects in inflammatory settings, such as delayed wound healing (12), decreased myocardial infarction recovery (13), and impaired response to LPS injection (14).
We find that in Sdc4−/− mice and Sdc4−/− endothelial cells, PGE2-induced ERK activation is decreased. We further show that PGE2 induces ERK activation by activating PKCα in an Sdc4-dependent manner and that activation of Sdc4-PKCα-ERK is involved in modulation of PGE2-dependent proangiogenic effects (i.e. migration and endothelial cord formation). In agreement with these results, Sdc4−/− and PKCα−/− mice show a reduction in PGE2-induced angiogenesis in vivo. Thus, Sdc4 plays a key role in mediation of PGE2 angiogenic signaling.
EXPERIMENTAL PROCEDURES
Cell Cultures
Human umbilical vein endothelial cells (HUVEC) were obtained from the Yale tissue culture core laboratory at passage 1 and maintained in complete M199 medium (Invitrogen), which contains 20% FBS, penicillin (100 units/ml), streptomycin (100 μg/ml), non-essential amino acid (1×), gentamicin (100 μg/ml), amphotericin B (2.5 μg/ml), heparin (100 μg/ml), and endothelial cell growth supplement (100 μg/ml) (Biomedical Technologies). HUVEC were used for experiments between P2 and P6.
Primary mouse EC were isolated as described previously (15). Briefly, organs (heart, lung, vein, and artery) of both wild-type and knockout mice were harvested, finely minced with scissors, and digested with 25 ml collagenase (2 mg/ml) at 37 °C for 45 min under gentle agitation. The crude preparation was triturated, passing it 12 times through a cannula needle, and then filtered on a 70-μm sterile cell strainer. The filtered preparation was spun at 400 × g, and the pellet was resuspended in 2 ml of 0.1% BSA. For EC selection, magnetic beads (Invitrogen) coated with anti-mouse CD31 (BD Biosciences) were added to the cell suspension and incubated with rotation at room temperature for 15 min. The bead-bound cells were recovered with a magnetic separator and washed with DMEM containing 20% FBS. Cells were suspended in 10 ml of complete DMEM and seeded on 10-cm plates. Lenti-XTM HEK 293T (Clontech) and HEK 293A (Invitrogen) cells were cultured in 10% FBS DMEM (Invitrogen) containing penicillin/streptomycin at the same concentration as indicated above.
Virus Generation
Lentivirus for stable shRNA integration into the host genome were generated as described previously (16). Briefly, packaging plasmids (Addgene) were mixed with shRNA plasmid (Mission® shRNA, Sigma-Aldrich) in Optimem® medium (Invitrogen) and Lipofectamine 2000 (Invitrogen) with the following ratios: 5 μg pMDL/pRRE, 2.5 μg pRSV-Rev, 2.5 μg pVCMV-VSG, and 10 μg shRNA. The mixture was transferred to 90% confluent 293T cells in 10-cm dishes for 6 h. The medium was replaced with regular DMEM 10% FBS and collected after 48 h. Medium containing virus was filtered through a 0.45-μm filter and used immediately for HUVEC transduction. The target sequence for human Sdc4 was 5′-CCGGGCCAGGTTCTTCTTGAGCTTTCTCGAGAAAGCTCAAGAAGAACCTGGCTTTTTG-3′.
Site-directed mutagenesis of Sdc4 was conducted using the QuikChange II XL site-directed mutagenesis kit (Stratagene) according to the instructions of the manufacturer. PCR reactions were assembled and performed under the following conditions: annealing, 60 °C, 50 s; extension, 68 °C, 7 min; and denaturation, 95 °C 50 s, 18 cycles. Then the DNA resulting from the PCR was digested to remove the template DNA, and the remaining plasmids were transformed into One Shot Top 10 Escherichia coli (Invitrogen). The mutated clones were selected and confirmed by sequencing. Adenoviruses expressing a rat full-length or mutated rat Sdc4 sequence were then generated as reported previously (16). Briefly, HA-tagged full-length or mutated Sdc4 coding sequences were subcloned into pENTR/D (Invitrogen) and then transferred into the adenovirus vector pAD/CMV/V5-DEST (Invitrogen). The adenovirus was generated by transfection of this plasmid into HEK 293A (Invitrogen) according to the instructions of the manufacturer.
Stable Knockdown and Silencing Experiments
To achieve a stable knockdown, HUVEC P2 were seeded on 10-cm plates and transduced at 70% confluence with freshly produced lentivirus carrying a Scrambled or Sdc4 shRNA sequence and expressing the puromycin-resistant gene. Cells were kept with virus-rich medium for 6 h, and then the medium was replaced with complete M199 medium (same ingredients reported in cell cultures paragraph). Forty-eight hours post-infection, puromycin (0.8 μg/ml) was added to cells, and selection was allowed for 3 days. Cells were used in the experiment or split for propagation. Selected cells were maintained in complete M199 medium with puromycin (0.4 μg/ml) and used for a maximum of two more passages after initial selection.
For PKCα silencing, HUVEC were seeded on 6-well plates and transfected at 70% confluence. PKCα or Scrambled siRNA (Origene) were resuspended in the provided buffer, and transfection was done using Lipofectamine RNAiMAX (Invitrogen) according to the instructions of the manufacturer. Cells were used for experiments 72 h post-transfection.
Western Blot Analysis
HUVEC or primary mouse EC were seeded onto 6-cm plates. Confluent cells were starved overnight (HUVEC) or 48 h (mouse EC) in 0.5% FBS and then stimulated with the indicated agent. For inhibition experiments, the PI3K inhibitor LY290042 (50 μm) and EP4 antagonist AH23848 (10 μm) (17) were preincubated for 30 min prior PGE2 treatment. Rescue experiments were carried out by infecting HUVEC with adenovirus (multiplicity of infection = 10) for 6 h and then starved for 18 h in 0.5% FBS. For cell stimulation, the PGE2 concentration (100 nm) was the same except were indicated. Following stimulation, cells were rapidly washed twice with ice-cold PBS and lysed with 200 μl of 0.1% TritonX-100 lysis buffer (Cell Signaling Technology, Inc.) containing protease inhibitor (Roche) and phosphatase inhibitor (Roche) mixtures. Total lysates were cleared with a 15,000 × g spin, and protein concentration was determined using the BCA method (Thermo-Scientific). The protein concentration of each lysate was adjusted accordingly, added to 1× reducing loading buffer, and boiled for 5 min. Samples were loaded on 4–15% gels for SDS-PAGE separation and then transferred to an Immobilon-P membrane (Millipore). Membranes were blocked 1 h with 5% fat dry milk in Tris-buffered saline containing 0.05% Tween20 (TBS-T) and then incubated overnight at 4 °C with primary antibody. Protein bands were visualized using HRP-conjugated secondary antibodies associated to enhanced chemiluminescence (ImmobilonTM Western, Millipore).
Densitometric Quantification
The signal from the chemiluminescence reaction was recorded in a digital acquisition system (G-Box by Syngene) equipped with a 1.4-megapixel charge-coupled device (CCD) camera with a “true” 1.4-megapixel resolution. The linear range is automatically calculated by the software and is displayed as a histogram with each acquired image. Multiple images of the same blot were acquired with incremental 1-min exposure. Images without band saturation were used for densitometric quantification. The total intensity of each band was determined with ImageJ software (18) as described, following published guidelines for background correction (19). For determination of phosphorylation levels, controls were always repeated in each experiment and loaded side-by-side with treated samples in the same gel. This allows each experiment to develop all sample signals in the same acquired image. Samples were probed with an antibody that recognizes the phosphorylated form (e.g. pERK) and with another one that recognizes both phosphorylated and non-phosphorylated form (e.g. tERK). After quantification (see above), the band intensity of the phosphorylated protein was normalized to the intensity of total protein in the same sample. These normalized values were used for calculation of the phosphorylation fold change in treated versus control samples. Fold change values were collected from replicated independent experiments (n = at least 3) and used for statistical analysis.
RNA Isolation and Real-time PCR
Cells were washed twice with PBS and homogenized with a QIAshredder kit (Qiagen). Total RNA was extracted with an RNeasy Plus mini kit (Qiagen), which allows DNA elimination. cDNA synthesis was performed with an iScript cDNA synthesis kit (Bio-Rad). Quantitative real-time PCR was performed in triplicate using an iQ SYBR Green Supermix kit and CFX96TM real-time system (Bio-Rad). Thermocycling condition were as follows: 95 °C for 3 min, followed by 45 cycles at 95 °C for 10 s and 60 °C for 30 s. Gene expression was normalized with the housekeeping gene (GAPDH), and relative expression was calculated using the ΔΔCt method. Primers for mouse EC were as follows: EP1, 5′-CATCCATCACTTCAGCCACA-3′ (sense) and 5-CGCTGCAGGGAGTTAGAGTT-3′ (antisense); EP2, 5′-AGAGGACTTCGATGGCAGAG-3′ (sense) and 5′-GGAGGTCCCACTTTTCCTTT-3′(antisense); EP3, 5′-GGATCATGTGTGTGCTGTCC-3′ (sense) and 5-CCCATCTGTGTCTTGCATTG-3′(antisense); EP4, 5′-TCTCTGGTGGTGCTCATCTG-3′ (sense) and 5′-ATGGGGTTCACAGAAGCAAT-3′ (antisense). Primers for HUVEC were as follows: first set, EP1, 5′-TTGTCGGTATCATGGTGGTG-3′ (sense) and 5′-ATGTACACCCAAGGGTCCAG-3′ (antisense); EP2, 5′-CCACCTCATTCTCCTGGCTA-3′ (sense) and 5′-TTCCTTTCGGGAAGAGGTTT-3′ (antisense); EP3, 5′-AGCTTATGGGGATCATGTGC-3′(sense) and 5′-TTTCTGCTTCTCCGTGTGTG-3′(antisense); EP4, 5′-TGCCGCGCCTCAGCGACTTTC-3′ (sense) and 5′-AATTCGGATGGCCTGCAAATCTGG-3′ (antisense). Primers for the second set were as follows: EP1, 5′-GGAAGAGGGAGGGAGGAAG-3′(sense) and 5′-GCAAGGGCTCATGTCAGG-3′ (antisense); EP2, 5′-GTCTGCTCCTTGCCTTTCAC-3′ (sense) and 5′-AACAGGAGGCCTAAGGATGG-3′ (antisense); EP3, 5′-GGTCTCCGCTCCTGATAATG-3′ (sense) and 5′-ACAGCAGGTAAACCCAAGGA-3′ (antisense); EP4, 5′-CGAGATCCAGATGGTCATCTTAC-3′ (sense) and 5′-TGGCTGATATAACTGGTTGACG-3′ (antisense). GAPDH primers were as follows: mouse, 5′-AACTTTGGCATTGTGGAAGG-3′ (sense) and 5′-ACACATTGGGGGTAGGAACA-3′ (antisense); and human, 5′-GAGTCAACGGATTTGGTCGT-3′ (sense) and 5′-GACAAGCTTCCCGTTCTCAG-3′ (antisense).
Scratch Assay
Cells were seeded on 6-well plates, allowed to reach confluence, and then starved overnight in 0.5% FBS. Each well was marked below the plate surface by drawing a vertical line. This allowed identification of the same scratched area to take consistent pictures. After overnight starving, five different scratches intercepting the marked line were done in each well using a 200-μl sterile tip. Picture of scratches were taken just before stimulation (time 0) and after 8 h (time 8). Except where indicated, the same concentration of PGE2 (100 nm) was used for the scratch assay. Migration was calculated using the scratch area at time 0 (At0) and the correspondent scratch area at time 8 (At8). The scratch area was measured using ImageJ software. Migration was expressed as the percentage of scratch closure after 8 h compared with the initial area according to the following formula: % closure = [(At0 - At8)/At0]×100. Migration in each well is defined by the mean closure of five different scratches.
In Vitro EC Cord Formation
Cells were starved overnight in 0.5% FBS, detached with trypsin, and seeded again in 12-well plates coated previously with 300 μl of reduced-growth factor Matrigel (BD Biosciences). 80,000 cells/well were seeded in 0.5% FBS with or without PGE2 (100 nm) or VEGF-A as a positive control. 7 h after cell seeding, five random pictures (×40 magnification) were taken in each well, and the total length of cord formation was quantified in each field. The mean of five total lengths/well represents an independent experimental point. Cord length in each random field was assessed using the NeuronJ plug-in (20) for ImageJ.
In Vivo Angiogenesis
In vivo angiogenesis was evaluated by Matrigel plug assay as reported previously (21) with modifications. Briefly, 8-week-old male mice were injected subcutaneously with reduced-growth factor Matrigel (BD Biosciences) in the flank area. Matrigel plugs containing either vehicle, PGE2 (10 μm), or VEGF-A (100 ng/ml + 5 units of heparin) were removed 7 days after injection, embedded in Optimum Cutting Temperature (O.C.T.) compound (Sakura Finetek), and allowed to solidify on a dry ice bed. Embedded plugs were cryosectioned (10-μm thickness) and processed by fixation and staining. Sections were stained with CD31 primary antibody (BD Biosciences) followed by fluorescence-conjugated secondary antibody. Four random images from each section were acquired and used for quantification of angiogenesis (see Fig. 5 for details).
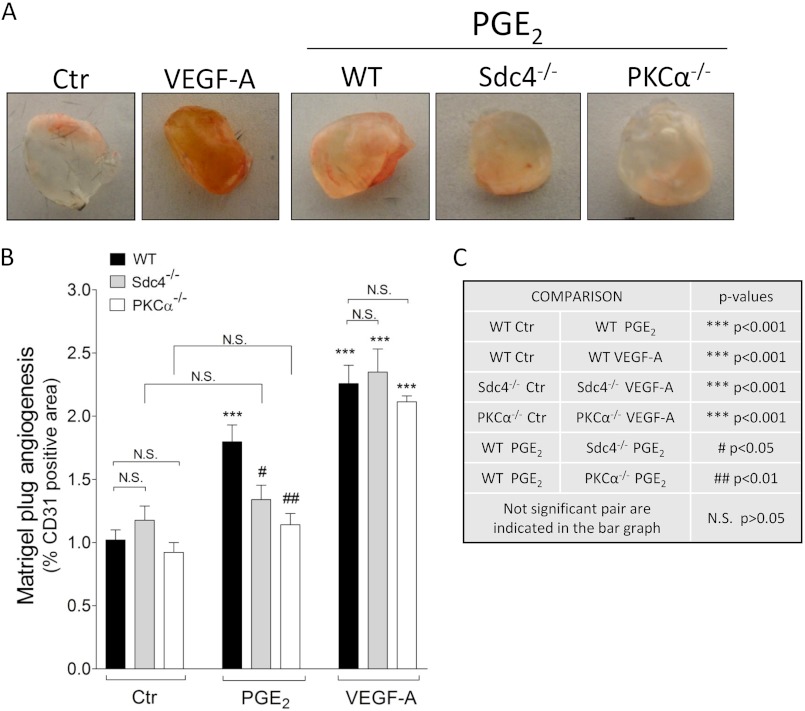
FIGURE 5.
Sdc4 and PKCα mediate PGE2-induced in vivo angiogenesis. Matrigel containing either vehicle, PGE2 (10 μm), or VEGF-A (100 ng/ml) was injected in the mouse flank area and excised 7 days later to evaluate angiogenesis. A, representative pictures of plugs extracted from WT, Sdc4−/−, or PKCα−/− mice. Ctr, control. B, quantification of Matrigel plug angiogenesis. Angiogenesis was evaluated by staining four different sections/plug and acquiring five random pictures in each section. Angiogenesis in each plug was calculated as mean of CD31-positive fluorescence pixels in the total pixel area. The diagram was generated by means derived from multiple plug experiments. Data are mean ± S.E., n = 4–12. C, explanation of the statistical symbols and p values in B.
Chemicals and Antibodies
Chemicals were purchased as follows: penicillin, streptomycin, gentamicin, and non-essential amino acid (100×) from Invitrogen; amphotericin B from Cellgro; heparin, FBS, and PGE2 from Sigma-Aldrich; LY290042 from Cell Signaling Technology, Inc.; AH28348 from Cayman Chemicals; and VEGF-A from R&D Systems. Antibodies were purchased as follows: phospho-p44/42 MAPK (pERK), p44/p42 MAPK (total ERK), phospho-AKT (Ser-473), AKT (total AKT), PKCα, and HA tag from Cell Signaling Technology, Inc.; EP4 (N-terminal region, extracellular domain) from Cayman Chemicals; tubulin (custom-made); Sdc4 (Ser-184 region, intracellular domain) from Abcam; and GFP from Santa Cruz Biotechnology, Inc.
Statistics
All statistical analyses were done using GraphPad Prism (GraphPad). Student's t test comparison was used in two-group data sets. One-way analysis of variance followed by Bonferroni or Dunnet post-tests was used in more than two-group data sets.
RESULTS
Sdc4 Regulates PGE2-induced ERK Activation
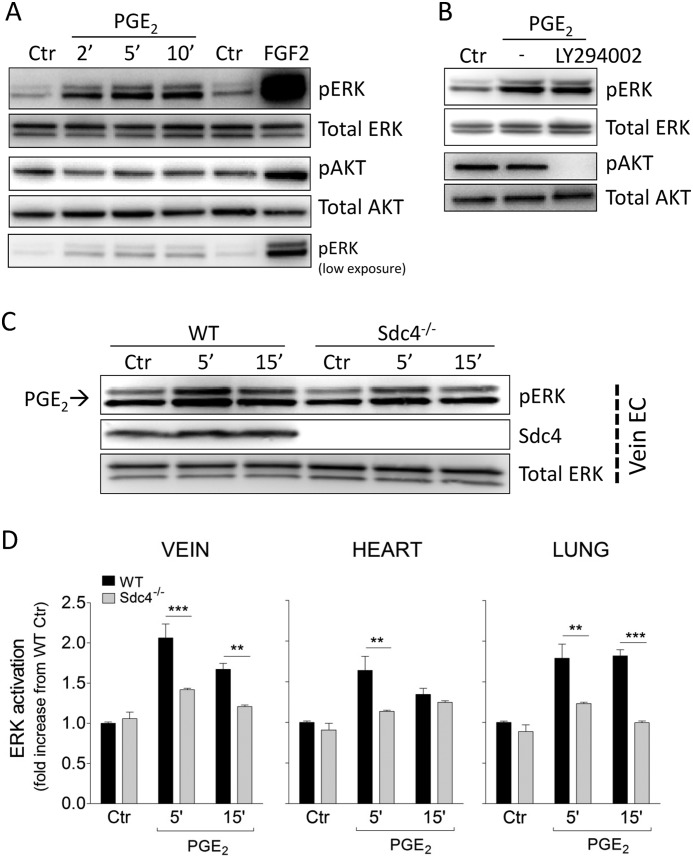
In agreement with previous studies that have shown that PGE2 induces ERK activation in EC, HUVEC displayed ERK activation as early as 2 min after PGE2 administration (Fig. 1A). Although in non-endothelial cell types PGE2-induced ERK activation has been shown to involve a PI3K-dependent mechanism (22, 23), in agreement with previous publications (24) we could not detect PGE2-induced AKT activation in HUVEC (Fig. 1A). Furthermore, PGE2-induced ERK activation could not be blocked with the PI3K inhibitor LY292000 (50 μm) (Fig. 1B).
FIGURE 1.
Sdc4 regulates PGE2-induced ERK activation. A, confluent HUVEC were serum-starved and then treated with PGE2 for the indicated times to assess ERK activation (pERK) and AKT activation (pAKT). Ctr, control. B, HUVEC were preincubated with the PI3K inhibitor LY294002 (50 μm) for 30 min before PGE2 stimulation for 5 min. C, primary mouse EC were isolated from WT or Sdc4−/− mice as described under “Experimental Procedures.” Confluent endothelial cells derived from the vein were serum-starved and then treated with PGE2 for the indicated times. Shown is one representative blot of three with similar results. D, pERK quantification in WT versus Sdc4−/− following treatment with PGE2 in EC isolated from different tissues (vein, lung, and heart). Each diagram is derived from three independent experiments (n = 3). Bars represent mean ± S.E. **, p < 0.01; ***, p < 0.001.
Because PGE2-induced ERK activation in EC has been shown to require FGFR1 transactivation (3), we next examined whether Sdc4, which can interact with and modulate FGFR signaling, plays a role in PGE2-induced ERK activation. To investigate this possibility, primary EC were isolated from WT or Sdc4−/− mice and treated with PGE2. Western blot analysis of vein EC stimulated with PGE2 demonstrated prompt activation in EC isolated from WT but not Sdc4−/− mice (Fig. 1C). Quantitative analysis of this blot as well as of blots of EC from hearts and lungs of wild-type and Sdc4−/− mice showed a 1.5- to 2-fold increase in ERK activation after 5-min stimulation with PGE2 in wild-type but not knockout EC (Fig. 1D).
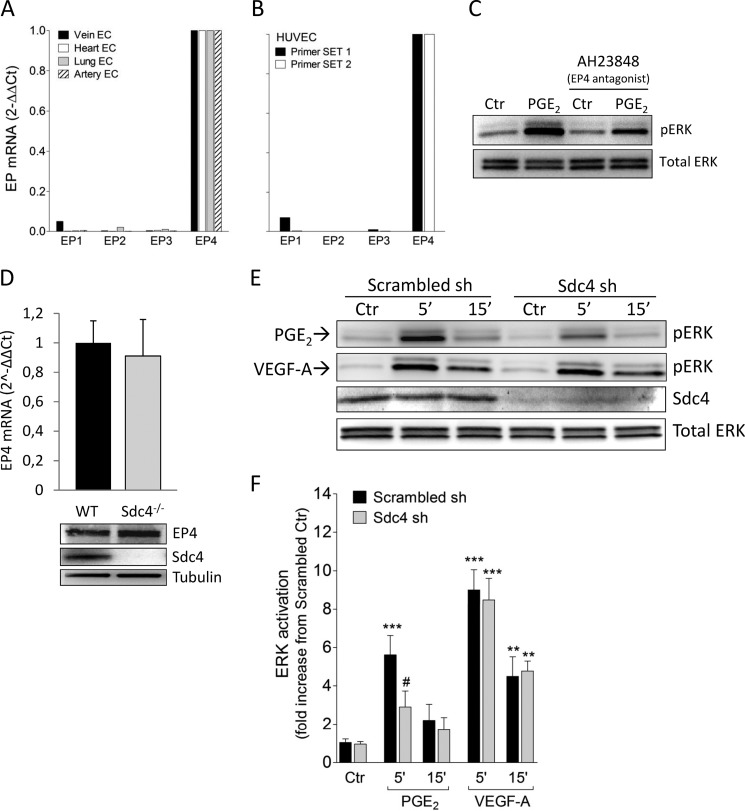
EP4 is thought to be the principal prostanoid receptor responsible for PGE2-induced ERK activation (24). In agreement with this, EP4 was the highest-expressed receptor in HUVEC (Fig. 2B) and in mouse EC isolated from blood vessel of four different tissues (A). Mean Ct (threshold cycle) values were as follows: hEP1, 27.0; hEP2, 35.1; hEP3, 29.1; hEP4, 22.4; mEP1, 27.5; mEP2, 31.0; mEP3, 30.5; and mEP4, 21.3. Furthermore, inhibition of PGE2-induced ERK activation was achieved in the presence of the EP4-selective antagonist AH23848 (10 μm) (Fig. 2C). One potential reason for decreased PGE2-induced ERK activation in Sdc4−/− EC could be reduced EP4 expression. However, both quantitative real-time PCR and Western blot analysis did not show differences in EP4 expression level between Sdc4−/− and WT EC (Fig. 2D).
FIGURE 2.
Sdc4 knockdown in HUVEC decreases PGE2-induced ERK activation. A, relative mRNA expression of EP isoforms in primary mouse EC isolated from blood vessels of four different tissues. B, EP expression in HUVEC using two different quantitative real-time PCR primer sets. C, HUVEC were pretreated with a selective EP4 antagonist, AH23848 (10 μm), for 30 min before PGE2 stimulation for 5 min. Ctr, control. D, mRNA (top panel) and protein level (bottom panel) of EP4 in EC isolated from WT or Sdc4−/− mice. E, Scrambled-infected (Scrambled sh) or Sdc4 knockdown HUVEC (Sdc4 sh) were treated with PGE2 or VEGF-A (50 ng/ml) for the indicated times. F, quantification of pERK (ERK activation) relative to the experimental protocol in E. The bars represent mean ± S.E. (n = 3–8). **, p < 0.01; ***, p < 0.001 from Ctr; #, p < 0.05 from Scrambled sh 5′ PGE2.
We then decided to use HUVEC to test whether human EC showed a similar effect as mouse EC with regard to the role of Sdc4 in PGE2 signaling. To this end, we knocked down Sdc4 in HUVEC using lentivirus carrying shRNA followed by stable selection with puromycin, achieving more that 80% Sdc4 knockdown, as shown by Western blot analysis (Fig. 2E). Knockdown of Sdc4 in HUVEC (Sdc4 sh) led to a similar decrease in PGE2-induced ERK activation (Fig. 2, E and F), as was observed in mouse Sdc4−/− cells. In contrast, ERK activation induced by VEGF-A was not affected by the knockdown of Sdc4 (Fig. 2, E and F).
PKCα Mediates PGE2-induced ERK Activation via Sdc4
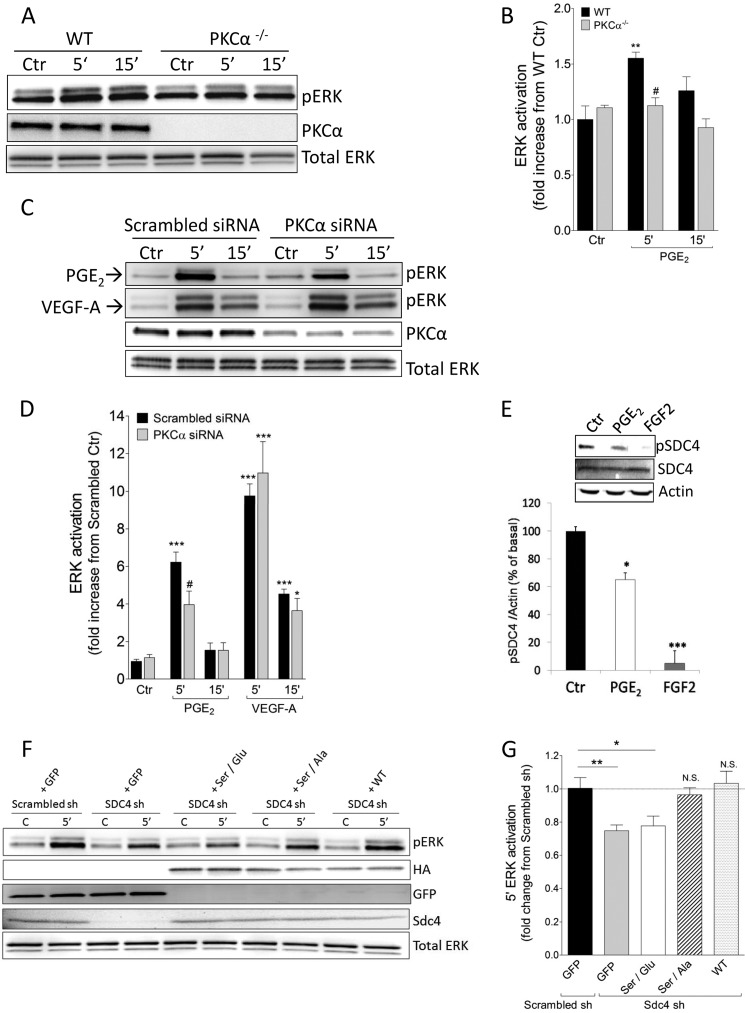
We next addressed the mechanism involved in Sdc4-dependent activation of ERK by PGE2. A key signaling event attributed to Sdc4 is recruitment and activation of PKCα in response to ligand-induced oligomerization (25, 26). To check whether PKCα is involved in PGE2-induced ERK activation, we first used mouse EC isolated from PKCα−/− mouse hearts. Compared with WT EC, PGE2-induced ERK activation was reduced significantly in PKCα−/− EC (Fig. 3, A and B). To verify this, we next knocked down PKCα in HUVEC (PKCα siRNA) and also observed a significant reduction in PGE2-induced ERK activation (Fig. 3, C and D). ERK activation induced by VEGF-A was not affected by the knockdown of PKCα in HUVEC (Fig. 3, C and D).
FIGURE 3.
PKCα mediates PGE2-induced ERK activation via Sdc4. A, primary mouse EC isolated from heart were treated with PGE2 for the indicated times. Ctr, control. B, pERK quantification in WT and PKCα−/− mice treated with PGE2. The bars represent mean ± S.E. (n = 3). **, p < 0.01 from WT Ctr; #, p < 0.05 from WT 5′ PGE2. C, Scrambled-transfected (Scrambled siRNA) or PKCα knockdown HUVEC (PKCα siRNA) were treated with PGE2 or VEGF-A (50 ng/ml) for the indicated times. D, pERK quantification relative to the experimental protocol of E. The bars represent means ± S.E. (n = 3–7). *, p < 0.05; ***, p < 0.01 from Ctr; #, p < 0.05 from Scrambled siRNA 5′ PGE2. E, HUVEC were treated with PGE2 or FGF2 (50 ng/ml) for 5 min, and Sdc4 phosphorylation at Ser-183 was evaluated by Western blot analysis. Top panel, representative Western blot analysis. Bottom panel, quantification of three independent experiments. *, p < 0.05; ***, p < 0.001. F, rescue experiment in Sdc4 knockdown HUVEC (Sdc4 sh) using a mutated rat Sdc4 sequence. Scrambled sh or Sdc4 sh HUVEC were infected with the indicated adenovirus for 6 h and then starved 18 h before stimulation with PGE2 for 5 min. C, control, no stimulation with PGE2. G, pERK quantification relative to the experimental protocol in F. The bars represent mean ± S.E. (n = 7–10). *, p < 0.05; **, p < 0.01; N.S., not significant from Scrambled sh GFP.
Prior studies have established that dephosphorylation of Ser-183 in Sdc4 cytoplasm is required for PKCα activation (27, 28). PGE2 treatment induced Ser-183 dephosphorylation in HUVEC (Fig. 3E) that is similar but not as extensive as dephosphorylation induced by FGF2, a known activator of this pathway (27). To verify that Sdc4-dependent activation is indeed central to PGE2-induced ERK activation, we mutated Ser-183 site to augment or inhibit Sdc4-dependent PKCα activation. Two mutants were used: a Ser-to-Ala mutation (S183A), which would be expected to favor PKCα activation, and S183E, which would be expected to inhibit it. Introduction of these mutant constructs in adenoviral vectors into Sdc4 sh HUVEC demonstrated that S183A (Ser/Ala) but not S183E (Ser/Glu) mutant rescued ERK activation by PGE2 (Fig. 3, F and G). Re-expression of full-length Sdc4 protein (WT) in Sdc4 sh HUVEC fully restored PGE2-dependent induction of ERK activation (Fig. 3, F and G). HA-tagged Sdc4 constructs used for rescue experiments achieved a similar protein expression after adenoviral infection (Fig. 3F). As a control for nonspecific infection effects, we used an adenovirus expressing GFP at an equal multiplicity of infection as the Sdc4 virus.
Sdc4 Regulates PGE2-induced the Angiogenic Phenotype in Vitro and in Vivo
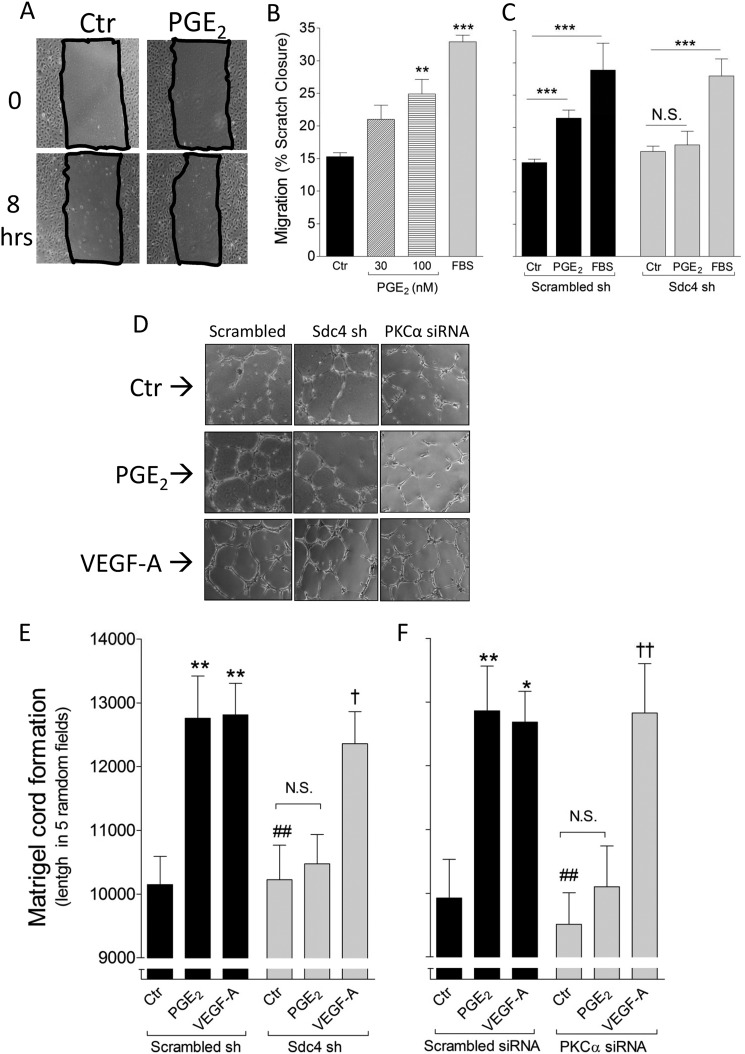
To demonstrate the biological relevance of Sdc4-dependent regulation of PGE2-induced ERK activation, we examined several aspects of EC behavior in vitro and in vivo. First, the effect of PGE2 on EC migration was examined using an in vitro scratch assay. PGE2 induced HUVEC migration in a concentration-dependent manner (Fig. 4, A and B). Next, the same assay was used to test the migration response of EC after knockdown of Sdc4. Although HUVEC infected with a scrambled shRNA increased their migration in response to PGE2, no such increase was observed in Sdc4 shRNA HUVEC (Fig. 4C). Sdc4 sh HUVEC were still able to respond to a general promigratory stimulus such 20% FBS (Fig. 4C). FBS was used as a positive control since it contains many factors that induce a strong promigratory response in HUVEC (29).
FIGURE 4.
Sdc4 mediates PGE2-induced proangiogenic effect in EC. A, representative pictures of scratch closure in HUVEC. Ctr, control. B, increasing PGE2 concentrations were tested for the ability to induce HUVEC migration. 20% FBS was used as a positive control. The bars represent mean of three independent experiments, each with a quantification of five scratched areas/group of treatment. **, p < 0.01; ***, p < 0.001. C, Scrambled sh or Sdc4 sh HUVEC were treated with PGE2 (100 nm) for 8 h, and the scratch closure was quantified. ***, p < 0.001; N.S., not significant. D, representative pictures of two-dimensional cord formation in the indicated cells in presence of PGE2 or VEGF-A (100 ng/ml). E, in vitro cord formation induced by PGE2 or VEGF-A in Scrambled versus Sdc4 sh HUVEC. The bars represent mean ± S.E. (n = 3). **, p < 0.01 from Scrambled sh Ctr; ##, p < 0.01 from Scrambled sh PGE2; †, p < 0.05 from Sdc4 sh Ctr. F, in vitro cord formation induced by PGE2 or VEGF-A (100 ng/ml) in Scrambled versus PKCα siRNA HUVEC. The bars represent mean ± S.E. (n = 3). *, p < 0.05; **, p < 0.01 from Scrambled siRNA Ctr; ##, p < 0.01 from Scrambled siRNA PGE2; ††, p < 0.01 from PKCα siRNA Ctr).
We then used an in vitro two-dimensional Matrigel assay to assess the effect of PGE2 on EC cord formation (Fig. 4D). Quantification of this assay showed that Sdc4 or PKCα knockdown in HUVEC inhibited their ability to form cords in response to PGE2 but not VEGF-A (Fig. 4, F and E). We next examined whether PGE2-induced angiogenesis is affected in Sdc4 or PKCα null mice. To investigate this we employed the Matrigel plug assay that has been used previously to test in vivo proangiogenic properties of PGE2 (21). Analysis of Matrigel plugs impregnated with PGE2 and implanted into WT, Sdc4−/−, and PKCα−/− mice after 7 days showed a significant increase in the CD31-positive area in WT mouse compared with WT control mouse, whereas no such increase was observed in either Sdc4−/− or PKCα−/− mice (Fig. 5, A and B). Of note, VEGF-induced in vivo angiogenesis was not affected in Sdc4−/− or PKCα−/− mice.
DISCUSSION
The proangiogenic properties of PGE2 have been widely investigated both in vivo and vitro (2). Administration of PGE2 to EC in vitro results in a series of “proangiogenic” events such as up-regulation of VEGF and FGF2 (30), increased migration and spreading via αVβ3 integrin-dependent Rac activation (31), and up-regulation of chemokine receptor CXCR4 (32). The proangiogenic phenotype promoted by PGE2 in EC has been associated with the activation of PGE2-EPs downstream signaling such cAMP/protein kinase A (33) and PI3K/AKT (34) pathways. However, given the complexity of PGE2 signaling, other pathways are likely to be involved. PGE2 is able to induce FGFR1 transactivation in EC via a cross-talk mechanism, promoting the proangiogenic phenotype in vitro (3).
ERK activation is a key event in the proangiogenic phenotype promoted by PGE2 in vitro (24), but the mechanism of this activation remains elusive. The results of this study show that PGE2 induces ERK activation in a Sdc4-dependent manner that, in turn, requires Sdc4-driven activation of PKCα. Furthermore, Sdc4 plays a critical role in the regulation of angiogenic activity of PGE2 both in vitro and in vivo. PGE2-induced migration and endothelial cord formation has been shown to be ERK-dependent in vitro (24). Our findings show that silencing of Sdc4 in HUVEC leads to reduced migration and cord formation in response to PGE2. In line with our in vitro data, we also observed impairment of the angiogenesis response in Sdc4−/− mice, demonstrating that failing to fully respond to PGE2, observed in vitro, has in vivo relevance.
PGE2 dependence on the Sdc4/PKCα pathway is demonstrated by a number of observations. First, PGE2 treatment of EC leads to dephosphorylation of the Ser-183 site in Sdc4 cytoplasmic tail, an event known to be required for PIP2-mediated PKCα binding to Sdc4 and its activation (27). Because the phosphatase responsible for this event is not known, we have not been able to determine how PGE2 achieves this effect. Second, PGE2 signaling is reduced similarly in Sdc4−/− and PKCα−/− mouse EC in vitro. Finally, PGE2-dependent ERK activation can be restored in Sdc4−/− EC by a WT Sdc4 construct but not by a construct unable to activate PKCα.
It is noteworthy that in cancer cells (23) and EP4-transfected HEK 293 cells (22), ERK activation mediated by EP4 requires PI3K involvement, but in EC EP4-induced ERK activation has been reported to be PI3K-independent (24). This study provides a mechanism for this observation, demonstrating that Sdc4/PKCα can activate ERK in response to PGE2 downstream of EP4. Furthermore, in agreement with a previous finding, we did not observe PGE2-induced AKT activation at early time points (2–15 min). This is also supported by the fact that ERK activation was unaffected by the PI3K inhibitor.
PKCα can promote ERK activation in a number of ways, including direct activation of RAF (36, 37) and RAS (38). ERK has been shown to be a key signal for PGE2-induced angiogenesis (3, 24). PKCα ability to stimulate angiogenesis both in vivo and in vitro has been well documented. Activation of PKCα but not other PKC isoforms is responsible for enhancing in vitro cord formation in HUVEC (39). This is mediated by up-regulation of VEGF that is evident after 6 h of PKCα activation (39). Interestingly, VEGF up-regulation induced by PMA (non selective PKC activator) can be abolished by a specific MEK inhibitor U0126 (40). Thus, PKCα-induced VEGF up-regulation appears to be ERK-dependent. In agreement with this, inhibition of Sdc4/PKCα pathway induced by overexpression of PKCδ leads to impaired in vitro EC cord formation (28). In vivo, PKCα knock-out mice have impaired neovascularization following myocardial infarction (35). PGE2-induced VEGF up-regulation requires ERK-dependent activation of JNK in EC (30) therefore PGE2/Sdc4/PKCα could represent the missing link in this pathway.
Sdc4−/− mice are viable and fertile but show an impaired response to insults that evoke inflammatory response such as wound healing (12) and myocardial infarction (16). Wound healing is a tightly regulated process driven and orchestrated by a plethora of inflammatory mediators (36) including PGE2 which among other effects can induce epithelial cell proliferation (37) and promote angiogenesis in the healing tissue (38). Our data showing that Sdc4−/− EC are unable to achieve a full angiogenic response following PGE2 administration can potentially explain some of the findings in Sdc4−/− mice including delayed angiogenesis in healing tissues leading to impaired wound healing (15) and larger infarct size (16). Furthermore, because Sdc4 is ubiquitously express in many cell types other than EC, including myofibroblasts, smooth muscle cells and epithelial cells among others, this pathway might be relevant in a number of other disease processes where inflammation and inflammatory response play an important role including atherosclerosis, certain chronic infections and cancer.
PGE2 is generally considered to have a potent tumor-promoting activity associated with a wide spectrum of effects which include cancer cell proliferation (39), angiogenesis (22) and metastasis (40). In tumor microenvironment the cyclooxygenase-2/PGE2 axis is highly up-regulated (5) and a direct role of PGE2 in tumorigenesis has been extensively showed in animal models (see (41) for Review). Finally, recent epidemiological studies (42–44) reported a marked reduction in cancer incidence/mortality and prevention of cancer onset in patients treated with low-dose aspirin or other non-steroidal anti-inflammatory drugs. This effect is believed to be associated with nonselective cyclooxygenase enzyme inhibition and reduction in prostanoid metabolites, including PGE2 levels. Our study unveiled a novel PGE2 signaling pathway that may provide a therapeutic opportunity to control tumor growth.
Acknowledgments
We thank Sandra Donnini for guidance and helpful suggestions, the Società Italiana di Farmacologia (SIF) for initial support for this project, Jiasheng Zhang for technical help in endothelial cell preparation, John Rhodes and Rong Ju for discussion and technical help, and Antony Lanahan for helpful suggestions during manuscript revision.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL062289 (to M. S.). This work was also supported by the Fondazione Istituto Toscano Tumori (to F. C.) and the Associazione Italiana per la Ricerca sul Cancro (AIRC) (to M. Z.).
- PGE2
- prostaglandin E2
- EP
- PGE2 receptor
- PKCα
- protein kinase Cα
- HUVEC
- human umbilical vein endothelial cell(s)
- EC
- endothelial cell(s)
- P
- cell passage number.
REFERENCES
- 1. Aoki T., Narumiya S. (2012) Prostaglandins and chronic inflammation. Trends Pharmacol. Sci. 33, 304–311 [DOI] [PubMed] [Google Scholar]
- 2. Wang D., Dubois R. N. (2010) Eicosanoids and cancer. Nat. Rev. Cancer 10, 181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Finetti F., Solito R., Morbidelli L., Giachetti A., Ziche M., Donnini S. (2008) Prostaglandin E2 regulates angiogenesis via activation of fibroblast growth factor receptor-1. J. Biol. Chem. 283, 2139–2146 [DOI] [PubMed] [Google Scholar]
- 4. Buchanan F. G., Gorden D. L., Matta P., Shi Q., Matrisian L. M., DuBois R. N. (2006) Role of β-arrestin 1 in the metastatic progression of colorectal cancer. Proc. Natl. Acad. Sci. U.S.A. 103, 1492–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liang X., Lin L., Woodling N. S., Wang Q., Anacker C., Pan T., Merchant M., Andreasson K. (2011) Signaling via the prostaglandin E(2) receptor EP4 exerts neuronal and vascular protection in a mouse model of cerebral ischemia. J. Clin. Invest. 121, 4362–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xin X., Majumder M., Girish G. V., Mohindra V., Maruyama T., Lala P. K. (2012) Targeting COX-2 and EP4 to control tumor growth, angiogenesis, lymphangiogenesis and metastasis to the lungs and lymph nodes in a breast cancer model. Lab. Invest. 92, 1115–1128 [DOI] [PubMed] [Google Scholar]
- 7. Tkachenko E., Rhodes J. M., Simons M. (2005) Syndecans. New kids on the signaling block. Circ. Res. 96, 488–500 [DOI] [PubMed] [Google Scholar]
- 8. Volk R., Schwartz J. J., Li J., Rosenberg R. D., Simons M. (1999) The role of syndecan cytoplasmic domain in basic fibroblast growth factor-dependent signal transduction. J. Biol. Chem. 274, 24417–24424 [DOI] [PubMed] [Google Scholar]
- 9. Chua C. C., Rahimi N., Forsten-Williams K., Nugent M. A. (2004) Heparan sulfate proteoglycans function as receptors for fibroblast growth factor-2 activation of extracellular signal-regulated kinases 1 and 2. Circ. Res. 94, 316–323 [DOI] [PubMed] [Google Scholar]
- 10. Elfenbein A., Rhodes J. M., Meller J., Schwartz M. A., Matsuda M., Simons M. (2009) Suppression of RhoG activity is mediated by a syndecan 4-synectin-RhoGDI1 complex and is reversed by PKCα in a Rac1 activation pathway. J. Cell Biol. 186, 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horowitz A., Simons M. (1998) Phosphorylation of the cytoplasmic tail of syndecan-4 regulates activation of protein kinase Cα. J. Biol. Chem. 273, 25548–25551 [DOI] [PubMed] [Google Scholar]
- 12. Echtermeyer F., Streit M., Wilcox-Adelman S., Saoncella S., Denhez F., Detmar M., Goetinck P. (2001) Delayed wound repair and impaired angiogenesis in mice lacking syndecan-4. J. Clin. Invest. 107, R9-R14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsui Y., Ikesue M., Danzaki K., Morimoto J., Sato M., Tanaka S., Kojima T., Tsutsui H., Uede T. (2011) Syndecan-4 prevents cardiac rupture and dysfunction after myocardial infarction. Circ. Res. 108, 1328–1339 [DOI] [PubMed] [Google Scholar]
- 14. Ishiguro K., Kadomatsu K., Kojima T., Muramatsu H., Iwase M., Yoshikai Y., Yanada M., Yamamoto K., Matsushita T., Nishimura M., Kusugami K., Saito H., Muramatsu T. (2001) Syndecan-4 deficiency leads to high mortality of lipopolysaccharide-injected mice. J. Biol. Chem. 276, 47483–47488 [DOI] [PubMed] [Google Scholar]
- 15. Allport J. R., Lim Y. C., Shipley J. M., Senior R. M., Shapiro S. D., Matsuyoshi N., Vestweber D., Luscinskas F. W. (2002) Neutrophils from MMP-9- or neutrophil elastase-deficient mice show no defect in transendothelial migration under flow in vitro. J. Leukocyte Biol. 71, 821–828 [PubMed] [Google Scholar]
- 16. Partovian C., Ju R., Zhuang Z. W., Martin K. A., Simons M. (2008) Syndecan-4 regulates subcellular localization of mTOR complex 2 and Akt activation in a PKCα-dependent manner in endothelial cells. Mol. Cell 32, 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coleman R. A., Grix S. P., Head S. A., Louttit J. B., Mallett A., Sheldrick R. L. (1994) A novel inhibitory prostanoid receptor in piglet saphenous vein. Prostaglandins 47, 151–168 [DOI] [PubMed] [Google Scholar]
- 18. Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Meth. 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gassmann M., Grenacher B., Rohde B., Vogel J. (2009) Quantifying Western blots. Pitfalls of densitometry. Electrophoresis 30, 1845–1855 [DOI] [PubMed] [Google Scholar]
- 20. Meijering E., Jacob M., Sarria J. C., Steiner P., Hirling H., Unser M. (2004) Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58, 167–176 [DOI] [PubMed] [Google Scholar]
- 21. Finetti F., Donnini S., Giachetti A., Morbidelli L., Ziche M. (2009) Prostaglandin E(2) primes the angiogenic switch via a synergic interaction with the fibroblast growth factor-2 pathway. Circ. Res. 105, 657–666 [DOI] [PubMed] [Google Scholar]
- 22. Fujino H., Xu W., Regan J. W. (2003) Prostaglandin E2 induced functional expression of early growth response factor-1 by EP4, but not EP2, prostanoid receptors via the phosphatidylinositol 3-kinase and extracellular signal-regulated kinases. J. Biol. Chem. 278, 12151–12156 [DOI] [PubMed] [Google Scholar]
- 23. Pozzi A., Yan X., Macias-Perez I., Wei S., Hata A. N., Breyer R. M., Morrow J. D., Capdevila J. H. (2004) Colon carcinoma cell growth is associated with prostaglandin E2/EP4 receptor-evoked ERK activation. J. Biol. Chem. 279, 29797–29804 [DOI] [PubMed] [Google Scholar]
- 24. Rao R., Redha R., Macias-Perez I., Su Y., Hao C., Zent R., Breyer M. D., Pozzi A. (2007) Prostaglandin E2-EP4 receptor promotes endothelial cell migration via ERK activation and angiogenesis in vivo. J. Biol. Chem. 282, 16959–16968 [DOI] [PubMed] [Google Scholar]
- 25. Horowitz A., Tkachenko E., Simons M. (2002) Fibroblast growth factor-specific modulation of cellular response by syndecan-4. J. Cell Biol. 157, 715–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tkachenko E., Simons M. (2002) Clustering induces redistribution of syndecan-4 core protein into raft membrane domains. J. Biol. Chem. 277, 19946–19951 [DOI] [PubMed] [Google Scholar]
- 27. Horowitz A., Simons M. (1998) Regulation of syndecan-4 phosphorylation in vivo. J. Biol. Chem. 273, 10914–10918 [DOI] [PubMed] [Google Scholar]
- 28. Murakami M., Horowitz A., Tang S., Ware J. A., Simons M. (2002) Protein kinase C (PKC) Δ regulates PKCα activity in a Syndecan-4-dependent manner. J. Biol. Chem. 277, 20367–20371 [DOI] [PubMed] [Google Scholar]
- 29. Mastyugin V., McWhinnie E., Labow M., Buxton F. (2004) A quantitative high-throughput endothelial cell migration assay. J. Biomol. Screen. 9, 712–718 [DOI] [PubMed] [Google Scholar]
- 30. Pai R., Szabo I. L., Soreghan B. A., Atay S., Kawanaka H., Tarnawski A. S. (2001) PGE(2) stimulates VEGF expression in endothelial cells via ERK2/JNK1 signaling pathways. Biochem. Biophys. Res. Commun. 286, 923–928 [DOI] [PubMed] [Google Scholar]
- 31. Dormond O., Bezzi M., Mariotti A., Ruegg C. (2002) Prostaglandin E2 promotes integrin α Vβ 3-dependent endothelial cell adhesion, Rac-activation, and spreading through cAMP/PKA-dependent signaling. J. Biol. Chem. 277, 45838–45846 [DOI] [PubMed] [Google Scholar]
- 32. Salcedo R., Zhang X., Young H. A., Michael N., Wasserman K., Ma W. H., Martins-Green M., Murphy W. J., Oppenheim J. J. (2003) Angiogenic effects of prostaglandin E2 are mediated by up-regulation of CXCR4 on human microvascular endothelial cells. Blood 102, 1966–1977 [DOI] [PubMed] [Google Scholar]
- 33. Zhang Y., Daaka Y. (2011) PGE2 promotes angiogenesis through EP4 and PKA Cγ pathway. Blood 118, 5355–5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dada S., Demartines N., Dormond O. (2008) mTORC2 regulates PGE2-mediated endothelial cell survival and migration. Biochem. Biophys. Res. Commun. 372, 875–879 [DOI] [PubMed] [Google Scholar]
- 35. Wang A., Nomura M., Patan S., Ware J. A. (2002) Inhibition of protein kinase Cα prevents endothelial cell migration and vascular tube formation in vitro and myocardial neovascularization in vivo. Circ. Res. 90, 609–616 [DOI] [PubMed] [Google Scholar]
- 36. Kolch W., Heidecker G., Kochs G., Hummel R., Vahidi H., Mischak H., Finkenzeller G., Marmé D., Rapp U. R. (1993) Protein kinase C α activates RAF-1 by direct phosphorylation. Nature 364, 249–252 [DOI] [PubMed] [Google Scholar]
- 37. Marais R., Light Y., Mason C., Paterson H., Olson M. F., Marshall C. J. (1998) Requirement of Ras-GTP-Raf complexes for activation of Raf-1 by protein kinase C. Science 280, 109–112 [DOI] [PubMed] [Google Scholar]
- 38. Clark J. A., Black A. R., Leontieva O. V., Frey M. R., Pysz M. A., Kunneva L., Woloszynska-Read A., Roy D., Black J. D. (2004) Involvement of the ERK signaling cascade in protein kinase C-mediated cell cycle arrest in intestinal epithelial cells. J. Biol. Chem. 279, 9233–9247 [DOI] [PubMed] [Google Scholar]
- 39. Xu H., Czerwinski P., Hortmann M., Sohn H. Y., Förstermann U., Li H. (2008) Protein kinase C α promotes angiogenic activity of human endothelial cells via induction of vascular endothelial growth factor. Cardiovasc. Res. 78, 349–355 [DOI] [PubMed] [Google Scholar]
- 40. Favata M. F., Horiuchi K. Y., Manos E. J., Daulerio A. J., Stradley D. A., Feeser W. S., Van Dyk D. E., Pitts W. J., Earl R. A., Hobbs F., Copeland R. A., Magolda R. L., Scherle P. A., Trzaskos J. M. (1998) Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273, 18623–18632 [DOI] [PubMed] [Google Scholar]
- 41. Nakanishi M., Rosenberg D. W. (2013) Multifaced roles of PGE2 in inflammation and cancer. Sem. Immunopathol. 35, 123–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rothwell P. M., Price J. F., Fowkes F. G., Zanchetti A., Roncaglioni M. C., Tognoni G., Lee R., Belch J. F., Wilson M., Mehta Z., Meade T. W. (2012) Short-term effects of daily aspirin on cancer incidence, mortality, and non-vascular death. Analysis of the time course of risks and benefits in 51 randomised controlled trials. Lancet 379, 1602–1612 [DOI] [PubMed] [Google Scholar]
- 43. Rothwell P. M., Wilson M., Price J. F., Belch J. F., Meade T. W., Mehta Z. (2012) Effect of daily aspirin on risk of cancer metastasis. A study of incident cancers during randomised controlled trials. Lancet 379, 1591–1601 [DOI] [PubMed] [Google Scholar]
- 44. Harris R. E. (2009) Cyclooxygenase-2 (cox-2) blockade in the chemoprevention of cancers of the colon, breast, prostate, and lung. Inflammopharmacology 17, 55–67 [DOI] [PubMed] [Google Scholar]