Background: The anti-apoptotic protein c-FLIP inhibits cell death induced by TNF family cytokines, including TRAIL.
Results: Reactive oxygen species (ROS)-dependent phosphorylation and ubiquitination of the c-FLIP protein causes its proteasome-mediated degradation, thus sensitizing to TRAIL-induced cell death.
Conclusion: ROS-dependent post-translational modifications regulate c-FLIP protein stability.
Significance: Understanding how ROS mediate c-FLIP protein degradation may inform therapeutic strategies targeting cell death mechanisms.
Keywords: Apoptosis, Reactive Oxygen Species (ROS), Trail, Ubiquitin, Ubiquitination, Ubiquitylation, Post-translational Modification, ROS, c-FLIP
Abstract
The cytosolic protein c-FLIP (cellular Fas-associated death domain-like interleukin 1β-converting enzyme inhibitory protein) is an inhibitor of death receptor-mediated apoptosis that is up-regulated in a variety of cancers, contributing to apoptosis resistance. Several compounds found to restore sensitivity of cancer cells to TRAIL, a TNF family death ligand with promising therapeutic potential, act by targeting c-FLIP ubiquitination and degradation by the proteasome. The generation of reactive oxygen species (ROS) has been implicated in c-FLIP protein degradation. However, the mechanism by which ROS post-transcriptionally regulate c-FLIP protein levels is not well understood. We show here that treatment of prostate cancer PPC-1 cells with the superoxide generators menadione, paraquat, or buthionine sulfoximine down-regulates c-FLIP long (c-FLIPL) protein levels, which is prevented by the proteasome inhibitor MG132. Furthermore, pretreatment of PPC-1 cells with a ROS scavenger prevented ubiquitination and loss of c-FLIPL protein induced by menadione or paraquat. We identified lysine 167 as a novel ubiquitination site of c-FLIPL important for ROS-dependent degradation. We also identified threonine 166 as a novel phosphorylation site and demonstrate that Thr-166 phosphorylation is required for ROS-induced Lys-167 ubiquitination. The mutation of either Thr-166 or Lys-167 was sufficient to stabilize c-FLIP protein levels in PPC-1, HEK293T, and HeLa cancer cells treated with menadione or paraquat. Accordingly, expression of c-FLIP T166A or K167R mutants protected cells from ROS-mediated sensitization to TRAIL-induced cell death. Our findings reveal novel ROS-dependent post-translational modifications of the c-FLIP protein that regulate its stability, thus impacting sensitivity of cancer cells to TRAIL.
Introduction
Apoptosis is a highly regulated form of cell death that is fundamental during embryonic development and in maintaining tissue homeostasis (1). Two major pathways of apoptosis have been extensively characterized: 1) the death receptor (DR)2-induced (extrinsic) pathway and 2) the mitochondria-mediated (intrinsic) pathway (reviewed in Refs. 2–4). In the extrinsic pathway, extracellular ligands of the tumor necrosis factor (TNF) superfamily activate death receptors at the cell surface causing their oligomerization and association with the adaptor protein FADD (Fas-associated protein with death domain) via interaction of their death domains. FADD then recruits the protease pro-caspase-8 (and pro-caspase-10 in humans) via interactions of their death effector domains (DEDs) to form the death-inducing signaling complex (DISC) (5). Under these conditions, initiator caspases dimerize, resulting in their proteolytic activation. Active caspase-8 can then directly cleave and activate caspases-3 and -7, whose proteolytic activity drives the organized demise of the cell by apoptosis.
A key inhibitor of the extrinsic apoptotic pathway is the protein c-FLIP (cellular Fas-associated death domain-like interleukin 1β-converting enzyme inhibitory protein). The widely expressed long isoform of c-FLIP (c-FLIPL) contains a tandem pair of DEDs and is similar in length and amino acid sequence to caspase-8. However, its caspase-like domain is enzymatically inactive (6). The c-FLIP protein competes with caspase-8 for binding to FADD at the DISC and it also forms non-apoptotic heterodimers with caspase-8 (7). This prevents further recruitment, proteolytic processing, and the release of active caspase-8 from the DISC, thus inhibiting execution of the apoptotic program (8).
Defects in apoptosis regulation contribute to anti-cancer drug resistance in numerous types of malignant cells (9). One such mechanism is the elevated expression of anti-apoptotic proteins. c-FLIP expression is known to be aberrantly up-regulated in several cancers including prostate, breast, ovarian, pancreatic, colorectal, gastric, melanoma, glioblastoma, and Birkett and non-Hodgkin lymphoma (reviewed in Ref. 9). c-FLIP protein expression is also elevated in castrate-resistant prostate cancer, which has been implicated in promoting tumor survival and reducing sensitivity to hormone ablation therapy (10). Elevated expression of c-FLIP protein correlates with resistance to cytokines and agonistic antibodies that stimulate DR-mediated apoptosis. Additionally, high levels of c-FLIP are also known to activate signal transduction pathways that promote cell survival and enhance cell proliferation (11, 12). These two actions in concert may provide circumstances that promote unrestrained tumor growth and resilience, making this anti-apoptotic protein a promising target for strategies aimed at killing cancer cells by sensitizing them to DR agonists.
The tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a promising anti-cancer reagent due to its ability to preferentially induce apoptosis in tumor cells with little toxicity to normal cells (13). Unfortunately, most cancer cells are resistant to TRAIL, even though they display receptors for TRAIL on their surface. A variety of studies exploring strategies for overcoming TRAIL resistance have reported that certain chemical compounds can sensitize cancer cells to TRAIL-induced apoptosis by targeting c-FLIP protein for degradation (14–22). Typically, this degradation is achieved by the ubiquitin-proteasome pathway. Post-translational modifications (PTM) have emerged as important regulators of protein stability. However, the specific details and the biological significance of PTMs for c-FLIP remain unclear and the exact sites of c-FLIP ubiquitination have not been fully elucidated.
Compounds that induce c-FLIP ubiquitination and degradation such as the triterpenoid 2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid (CDDO) and berberine have also been shown to generate reactive oxygen species (ROS) such as superoxide and hydrogen peroxide within cells. ROS generation by these compounds was shown to be critical for apoptosis induction (21, 23, 24). Moreover, apoptosis of cancer epithelial cells by Fas ligand (FasL) reported down-regulation of c-FLIP via a ROS-dependent ubiquitin-proteasomal degradation process (25). These findings suggest that c-FLIP could be a redox-sensitive protein.
Here we studied the effects of ROS on the post-translational regulation of c-FLIP protein levels and on TRAIL-induced apoptosis in cancer cell lines. We identified novel phosphorylation and ubiquitination sites that mediate ROS-dependent proteasomal degradation of c-FLIP and concomitant sensitization of cancer cells to TRAIL-induced apoptosis. Our findings suggest that ROS-dependent PTM of c-FLIP contributes to the sensitization of cancer cells to TRAIL.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Menadione was purchased from Sigma and paraquat was from ChemService. MG132 was from Calbiochem and tetramethylpiperidine-N-oxyl (TEMPO) was from MP Biomedicals. TRAIL was from ENZO Life Sciences and Lipofectamine 2000 was from Invitrogen. Protease inhibitor mixture and phosphatase inhibitor mixture were from Roche Applied Science. BCA protein assay was from Pierce, BSA and imidazole were from Fischer Scientific, and Ni-NTA-agarose was from Qiagen. Protein G-Sepharose beads, Trypan blue, ×10 annexin V binding buffer, and propidium iodide were from Invitrogen. Annexin V-APC was from eBioscience. 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) was from Molecular Probes. ECL Western blotting detection reagents were from GE Healthcare. We used the following antibodies: rabbit antibodies to c-FLIP (F-9800, Sigma), and FADD (number 06-711, Millipore); mouse antibodies to Ubiquitin (P4D1) (number 3936, Cell Signaling Technology, and number MMS-257P, Covance), the HA epitope (number 11583816001, Roche Applied Science), c-FLIP (sc-5276, Santa Cruz Biotechnology), and GFP (sc-9996, Santa Cruz Biotechnology); and rat antibody to HA (number 11867431001, Roche Applied Science). The following secondary antibodies were used: HRP-conjugated anti-mouse (NA931V, GE Healthcare) and HRP-conjugated anti-rabbit (NA934, GE Healthcare).
DNA Constructs
A cDNA comprising the open reading frame encoding c-FLIP was subcloned into various plasmids to produce the pcDNA3.1His6-c-FLIPL and pcDNA3-HA-c-FLIPL expression vectors. Site-directed mutagenesis of FLIPL was performed to generate the single T166A and K167R substitution mutations as well as the T166A,K167R double substitution mutations using the QuikChangeTM Site-directed Mutagenesis kit (Stratagene) as per the manufacturer's protocol, with pcDNA3-HA-c-FLIPL or pcDNA3.1His6-c-FLIPL plasmids as DNA template and the mutagenic primers: (a) for T166A, 5′-CACAGAATAGACCTGAAGGCAAAAATCCAGAAGTACAAG-3′ and 5′-CTTGTACTTCTGGATTTTTGCCTTCAGGTCTATTCTGTG-3′; (b) for K167R, 5′-CAGAATAGACCTGAAGACACGAATCCAGAAGTACAAGCAG-3′ and 5′-CTGCTTGTACTTCTGGATTCGTGTCTTCAGGTCTATTCTG-3′; (c) for T166A,K167R, 5′-CCACAGAATAGACCTGAAGGCACGAATCCAGAAGTACAAGCAG-3′ and 5′-CTGCTTGTACTTCTGGATTCGTGCCTTCAGGTCTATTCTGTGG-3′ and verified by DNA sequencing. Full-length human ubiquitin cDNA was subcloned into pEGFP2-C2 (BioSignal Packard) in-frame with GFP2 using PCR to produce the EGFP-C2-Ubiquitin expression vector.
Cell Culture and Transfections
Human prostate cancer, PPC-1 cells were cultured in RPMI 1640 medium (Mediatech) supplemented with 10% fetal bovine serum (FBS) (HyClone), penicillin (100 IU), and streptomycin (100 μg/ml) at 37 °C in 5% CO2, 95% air. Human embryonic kidney cancer cell line (HEK293T) and HeLa cervical cancer cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Mediatech) with 10% FBS, penicillin (100 IU), and streptomycin (100 μg/ml) at 37 °C in 5% CO2, 95% air. Transient transfections were performed using Lipofectamine 2000.
For mass spectrometry analysis, the His6-c-FLIPL plasmid (5 μg of DNA/10-cm dish) was transfected into PPC-1 cells and cultured for 16 h in complete media. Cells were then treated with 5 μm menadione in the presence of 1 μm MG132 for 10 h in RPMI media supplemented with 0.5% FBS. Multiple 10-cm dishes with identical treatments were pooled before lysis to achieve a highly concentrated sample for optimal mass spectrometry analysis.
For immunoprecipitation and immunoblot assays, PPC-1 cells were transfected with various His6-tagged or HA-tagged c-FLIP plasmids (wild-type (WT), T166A, K167R, and T166A,K167R double mutant) (5 μg of DNA/10-cm dish). Cells were cultured for 16 h in complete media and then treated with 5 μm menadione with or without 0.5 μm MG132 for 8 h in RPMI media supplemented with 0.5% FBS. For paraquat treatments, PPC-1, HEK293T, or HeLa cells were co-transfected with GFP-Ubiquitin plasmid (10 μg of DNA/10-cm dish) together with either HA-FLIP-WT or mutant c-FLIP plasmids (T166A, K167R, and T166A,K167R double mutant) (5 μg of DNA/10-cm dish). Cells were cultured for 16 h in complete media prior to 2 mm paraquat treatment with or without 0.5 μm MG132 for 8 h in RPMI media supplemented with 0.5% FBS.
For cycloheximide chase experiments, HA-FLIP-WT or mutant c-FLIP plasmids (T166A, K167R, and T166A,K167R double mutant) (2 μg/6-well plate) were transfected into PPC-1 cells and cultured for 16 h in complete media. Cells were then treated with 25 μg/ml of cycloheximide in the presence of 5 μm menadione with or without MG132 (0.5 μm) for 12 h.
For cell titer glow (Promega) and trypan blue exclusion assays, HA-FLIP-WT or mutant c-FLIP plasmids (T166A, K167R, and T166A,K167R double mutant) (5 μg of DNA/10-cm dish) were transfected into PPC-1 cells. Cells were cultured for 16 h in complete media. For cell titer glow experiments, 2.5 × 104 cells/well were transferred into 96-well tissue culture plates. For trypan blue exclusion experiments, 7 × 104 cells/well were transferred into 24-well tissue culture plates and then incubated for a further 8 h in complete media. Cells were then treated with various menadione concentrations with or without TRAIL for the indicated times.
For Annexin-V staining by FACS analysis, PPC-1 cells were co-transfected with HA-FLIP-WT or mutant c-FLIP plasmids (T166A, K167R, and T166A,K167R double mutant) (1 μg of DNA/6-well plate) along with EGFP-C2 plasmid (0.2 μg of DNA/6-well plate) and cultured for 16 h in complete media. Cells were then treated with 10 μm menadione with or without 25 ng/ml of TRAIL for 16 h.
Immunoprecipitation and Protein Analysis
HA-tagged cFLIP-transfected cells in 10-cm plates were lysed 8 h post-treatment with menadione or paraquat with or without MG132 or TEMPO in 1 ml of lysis buffer (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 2 mm Na4VO3) containing a protease and phosphatase inhibitor mixture at 4 °C for 45 min on a wheel rotor. The lysates were cleared of cell debris by centrifugation at 10,000 × g for 15 min and the total protein content was quantified by a BCA assay. To reduce nonspecific binding the supernatants were preincubated with protein G-Sepharose beads at 4 °C for 1 h. Following a brief centrifugation, the supernatants were incubated with anti-HA rat-specific antibody with gentle agitation overnight at 4 °C, followed by incubation with protein G-Sepharose beads at 4 °C for 2 h. After a brief centrifugation the beads were washed three times in lysis buffer, and resuspended in Laemmli sample buffer containing no β-mercaptoethanol and boiled for 5 min to release bound proteins. Proteins were loaded onto SDS-PAGE (10, 12, or 4–20% gels) and subjected to immunoblotting. For immunoblot and mass spectrometry analysis, His6-tagged c-FLIP-transfected cells in 10-cm dishes were lysed in 1 ml of 50 mm NaH2PO4, pH 8.0, containing 300 mm NaCl, 10 mm imidazole, 0.05% Tween 20, and a protease and phosphatase inhibitor mixture at 4 °C for 45 min on a wheel rotor and complemented with sonication (6 pulses) on ice. The lysates were centrifuged at 10,000 × g for 15 min, then the resulting supernatants were incubated with Ni-NTA-agarose beads at 4 °C for 2 h. After a brief centrifugation, the beads were washed three times in washing buffer (50 mm NaH2PO4, pH 8.0, 300 mm NaCl, 20 mm imidazole, 0.05% Tween 20) and the bound proteins were released in elution buffer (50 mm NaH2PO4, pH 8.0, 300 mm NaCl, 250 mm imidazole, 0.05% Tween 20) and combined with Laemmli sample buffer; proteins were separated by (11%) SDS-PAGE before being digested with trypsin for mass spectrometry analysis. For direct immunoblot analysis using cell lysates, cells were lysed with RIPA buffer (50 mm Tris-Cl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 0.1% SDS, 0.5% deoxycholic acid, 1% Triton X-100) containing a protease and phosphatase inhibitor mixture at 4 °C for 30 min on a wheel rotor. The lysates were centrifuged at 10,000 × g for 15 min. FLIP levels were quantified by densitometry using ImageJ software. For sequential probing with multiple antibodies, the blots were stripped using 0.1 m glycine, pH 2.5, with 0.1% Tween at 37 °C for 30–60 min and washed with Tris-buffered saline containing 0.1% Tween 20 (TBST20).
Measurement of Cellular Reactive Oxygen Species
After treatment with menadione, cells were washed twice with phosphate-buffered saline (PBS) and combined with 10 μm H2DCFDA in PBS solution. Cultures were incubated at 37 °C under 5% CO2 for 30 min. Any unbound dye was removed by washing with PBS, then the cells were trypsinized and cell pellets were washed and resuspended in PBS. Fluorescence (excitation 485 nm, emission 530 nm) was measured by fluorescence-activated cell scanning (FACS; BD Biosciences).
FLIP mRNA Analysis
Following the treatment of cells with menadione or paraquat, total RNA was extracted from cells using the RNeasy mini kit (Qiagen). For quantitative real-time PCR (RT-PCR) analysis of c-FLIP mRNA, cDNA was generated using the SuperScript III First-strand synthesis system (Invitrogen). The primers used for PCR-mediated c-FLIP cDNA amplification were: 5′-CTTCTTCTGGAGCCTGTGTACTG-3′ and 5′-TCTTGTCTCAGTTTCTGGGAGAG-3′. Thermal DNA melting experiments showed single melting peaks for the PCR products generated. Quantitative PCR was performed using the Mx3000P Q-PCR system (Stratagene) with SYBR Green PCR master mixture (Qiagen) and analyzed with the MxPro software (Stratagene).
Tandem Mass Spectrometry Analysis of FLIP Post-translational Modifications
His-FLIP-WT protein was captured from transfected PPC-1 cells following a 10-h treatment with 5 μm menadione in the presence of 1 μm MG132 by Ni-NTA-agarose. The whole lane of FLIP protein bands were excised from the SDS-PAGE gel, reduced, alkylated, and digested by trypsin. The C18 ZipTip (Millipore)-cleaned peptides were subjected to LC/MS-MS analysis using capillary reverse-phase column chromatography (15-cm Magic C18 AQ resin (Michrom)) using a Michrom Paradigm MS4 HPLC.HTC-PAL autosampler (Bruker-Michrom, Aubrun, CA) and an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, San Jose, CA). The RP-HPLC gradient was run from 2.0% solvent B to 35.0% B from 0.0 to 120.0 min, 80.0% B from 120.1 to 124.0 min, and 2.0% B from 124.1 to 134.0 min (solvent A = 0.1% formic acid, solvent B = acetonitrile). The mass spectrometer was programmed to scan precursors in the Orbitrap at 60,000 resolution followed by data-dependent, top 4 MS/MS scan fragment ions, obtained by collision-induced dissociation, of the most abundant precursors in the linear ion trap. Charge state screening and monoisotopic precursor selection was enabled. Dynamic exclusion was enabled for 120 s with a repeat count of 2. A Sorcerer-SEQUEST version 3.10.7 search (SorcererTM-SEQUEST®, SageN Research, Milipitas, CA) was performed on raw MS/MS data. SEQUEST was set up to search the target-decoy ipi.Human.v.3.22 protein database containing protein sequences using trypsin for enzyme with the allowance of up to 2 missed cleavages, full tryptic search, and precursor mass tolerance of 10 ppm. Differential search includes 16 Da for methionine oxidation, 57 Da for cysteines to account for carboxyamidomethylation in case of alkylation of cysteines; serine, threonine, and tyrosine phosphorylation (79.9 Da), lysine ubiquitination with GG tag (114 Da), LRGG tag (382 Da), proline hydroxylation (16 Da), and lysine acetylation (42 Da) to identify these modifications. The search results were viewed, sorted, and filtered using comprehensive proteomics data analysis software (Peptide/Protein prophet version 3.3.0 (Institute for Systems Biology, Seattle, WA)). In this study, we used the following two data filtering criteria. First, the minimum trans-proteomic pipeline probability score for proteins were set to 0.99, to assure very low error identification (much less than FDR 2%) with reasonably good sensitivity. Second, we set up a threshold of cross-correlation (Xcorr) scores set for filtered peptides to 1.5, 2.0, and 2.5 for 1, 2, and 3 charged fully digested tryptic peptides, respectively.
Cell Death Assays
Cellular ATP concentrations were assessed using the CellTiter-Glo luminescent cell viability assay (Promega) following the manufacturer's protocol. In brief, 2.5 × 104 transfected cells expressing WT or mutant c-FLIP proteins were seeded in 96-well plates and cultured for 8 h prior to treatment. Cells were then treated with increasing concentrations of menadione or paraquat with or without 10 ng/ml of TRAIL for 24 h. Luminescence was measured using the Luminoskan Ascent (Thermo Electron Corp.) at 1-s integration time per sample. For trypan blue exclusion assays 7 × 104 transfected PPC-1 cells expressing WT or mutant c-FLIP proteins were seeded in 24-well plates and cultured for 8 h prior to treatment. Cells were then treated with 15 μm menadione with or without TRAIL (10 ng/ml) for 24 h. Cell viability was assessed by exclusion of trypan blue using the Countess automated cell counter (Invitrogen). For FACS analysis, PPC-1 cells were co-transfected with either WT or mutant c-FLIP proteins together with GFP plasmid, then treated with 10 μm menadione with or without 25 ng/ml of TRAIL for 16 h. Apoptosis was assessed by staining with Annexin V-fluorescence isothiocynate (FITC) using a kit (Invitrogen) per the manufacturer's instructions, except that annexin V-FITC was substituted with annexin V-APC (allophycocyanin). Flow cytometry was performed with a FACSVantageSE DiVa (BD Biosciences) and the data analyzed using FACSDiva software (BD Biosciences).
RESULTS
Menadione Reduces c-FLIP Protein Levels through a ROS-dependent Ubiquitination Mechanism
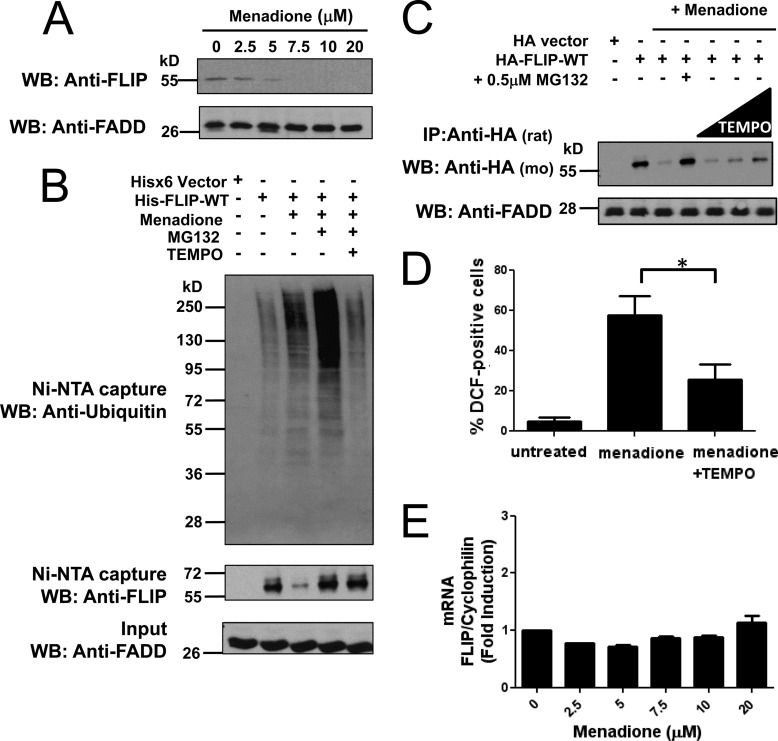
We studied the effect of the ROS-generating compound menadione on c-FLIP ubiquitination and degradation. Treatment of prostate cancer cell line PPC-1 with increasing concentrations of menadione caused striking reductions in the levels of endogenous c-FLIP protein in a dose-dependent manner (Fig. 1A.). We expressed His6-tagged wild-type FLIP (His-FLIP-WT) in PPC-1 cells for 16 h followed by 10 h treatment with menadione. Multiple dishes with identical treatments were pooled to concentrate samples for further mass spectrometry analysis. Cells were lysed, c-FLIP was isolated by nickel-agarose, and c-FLIP protein levels was analyzed by immunoblotting with anti-FLIP antibody, whereas ubiquitination of His6-FLIP was analyzed by anti-ubiquitin antibody. Consistent with endogenous c-FLIP protein, His6-tagged c-FLIP was similarly reduced following treatment with menadione (Fig. 1B, middle panel). Inhibition of the proteasome by MG132 blocked the loss of c-FLIP protein and greatly enhanced the level of c-FLIP ubiquitination (Fig. 1B, upper panel). Culturing cells with the ROS scavenger TEMPO reduced mendione-induced ubiquitination of the His6-FLIP protein, which was measured in cells treated with MG132 (Fig. 1B, upper panel). To further examine the effects of ROS in menadione-treated cells, we also monitored HA-tagged c-FLIP in transfected PPC-1 cells. Treatment of PPC-1 cells with TEMPO prevented menadione-induced reductions in HA-FLIP protein levels, even without proteasomal inhibition (Fig. 1C).
FIGURE 1.
ROS-dependent ubiquitination and degradation of FLIP. A, PPC1 cells were treated with increasing concentrations of menadione for 8 h. Total cell lysates were analyzed by immunoblotting using a specific mouse anti-FLIP antibody. FADD served as a loading control. B, PPC1 cells were transfected with pcDNA3.1-His6 vector (first lane) or plasmid encoding His-FLIP (second to fifth lanes) for 16 h. Cells were treated with menadione (5 μm) in the presence of MG132 (1 μm) or TEMPO (1 mm) as indicated for 10 h. Multiple dishes with identical treatments were pooled before lysis. His-tagged FLIP proteins were captured by Ni-NTA resin and eluted by imidazole solution. The inputs (1/20 of lysates used for FLIP protein capture) and the Ni-NTA-purified proteins were analyzed by immunoblotting using mouse anti-FLIP, anti-Ubiquitin, and rabbit anti-FADD antibodies. C, PPC1 cells were transfected with either pcDNA3-HA vector (first lane) or HA-tagged FLIP-WT plasmid (second to seventh lanes) for 16 h. Cells were then treated with menadione (5 μm) in the presence of MG132 (0.5 μm) or increasing concentrations of TEMPO (500 μm, 1 mm, and 2 mm) for 8 h. Cell lysates were immunoprecipitated (IP) using rat anti-HA antibody and the immunoprecipitated proteins were analyzed by immunoblotting using mouse anti-HA to detect FLIP. The inputs (1/10 of lysates used for immunoprecipitation) were analyzed by immunoblotting using rabbit anti-FADD as loading control. D, PPC1 cells were treated with menadione (30 μm) with or without TEMPO (1 mm) for 8 h. The adherent cells were incubated with H2DCFDA for 30 min at 37 °C, washed, collected, and analyzed by FACS. Statistical significance (mean ± S.E.; n = 4) was determined by one-way analysis of variance and Tukey post-test. *, indicates the p value is p < 0.05. E, PPC1 cells were treated with increasing concentrations of menadione for 8 h and then total RNA was extracted from cells. Relative levels of endogenous FLIP mRNA were assessed by quantitative RT-PCR.
The generation of ROS by menadione was confirmed in PPC-1 cells by measuring DCF fluorescence by flow cytometry. Addition of the ROS scavenger TEMPO, even at moderate concentrations, significantly reduced the amount of ROS generated within cells by menadione (Fig. 1D). To assess whether the loss of FLIP protein was solely due to post-translational mechanisms, levels of c-FLIP mRNA were quantified by quantitative RT-PCR. No significant changes in c-FLIP mRNA levels were detected during the 8-h treatment with menadione at varying concentrations (Fig. 1E). Taken together, these experiments suggest that menadione-generated ROS cause c-FLIP ubiquitination and subsequent proteasome-dependent degradation.
ROS-dependent Degradation of c-FLIP Induced by Paraquat
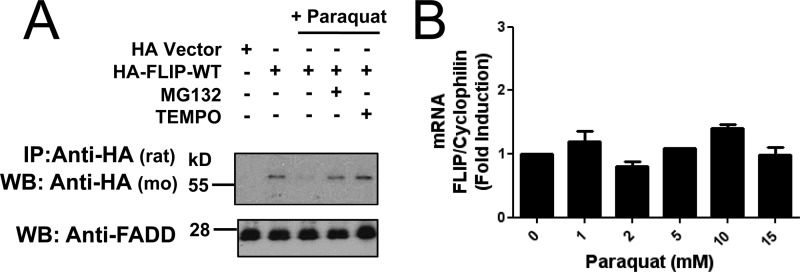
To explore whether this ROS-dependent degradation of c-FLIP was specific to menadione treatment or attributed to a more general response to ROS exposure, we tested the effect on c-FLIP degradation of paraquat, another compound that generates ROS by a similar mechanism as menadione. Treatment of PPC-1 cells with paraquat induced a reduction in HA-FLIP protein that was blocked by the presence of MG132 (Fig. 2A). Moreover, when ROS scavenger TEMPO was added to cultures, HA-FLIP protein was no longer degraded. No significant changes in c-FLIP mRNA levels were observed during the paraquat treatments concluding that paraquat likewise reduces c-FLIP protein expression through a post-transcriptional mechanism (Fig. 2B).
FIGURE 2.
ROS-dependent degradation of FLIP following paraquat treatment. A, PPC1 cells were transfected with either pcDNA3-HA vector (first lane) or HA-tagged FLIP-WT plasmid (second to fifth lanes) for 16 h. Cells were then treated with paraquat (2 mm) in the presence of MG132 (0.5 μm) or TEMPO (500 μm) for 8 h. Cell lysates were immunoprecipitated (IP) using rat anti-HA antibody and the immunoprecipitated proteins were analyzed by immunoblotting using mouse anti-HA to detect FLIP. The inputs (1/10 of lysates used for immunoprecipitation) were analyzed by immunoblotting using rabbit anti-FADD as loading control. B, PPC1 cells were treated with increasing concentrations of paraquat for 8 h and then total RNA was extracted from the cells. Relative levels of endogenous FLIP mRNA were assessed by quantitative RT-PCR.
An alternative method of ROS generation was also tested using buthionine sulfoximine, which depletes cellular glutathione levels. Exposure of PPC-1 cells to buthionine sulfoximine reduced HA-FLIP protein levels, which was prevented by the addition of MG132 or TEMPO (supplemental Fig. S1). Consistent with observations for epitope-tagged FLIP, ROS-dependent proteasomal degradation of the endogenous c-FLIPL protein was verified by all three inducers of ROS tested here: menadione, paraquat, and buthionine sulfoximine (supplemental Fig. S2).
Phosphorylation of Threonine 166 and Ubiquitination of Lysine 167 Are Required for ROS-dependent Degradation of c-FLIP
We assessed ROS-induced PTMs of c-FLIP by mass spectrometry proteomics analysis. PPC-1 cells overexpressing His6-FLIP-WT were treated with menadione in the presence of MG132 for 10 h. Cells were lysed, His6-FLIP-WT protein was isolated by nickel-agarose, and His6-FLIP protein was visualized by Sypro Ruby staining. The His6-FLIP protein bands were excised, digested with trypsin, and analyzed by mass spectrometry (supplemental Table S1). Two PTMs were identified, phosphorylation of threonine 166 and ubiquitination of the adjacent lysine 167 (Table 1). Fragmentation mass spectra of the FLIP peptide containing both PTMs of Thr-166 and Lys-167 indicate good coverage for the PTM sites identified (supplemental Fig. S3). These residues are located near the end of the second DED (supplemental Fig. S4).
TABLE 1.
Assignments for peptide sequences identified from trypsin digestion of captured FLIP protein bands
The whole lane of c-FLIP protein bands were excised from the SDS-PAGE gel, reduced, alkylated, digested by trypsin, and subjected to LC/MS-MS analysis using the Michrom Paradigm MS4 HPLC.HTC-PAL autosampler/LTQ Orbitrap XL mass spectrometery for the analyses. The first column indicates the number of the first and last amino acid of the identified FLIP peptides, whereas the second column shows the corresponding amino acid sequences. TP or T(181 amu) indicates Thr mass + phospho group; KUb or K(242 amu) indicates Lys mass + ubiquitin GG tag; and MO or M(147 amu) indicates Met mass + oxidation.
| Residues | Sequence |
|---|---|
| 27–38 | DVAIDVVPPNVR |
| 39–45 | DLLDILR |
| 50–61 | LSVGDLAELLYR |
| 77–85 | KAVETHLLR |
| 95–106 | VLMOAEIGEDLDK |
| 141–154 | LNLVAPDQLDLLEK |
| 162–170 | IDLKTpKUbIQK |
| 213–226 | LKEQLGAQQEPVKK |
| 227–243 | SIQESEAFLPQSIPEER |
| 269–280 | DTFTSLGYEVQK |
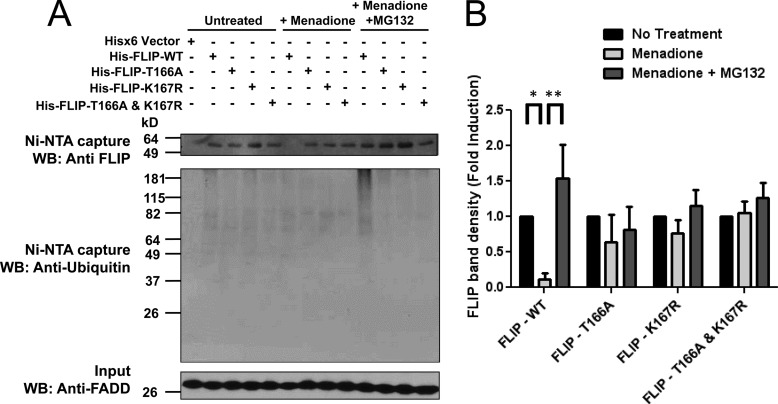
To verify phosphorylation on Thr-166, we performed a Phostag gel electrophoresis comparison of wild-type and T166A c-FLIP proteins. Accordingly, HA-tagged FLIP wild-type (HA-FLIP-WT) or phosphothreonine mutant (T166A) HA-FLIP proteins were expressed by transfection in PPC-1 cells, then the cells were treated with menadione and MG132. Cells were lysed after 8 h and HA-FLIP protein was immunoprecipitated with anti-HA rat antibody. The precipitates were separated by Phostag gel electrophoresis and migration of phosphorylated c-FLIP isoforms was analyzed by immunoblotting with anti-HA mouse antibody. Phostag gel analysis of the T166A c-FLIP mutant revealed loss of a band observed for the WT c-FLIP protein, presumably corresponding to phosphorylation on Thr-166 (supplemental Fig. S5), thus supporting the notion that Thr-166 is a site of phosphorylation in the c-FLIP protein. To ascertain the importance of these two PTMs on FLIP protein stability, we expressed His6-tagged c-FLIP wild-type (His-FLIP-WT), phosphothreonine mutant (His-FLIP-T166A), ubiquitin mutant (His-FLIP-K167R), and the double mutant where both the threonine and lysine residues were substituted (His-FLIP-T166A,K167R) in PPC-1 cells. The cells were then treated with or without menadione in the presence of MG132. Cells were lysed after 8 h and c-FLIP protein was isolated by nickel-agarose. The c-FLIP protein levels in the precipitates were analyzed by immunoblotting with anti-FLIP antibody and ubiquitination was analyzed with anti-ubiquitin antibody. WT c-FLIP was degraded upon menadione treatment and the polyubiquitinated WT c-FLIP protein accumulated when MG132 was present. In contrast, c-FLIP mutants T166A, K167R, or T166A,K167R were not susceptible to menadione-induced degradation (Fig. 3A). The FLIP protein band densities from multiple experiments were quantified, confirming that c-FLIP protein levels are stabilized when the identified phosphorylation and ubiquitination PTMs are ablated by site-directed mutagenesis (Fig. 3B).
FIGURE 3.
Phosphorylation of Thr-166 and ubiquitination of Lys-167 are required for ROS-dependent ubiquitination and degradation of FLIP following menadione treatment. A, PPC1 cells were transfected with pcDNA3.1-His6 vector (first lane) or various His-tagged FLIP plasmids (WT, T166A, K167R, and T166A,K167R double mutant) for 16 h. Cells were then treated with menadione (5 μm) in the presence of MG132 (1 μm) for 8 h. His-tagged FLIP proteins were captured by Ni-NTA resin and eluted by imidazole solution. The Ni-NTA-purified proteins were analyzed by immunoblotting using mouse anti-FLIP and anti-Ubiquitin antibodies. The inputs (1/20 of lysates used for FLIP protein capture) were analyzed by immunoblotting using rabbit anti-FADD as a loading control. B, levels of His-FLIP protein in A were quantified using scanning densitometry. Vector groups with no treatment were adjusted to 1. Statistical significance (mean ± S.E.; n = 3) was determined by two-way analysis of variance and Bonferroni post-test. *, indicates the p value is p < 0.05; **, indicates p < 0.001.
To further explore the requirement of phosphorylation for ROS-dependent ubiquitination and degradation, we generated phosphomimic mutants HA-FLIP-T166D and HA-FLIP-T166E, which were expressed by transfection in PPC-1 cells, followed by treatment of the cells with menadione and MG132. However, no increased ubiquitination or degradation was observed for either phosphorylation mimicking mutant of c-FLIP (supplemental Fig. S6). Therefore, these mutations may not effectively imitate the effect of phosphorylation at Thr-166 with respect to ROS-mediated ubiquitination and degradation.
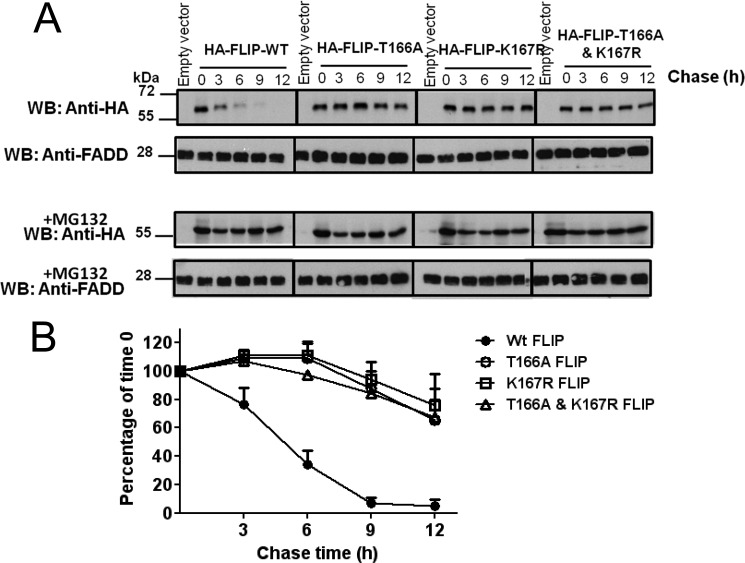
The effect of menadione treatment on FLIP protein stability was additionally examined by cycloheximide chase experiments. PPC-1 cells expressing HA-tagged FLIP-WT or PTM mutants were treated with cycloheximide to prevent further protein synthesis and with menadione to induce ROS production. Cells were cultured with or without MG132. Immunoblotting with anti-HA rat antibody monitored FLIP protein levels over a 12-h period (Fig. 4A). For HA-FLIP-WT, a half-life of ∼4.5 h was observed (Fig. 4B). In contrast, the c-FLIP mutants T166A, K167R, and the double mutant (where both post-translational sites were modified) showed increased stability, with more than 65% retention of c-FLIP protein levels following 12 h treatment (Fig. 4B). These results further support that menadione down-regulates c-FLIP protein levels in a post-translational manner and show that mutagenesis of the threonine 166 and lysine 167 residues stabilizes c-FLIP.
FIGURE 4.
Mutations of Thr-166 or Lys-167 stabilize the c-FLIP protein. A, PPC1 cells were transfected with pcDNA3-HA vector or various HA-tagged FLIP plasmids (WT, T166A, K167R, and T166A,K167R double mutant) for 16 h. Cells were treated with 25 μg/ml of cycloheximide, menadione (5 μm), MG132 (0.5 μm), or various combinations of these reagents, then harvested at the indicated times. An equal amount of protein from each cell lysate was analyzed by immunoblotting using rat anti-HA antibody. FADD served as a loading control. B, levels of HA-FLIP protein in A were quantified using scanning densitometry and expressed as a percentage of FLIP expression at 0 h for each vector group (mean ± S.E.; n = 3).
Other studies were performed to explore whether nitrosylative PTMs occur on c-FLIP in response to menadione treatment. Following treatment with menadione and MG132, the His6-FLIP-WT protein was isolated from cell lysates by nickel-agarose and reacted with 2,3-diaminonaphthalene reagent. The conversion of 2,3-diaminonaphthalene to the fluorescent compound 2,3-napthyltriazole by nitrosothiols was measured on a fluorometric plate reader. BSA nitrosylation by S-nitrocysteine served as a positive control. No evidence of c-FLIP nitrosylation was detected (supplemental Fig. S7).
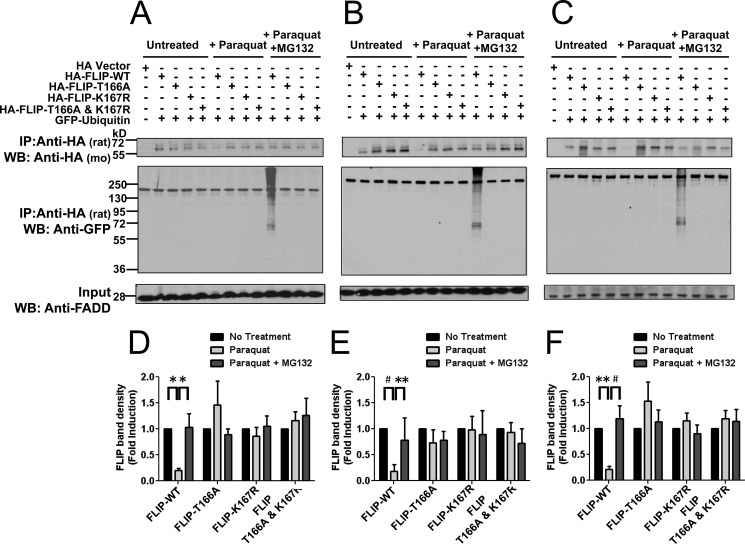
The resistance of phosphorylation and ubiquitination c-FLIP mutants to menadione-induced degradation was also explored in PPC-1 cells treated with the alternative ROS-generating compound, paraquat. In these experiments, HA-tagged c-FLIP WT and the various c-FLIP mutants were co-expressed with GFP-ubiquitin to enhance the ability to detect c-FLIP ubiquitination. Following 8 h treatment with or without paraquat in the presence of MG132, the cells were lysed and c-FLIP protein was immunoprecipitated with anti-HA rat antibody. The levels of c-FLIP protein in immunoprecipitates were analyzed by immunoblotting with anti-HA mouse antibody and ubiquitination was analyzed with anti-GFP antibody. Analogous to observations with menadione, the WT c-FLIP protein was degraded following exposure to paraquat and this decline in c-FLIP protein was prevented by inhibition of the proteasome (Fig. 5, A and D). Furthermore, T166A and K167R single and double mutants showed enhanced protein stability following paraquat treatment. Moreover, in the presence of proteasomal inhibitor (MG132), no c-FLIP ubiquitination of these mutants was apparent (Fig. 5, A and D). Very similar results were obtained in HeLa (Fig. 5, B and E) and 293T (Fig. 5, C and F) cells, showing that mutation of Thr-166 or Lys-167 ablates ROS-dependent c-FLIP ubiquitination and degradation.
FIGURE 5.
Phosphorylation of Thr-166 and ubiquitination of Lys-167 are required for ROS-dependent ubiquitination and degradation of FLIP following paraquat treatment. A-C, PPC1 (A), HeLa (B), or 293T (C) cells were co-transfected with EGFP-C2-ubiquitin and either pcDNA3-HA vector (first lane) or various HA-tagged FLIP plasmids (WT, T166A, K167R, and T166A,K167R double mutant) for 16 h. Cells were then treated with paraquat (2 mm) in the presence of MG132 (0.5 μm) for 8 h. Cell lysates were immunoprecipitated (IP) using rat anti-HA antibody and the immunoprecipitated proteins were analyzed by immunoblotting using mouse anti-HA to detect FLIP and anti-GFP to detect ubiquitination. The inputs (1/10 of lysates used for immunoprecipitation) were analyzed by immunoblotting using rabbit anti-FADD antibody as a loading control. D–F, levels of HA-FLIP protein in A–C, respectively, were quantified using scanning densitometry. Vector groups with no treatment were adjusted to 1. Statistical significance (mean ± S.E.; n = 6 PPC1 and 293T; n = 4 HeLa) was determined by two-way analysis of variance and Bonferroni post-test. *, indicates the p value is p < 0.05; **, indicates p < 0.01; #, indicates p < 0.001.
Menadione and Paraquat Increase Sensitivity of PPC-1 Cells to TRAIL
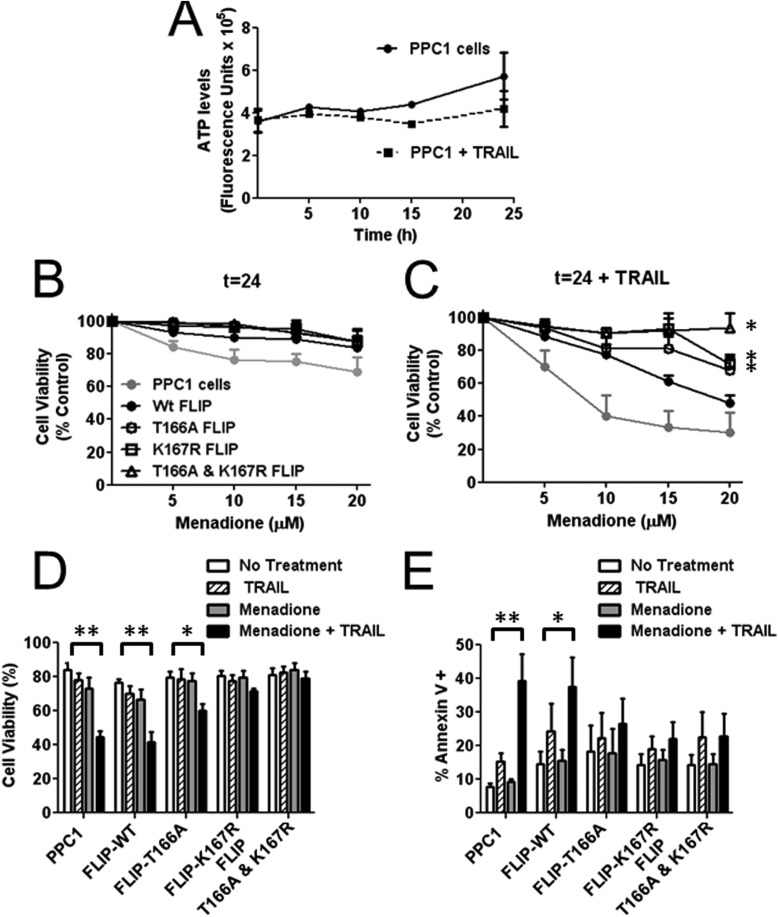
The sensitization of tumor cell lines to TRAIL has been attributed in several contexts to the loss of c-FLIP protein levels. We therefore determined the effect of ROS-generating compounds menadione and paraquat on the viability of cells treated with TRAIL. Addition of TRAIL by itself had only a minor effect on PPC-1 cell viability and growth over 24 h, as assessed by assays measuring cellular ATP levels as a surrogate indicator of cell viability (Fig. 6A). Menadione treatment of control PPC-1 cells or PPC-1 cells overexpressing WT or mutant c-FLIP variants had little effect on cell viability even at doses as high as 20 μm (Fig. 6B). However, in combination with TRAIL, menadione treatment resulted in concentration-dependent reductions in cell viability (EC50 < 10 μm) (Fig. 6C). PPC-1 cells overexpressing HA-FLIP-WT showed an increase in cell viability at low concentrations of menadione but these cells were still sensitive to TRAIL with increasing concentrations of menadione. In contrast, the combination of menadione and TRAIL was essentially not cytotoxic to PPC-1 cells overexpressing the phosphorylation or ubiquitination single site c-FLIP mutants. In particular, the c-FLIP double mutant provided protection even against the highest concentration of menadione tested (20 μm) in combination with TRAIL (Fig. 6C).
FIGURE 6.
Expression of FLIP mutants rescues PPC1 cells from menadione-mediated sensitization to TRAIL-induced cell death. A, PPC1 cells were treated with or without TRAIL (10 ng/ml) for 24 h. At various time points cells were lysed and ATP levels were measured using cell titer glow to assess cell viability. B, PPC1 cells were transfected with various HA-tagged FLIP plasmids (WT, T166A, K167R, and T166A,K167R double mutant) for 16 h and then treated with increasing concentrations of menadione alone or (C) with 10 ng/ml of TRAIL for 24 h. Cells were then lysed and ATP levels were measured using cell titer glow to assess cell viability. Vector groups with no treatment were adjusted to 100%. Statistical significance (mean ± S.E.; n = 3) was determined by repeated measures analysis of variance and Tukey post-test. *, indicates the p value is p < 0.01 for a comparison with control PPC1 cells. D, PPC1 cells were transfected with various HA-tagged FLIP plasmids (WT, T166A, K167R, and T166A,K167R double mutant) for 16 h and then treated with 15 μm menadione with or without TRAIL (10 ng/ml) for 24 h. Cell viability was assessed by exclusion of trypan blue. Statistical significance (mean ± S.E.; n = 4) was determined by two-way analysis of variance and Bonferroni post-test. *, indicates p < 0.05 and **, indicates p < 0.001. E, PPC1 cells were co-transfected with various HA-tagged FLIP plasmids (WT, T166A, K167R, and T166A,K167R double mutant) each with EGFP-C2 for 16 h and then treated with 10 μm menadione with or without TRAIL (25 ng/ml) for 16 h. To assess cell death, cells were stained with annexin V-APC and propidium iodide and the percentage of annexin V positive cells was determined by FACS. Statistical significance (mean ± S.E.; n = 3) was determined by two-way analysis of variance and Bonferroni post-test. *, indicates p < 0.05 and **, indicates p < 0.01.
Trypan blue exclusion was assessed as a second measure of cell viability, confirming the results (Fig. 6D). Cell death assessments by flow cytometry analysis measuring annexin V-APC and propidium iodide staining showed a significant increase in the percentage of apoptotic control PPC-1 cells or PPC-1 cells expressing HA-FLIP-WT following exposure to menadione with TRAIL (Fig. 6E). No significant induction of apoptosis was detected in PPC-1 cells during the time frame of these experiments when T166A, K167R, or the T166A,K167R double mutant c-FLIP protein was expressed in cells treated with a combination of menadione and TRAIL (Fig. 6E).
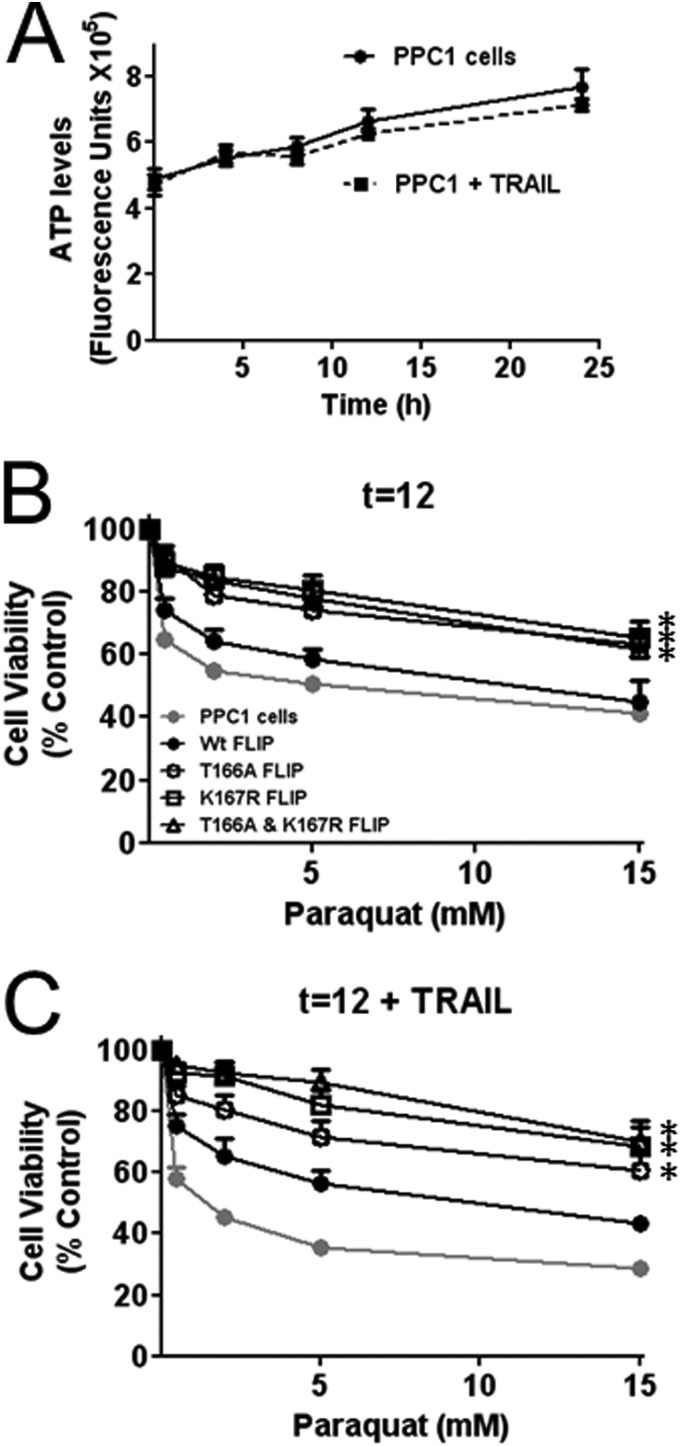
In comparison to menadione, treatment of PPC-1 cells with paraquat alone resulted in significant cell death (Fig. 7B). PPC-1 cells expressing HA-FLIP-WT were equally susceptible to this paraquat-induced cell death. In contrast, expression of phosphorylation and ubiquitination c-FLIP mutants improved survival of paraquat-treated cells (Fig. 7B). The addition of TRAIL augmented cell death in cultures of paraquat-treated PPC-1 control cells (Fig. 7C). In contrast, expression of HA-FLIP-T166A, K167R, or the phosphorylation and ubiquitination c-FLIP double mutant (HA-FLIP-T166A,K167R) protected the PPC-1 cells from the incremental cell death observed upon addition of TRAIL to paraquat-treated cultures (Fig. 7C).
FIGURE 7.
Expression of FLIP PTM mutants rescues PPC1 cells from paraquat-induced cell death and sensitization to TRAIL. A, PPC1 cells were treated with or without TRAIL (10 ng/ml) for 24 h. At various times, cells were lysed and ATP levels were measured using cell titer glow to assess cell viability. B, PPC1 cells were transfected with various HA-tagged FLIP plasmids (WT, T166A, K167R, and T166A,K167R double mutant) for 16 h and then treated with increasing concentrations of paraquat alone or (C) with 10 ng/ml of TRAIL for 12 h. Cells were then lysed and ATP levels were measured using cell titer glow to assess cell viability. Vector groups with no treatment were adjusted to 100%. Statistical significance (mean ± S.E.; n = 4) was determined by repeated measures analysis of variance and Tukey post-test. *, indicates p < 0.01 for a comparison with control PPC1 cells.
DISCUSSION
The protein c-FLIP is an important inhibitor of DR-mediated apoptosis (6). Numerous studies have demonstrated c-FLIP up-regulation in a variety of cancers, contributing to the resistance of tumor cells to DR-induced apoptosis (9). The ubiquitin-proteasome pathway has emerged as a central regulator of c-FLIP expression that conceivably could be exploited to effectively re-sensitize cancer cells to DR-mediated apoptosis. Although c-FLIP ubiquitination and degradation have been extensively reported in response to a variety of chemical compounds and cellular circumstances, the exact sites of ubiquitination or other PTMs involved have not been elucidated. ROS have recently been implicated in the down-regulation of c-FLIP but the underlying mechanism is not well defined. Our findings indicate that ROS-induced proteasomal degradation of c-FLIP is mediated through PTMs that include phosphorylation and ubiquitination. The specific phosphorylation and ubiquitination sites identified that confer c-FLIP protein instability and degradation following exposure to ROS reside at the end of the second DED domain. Mutation of either the phosphorylation site at Thr-166 or the ubiquitination site at Lys-167 was sufficient to confer stability on the c-FLIP protein in the face of ROS challenge, with ablation of both Thr-166 and Lys-167 sites simultaneously being slightly more effective than either site alone. This supports the concept that FLIP is a redox-sensitive protein whereby both of these PTMs are involved for mediating ROS-dependent degradation of FLIP, and thus important determinants of ROS-dependent sensitization of cells to DR agonists such as TRAIL.
The residues corresponding to the Thr-166 phosphorylation site and the Lys-167 ubiquitination site are highly conserved in c-FLIP across species. However, they are not present in other DED domain-containing proteins. Interestingly, two phosphorylation sites have previously been characterized for c-FLIP. In this regard, c-FLIP is phosphorylated at serine 273 by Akt in macrophages stimulated with TNFα or lipopolysaccharide (LPS) (26). Expression of a S273A mutant was less susceptible to LPS-stimulated reductions of c-FLIP compared with the WT c-FLIP protein, although reduction was still evident. In our analysis of menadione-treated PPC-1 cells, serine 273 phosphorylation was not observed. This disparity could be due to differences in the stimulus and cell-type used. Second, phosphorylation on serine 193 by protein kinase C (PKC) reportedly blocks ubiquitination of the short isoform of c-FLIP (c-FLIPS), increasing its stability in erythroleukemia cells (27). Conversely, PKC-mediated phosphorylation of c-FLIPL had no effect on its ubiquitination and degradation. Our mass spectrometry analysis did not cover the region of c-FLIP protein where serine 193 is located. However, this phosphorylation site could potentially correspond to the unknown phosphorylation band observed in phostag gel separation analysis (supplemental Fig. S5). As this PTM is observed on the WT c-FLIP protein (which undergoes ROS-induced degradation), our data are in agreement with Eriksson and colleagues (27), who have suggested that phosphorylation of Ser-193 is not important for regulating ROS-mediated degradation of c-FLIPL. Irrespective of effects on protein degradation, phosphorylation of c-FLIP has also been reported to affect the sensitivity of cells to DR-mediated apoptosis. For example, PKC-mediated phosphorylation of c-FLIP reportedly interferes with binding to FADD, thus enhancing TRAIL-mediated apoptosis (28). In addition, threonine phosphorylation of c-FLIP by calcium/calmodulin-dependent protein kinase II (CMKII) reportedly directs c-FLIP to the DISC complex, inhibiting execution of apoptosis in Fas and TRAIL-resistant malignant cells (29–31). We show here that mutagenesis of the phosphorylation site at threonine 166 stabilizes c-FLIP protein levels in the face of ROS challenge, whereas conferring protection of cells against ROS-mediated sensitization to apoptosis induced by TRAIL. Furthermore, mutation of Thr-166 also prevented ubiquitination of the adjacent lysine suggesting that this phosphorylation site tags c-FLIP for polyubiquitination and proteasomal degradation. For protein-protein interactions, examples have been elucidated where the bulkiness of the phosphate group or protein conformational changes induced by phosphorylation account for the actions of phosphorylation as opposed to its negative charge (32). This may explain why the T166D or T166E mutants could not mimic phosphorylation with respect to ROS-mediated ubiquitination and degradation.
Although ubiquitin-mediated proteasomal degradation of c-FLIP has been reported in response to a number of chemical compounds (14, 16–22), our mutagenesis experiments support the novel identification of lysine 167 as a key ROS-dependent ubiquitination site that is central for proteasomal degradation of FLIP. Prior to this study, only one other c-FLIP ubiquitination site, lysine 192, was reported in a proteome-wide mass spectrometry analysis of endogenous ubiquitination sites in untreated HEK293T (33). This region of the FLIP protein was not covered by our peptides analyzed by mass spectrometry. As the previous study was performed on resting untreated cells, this ubiquitination site may be involved in the basal turnover of c-FLIP protein and the background level of ubiquitination observed in some of our immunoblots may correlate to such an ubiquitination site. Nitrosylation of the c-FLIP protein has also previously been shown to stabilize the protein and interfere with degradation via the ubiquitin-proteasome pathway (34). The implication that ROS generation and oxidative stress can also increase nitric oxide (NO) within cells raised the possibility of nitrosylation PTMs occurring on c-FLIP following treatment with agents such as menadione and paraquat. However, we obtained no evidence for FLIP nitrosylation in menadione-treated cells (supplemental Fig. S7). In support, it has been demonstrated that superoxide can react with NO thereby reducing the available pool of NO present to react with c-FLIP (25).
The conjugation of ubiquitin to target proteins requires the activity of an E3 ligase (35). Karin and colleagues (36) demonstrated that Itch was the E3 ligase responsible for ubiquitination and degradation of c-FLIP during Fas-mediated apoptosis involving the Jun kinase (JNK) signaling pathway. They showed JNK acted not by directly phosphorylating c-FLIP but by phosphorylating and activating Itch. JNK activity was also required to trigger c-FLIP degradation in breast tumor cells treated with antimicrotubule agents and TRAIL (37) and treatment of TRAIL with anti-DR5 antibody induced ROS generation that stimulated JNK phosphorylation and down-regulation of c-FLIP in HIV-infected macrophages (38). Moreover, Itch-dependent ubiquitination and proteasomal degradation of c-FLIP were reported in cisplatin-treated ovarian cancer cells (39). Itch specifically ubiquitinates FLIPL by binding to the caspase domain (36). The ubiquitination site Lys-167 identified in our studies is located in the highly conserved DED region. Therefore it is possible that JNK/Itch-mediated proteasomal degradation may not be involved in the degradation of c-FLIP protein induced by ROS challenge of cancer cells. Importantly, Itch and JNK-independent c-FLIP ubiquitination have been reported (20, 22, 26, 40). For example, proteasomal degradation of c-FLIP still occurs in breast tumor cells treated with the histone deacetylase inhibitor suberoylanilide hydroxamic acid under circumstances where Itch had been depleted by siRNA, suggesting proteasomal targeting of c-FLIP through a pathway independent of the E3 ubiquitin ligase Itch (22). An alternative ligase was not identified by the group. Pope and colleagues (26) point to the phosphatidylinositol 3-kinase/Akt pathway in the c-FLIP degradation observed in macrophages stimulated with TNFα or LPS. The ataxia telangiectasia-mutated kinase was also found to be required for c-FLIP down-regulation and sensitization to TRAIL by the DNA-damaging agents neocarzinostatin and 5-fluorouracil in human hepatocellular carcinoma cell lines (40). Therefore, it is clear that different kinases and ubiquitin ligases are involved in c-FLIP down-regulation, depending on the cell type and circumstances. Computational analysis of the amino acid sequences surrounding Thr-166 by GPS (Group-base Prediction System) version 2.1 (41), NetPhosK 1.0 (42), and PhosphoMotif Finder (43) databases predict Thr-166 phosphorylation by MAP3K11 (MLK3), PKC, and casein kinase 1, respectively. Oxidative stress is known to activate PKC and MLK3 activity (44, 45). Moreover, apoptosis can be stimulated by MLK3-mediated JNK and p38 signaling (46). Further studies are required to clarify the specific kinase and ligase important for c-FLIP proteasomal degradation in response to ROS.
ROS can elicit a wide spectrum of biological responses from proliferation to cell death (47). Almost a decade ago, studies of cardiac myocytes showed a requirement for ROS generation by doxorubicin to trigger a loss of c-FLIP protein and subsequent sensitization of cells to Fas-induced apoptosis (48). A later study demonstrated that Fas ligand generated ROS in normal lung epithelial cells through activation of Rac1 and the NADPH oxidase. These ROS specifically mediated the ubiquitination and degradation of c-FLIP to increase cell sensitivity to Fas-induced cell death (25). More recently, an investigation of the natural alkaloid berberine also attributed the observed ROS-dependent loss of c-FLIP to proteasomal degradation (21). In addition, targeting of c-FLIP to the proteasome was similarly shown to be a route by which cells normally are resistant to TRAIL-induced apoptosis can be rendered susceptible to cell death. In our present study, treatment with the ROS-generating compounds menadione or paraquat resulted in decreased c-FLIP expression that was prevented by pretreatment of cells with the ROS scavenger TEMPO or proteasome inhibitor MG132. Consistent with other studies the loss of c-FLIP protein was solely due to the post-translational regulation as the level of c-FLIP mRNA transcripts were unaffected by ROS generation. We also observed that chemical inducers of ROS can sensitize cancer cells to TRAIL. In contrast, overexpression of c-FLIP mutants lacking Thr-166 or Lys-167 demonstrated increased protein stability and protected cells from ROS-mediated sensitization to TRAIL-induced cell death.
Recently, hepatocytes from mice with liver-specific deletion of genes encoding c-FLIP were reported to be hypersensitive to low concentrations of menadione and vulnerable to ROS-induced apoptosis (49). Interestingly, viability differences between WT and c-FLIP knock-out cells were lost at higher concentrations of menadione, indicating that c-FLIP-mediated protection is overwhelmed in the face of massive ROS exposure. Treatment of hepatocytes with high menadione concentrations also activated JNK (49, 50) and JNK phosphorylation was augmented in c-FLIP knock-out hepatocytes (49) but reversed in the presence of superoxide scavengers (50). In conditions where NF-κB is attenuated, TNF-α-induced ROS were shown to oxidize and inhibit JNK-inactivating phosphatases, thus resulting in sustained JNK activation and execution of cell death (51). Moreover, c-FLIP reportedly binds to MAP kinase kinase-7 (MKK7) and suppresses JNK signaling (52). Therefore, when NF-κB is inhibited, ROS promote JNK activation as well as c-FLIP degradation thus alleviating the brake that FLIP has on the pro-apoptotic signaling cascade.
Menadione and paraquat treatment of cells up-regulate GRP94, GRP78, CHOP, and induce phosphorylation of eIF2, IRE1, and JNK, indicative of endoplasmic reticulum (ER) stress (53–57). In this regard, ER stress up-regulates expression of DR5 (TRAIL-R2) via a CHOP-dependent mechanism, stimulating caspase-8-dependent apoptosis in some cellular contexts (58–62). Moreover, c-FLIP down-regulation in breast cancer cells undergoing ER stress contributes to sensitization to TRAIL-induced apoptosis (63). Thus, the combined use of agents that induce ER stress with TRAIL has received attention as potential anti-cancer therapies. In this regard, α-TEA, an analog of vitamin E, induces c-FLIP down-regulation through the ER stress JNK/CHOP/DR5 pathway via Itch-mediated ubiquitination and loss of c-FLIP (59). However, a study using multiple chemical inducers of CHOP expression did not find a role for JNK or Itch in mediating CHOP-induced c-FLIP ubiquitination and degradation (64). Further experiments are needed to determine whether the oxidative stress caused by menadione or paraquat treatment of cells at the concentrations used in our study induces the ER stress response and whether the ER stress signal transduction pathway is involved in the ROS-mediated down-regulation of c-FLIP observed in our experiments.
In summary, we identified the specific sites of phosphorylation and ubiquitination on c-FLIP that mediate ROS-dependent degradation and subsequent sensitization of cancer cells to TRAIL. Future identification of the specific kinase(s) and ubiquitin ligase(s) involved in ROS-mediated c-FLIP degradation may provide attractive targets for developing novel therapeutic strategies that either seek to (a) preserve c-FLIP expression and promote cell survival in the face of ROS challenge, such as in cardiomyocytes during ischemia-reperfusion injury, or alternatively to (b) stimulate c-FLIP degradation for sensitizing unwanted cells such as cancers and autoimmune lymphocytes to DR-mediated apoptosis.
Acknowledgments
We thank Dr. Ranxin Shi for His6-c-FLIPL plasmid and Dr. Charitha Madiraju and Michael Cuddy for GFP-Ubiquitin plasmid. We thank Drs. Khatereh Motamedchaboki and Laurence M. Brill for mass spectrometry-based proteomic analysis, Jonna Hurtado for technical assistance with FACS analysis, Dr. Paul Diaz for helpful discussion, and Melanie Hanaii for manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-CA-163743.

This article contains supplemental Table S1 and Figs. S1–S7.
- DR
- death receptor
- FADD
- Fas-associated protein with death domain
- DED
- death effector domain
- DISC
- death-inducing signaling complex
- c-FLIP
- cellular Fas-associated death domain-like interleukin 1β-converting enzyme inhibitory protein
- TRAIL
- tumor necrosis factor-related apoptosis-inducing ligand
- PTM
- Post-translational modification
- ROS
- reactive oxygen species
- TEMPO
- tetramethylpiperidine-N-oxyl
- Ni-NTA
- nickel-nitrilotriacetic acid
- H2DCFDA
- 2′,7′-dichlorodihydrofluorescein diacetate
- APC
- allophycocyanin
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Kerr J. F., Wyllie A. H., Currie A. R. (1972) Apoptosis. A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Budihardjo I., Oliver H., Lutter M., Luo X., Wang X. (1999) Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 15, 269–290 [DOI] [PubMed] [Google Scholar]
- 3. Hengartner M. O. (2000) The biochemistry of apoptosis. Nature 407, 770–776 [DOI] [PubMed] [Google Scholar]
- 4. Riedl S. J., Salvesen G. S. (2007) The apoptosome. Signalling platform of cell death. Nat. Rev. Mol. Cell Biol. 8, 405–413 [DOI] [PubMed] [Google Scholar]
- 5. Nagata S. (1999) Fas ligand-induced apoptosis. Annu. Rev. Genet. 33, 29–55 [DOI] [PubMed] [Google Scholar]
- 6. Irmler M., Thome M., Hahne M., Schneider P., Hofmann K., Steiner V., Bodmer J.-L., Schröter M., Burns K., Mattmann C., Rimoldi D., French L. E., Tschopp J. (1997) Inhibition of death receptor signals by cellular FLIP. Nature 388, 190–195 [DOI] [PubMed] [Google Scholar]
- 7. Oberst A., Green D. R. (2011) It cuts both ways. Reconciling the dual roles of caspase 8 in cell death and survival. Nat. Rev. Mol. Cell Biol. 12, 757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roth W., Reed J. C. (2004) FLIP protein and TRAIL-induced apoptosis (review). Vitam. Horm. 67, 189–206 [DOI] [PubMed] [Google Scholar]
- 9. Safa A. R., Pollok K. E. (2011) Targeting the anti-apoptotic protein c-FLIP for cancer therapy. Cancers 3, 1639–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCourt C., Maxwell P., Mazzucchelli R., Montironi R., Scarpelli M., Salto-Tellez M., O'Sullivan J. M., Longley D. B., Waugh D. J. (2012) Elevation of c-FLIP in castrate-resistant prostate cancer antagonizes therapeutic response to androgen receptor-targeted therapy. Clin. Cancer Res. 18, 3822–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kataoka T., Budd R. C., Holler N., Thome M., Martinon F., Irmler M., Burns K., Hahne M., Kennedy N., Kovacsovics M., Tschopp J. (2000) The caspase-8 inhibitor FLIP promotes activation of NF-κB and Erk signaling pathways. Curr. Biol. 10, 640–648 [DOI] [PubMed] [Google Scholar]
- 12. Yu J. W., Shi Y. (2008) FLIP and the death effector domain family. Oncogene 27, 6216–6227 [DOI] [PubMed] [Google Scholar]
- 13. Johnstone R. W., Frew A. J., Smyth M. J. (2008) The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat. Rev. Cancer 8, 782–798 [DOI] [PubMed] [Google Scholar]
- 14. Kim Y., Suh N., Sporn M., Reed J. C. (2002) An inducible pathway for degradation of FLIP protein sensitizes tumor cells to TRAIL-induced apoptosis. J. Biol. Chem. 277, 22320–22329 [DOI] [PubMed] [Google Scholar]
- 15. Suh W.-S., Kim Y. S., Schimmer A. D., Kitada S., Minden M., Andreeff M., Suh N., Sporn M., Reed J. C. (2003) Synthetic triterpenoids activate a pathway for apoptosis in AML cells involving down-regulation of FLIP and sensitization to TRAIL. Leukemia 17, 2122–2129 [DOI] [PubMed] [Google Scholar]
- 16. Zhang S., Shen H. M., Ong C. N. (2005) Down-regulation of c-FLIP contributes to the sensitization effect of 3,3′-diindolylmethane on TRAIL-induced apoptosis in cancer cells. Mol. Cancer Ther. 4, 1972–1981 [DOI] [PubMed] [Google Scholar]
- 17. Palacios C., Yerbes R., López-Rivas A. (2006) Flavopiridol induces cellular FLICE-inhibitory protein degradation by the proteasome and promotes TRAIL-induced early signaling and apoptosis in breast tumor cells. Cancer Res. 66, 8858–8869 [DOI] [PubMed] [Google Scholar]
- 18. Chen S., Liu X., Yue P., Schönthal A. H., Khuri F. R., Sun S. Y. (2007) CCAAT/enhancer binding protein homologous protein-dependent death receptor 5 induction and ubiquitin/proteasome-mediated cellular FLICE-inhibitory protein down-regulation contribute to enhancement of tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by dimethylcelecoxib in human non-small-cell lung cancer cells. Mol. Pharmacol. 72, 1269–1279 [DOI] [PubMed] [Google Scholar]
- 19. Frew A. J., Lindemann R. K., Martin B. P., Clarke C. J., Sharkey J., Anthony D. A., Banks K. M., Haynes N. M., Gangatirkar P., Stanley K., Bolden J. E., Takeda K., Yagita H., Secrist J. P., Smyth M. J., Johnstone R. W. (2008) Combination therapy of established cancer using a histone deacetylase inhibitor and a TRAIL receptor agonist. Proc. Natl. Acad. Sci. U.S.A. 105, 11317–11322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin Y., Liu X., Yue P., Benbrook D. M., Berlin K. D., Khuri F. R., Sun S. Y. (2008) Involvement of c-FLIP and survivin down-regulation in flexible heteroarotinoid-induced apoptosis and enhancement of TRAIL-initiated apoptosis in lung cancer cells. Mol. Cancer Ther. 7, 3556–3565 [DOI] [PubMed] [Google Scholar]
- 21. Lee S. J., Noh H. J., Sung E. G., Song I. H., Kim J. Y., Kwon T. K., Lee T. J. (2011) Berberine sensitizes TRAIL-induced apoptosis through proteasome-mediated down-regulation of c-FLIP and Mcl-1 proteins. Int. J. Oncol. 38, 485–492 [DOI] [PubMed] [Google Scholar]
- 22. Yerbes R., López-Rivas A. (2012) Itch/AIP4-independent proteasomal degradation of cFLIP induced by the histone deacetylase inhibitor SAHA sensitizes breast tumour cells to TRAIL. Invest. New Drugs 30, 541–547 [DOI] [PubMed] [Google Scholar]
- 23. Ikeda T., Sporn M., Honda T., Gribble G. W., Kufe D. (2003) The novel triterpenoid CDDO and its derivatives induce apoptosis by disruption of intracellular redox balance. Cancer Res. 63, 5551–5558 [PubMed] [Google Scholar]
- 24. Deeb D., Gao X., Jiang H., Janic B., Arbab A. S., Rojanasakul Y., Dulchavsky S. A., Gautam S. C. (2010) Oleanane triterpenoid CDDO-Me inhibits growth and induces apoptosis in prostate cancer cells through a ROS-dependent mechanism. Biochem. Pharmacol. 79, 350–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang L., Azad N., Kongkaneramit L., Chen F., Lu Y., Jiang B. H., Rojanasakul Y. (2008) The Fas death signaling pathway connecting reactive oxygen species generation and FLICE inhibitory protein down-regulation. J. Immunol. 180, 3072–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi B., Tran T., Sobkoviak R., Pope R. M. (2009) Activation-induced degradation of FLIPL is mediated via the phosphatidylinositol 3-kinase/Akt signaling pathway in macrophages. J. Biol. Chem. 284, 14513–14523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaunisto A., Kochin V., Asaoka T., Mikhailov A., Poukkula M., Meinander A., Eriksson J. E. (2009) PKC-mediated phosphorylation regulates c-FLIP ubiquitylation and stability. Cell Death Differ. 16, 1215–1226 [DOI] [PubMed] [Google Scholar]
- 28. Higuchi H., Yoon J. H., Grambihler A., Werneburg N., Bronk S. F., Gores G. J. (2003) Bile acids stimulate cFLIP phosphorylation enhancing TRAIL-mediated apoptosis. J. Biol. Chem. 278, 454–461 [DOI] [PubMed] [Google Scholar]
- 29. Yang B. F., Xiao C., Roa W. H., Krammer P. H., Hao C. (2003) Calcium/calmodulin-dependent protein kinase II regulation of c-FLIP expression and phosphorylation in modulation of Fas-mediated signaling in malignant glioma cells. J. Biol. Chem. 278, 7043–7050 [DOI] [PubMed] [Google Scholar]
- 30. Xiao C., Yang B. F., Song J. H., Schulman H., Li L., Hao C. (2005) Inhibition of CaMKII-mediated c-FLIP expression sensitizes malignant melanoma cells to TRAIL-induced apoptosis. Exp. Cell Res. 304, 244–255 [DOI] [PubMed] [Google Scholar]
- 31. Yang B. F., Xiao C., Li H., Yang S. J. (2007) Resistance to Fas-mediated apoptosis in malignant tumours is rescued by KN-93 and cisplatin via down-regulation of c-FLIP expression and phosphorylation. Clin. Exp. Pharmacol. Physiol. 34, 1245–1251 [DOI] [PubMed] [Google Scholar]
- 32. Paleologou K. E., Schmid A. W., Rospigliosi C. C., Kim H. Y., Lamberto G. R., Fredenburg R. A., Lansbury P. T., Jr., Fernandez C. O., Eliezer D., Zweckstetter M., Lashuel H. A. (2008) Phosphorylation at Ser-129 but not the phosphomimics S129E/D inhibits the fibrillation of α-synuclein. J. Biol. Chem. 283, 16895–16905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chanvorachote P., Nimmannit U., Wang L., Stehlik C., Lu B., Azad N., Rojanasakul Y. (2005) Nitric oxide negatively regulates Fas (CD95)-induced apoptosis through inhibition of ubiquitin-proteasome mediated degradation of FLIP. J. Biol. Chem. 280, 42044–42050 [DOI] [PubMed] [Google Scholar]
- 35. Hershko A., Heller H., Elias S., Ciechanover A. (1983) Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 258, 8206–8214 [PubMed] [Google Scholar]
- 36. Chang L., Kamata H., Solinas G., Luo J. L., Maeda S., Venuprasad K., Liu Y. C., Karin M. (2006) The E3 ubiquitin ligase itch couples JNK activation to TNFα-induced cell death by inducing c-FLIPL turnover. Cell 124, 601–613 [DOI] [PubMed] [Google Scholar]
- 37. Sánchez-Pérez T., Ortiz-Ferrón G., López-Rivas A. (2010) Mitotic arrest and JNK-induced proteasomal degradation of FLIP and Mcl-1 are key events in the sensitization of breast tumor cells to TRAIL by antimicrotubule agents. Cell Death Differ. 17, 883–894 [DOI] [PubMed] [Google Scholar]
- 38. Zhu D. M., Shi J., Liu S., Liu Y., Zheng D. (2011) HIV infection enhances TRAIL-induced cell death in macrophage by down-regulating decoy receptor expression and generation of reactive oxygen species. PLoS ONE 6, e18291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abedini M. R., Muller E. J., Brun J., Bergeron R., Gray D. A., Tsang B. K. (2008) Cisplatin induces p53-dependent FLICE-like inhibitory protein ubiquitination in ovarian cancer cells. Cancer Res. 68, 4511–4517 [DOI] [PubMed] [Google Scholar]
- 40. Stagni V., Mingardi M., Santini S., Giaccari D., Barilà D. (2010) ATM kinase activity modulates cFLIP protein levels. Potential interplay between DNA damage signalling and TRAIL-induced apoptosis. Carcinogenesis 31, 1956–1963 [DOI] [PubMed] [Google Scholar]
- 41. Xue Y., Ren J., Gao X., Jin C., Wen L., Yao X. (2008) GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell Proteomics 7, 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649 [DOI] [PubMed] [Google Scholar]
- 43. Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S. G., Pandey A. (2007) A curated compendium of phosphorylation motifs. Nat. Biotechnol. 25, 285–286 [DOI] [PubMed] [Google Scholar]
- 44. Gopalakrishna R., Jaken S. (2000) Protein kinase C signaling and oxidative stress. Free Radic. Biol. Med. 28, 1349–1361 [DOI] [PubMed] [Google Scholar]
- 45. Sharma M., Gadang V., Jaeschke A. (2012) Critical role for mixed-lineage kinase 3 in acetaminophen-induced hepatotoxicity. Mol. Pharmacol. 82, 1001–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gallo K. A., Johnson G. L. (2002) Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat. Rev. Mol. Cell Biol. 3, 663–672 [DOI] [PubMed] [Google Scholar]
- 47. Martindale J. L., Holbrook N. J. (2002) Cellular response to oxidative stress. Signaling for suicide and survival. J. Cell Physiol. 192, 1–15 [DOI] [PubMed] [Google Scholar]
- 48. Nitobe J., Yamaguchi S., Okuyama M., Nozaki N., Sata M., Miyamoto T., Takeishi Y., Kubota I., Tomoike H. (2003) Reactive oxygen species regulate FLICE inhibitory protein (FLIP) and susceptibility to Fas-mediated apoptosis in cardiac myocytes. Cardiovasc. Res. 57, 119–128 [DOI] [PubMed] [Google Scholar]
- 49. Schattenberg J. M., Wörns M. A., Zimmermann T., He Y. W., Galle P. R., Schuchmann M. (2012) The role of death effector domain-containing proteins in acute oxidative cell injury in hepatocytes. Free Radic. Biol. Med. 52, 1911–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Conde de la Rosa L., Schoemaker M. H., Vrenken T. E., Buist-Homan M., Havinga R., Jansen P. L., Moshage H. (2006) Superoxide anions and hydrogen peroxide induce hepatocyte death by different mechanisms. Involvement of JNK and ERK MAP kinases. J. Hepatol. 44, 918–929 [DOI] [PubMed] [Google Scholar]
- 51. Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. (2005) Reactive oxygen species promote TNFα-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell 120, 649–661 [DOI] [PubMed] [Google Scholar]
- 52. Nakajima A., Komazawa-Sakon S., Takekawa M., Sasazuki T., Yeh W. C., Yagita H., Okumura K., Nakano H. (2006) An antiapoptotic protein, c-FLIPL, directly binds to MKK7 and inhibits the JNK pathway. EMBO J. 25, 5549–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Muruganandan S., Cribb A. E. (2006) Calpain-induced endoplasmic reticulum stress and cell death following cytotoxic damage to renal cells. Toxicol. Sci. 94, 118–128 [DOI] [PubMed] [Google Scholar]
- 54. Dejeans N., Tajeddine N., Beck R., Verrax J., Taper H., Gailly P., Calderon P. B. (2010) Endoplasmic reticulum calcium release potentiates the ER stress and cell death caused by an oxidative stress in MCF-7 cells. Biochem. Pharmacol. 79, 1221–1230 [DOI] [PubMed] [Google Scholar]
- 55. Yang W., Tiffany-Castiglioni E., Koh H. C., Son I. H. (2009) Paraquat activates the IRE1/ASK1/JNK cascade associated with apoptosis in human neuroblastoma SH-SY5Y cells. Toxicol. Lett. 191, 203–210 [DOI] [PubMed] [Google Scholar]
- 56. Ge W., Ge W., Zhang Y., Han X., Ren J. (2010) Cardiac-specific overexpression of catalase attenuates paraquat-induced myocardial geometric and contractile alteration. Role of ER stress. Free Radic. Biol. Med. 49, 2068–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen Y. W., Yang Y. T., Hung D. Z., Su C. C., Chen K. L. (2012) Paraquat induces lung alveolar epithelial cell apoptosis via Nrf-2-regulated mitochondrial dysfunction and ER stress. Arch. Toxicol. 86, 1547–1558 [DOI] [PubMed] [Google Scholar]
- 58. Ravanan P., Sano R., Talwar P., Ogasawara S., Matsuzawa S., Cuddy M., Singh S. K., Rao G. S., Kondaiah P., Reed J. C. (2011) Synthetic triterpenoid cyano enone of methyl boswellate activates intrinsic, extrinsic, and endoplasmic reticulum stress cell death pathways in tumor cell lines. Mol. Cancer Ther. 10, 1635–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tiwary R., Yu W., Li J., Park S. K., Sanders B. G., Kline K. (2010) Role of endoplasmic reticulum stress in α-TEA mediated TRAIL/DR5 death receptor dependent apoptosis. PLoS ONE 5, e11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Park S. K., Sanders B. G., Kline K. (2010) Tocotrienols induce apoptosis in breast cancer cell lines via an endoplasmic reticulum stress-dependent increase in extrinsic death receptor signaling. Breast Cancer Res. Treat. 124, 361–375 [DOI] [PubMed] [Google Scholar]
- 61. Cazanave S. C., Mott J. L., Bronk S. F., Werneburg N. W., Fingas C. D., Meng X. W., Finnberg N., El-Deiry W. S., Kaufmann S. H., Gores G. J. (2011) Death receptor 5 signaling promotes hepatocyte lipoapoptosis. J. Biol. Chem. 286, 39336–39348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang J. F., Cao J. G., Tian L., Liu F. (2012) 5,7-Dimethoxyflavone sensitizes TRAIL-induced apoptosis through DR5 up-regulation in hepatocellular carcinoma cells. Cancer Chemother. Pharmacol. 69, 195–206 [DOI] [PubMed] [Google Scholar]
- 63. Martín-Pérez R., Niwa M., López-Rivas A. (2012) ER stress sensitizes cells to TRAIL through down-regulation of FLIP and Mcl-1 and PERK-dependent up-regulation of TRAIL-R2. Apoptosis 17, 349–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Noh H. J., Lee S. J., Sung E. G., Song I. H., Kim J. Y., Woo C. H., Kwon T. K., Lee T. J. (2012) CHOP down-regulates cFLIP(L) expression by promoting ubiquitin/proteasome-mediated cFLIP(L) degradation. J. Cell Biochem. 113, 3692–3700 [DOI] [PubMed] [Google Scholar]