Background: It remains largely unexplored how post-translational modifications regulate reprogramming of somatic cells into induced pluripotent stem (iPS) cells.
Results: Substitution of the sole sumoylation site of the well known reprogramming factor KLF4 promotes iPS cell formation.
Conclusion: KLF4 sumoylation inhibits iPS cell induction but stimulates adipocyte differentiation.
Significance: The study highlights the importance of KLF4 sumoylation in regulating pluripotency and cell fate determination.
Keywords: Adipocyte, Cell Differentiation, Reprogramming, Sumoylation, Transcription Factors, iPS Cells, KLF2, KLF4, KLF5
Abstract
Ectopic expression of transcription factors has been shown to reprogram somatic cells into induced pluripotent stem (iPS) cells. It remains largely unexplored how this process is regulated by post-translational modifications. Several reprogramming factors possess conserved sumoylation sites, so we investigated whether and how this modification regulates reprogramming of fibroblasts into iPS cells. Substitution of the sole sumoylation site of the Krüppel-like factor (KLF4), a well known reprogramming factor, promoted iPS cell formation. In comparison, much smaller effects on reprogramming were observed for sumoylation-deficient mutants of SOX2 and OCT4, two other classical reprogramming factors. We also analyzed KLF2, a KLF4 homolog and a member of the KLF family of transcription factors with a known role in reprogramming. KLF2 was sumoylated at two conserved neighboring motifs, but substitution of the key lysine residues only stimulated reprogramming slightly. KLF5 is another KLF member with an established link to embryonic stem cell pluripotency. Interestingly, although it was much more efficiently sumoylated than either KLF2 or KLF4, KLF5 was inactive in reprogramming, and its sumoylation was not responsible for this deficiency. Furthermore, sumoylation of KLF4 but not KLF2 or KLF5 stimulated adipocyte differentiation. These results thus demonstrate the importance KLF4 sumoylation in regulating pluripotency and adipocyte differentiation.
Introduction
Post-translational modification is essential for regulating protein functions in diverse organisms. Rather than attachment of small chemical moieties such as phospho, acetyl, and methyl groups, sumoylation adds an ∼10-kDa small ubiquitin-like modifier (SUMO)5 polypeptide to the ϵ-amino group of lysine residues (1–3). Sumoylation exists in all eukaryotes and is essential for viability (4). In humans, there are four different SUMO proteins, SUMO1, -2, -3, and -4. SUMO2 and SUMO3 are highly homologous (95% identical) and allow both mono- and polysumoylation (5). SUMO1 is 47% identical to SUMO2 and -3, but it lacks the key lysine residue for polysumoylation, thereby conferring only monosumoylation (2, 3). The functional relevance of SUMO4 remains unclear. SUMOs are ∼18% identical to ubiquitin at the sequence level and have three-dimensional structural folds similar to that of ubiquitin (2, 3). Like ubiquitination, a conserved enzymatic cascade catalyzes sumoylation, including a heterodimeric E1 activating enzyme (SAE1/SAE2), an E2 conjugating enzyme (UBC9), and multiple E3 ligases (1, 3). Sentrin-specific proteases remove SUMO polypeptides from target proteins, rendering sumoylation dynamic and reversible (5).
Numerous proteins have been found to be sumoylated in diverse species from yeast to human, and ∼50% of known sumoylation sites conform to the classical consensus sequence ψKXE, where ψ is a bulky hydrophobic amino acid (such as Ile, Leu, and Val), and X is any residue (1, 5, 6). A subgroup of known sumoylation sites contains one or a few acidic residues located two residues C-terminal from the core motif ψKXE and forms a negatively charged amino acid-dependent sumoylation motif (7). The negative charge enhances the affinity for Ubc9 through binding to its positively charged surface close to the sumoylation pocket (7, 8). Similar to the negatively charged motif is the phosphorylation-dependent sumoylation motif ψKXEXX(S/T), shared by HSF1, PPARγ, MEF2, estrogen-related receptors (ERRs), and others (9–12). Signal-dependent phosphorylation of the Ser/Thr residue promotes sumoylation and provides a unique mechanism for phosphorylation-dependent sumoylation in response to different signaling pathways (13). The negative charge resulting from phosphorylation enhances the affinity for a positively charged surface on Ubc9 and promotes sumoylation (8).
We searched sequence databases with the extended motif ψKXEXX(S/T) and identified additional targets that are potentially subject to phosphorylation-dependent sumoylation (13). Two of them are KLF4 and SOX2, both of which are sumoylated (14–16), raising the question whether neighboring phosphorylation regulates sumoylation. Strikingly, both are among the four transcription factors initially found to reprogram mouse fibroblasts to iPS cells (17). The other two reprogramming factors are OCT4 (also known as Pou5f1) and c-MYC. Interestingly, OCT4 also contains a sumoylation site (18, 19), so three of the four reprogramming factors have been shown to be sumoylated. In addition, two KLF4 homologs, KLF2 and KLF5, play a role in reprogramming (20). Although KLF5 is known to be sumoylated (21, 22), KLF2 possesses two putative sumoylation sites awaiting characterization. Furthermore, ERRs are subject to phosphorylation-dependent sumoylation and have a role in reprogramming (12, 23). These observations suggest the intriguing possibility that sumoylation regulates iPS cell generation and pluripotency.
Since the initial description of iPS cell formation by ectopic expression of only four transcription factors (17), there have been intensive research efforts to apply this technology to disease modeling and autologous cell therapy (24–26). A better understanding of the underlying molecular and cellular mechanisms is important for further improvement and eventual optimization of this powerful technology. Although post-translational modifications such as sumoylation are crucial for various transcription factors to function (27–29), it remains not so clear how such modifications may affect iPS cell generation. We thus analyzed how sumoylation of KLF4, SOX2, and OCT4 may regulate iPS cell formation. Here, we report that sumoylation of these factors inhibits iPS cell induction. Interestingly, KLF4 sumoylation appeared to stimulate adipocyte differentiation. In addition, we analyzed KLF2 and KLF5. Like KLF5, KLF2 was sumoylated at two sumoylation sites. However, substitution of the sumoylation sites on KLF2 or KLF5 had minimal effects on reprogramming or adipocyte differentiation. These findings support the importance of KLF4 sumoylation in pluripotency induction and adipocyte differentiation.
MATERIALS AND METHODS
Cell Culture and Animal Care
HEK293 and 3T3-L1 were cultured and expanded in DMEM, 10% fetal bovine serum (FBS), and penicillin and streptomycin (50 μg/ml each). Mouse embryonic fibroblasts (MEFs) were derived from 13.5-day post-coitus mouse embryos as described previously (30). Briefly, a 13.5-day post-coitus pregnant mouse (FVB strain, Jax) was sacrificed by cervical dislocation under sterile conditions, and the embryos were surgically removed from the uterus. After the head, forelimb, tail, and abdomen were surgically removed, the remaining embryos were nipped into pieces and incubated in 0.25% trypsin for 30 min. Trypsinization was repeated twice, and after each time, trypsinized cells were separated from the tissue pieces and plated onto a 10-cm culture dish. This was considered as passage 0, and MEFs were expanded for 3 to 4 passages. MEFs at passages 2–3 were used for iPS cell generation. For preparation of feeder layers, MEFs were irradiated at 6000 rads or treated with 10 μg/ml mitomycin C (Sigma) for 3 h. FVB mice were maintained, bred, and sacrificed according to an animal use protocol approved by McGill University Animal Care Committee.
For MEF preparation, we encountered mysterious contamination by “black swimming dots,” which appeared to be very similar to what was reported to be Achromobacter (31). In light of the similarity, ciprofloxacin and piperacillin (10 μg/ml each; Sigma, Cat. nos. 17850 and P8396, respectively) were included in the MEF medium. The former is an acid and was prepared in 30 mm NaOH for a 10 mg/ml stock, prior to sterilization by filtration, whereas the latter is a salt and was prepared in water or PBS, with all stocks stored as aliquots at −20 °C). This remedy was effective in eradiating and preventing the contamination. Such contamination has occurred in many other laboratories (31).
Construction of Expression Plasmids
The following expression plasmids or cDNA constructs were purchased from Open Biosystems: KLF2 (BC071983); KLF4 (MHS1011-7509690); KLF5 (MHS1011-61504); OCT4 (MHS1768-98081221); SOX2 (MHS1011-169828), and c-Myc (MHS1010-9205764). The following lentivirus plasmids were obtained from Addgene: pLOVE (15948); pLOVE-Klf4 (15950); pLOVE-N-MYC (15951); pSin-EF2-SOX2-Pur (16577), and pSin-EF2-OCT4-Pur (16579). The plasmids for FLAG-tagged wild-type and mutant proteins were constructed by use of standard subcloning and mutagenesis protocols. Lentiviral shuttle vectors were prepared on pENTR11 for homologous recombination with pLOVE via the Gateway system (Invitrogen).
Antibodies
The following antibodies were purchased as specified: anti-Gal4 (Santa Cruz Biotechnology, RK5CI); anti-FLAG (Sigma, F3165); anti-HA (Babco/Covance); anti-mouse HRP IgG (Amersham Biosciences, NA93IV); goat anti-rabbit IgG (Fisher, AP307FMI); anti-Nanog (Bethyl Laboratories, BL1662); anti-KLF4 (Santa Cruz Biotechnology, H-180), and anti-Ssea-1 (Cell Signaling Technology, MC-480).
Sumoylation Assays
The procedure has been described previously (11). Briefly, an HA-tagged SUMO construct and a FLAG-tagged transcription factor construct were co-transfected into HEK293 cells by using the Superfect transfection reagent (Qiagen, 301307). After 48 h, the cells were lysed in buffer S (SDS sample buffer (0.15 m Tris-HCl, pH 6.7, 5% SDS, and 30% glycerol) diluted 1:10 in PBS containing 0.5% Nonidet P-40, and protease inhibitors), followed by 15 s of sonication three times at the power setting of 3.5 (Model Virsonic 100 sonicator) to break up chromatin and decrease the viscosity. Anti-FLAG M2 beads (Sigma, A2220) were used to immunoprecipitate FLAG-tagged proteins according to the manufacturer's instructions. Briefly, prewashed anti-FLAG M2-agarose was mixed with soluble extracts and rotated for 2 h at 4 °C. The agarose was collected by centrifugation at 400 × g for 1 min and washed three times with buffer R (PBS, 0.5% Nonidet P-40, 1 mm PMSF, 12.8 mm β-mercaptoethanol, and protease inhibitors). For elution, the agarose was incubated with buffer R containing 0.2 mg/ml FLAG peptide for 30 min on a rotator at 4 °C. After a brief spin, the supernatant was collected for SDS-PAGE and Western blotting.
Reporter Gene Assays
On the day before transfection, 4 × 104 HEK293 cells or 2 × 104 MEFs were seeded per well onto a 12-well plate. 200 ng of the luciferase reporter Gal4-tk-luc or Nanog-luc (pGL3-Nanog(−2342 to +50), obtained from Takashi Tada, Kyoto University (32)) were transfected along with 200 ng of expression plasmids. The β-galactosidase expression plasmid CMV-β-Gal (50 ng) was co-transfected as an internal control. The transfection reagents Superfect (Qiagen, catalog no. 301307) and Lipofectamine 2000 (Invitrogen, catalog no. 11668-019) were employed for transfection of HEK293 cells and MEFs, respectively. 48 h post-transfection, the cells were lysed in situ, and soluble extracts were prepared for measurement of luciferase and β-galactosidase activities with a 96-well plate luminometer (Dynex). d-(−)-Luciferin (Roche Applied Science) and Galacto-Light Plus (Tropix) were used as the substrates for luciferase and β-galactosidase, respectively.
Chromatin Immunoprecipitation (ChIP)
The procedure has been described previously (33). On irradiated MEF feeders precultured on three 10-cm gelatinized culture dishes, mouse R1 ES cells were grown to ∼90% confluency, resulting in ∼1.5 × 107 cells for ChIP. 0.27 ml of 37% formaldehyde was added directly to each dish to achieve a final concentration of 1%. The three dishes were transferred to an orbital shaker and shaken for 10 min at room temperature. This cross-linking reaction was stopped by addition of glycine to a final concentration of 0.125 m, which was followed by a 5-min incubation at room temperature. The dishes were washed twice with ice-cold PBS and scraped on ice to harvest the cells in some residual PBS. The cell suspension was combined into one Falcon tube and centrifuged at 220 × g for 10 min at 4 °C. The cell pellet was lysed in 150 μl of the lysis buffer (50 mm Tris-HCl, pH 8.0, 1% SDS, 10 mm EDTA, and protease inhibitors) and sonicated three times on ice for 20 s each at setting 6 (Virsonic 100 sonicator). The cell lysate was centrifuged at 16,000 × g for 10 min at 4 °C. The resulting supernatant was diluted in 1.2 ml of ChIP dilution buffer (20 mm Tris-HCl, pH 8.0, 1% Triton X-100, 2 mm EDTA, and 150 mm NaCl). Sheared salmon sperm DNA was mixed with protein A-agarose (Upstate) and incubated with the diluted lysate for pre-clearance. The suspension was rotated for 1 h at 4 °C. After brief centrifugation, the supernatant was incubated with the primary antibody overnight at 4 °C. The next day, 40 μl of salmon sperm DNA/protein A-agarose bead suspension was added into the lysate/antibody mixture and incubated for 2 h at 4 °C. The beads were then separated from the lysate by centrifugation at 750 × g for 1 min, resuspended in 1 ml of wash buffer 1 (low salt buffer: 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.1% SDS, 1% Triton, and 2 mm EDTA), and rotated for 10 min. The beads were separated from buffer 1 by brief centrifugation and resuspended in 1 ml of buffer 2 (high salt buffer: 20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 0.1% SDS, 1% Triton, and 2 mm EDTA). The same washing step was repeated in freshly prepared buffer 3 (LiCl buffer: 10 mm Tris-HCl, pH 8.0, 0.25 m LiCl, 1% Nonidet P-40, 1 mm EDTA, and 1% sodium deoxycholate) and cold buffer TE (10 mm Tris-HCl, pH 7.5, and 1 mm EDTA). 150 μl of de-crosslink buffer (0.1 m NaHCO3 and 1% SDS) was added onto the beads and incubated at 65 °C for 6–18 h. The supernatant was mixed with 5 μl of proteinase K (1 mg/ml) and incubated for 1 h at 55 °C. Afterward, DNA was purified with the QIAQuick PCR purification column kit (Qiagen) for PCR. For the Nanog promoter, the PCR primers were 5′-GTGAAATGAGGTAAAGCCTCTTTT-3′ and 5′-AAGGCCAACGGCTCAAGGCGATAG-3′. For the Lefty promoter, the primers 5′-AAGCTGCAGACTTCATTCCA-3′ and 5′-CGGGGGATAGATGAAGAAAC-3′ were used (34).
Immunofluorescence Microscopy
Coverslips were flamed and put onto wells of 12- or 24-well dishes for culturing cells. Coverslips containing cultured cells were washed twice with PBS prior to fixation with 2% paraformaldehyde for 20 min at room temperature. Cells were then washed three times with PBS and permeabilized with 0.2% Triton X-100 solution/PBS for 10 min. Afterward, the coverslips were rinsed three times with 100 mm glycine/PBS and blocked in the blocking solution (2% BSA prepared in the IF buffer (PBS, 0.2% Triton X-100 and 0.05% Tween 20)). After 30 min, the coverslips were incubated for 1 h at room temperature with primary antibodies diluted 1:100 or 1:200 in the blocking solution. Cells were then washed three times with the IF buffer and incubated with fluorophore-conjugated secondary antibodies (diluted 1:500 to 1:1000) for 45 min. Afterward, the coverslips were rinsed three times with the IF buffer, briefly exposed to DAPI or Hoechst 33258 to stain the nuclei, and mounted for examination under a Zeiss Axiovert 135 fluorescence microscope.
Lentivirus Preparation
293FT cells (Invitrogen) were maintained in DMEM/10% FBS medium containing 400 μg/ml neomycin (geneticin, Invitrogen, catalog no. 11811-098) per the manufacturer's instructions and incubated in the antibiotic-free medium for at least 8 h prior to transfection with Lipofectamine 2000. 10 μg of expression plasmid was mixed with 6.5 μg of psPAX2, 3.5 μg of pMD2.G, 50 μl of Lipofectamine 2000, and 1 ml of Opti-MEM to transfect 8 × 106 293FT cells in a 10-cm dish according to the manufacturer's instructions (Invitrogen). 24 h post-transfection, the medium was collected as the viral supernatant everyday for 3 days and was subjected to centrifugation at 76,000 × g for 1.5 h. The viral pellet was then suspended in DMEM, 10% heat-inactivated FBS and rotated overnight at 4 °C. The resulting virus supernatant was used to infect cells directly or was flash-frozen in aliquots on dry ice for long term storage at −80 °C.
iPS Cell Induction
The procedure for drug selection-free iPS cell induction has been described previously (35). Briefly, the day before virus infection, MEFs (passage 2 or 3) were plated at 0.8–1 × 105 cells per well for a 12-well plate or 3 × 105 cells per well of a 6-well plate and incubated overnight in DMEM containing 10% FBS and penicillin/streptomycin (50 μg/ml each) inside a 37 °C CO2 incubator. The next day, the cells were washed once with PBS or plain DMEM and refed with DMEM, 10% heat-inactivated FBS, 8 μg/ml Polybrene. After addition of the concentrated virus stock (suspended in DMEM, 10% heat-inactivated FBS, see above) to the medium, the cells were incubated in a 37 °C CO2 incubator for 2 days. The virus-containing medium was then removed, and the cells were then washed once with PBS or plain DMEM for culturing in the mESC medium (DMEM high glucose, 1% nonessential amino acids (100× stock, Invitrogen), 1% sodium pyruvate (100× stock, Invitrogen), 0.1 mm β-mercaptoethanol, 15% FBS, penicillin, and streptomycin (50 μg/ml each), and 1000 units of murine leukemia inhibitory factor/ml (Millipore, ESGRO®)). The medium was changed every 2 days. iPS colonies appeared 5–6 days after infection. For alkaline phosphatase staining, a detection kit (Millipore, SCR004) was used according to the manufacturer's instructions.
RT-PCR
cDNA synthesis was carried out with Expand Reverse Transcriptase (Roche Applied Science) and PCR was performed according to the manufacturer's instructions. The PCR primers for the ES markers Nanog, Eras, Rex1, and Dax1 have been described previously (17).
Adipocyte Differentiation and Oil Red O Staining
3T3-L1 preadipocytes (36) and MEFs were cultured in DMEM containing 10% FBS, 50 μg/ml penicillin, and 50 μg/ml streptomycin until reaching full confluency. Two days later (day 2), differentiation was induced by addition of insulin (5 μg/ml; Sigma, catalog no. 19278), dexamethasone (1 μm; Sigma, catalog no. D8893), and rosiglitazone (0.1 to 1 μm) (or isobutyl-1-methylxanthine (0.05 to 0.5 mm; Sigma catalog no. 178018). On day 4, the medium was replaced with the same medium containing 5 μg/ml insulin only. This medium was changed every 2 days until the end of differentiation. For Oil Red O (Sigma) staining, the medium was removed, and the cells were washed with PBS. The cells were then incubated with 10% formalin for 1 h and rinsed with 60% isopropyl alcohol. After air-drying, the fixed cells were incubated for 10 min with the Whatman No. 1 paper-filtered Oil Red O working solution: 3:2 dilution in Nano-pure H2O from the stock solution of 0.35% Oil Red O dissolved in isopropyl alcohol. The wells were rinsed five times with Nano-pure H2O prior to photographing or scanning. For quantification, Oil Red O was extracted with 100% isopropyl alcohol, and the absorbance was measured at 520 nm with a spectrophotometer (Thermo Spectronic) as described (37).
Statistical Analysis
Data are presented as means ± S.D. Unpaired one-tailed Student's t tests were performed to calculate p value. For experiments with more than two conditions, one-way analyses of variance were performed with a Bonferroni post-hoc test.
RESULTS
Sumoylation Inhibits Transcriptional Activity of KLF4
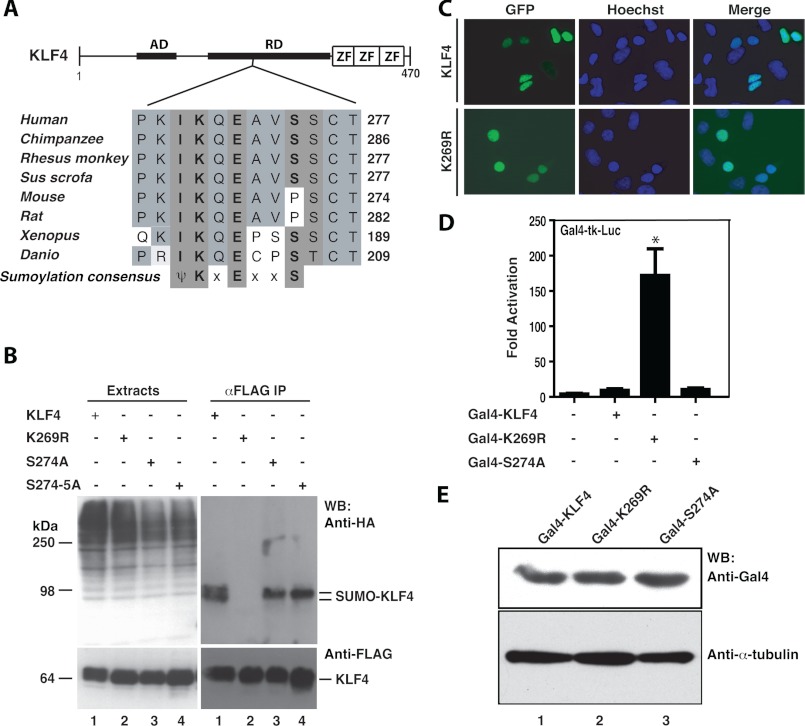
KLF4 belongs to the Krüppel-like factor (KLF)/SP family of zinc finger transcription factors and contains an activation domain at the N-terminal portion, a repression domain in the middle portion, and three Krüppel-like zinc fingers at the C-terminal part (Fig. 1A). Blast search and manual sequence inspection revealed a ψKXE motif, which is conserved from fish to humans and located within the transcriptional repression domain (Fig. 1A). A Ser/Thr cluster is a few residues C-terminal to the motif, suggesting a potential cross-talk between sumoylation and phosphorylation. We first examined whether conserved Lys-269 is indeed a sumoylation site. As shown in Fig. 1B (lanes 1 and 2), mutation K269R abolished KLF4 sumoylation, confirming that Lys-269 is the sole sumoylatable residue. Consistent with this, two recent studies showed that KLF4 is sumoylated at an equivalent site when different isoforms were used (14, 15). Next, we investigated how Ser-274 may control Lys-269 sumoylation. As shown in Fig. 1B, mutation of S274A reduced the sumoylated KLF4 double bands to a single band but only slightly decreased total sumoylation level (compare lanes 1 and 3). The doublet observed with wild-type KLF4 could be due to Ser-274 phosphorylation. The additional mutation S275A did not decrease sumoylation further (Fig. 1B, compare lanes 3 and 4). These results indicate that at least under the assay conditions employed, Ser-274 or Ser-275 had no major effects on sumoylation of human KLF4. Related to this, Ser-274 is not conserved in mouse and rat KLF4 (Fig. 1A).
FIGURE 1.
Sumoylation represses transcriptional activity of KLF4. A, domain organization of KLF4 illustrated with sequence alignment of a conserved sumoylation motif and adjacent residues in KLF4 proteins from zebrafish to human. AD, transcriptional activation domain; RD, repression domain; ZF, zinc finger; ψ, bulky hydrophobic residues such as Ile, Leu, or Val; x, any residue. B, sumoylation assays. Expression plasmids for HA-SUMO2 and FLAG-tagged wild-type and mutant KLF4 were co-transfected into HEK293 cells as indicated. 48 h post-transfection, cells were lysed in buffer S, and soluble extracts were prepared for immunoprecipitation (IP) on anti-FLAG M2-agarose and Western blotting (WB) with anti-HA and -FLAG antibodies. HA-SUMO1 was difficult to express, so SUMO2 was used instead. C, subcellular localization of GFP-KLF4 and -K269R after transient transfection of the corresponding expression plasmids into HEK293 cells. Cells were fixed and incubated with Hoechst 33258 for subsequent fluorescence microscopy to detect GFP expression (green) and nuclei (blue). D, reporter gene assays. The Gal4-tk-Luc construct contains five tandem binding sites for the yeast transcription factor Gal4 upstream from the thymidine kinase (tk) core promoters and the coding sequence of luciferase. This construct was transfected into HEK293 cells along with an expression plasmid for expression of fusion proteins containing the Gal4 DNA-binding domain (residues 1–147) fused to KLF4 or its point mutants as indicated. Luciferase activities were normalized to β-galactosidase activities expressed from the CMV-β-Gal expression plasmid that was co-transfected as the internal control. The results were based on three independent assays. *, p = 0.0006 when compared with the empty vector-transfected control. E, Western blotting analysis of Gal4 fusion protein expression. Expression plasmids for the indicated Gal4 fusion proteins (wild-type and mutant KLF4 proteins fused to the C-terminal end of the N-terminal 147 residues of yeast Gal4) were transfected into HEK293 cells, and 2 days later, soluble extracts were prepared for immunoblotting with anti-Gal4 antibody (top) and anti-α-tubulin (bottom) antibodies.
As for the functional consequence of sumoylation, protein levels of wild-type KLF4 and mutants were comparable (Fig. 1B), indicating that sumoylation does not affect protein stability. We then examined potential effects on subcellular localization using fluorescence microscopy, and no difference was found between wild-type KLF4 and mutant K269R (Fig. 1C). Then the wild-type and mutant forms of KLF4 were expressed as proteins fused to the C-terminal end of the DNA-binding domain of the yeast transcription factor Gal4. Reporter gene assays with these constructs revealed that mutation K269R but not S274A dramatically activated the transcriptional activity of KLF4 (Fig. 1D). These fusion proteins were expressed to similar levels (Fig. 1E), further attesting to the conclusion that sumoylation does not affect KLF4 stability. These results indicate that Lys-269 sumoylation inhibits KLF4-dependent transcription.
Sumoylation Inhibits Transcriptional Activity of Sox2
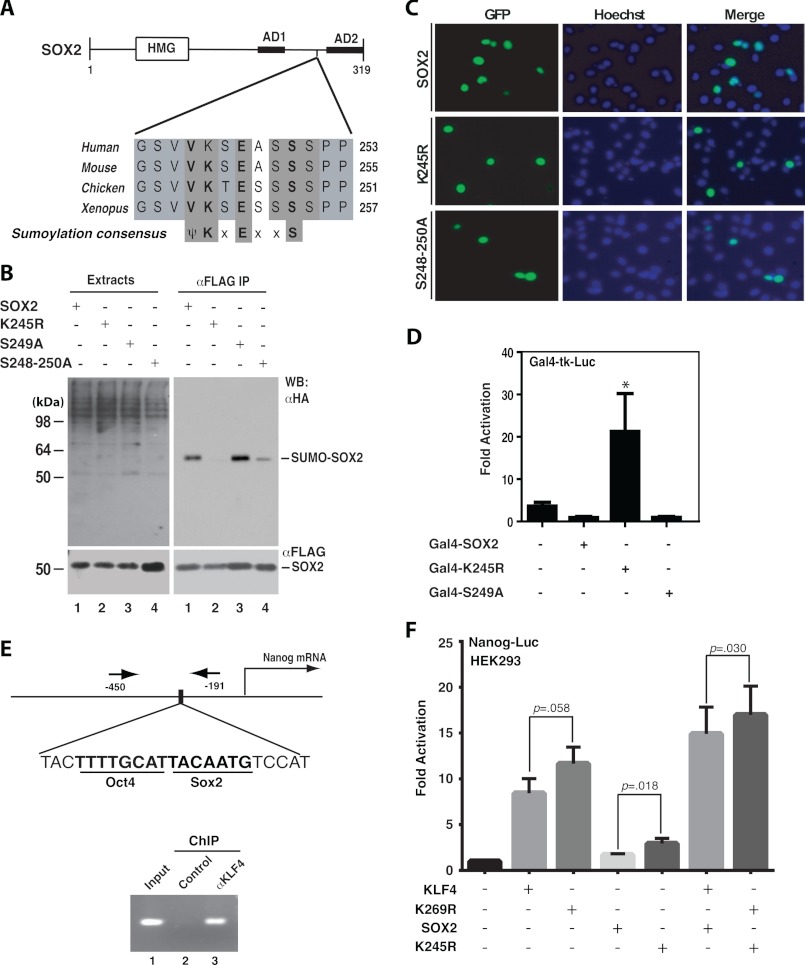
Sox2 is a member of the Sox ((Sex determining region Y)-box) family of transcription factors containing an high mobility group DNA-binding domain at the N-terminal portion and two activation domains at the C-terminal part (Fig. 2A). We previously reported that SOX2 contains a ψKXEXXS motif for potential phosphorylation-dependent sumoylation (13). This motif is conserved from Xenopus to humans and located between the two transcriptional activation domains (Fig. 2A). Lys-247 of mouse Sox2 has been shown to be sumoylated, and the modification was found to inhibit DNA binding (16). As shown in Fig. 2B, the mutation K245R dramatically reduced sumoylation of human SOX2 (compare lanes 1 and 2), confirming that Lys-245 of human SOX2 is a major sumoylation site (equivalent to Lys-247 of mouse Sox2, see Fig. 2A). This mutation had no effects on the subcellular localization (Fig. 2C) but promoted the transcriptional activity of SOX2 (Fig. 2D). Then we investigated how the serine cluster (Ser-248 to Ser-250) may regulate Lys-245 sumoylation. The mutation S249A had minimal impact on sumoylation (Fig. 2B, compares lanes 1 and 3) or transcription (Fig. 2D), but substitution of all three serine residues with alanine reduced sumoylation (Fig. 2B, compare lanes 1 and 4), indicating that the three serine residues are required for optimal sumoylation. In subcellular localization, the triple mutant behaved similarly as the wild-type and K245R mutant of SOX2 (Fig. 2C). It was somewhat surprising that the triple mutation S248A/S249A/S250A decreased sumoylation (Fig. 2B, compares lanes 1 and 4) but had little impact on transcription (data not shown). One explanation is that the residual amount of sumoylation is sufficient to repress transcription. To determine whether phosphorylation plays a role and can be mimicked by aspartate, we analyzed mutants S249D (Asp substitution of Ser-249) and S248D/S249D/S50D (Asp substitution of Ser-248, Ser-249, and Ser-250). As shown in supplemental Fig. S1, neither mutant promoted sumoylation, suggesting that negative charge per se is insufficient to stimulate sumoylation.
FIGURE 2.
Sumoylation inhibits transcriptional activity of SOX2. A, domain organization of mouse Sox2 shown with sequence alignment of a conserved sumoylation motif and adjacent residues in Sox2 proteins from Xenopus to humans. HMG, high mobility group DNA binding domain; AD1 and AD2, transcriptional activation domains 1 and 2, respectively. B, sumoylation assays. Expression plasmids for HA-SUMO2 and FLAG-tagged wild-type human SOX2 and mutants were co-transfected into HEK293 cells. Extracts were prepared as in Fig. 1B for IP on anti-FLAG M2-agarose and subsequent immunoblotting with anti-HA and -FLAG antibodies as indicated. C, subcellular localization of GFP-tagged wild-type and mutant SOX2 proteins. After transient transfection with the corresponding expression plasmids, HEK293 cells were fixed and incubated with Hoechst 33258 for fluorescence microscopy to detect GFP expression (green) and nuclei (blue). D, reporter gene assays. The assays were performed as for Fig. 1D except that expression plasmids for Sox2 and mutants were analyzed. The results were calculated from three independent assays. *, p < 0.05 when compared with the empty vector-transfected control. E, ChIP analysis. Formaldehyde was used to fix mouse ES cells, and after brief sonication, soluble chromatin was prepared for IP with the α-KLF4 antibody (lane 3) or control IgG (lane 2). The immunoprecipitates were used for PCR to amplify the fragment corresponding to nucleotides −450 to −191 of the mouse Nanog promoter, indicated by arrows (top). F, reporter gene assays. The assays were performed as for Fig. 1D except that the reporter construct contains a mouse Nanog promoter fragment (−2340 to +150) controlling luciferase expression. The expression plasmids for FLAG-tagged KLF4 and SOX2 were transfected as specified. The results were calculated from five different sets of data, with p values indicated.
Sumoylation of KLF4 and Sox2 Inhibits Nanog Promoter Activity
To complement the Gal4-based reporter gene assays (Fig. 1D and Fig. 2D), we sought to use a native promoter fragment. For this, we analyzed a promoter fragment of the Nanog gene because Nanog is an established embryonic stem (ES) cell-specific marker with a central role in regulating pluripotency and self-renewal of ES cells (38, 39). Its unexpected absence among the four reprogramming factors that were initially identified (17) suggests that its activation is secondary to expression of other factors. Consistent with this, OCT4 and SOX2 bind to a composite site at the mouse Nanog promoter and activate its transcription (Fig. 2E, top) (32, 40). In addition, two KLF4-binding loci are at the Nanog promoter, and one of them overlaps with the OCT4-SOX2-binding site (41). We thus wondered whether KLF4 sumoylation regulates Nanog transcription. To test this, we performed ChIP to verify KLF4 binding to the Nanog promoter in mouse ES cells. As shown in Fig. 2E (bottom), ChIP revealed that KLF4 binds to the Nanog promoter, and the binding site overlaps with the OCT4-SOX2 composite element. In addition, we analyzed the promoter of Lefty, another ES cell marker. This promoter also contains a composite OCT4-SOX2-binding site close to the KLF4-binding site (supplemental Fig. S2A) (34). As shown in supplemental Fig. S2B, ChIP revealed that KLF4 also binds to the OCT4-SOX2-binding site on the Lefty promoter. These observations suggest that KLF4 may interact with OCT4 and SOX2 to occupy Nanog and Lefty promoters. To substantiate this, we performed co-IP. As shown in supplemental Fig. S2C, FLAG-tagged KLF4 precipitated HA-tagged OCT4 and SOX2. Consistent with this, these proteins co-localized in the nucleus (supplemental Fig. S2D). Of relevance, it has been reported that KLF4 interacts with Oct4 and Sox2 (42).
Co-occupancy of KLF4, OCT4, and SOX2 at the Nanog promoter and the presence of sumoylation motifs in all of them suggest that sumoylation might regulate reprogramming. To test this, we first compared wild-type KLF4 and mutant K269R in activating the Nanog promoter. We used the reporter construct pGL3-Nanog(−2342 to +50), which contains a mouse Nanog promoter fragment driving luciferase expression (32). Both wild-type KLF4 and mutant K269R activated the Nanog-luc reporter, and the latter was more active (Fig. 2F). Although wild-type SOX2 and mutant K245R activated the reporter only slightly, they synergized with the KLF4 proteins (Fig. 2F). These findings suggest that sumoylation inhibits the ability of KLF4 and Sox2 to activate the Nanog promoter.
KLF4 Sumoylation Inhibits iPS Cell Induction
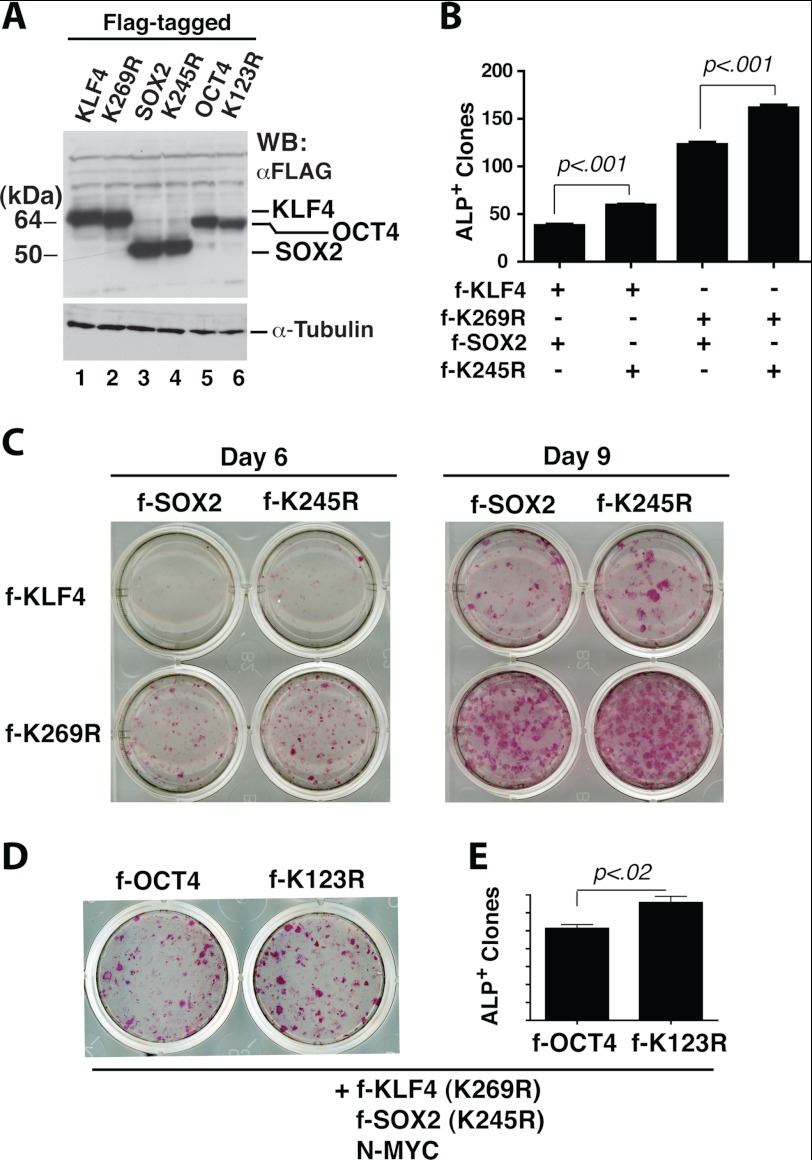
We next analyzed how the wild-type and sumoylation-deficient mutant of KLF4 might reprogram MEFs into iPS cells. For easy comparison of expression levels of the wild-type and mutant proteins, we generated lentiviruses expressing FLAG-tagged KLF4 proteins (Fig. 3A, lanes 1 and 2). MEFs were infected with lentiviruses expressing the four reprogramming factors (or mutant counterparts), and after 6–10 days, alkaline phosphatase staining was performed to determine reprogramming efficiency. Replacing wild-type KLF4 with mutant K269R enhanced reprogramming efficiency by ∼3.3-fold, and ALP+ colonies also appeared 1–2 days earlier (Fig. 3, B and C).
FIGURE 3.
Sumoylation of KLF4 and SOX2 down-regulates iPS cell induction. A, Western blotting analysis. HEK293 cell lysates were analyzed 48 h after transduction with lentivirus expressing FLAG-tagged KLF4, SOX2, or OCT4 as indicated. B, quantification of alkaline phosphatase (ALP)-positive colonies 9 days after MEFs were transduced with lentiviruses expressing OCT4, N-MYC (both untagged), and FLAG-tagged (f-) KLF4 and SOX2 proteins as indicated. The p value is shown (n = 3). C, alkaline phosphatase staining of primary iPS colonies 6 or 9 days after infection of MEFs with a mixture of lentiviruses expressing untagged OCT4 and N-MYC, along with lentiviruses for FLAG-tagged KLF4 and SOX2 as indicated. D, alkaline phosphatase staining of primary iPS colonies 6 days after MEFs were infected with the lentivirus expressing FLAG-tagged OCT4 or mutant K123R, along with lentiviruses expressing N-MYC and FLAG-tagged sumoylation-deficient mutants of KLF4 and SOX2. E, quantification of experiments performed as in D.
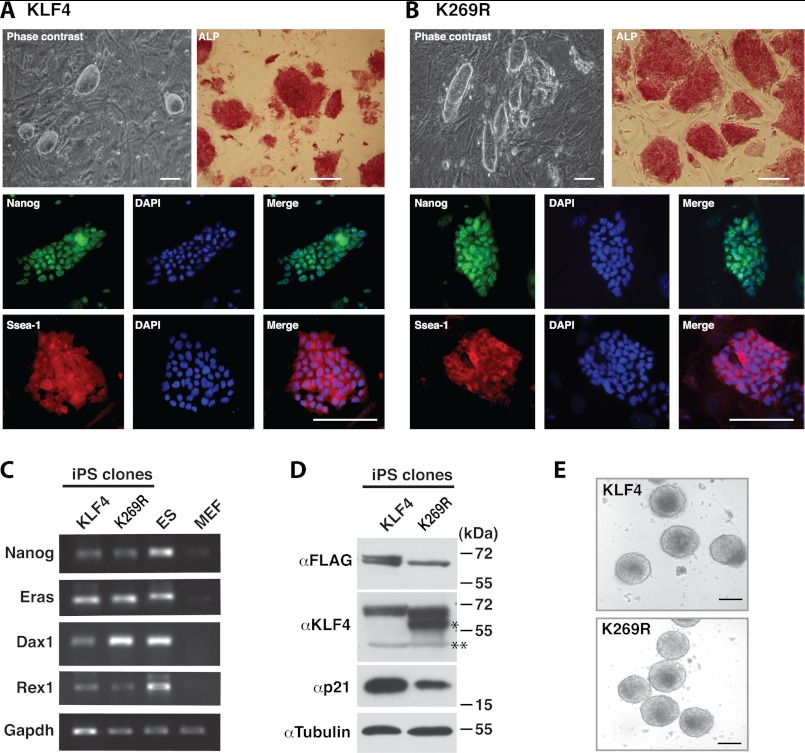
Several colonies were selected and expanded. They survived trypsinization and exhibited the characteristic morphology of mouse ES cell colonies (Fig. 4, A and B, phase contrast). RT-PCR analysis revealed expression of mouse ES cell markers, including Nanog, Eras, Dax1, and Rex1 (Fig. 4C). Immunofluorescence microscopy confirmed that the expanded iPS cell clones exhibited almost homogeneous expression of Nanog and Ssea-1, another mouse ES cell marker (Fig. 4, A and B, fluorescent images). Immunoblotting indicated that FLAG-KLF4 and -K269R were expressed in the iPS cell clones, and a higher level of endogenous KLF4 but a lower level of p21 were detected in extracts from the K269R iPS cell clone (Fig. 4D), indicating that K269R colonies were more successful in reactivating the endogenous KLF4 promoter. About p21, we compared wild-type and mutant KLF4 in activating its transcription using a luciferase reporter controlled by a p21 promoter fragment, but no difference was detected (data not shown). Stable expression of the mutant stimulated p21 expression as the wild-type (data not shown), suggesting that the decreased level of p21 in the K269R iPS cell clone was due to indirect effects. To determine the differentiation potential of the iPS cell clones, we analyzed embryoid body formation. After a 2-day culture in hanging drops, the wild-type KLF4 and mutant K269R iPS cell clones formed well shaped embryoid bodies (Fig. 4E). These results indicate that although KLF4 sumoylation inhibits MEF-to-iPS cell reprogramming, it does not affect the quality of the resulting iPS cell clones.
FIGURE 4.
Characterization of iPS cell clones expressing wild-type or mutant KLF4. A, phase contrast, alkaline phosphatase staining, and immunostaining of a representative iPS cell clone derived from MEFs infected with lentiviruses expressing KLF4 (FLAG-tagged), untagged OCT4, SOX2, and N-MYC. B, phase contrast, alkaline phosphatase staining, and immunostaining of a representative iPS cell clone derived from MEFs infected with lentivirus expressing K269R (FLAG-tagged), untagged OCT4, SOX2, and N-MYC. C, RT-PCR analysis of different ES cell markers in the two representative clones described in A and B. Mouse ES cells and MEFs were used as positive and negative controls, respectively. D, Western blotting analysis of the two representative iPS cell clones described in A and B. Top blot, soluble extracts were used for IP on anti-FLAG M2-agarose, and the eluted proteins were analyzed by immunoblotting with the anti-FLAG antibody. Perhaps due to different sites of integration, mutant K269R was expressed to a slightly lower level than that of the wild type. The doublet on lane 1 is perhaps due to phosphorylation. A similar doublet was observed elsewhere (e.g. Fig. 1B, top right). Lower three blots, soluble extracts were directly used for immunoblotting with the indicated antibodies. On the anti-KLF4 blot, the single asterisk labels a lower mysterious band perhaps related to endogenous KLF4, and the double asterisks mark the position of nonspecific bands. E, embryoid body formation of cells from the two representative iPS cell clones described in A and B.
Synergistic Inhibition of Reprogramming by Sumoylation of KLF4, SOX2, and OCT4
Reporter gene assays revealed a synergy of the KLF4 mutant K269R with the sumoylation-deficient SOX2 mutant K245R in activating the Nanog promoter (Fig. 2F), so an interesting question is whether these mutants synergize each other in MEF reprogramming to iPS cells. As it was unclear how sumoylation of Sox2 plays a role in this process, we first compared wild-type SOX2 with its sumoylation-deficient mutant. Both were expressed to similar levels (Fig. 3A, lanes 3 and 4). Compared with the wild-type, mutant K245R was slightly more active in inducing formation of ALP+ colonies (Fig. 3, B–D, and supplemental Fig. S3). We also compared K245R with mutants S249D and S248D/S249D/S250D. Consistent with the results from sumoylation assays (Fig. 2 and supplemental Fig. S1), these two mutants behaved similarly to the wild type (supplemental Fig. S3). Interestingly, when the KLF4 mutant K269R was co-expressed, the SOX2 mutant K245R synergized with this KLF4 mutant in promoting formation of ALP+ colonies (Fig. 3, B and C). Therefore, SOX2 sumoylation synergizes with KLF4 sumoylation to inhibit reprogramming.
As it was unclear how sumoylation of OCT4 plays a role in the process, we compared wild-type OCT4 with its sumoylation-deficient mutant K123R (18, 19). As shown in Fig. 3A (lanes 5 and 6), the wild-type and mutant proteins were expressed to similar levels. Compared with the wild type, K123R was slightly more active in promoting formation of ALP+ colonies (Fig. 3, D and E), suggesting that OCT4 sumoylation inhibits reprogramming. Interestingly, when the KLF4 mutant K269R and/or the SOX2 mutant K245R were co-expressed, the OCT4 mutant K123R slightly synergized with these two mutants in promoting formation of ALP+ colonies (Fig. 2, D and E), supporting synergistic inhibition of MEF reprogramming by sumoylation of KLF4, SOX2, and OCT4.
Effects of KLF2 and KLF5 Sumoylation on Reprogramming
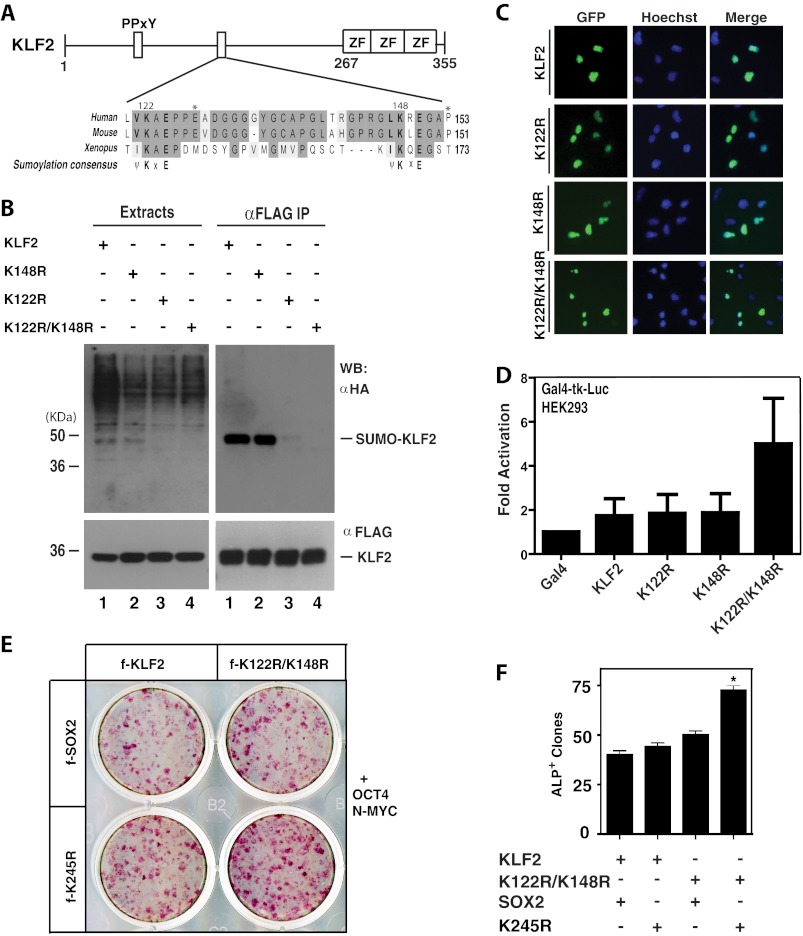
KLF2 and KLF5 are two members of the KLF family shown to replace KLF4 in iPS cell generation (20). Both contain a pair of conserved sumoylation motifs (Figs. 5A and 6A and supplemental Fig. S4). These two motifs of KLF2 are conserved among KLF2 proteins from different species but are located within the N-terminal region that is divergent in different members of the KLF family (supplemental Fig. S4), suggesting the unique function of these motifs. As sumoylation of KLF2 has not been reported, we first examined this. Mutant K122R was sumoylated to a residual level, and the double mutant K122R/K148R exhibited no detectable levels of sumoylation (Fig. 5B), indicating that Lys-122 is a major sumoylation site, whereas Lys-148 is a minor one. The double mutant K122R/K148R was similarly localized to the nucleus as the wild type (Fig. 5C), but it was slightly more active in stimulating transcription (Fig. 5D). We next compared wild-type KLF2 with mutant K122R/K148R in MEF reprogramming. As shown in Fig. 5, E and F, mutant K122R/K148R was only slightly more active than wild-type KLF2. These results suggest that KLF2 sumoylation plays a very minor role in regulating the transcriptional activity and controlling the ability to induce pluripotency.
FIGURE 5.
Sumoylation of KLF2 inhibits transcription and reprogramming. A, domain organization of KLF2 shown with sequence alignment of conserved sumoylation motifs and adjacent residues from human, mouse, and Xenopus KLF2 proteins. See supplemental Fig. S4 for the entire sequence alignment. PPXY, a motif for interaction with WW domains such as those in two nuclear targets in hippo signaling and organ size control (37, 47, 48); ZF, zinc finger. B, sumoylation assays. Expression plasmids for HA-SUMO2 and FLAG-tagged wild-type and mutant KLF2 were co-transfected into HEK293 cells as indicated. Extracts were prepared 48 h later for IP on anti-FLAG M2-agarose and subsequent immunoblotting analysis with anti-HA and -FLAG antibodies. WB, Western blot. C, subcellular localization of GFP-tagged wild-type and mutants of KLF2 after transient transfection of the corresponding expression plasmids into HEK293 cells. Cells were fixed and incubated with Hoechst 33258 for fluorescence microscopy to detect GFP expression (green) and nuclei (blue). D, reporter gene assays. The Gal4-tk-Luc construct was transfected into HEK293 cells along with expression plasmids for expression of the Gal4 fusion protein containing the Gal4 DNA-binding domain fused to the wild-type and point mutants of KLF2 as indicated. Luciferase activities were normalized to β-galactosidase activities expressed from CMV-β-Gal co-transfected as the internal control. E, alkaline phosphatase staining of primary iPS colonies 9 days after infection of MEFs with a mixture of lentiviruses expressing untagged Oct4 and N-MYC, along with lentiviruses for FLAG-tagged KLF2 and Sox2 as indicated. F, quantification of alkaline phosphatase staining as performed in E. The quantification was based on two sets of independent experiments; *, p = 0.0048.
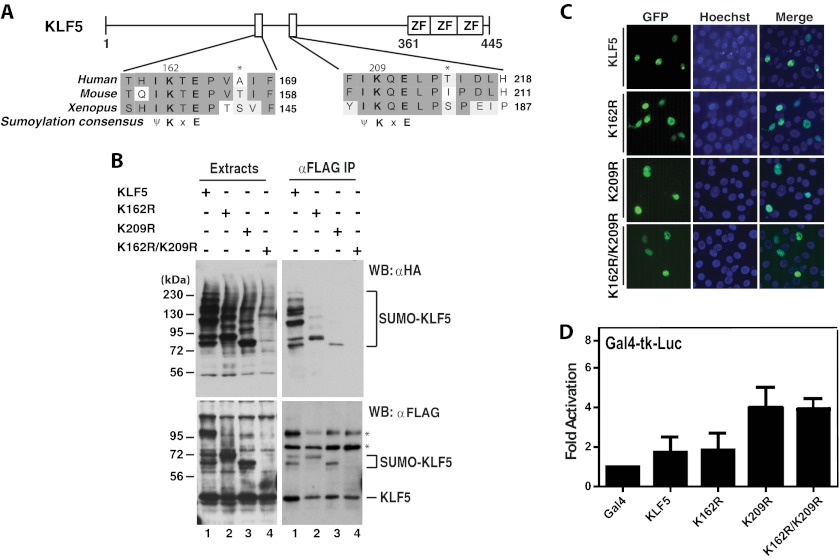
FIGURE 6.
KLF5 sumoylation and the effects on subcellular localization and transcription. A, domain organization of KLF5 illustrated with sequence alignment of two conserved sumoylation motifs and adjacent residues in human, mouse, and Xenopus KLF5 proteins. ZF, zinc finger. B, sumoylation assays. Expression plasmids for HA-SUMO2 and FLAG-tagged wild-type and mutant KLF5 were co-transfected into HEK293 cells as indicated. 48 h later, extracts were prepared for IP on anti-FLAG M2-agarose and immunoblotting with anti-HA and -FLAG antibodies. Asterisks denote nonspecific bands. WB, Western blot. C, subcellular localization of GFP-tagged wild-type and mutants of KLF5 and after transient transfection of the corresponding expression plasmids into HEK293 cells. Cells were fixed and incubated with Hoechst 33258 for fluorescence microscopy to detect GFP expression (green) and nuclei (blue). D, reporter gene assays. The Gal4-tk-Luc construct was transfected into HEK293 cells along with expression plasmids for expression of the Gal4 fusion proteins containing the Gal4 DNA-binding domain fused to the wild-type and point mutants of KLF5 as indicated. Luciferase activities were normalized to β-galactosidase activities expressed from CMV-β-Gal co-transfected as the internal control.
As was reported previously (21, 22), KLF5 was sumoylated at Lys-162 and Lys-209 (Fig. 6, A and B). Interestingly, these two sites appeared to cross-talk with each other and synergize the polysumoylation. As shown in Fig. 6C, no effect of mutation K162R/K209R on subcellular localization was observed, which is different from a previous report on mouse KLF5 (21). Although sumoylation was highly efficient (>20%, Fig. 6B; much more efficient than sumoylation of KLF2 or KLF4), the impact of double mutations K162R/K209R on KLF5 transcriptional activity was small (Fig. 6D). It is known that compared with KLF2, KLF5 was much less efficient to replace KLF4 in iPS cell generation (20). In our MEF reprogramming assays, we could not detect any reproducible effects when either the wild-type or mutant KLF5 was co-expressed with OCT4, SOX2, and N-MYC (data not shown), confirming that KLF5 is not as potent as KLF2 and KLF4 in inducing reprogramming (20). As the sumoylation-deficient mutant behaved similarly as the wild type (data not shown), sumoylation per se is not that reason for this difference. These results suggest that, compared with KLF4, sumoylation of KLF2 or KLF5 is much less important for reprogramming.
KLF4 Sumoylation Promotes Adipocyte Differentiation
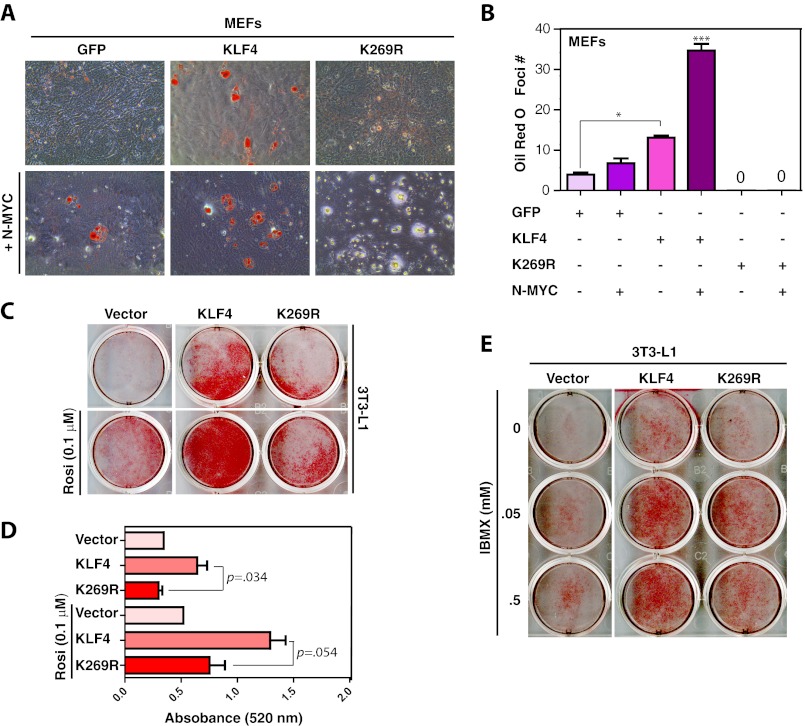
Because of skin defects and massive loss of body fluid, KLF4 knock-out mice die within a few hours after birth (43). Failure of secretion and deposition of lipids in keratinocytes has been suggested as one of the factors that break the skin barrier in these mice (43). In addition, KLF4 activates the CCAAT/enhancer-binding protein β promoter and regulates early stages of adipogenesis (44). During the process of reprogramming, we noticed that after culturing for 10–14 days under the reprogramming conditions, a low percentage of MEFs differentiated to adipocytes and accumulated oil droplets within the cytoplasmic region (data not shown). Such adipocyte-like cells were much fewer in MEFs expressing mutant K269R than those expressing wild-type KLF4 (data not shown). We thus hypothesized that wild-type KLF4 is more active than K269R in inducing adipocyte differentiation. To test this, we expressed wild-type and mutant KLF4 in MEFs, induced adipocyte differentiation for 14 days, and stained them with Oil Red O. Consistent with the above observation, wild-type KLF4 was more effective than K269R in inducing the differentiation (Fig. 7, A and B). During iPS cell generation, we also noticed that N-MYC had a positive effect on formation of adipocytes (data not shown), so we investigated the effect of N-MYC co-expression. As shown in Fig. 7, A and B, co-expression of N-MYC with KLF4 stimulated adipocyte differentiation further, whereas co-expression of mutant K269R with N-Myc appeared to transform MEFs.
FIGURE 7.
Defects of the KLF4 mutant K269R in promoting adipocyte differentiation. A, adipocyte differentiation from MEFs. The cells were infected with lentivirus expressing GFP, FLAG-KLF4, or FLAG-K269R with (bottom) or without (top) lentivirus for N-MYC. Two days after infection, adipocyte differentiation was initiated with the differentiation medium containing 5 μg/ml insulin, 1 μm dexamethasone, and 0.1 μm rosiglitazone. After 2 days, cells were switched to the medium containing 5 μg/ml insulin only and maintained there for the rest of the experiment. Oil Red O staining was performed on day 14; stained dishes were examined under a light microscope, and images of representative areas were taken. B, quantification of Oil Red O staining assays performed in A. The quantification was based on two sets of independent experiments. *, p < 0.05; ***, p < 0.001. C, adipocyte differentiation from 3T3-L1 preadipocytes. The cells were infected with lentivirus expressing GFP, FLAG-KLF4, or FLAG-K269R. Two days after infection, the cells were switched to the differentiation medium containing 5 μg/ml insulin, 1 μm dexamethasone in the absence (top) or presence (bottom) of 0.1 μm rosiglitazone. Oil Red O staining was performed 8 days later, and images of stained dishes were taken afterward. D, quantification of adipocyte differentiation. Isopropyl alcohol was used to extract Oil-Red O stain from stained cells in the dishes (C) for measurement of absorbance at 520 nm. The quantification was based on two sets of independent experiments, with the p values shown. E, adipocyte differentiation from 3T3-L1 preadipocytes. The cells were infected with lentivirus as in C. Two days later, adipocyte differentiation was initiated with the medium containing 5 μg/ml insulin, 1 μm dexamethasone, and isobutyl-1-methylxanthine at the indicated concentrations. Oil Red O staining was performed on day 8, and images of stained dishes were taken afterward.
To further investigate the role of KLF4 sumoylation in the process, we transduced the 3T3-L1 preadipocytes, a well established model for in vitro adipocyte differentiation (36), with lentiviruses expressing wild-type KLF4 or mutant K269R. 48 h after transduction, adipocyte differentiation was initiated in the presence or absence of rosiglitazone. Differentiated cells were stained with Oil Red O 8 days post-transduction. To quantify adipogenesis, we extracted Oil Red O from the stained cells and measured the absorbance at 520 nm. As shown in Fig. 7, C and D, mutant K269R was less active in promoting 3T3-L1 differentiation. When rosiglitazone was replaced with different concentrations of isobutyl-1-methylxanthine, we observed a similar difference between wild-type KLF4 and mutant K269R (Fig. 7E). Together, these results indicate that the K269R mutation inhibits the ability of KLF4 to induce adipocyte differentiation. KLF2 and KLF5 are known to inhibit and stimulate the process, respectively (45, 46), so we analyzed their sumoylation-deficient mutants in regulating 3T3-L1 differentiation. No major differences were observed when compared with the wild-type proteins (supplemental Fig. S5), indicating that sumoylation of the three KLF proteins has distinct effects on adipocyte differentiation.
DISCUSSION
Since iPS cells were first reported in 2006, there have been numerous studies on this unexpected technology (24–26). Although a majority of the studies focus on the potential application, it is important to explore and understand the underlying molecular and cellular mechanisms. This will not only provide vital information on how to refine and optimize this technology but also shed novel light on related cellular and developmental processes in normal and pathological states. One important aspect about the underlying mechanisms is how post-translational modifications regulate iPS cell induction. Related to this, the results described herein indicate that sumoylation of KLF4 repressed the Nanog promoter activity (Fig. 2F) and inhibited reprogramming (Figs. 3 and 4). Consistent with our results (Fig. 1), two other groups have found that KLF4 is conjugated by SUMO1 and the modification inhibits transcription (14, 15). Like KLF4, KLF2 was also sumoylated (Fig. 5 and supplemental Fig. S4), but the modification had a relatively smaller effect (Fig. 5). Although sumoylation of KLF5 was more efficient than that of KLF4 (Fig. 6), KLF5 played a much less obvious role in reprogramming, and sumoylation did not appear to contribute to this deficiency (data not shown). Thus, how sumoylation regulates functions of KLF2 and KLF5 awaits further investigation. Related to KLF2, it is noteworthy that N-terminal to the sumoylation sites is a conserved PPXY motif (Fig. 5A and supplemental Fig. S4); similar motifs are known to play important roles in regulation of several other proteins (37, 47, 48). Whether the PPXY motif interplays with the two sumoylation motifs is worthy of investigation.
Sumoylation of the nuclear receptor Nr5a2 was recently shown to inhibit reprogramming (49). Moreover, ERRγ can replace KLF4 in iPS cell generation (23), and sumoylation inhibits the transcriptional activity of ERRs (12), suggesting that ERRγ sumoylation may inhibit reprogramming. Thus, sumoylation of these known reprogramming factors plays a negative role. However, the sole SUMO E2 enzyme UBC9 is required for reprogramming,6 so sumoylation of an unidentified factor(s) may promote this process.
Our results reveal a sumoylation-dependent synergism among KLF4, Sox2, and Oct4 in activating the Nanog promoter (Fig. 2F and supplemental Fig. S2). Of relevance, another group also found that sumoylation of OCT4 and SOX2 represses the Nanog promoter activity (50). In parallel to the synergism at the Nanog promoter, we found that sumoylation-deficient mutants of KLF4, SOX2, and OCT4 synergized with each other and enhanced reprogramming efficiency (Fig. 3). This synergistic effect is reminiscent of the synergy control motif in other transcription factors (51, 52). Proximity of binding sites of these transcription factors and location of sumoylation motifs in the inhibitory domain of KLF4 (Fig. 1A) and between activation domains of SOX2 (Fig. 2A) further support this possibility.
An important issue is how cell signaling regulates sumoylation of these factors. Consistent with phosphorylation dynamics in ES cells (53), Ser-248, Ser-249, and Ser-250 of SOX2 were required for optimal sumoylation of Lys-245 (Fig. 2, A and B). But no major impact on reprogramming was detected with Asp substitution (supplemental Fig. S3). Mutant S274A did not exhibit a doublet as wild-type KLF4 (Fig. 1B, compare lanes 1 and 4, top right panel), suggesting that Ser-274 may be phosphorylated, but this does not affect sumoylation or transcription (Fig. 1). Thus, although we initially set out to study phosphorylation-dependent sumoylation of KLF4 and SOX2, such cross-talks do not play a major role in reprogramming. TGFβ signaling inhibits reprogramming (54–56). The SUMO E3 ligase PIAS1 mediates TGFβ signaling to activate α-actin expression in smooth muscles and inhibit KLF4-repression of the actin promoter by sumoylation (14). Whether similar mechanisms regulate KLF4 activity in Nanog expression and iPS cell induction is interesting for further investigation.
One implication of the findings is to express sumoylation-deficient mutants of reprogramming factors for iPS cell induction. This applies not only to the frequently used DNA-based methods (25, 26) but also to the new development using capped mRNAs (57). Only MEFs have been analyzed here, so it will be important to verify whether the obtained results can be extended to other cells and species, especially those of human origin. In addition, the four classical reprogramming factors also convert fibroblasts to epiblast stem cells (58), and Oct4 alone is sufficient to reprogram fibroblasts into multilineage blood progenitors (59). Ectopic expression of other transcription factors promotes trans-differentiation of adult pancreatic exocrine cells into β-cells (60) and conversion of fibroblasts to macrophage-like cells (61), neurons (62), cardiomyocytes (63), and multilineage blood progenitors (59). Effects of sumoylation suggest that it may be important to investigate whether post-translational modifications regulate these types of reprogramming.
We also investigated how sumoylation of KLF2, KLF4, and KLF5 regulates adipocyte differentiation. Although sumoylation of KLF2 has not been reported before (Fig. 5), KLF5 sumoylation (Fig. 6) is known to regulate expression of adipocyte-specific genes (21, 22). Sumoylation of KLF4 but not KLF2 or KLF5 was required for adipocyte differentiation (Fig. 7 and supplemental Fig. S5). Many members of KLF/SP family of transcription factors contain sumoylation motifs, and some of them are sumoylated. The modification inhibits activities of KLF1 (EKLF) (64), KLF3 (BKLF) (65), KLF8 (66), and SP3 (67). Moreover, concerted action of KLF family members regulates axon regeneration, with some inducing axonal growth and the others suppressing it (68). In addition to adipogenesis and axonal regeneration, KLF proteins are important in ES cell biology, leukocyte development, endothelial biology, and cancer development (41, 69–72). Overall, our findings provide new evidence that sumoylation serves as an additional layer of control over roles of KLFs in diverse cellular programs.
Acknowledgments
A. Sorisky and A. Gagnon provided 3T3-L1 preadipocytes and valuable advice on the use of these cells.
This work was supported in part by operating grants from Natural Sciences and Engineering Research Council of Canada and Ministère du Développement Économique, Innovation et Exportation du Québec (to X. J. Y.).

This article contains supplemental Figs. S1–S5.
S. Tahmasebi, M. Ghorbani, and X.-J. Yang, submitted for publication.
- SUMO
- small ubiquitin-like modifier
- iPS
- induced pluripotent stem
- mEF
- mouse embryonic fibroblast
- ERR
- estrogen-related receptor
- IP
- immunoprecipitation
- KLF
- Krüppel-like factor.
REFERENCES
- 1. Gill G. (2004) SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18, 2046–2059 [DOI] [PubMed] [Google Scholar]
- 2. Hochstrasser M. (2009) Origin and function of ubiquitin-like proteins. Nature 458, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 4. Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C., Pandolfi P. P., Dejean A. (2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9, 769–779 [DOI] [PubMed] [Google Scholar]
- 5. Hay R. T. (2005) SUMO: a history of modification. Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 6. Blomster H. A., Hietakangas V., Wu J., Kouvonen P., Hautaniemi S., Sistonen L. (2009) Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites. Mol. Cell. Proteomics 8, 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang S. H., Galanis A., Witty J., Sharrocks A. D. (2006) An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 25, 5083–5093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mohideen F., Capili A. D., Bilimoria P. M., Yamada T., Bonni A., Lima C. D. (2009) A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nat. Struct. Mol. Biol. 16, 945–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hietakangas V., Anckar J., Blomster H. A., Fujimoto M., Palvimo J. J., Nakai A., Sistonen L. (2006) PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. U.S.A. 103, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shalizi A., Gaudillière B., Yuan Z., Stegmüller J., Shirogane T., Ge Q., Tan Y., Schulman B., Harper J. W., Bonni A. (2006) A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 311, 1012–1017 [DOI] [PubMed] [Google Scholar]
- 11. Grégoire S., Tremblay A. M., Xiao L., Yang Q., Ma K., Nie J., Mao Z., Wu Z., Giguère V., Yang X. J. (2006) Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J. Biol. Chem. 281, 4423–4433 [DOI] [PubMed] [Google Scholar]
- 12. Tremblay A. M., Wilson B. J., Yang X. J., Giguère V. (2008) Phosphorylation-dependent sumoylation regulates estrogen-related receptor-α and -γ transcriptional activity through a synergy control motif. Mol. Endocrinol. 22, 570–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang X. J., Grégoire S. (2006) A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol. Cell 23, 779–786 [DOI] [PubMed] [Google Scholar]
- 14. Kawai-Kowase K., Ohshima T., Matsui H., Tanaka T., Shimizu T., Iso T., Arai M., Owens G. K., Kurabayashi M. (2009) PIAS1 mediates TGFβ-induced SM α-actin gene expression through inhibition of KLF4 function-expression by protein sumoylation. Arterioscler. Thromb. Vasc. Biol. 29, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Du J. X., McConnell B. B., Yang V. W. (2010) A small ubiquitin-related modifier-interacting motif functions as the transcriptional activation domain of Kruppel-like factor 4. J. Biol. Chem. 285, 28298–28308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tsuruzoe S., Ishihara K., Uchimura Y., Watanabe S., Sekita Y., Aoto T., Saitoh H., Yuasa Y., Niwa H., Kawasuji M., Baba H., Nakao M. (2006) Inhibition of DNA binding of Sox2 by the SUMO conjugation. Biochem. Biophys. Res. Commun. 351, 920–926 [DOI] [PubMed] [Google Scholar]
- 17. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 18. Wei F., Schöler H. R., Atchison M. L. (2007) Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation. J. Biol. Chem. 282, 21551–21560 [DOI] [PubMed] [Google Scholar]
- 19. Tolkunova E., Malashicheva A., Parfenov V. N., Sustmann C., Grosschedl R., Tomilin A. (2007) PIAS proteins as repressors of Oct4 function. J. Mol. Biol. 374, 1200–1212 [DOI] [PubMed] [Google Scholar]
- 20. Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. (2008) Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26, 101–106 [DOI] [PubMed] [Google Scholar]
- 21. Du J. X., Bialkowska A. B., McConnell B. B., Yang V. W. (2008) SUMOylation regulates nuclear localization of Kruppel-like factor 5. J. Biol. Chem. 283, 31991–32002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oishi Y., Manabe I., Tobe K., Ohsugi M., Kubota T., Fujiu K., Maemura K., Kubota N., Kadowaki T., Nagai R. (2008) SUMOylation of Kruppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-δ. Nat. Med. 14, 656–666 [DOI] [PubMed] [Google Scholar]
- 23. Feng B., Jiang J., Kraus P., Ng J. H., Heng J. C., Chan Y. S., Yaw L. P., Zhang W., Loh Y. H., Han J., Vega V. B., Cacheux-Rataboul V., Lim B., Lufkin T., Ng H. H. (2009) Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat. Cell Biol. 11, 197–203 [DOI] [PubMed] [Google Scholar]
- 24. Jaenisch R., Young R. (2008) Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132, 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamanaka S., Blau H. M. (2010) Nuclear reprogramming to a pluripotent state by three approaches. Nature 465, 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stadtfeld M., Hochedlinger K. (2010) Induced pluripotency: history, mechanisms, and applications. Genes Dev. 24, 2239–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holmberg C. I., Tran S. E., Eriksson J. E., Sistonen L. (2002) Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem. Sci. 27, 619–627 [DOI] [PubMed] [Google Scholar]
- 28. Latham J. A., Dent S. Y. (2007) Cross-regulation of histone modifications. Nat. Struct. Mol. Biol. 14, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 29. Yang X. J., Seto E. (2008) Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell 31, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 31. Gray J. S., Birmingham J. M., Fenton J. I. (2010) Got black swimming dots in your cell culture? Identification of Achromobacter as a novel cell culture contaminant. Biologicals 38, 273–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuroda T., Tada M., Kubota H., Kimura H., Hatano S. Y., Suemori H., Nakatsuji N., Tada T. (2005) Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 25, 2475–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dufour C. R., Wilson B. J., Huss J. M., Kelly D. P., Alaynick W. A., Downes M., Evans R. M., Blanchette M., Giguère V. (2007) Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRα and -γ. Cell Metab. 5, 345–356 [DOI] [PubMed] [Google Scholar]
- 34. Nakatake Y., Fukui N., Iwamatsu Y., Masui S., Takahashi K., Yagi R., Yagi K., Miyazaki J., Matoba R., Ko M. S., Niwa H. (2006) Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol. Cell. Biol. 26, 7772–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blelloch R., Venere M., Yen J., Ramalho-Santos M. (2007) Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell 1, 245–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gagnon A., Sorisky A. (1998) The effect of glucose concentration on insulin-induced 3T3-L1 adipose cell differentiation. Obes. Res. 6, 157–163 [DOI] [PubMed] [Google Scholar]
- 37. Hong J. H., Hwang E. S., McManus M. T., Amsterdam A., Tian Y., Kalmukova R., Mueller E., Benjamin T., Spiegelman B. M., Sharp P. A., Hopkins N., Yaffe M. B. (2005) TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 309, 1074–1078 [DOI] [PubMed] [Google Scholar]
- 38. Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 [DOI] [PubMed] [Google Scholar]
- 39. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodda D. J., Chew J. L., Lim L. H., Loh Y. H., Wang B., Ng H. H., Robson P. (2005) Transcriptional regulation of Nanog by OCT4 and SOX2. J. Biol. Chem. 280, 24731–24737 [DOI] [PubMed] [Google Scholar]
- 41. Jiang J., Chan Y. S., Loh Y. H., Cai J., Tong G. Q., Lim C. A., Robson P., Zhong S., Ng H. H. (2008) A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 10, 353–360 [DOI] [PubMed] [Google Scholar]
- 42. Wei Z., Yang Y., Zhang P., Andrianakos R., Hasegawa K., Lyu J., Chen X., Bai G., Liu C., Pera M., Lu W. (2009) Klf4 interacts directly with Oct4 and Sox2 to promote reprogramming. Stem Cells 27, 2969–2978 [DOI] [PubMed] [Google Scholar]
- 43. Segre J. A., Bauer C., Fuchs E. (1999) Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 22, 356–360 [DOI] [PubMed] [Google Scholar]
- 44. Birsoy K., Chen Z., Friedman J. (2008) Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 7, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Banerjee S. S., Feinberg M. W., Watanabe M., Gray S., Haspel R. L., Denkinger D. J., Kawahara R., Hauner H., Jain M. K. (2003) The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-γ expression and adipogenesis. J. Biol. Chem. 278, 2581–2584 [DOI] [PubMed] [Google Scholar]
- 46. Oishi Y., Manabe I., Tobe K., Tsushima K., Shindo T., Fujiu K., Nishimura G., Maemura K., Yamauchi T., Kubota N., Suzuki R., Kitamura T., Akira S., Kadowaki T., Nagai R. (2005) Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 1, 27–39 [DOI] [PubMed] [Google Scholar]
- 47. Wang K., Degerny C., Xu M., Yang X. J. (2009) YAP, TAZ, and Yorkie: a conserved family of signal-responsive transcriptional coregulators in animal development and human disease. Biochem. Cell Biol. 87, 77–91 [DOI] [PubMed] [Google Scholar]
- 48. Pan D. (2010) The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heng J. C., Feng B., Han J., Jiang J., Kraus P., Ng J. H., Orlov Y. L., Huss M., Yang L., Lufkin T., Lim B., Ng H. H. (2010) The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell 6, 167–174 [DOI] [PubMed] [Google Scholar]
- 50. Wu Y., Guo Z., Wu H., Wang X., Yang L., Shi X., Du J., Tang B., Li W., Yang L., Zhang Y. (2012) SUMOylation represses Nanog expression via modulating transcription factors Oct4 and Sox2. PLoS One 7, e39606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Iñiguez-Lluhí J. A., Pearce D. (2000) A common motif within the negative regulatory regions of multiple factors inhibits their transcriptional synergy. Mol. Cell. Biol. 20, 6040–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Komatsu T., Mizusaki H., Mukai T., Ogawa H., Baba D., Shirakawa M., Hatakeyama S., Nakayama K. I., Yamamoto H., Kikuchi A., Morohashi K. (2004) Small ubiquitin-like modifier 1 (SUMO-1) modification of the synergy control motif of Ad4 binding protein/steroidogenic factor 1 (Ad4BP/SF-1) regulates synergistic transcription between Ad4BP/SF-1 and Sox9. Mol. Endocrinol. 18, 2451–2462 [DOI] [PubMed] [Google Scholar]
- 53. Van Hoof D., Muñoz J., Braam S. R., Pinkse M. W., Linding R., Heck A. J., Mummery C. L., Krijgsveld J. (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5, 214–226 [DOI] [PubMed] [Google Scholar]
- 54. Maherali N., Hochedlinger K. (2009) Tgfβ signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 19, 1718–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ichida J. K., Blanchard J., Lam K., Son E. Y., Chung J. E., Egli D., Loh K. M., Carter A. C., Di Giorgio F. P., Koszka K., Huangfu D., Akutsu H., Liu D. R., Rubin L. L., Eggan K. (2009) A small-molecule inhibitor of Tgf-β signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 5, 491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lin T., Ambasudhan R., Yuan X., Li W., Hilcove S., Abujarour R., Lin X., Hahm H. S., Hao E., Hayek A., Ding S. (2009) A chemical platform for improved induction of human iPSCs. Nat. Methods 6, 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Warren L., Manos P. D., Ahfeldt T., Loh Y. H., Li H., Lau F., Ebina W., Mandal P. K., Smith Z. D., Meissner A., Daley G. Q., Brack A. S., Collins J. J., Cowan C., Schlaeger T. M., Rossi D. J. (2010) Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Han D. W., Greber B., Wu G., Tapia N., Araúzo-Bravo M. J., Ko K., Bernemann C., Stehling M., Schöler H. R. (2011) Direct reprogramming of fibroblasts into epiblast stem cells. Nat. Cell Biol. 13, 66–71 [DOI] [PubMed] [Google Scholar]
- 59. Szabo E., Rampalli S., Risueño R. M., Schnerch A., Mitchell R., Fiebig-Comyn A., Levadoux-Martin M., Bhatia M. (2010) Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 468, 521–526 [DOI] [PubMed] [Google Scholar]
- 60. Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D. A. (2008) In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 455, 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Feng R., Desbordes S. C., Xie H., Tillo E. S., Pixley F., Stanley E. R., Graf T. (2008) PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc. Natl. Acad. Sci. U.S.A. 105, 6057–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vierbuchen T., Ostermeier A., Pang Z. P., Kokubu Y., Südhof T. C., Wernig M. (2010) Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ieda M., Fu J. D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., Srivastava D. (2010) Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Siatecka M., Xue L., Bieker J. J. (2007) Sumoylation of EKLF promotes transcriptional repression and is involved in inhibition of megakaryopoiesis. Mol. Cell. Biol. 27, 8547–8560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Perdomo J., Verger A., Turner J., Crossley M. (2005) Role for SUMO modification in facilitating transcriptional repression by BKLF. Mol. Cell. Biol. 25, 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wei H., Wang X., Gan B., Urvalek A. M., Melkoumian Z. K., Guan J. L., Zhao J. (2006) Sumoylation delimits KLF8 transcriptional activity associated with the cell cycle regulation. J. Biol. Chem. 281, 16664–16671 [DOI] [PubMed] [Google Scholar]
- 67. Ross S., Best J. L., Zon L. I., Gill G. (2002) SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 10, 831–842 [DOI] [PubMed] [Google Scholar]
- 68. Moore D. L., Blackmore M. G., Hu Y., Kaestner K. H., Bixby J. L., Lemmon V. P., Goldberg J. L. (2009) KLF family members regulate intrinsic axon regeneration ability. Science 326, 298–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brey C. W., Nelder M. P., Hailemariam T., Gaugler R., Hashmi S. (2009) Kruppel-like family of transcription factors: an emerging new frontier in fat biology. Int. J. Biol. Sci. 5, 622–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cao Z., Sun X., Icli B., Wara A. K., Feinberg M. W. (2010) Role of Kruppel-like factors in leukocyte development, function, and disease. Blood 116, 4404–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Atkins G. B., Jain M. K. (2007) Role of Kruppel-like transcription factors in endothelial biology. Circ. Res. 100, 1686–1695 [DOI] [PubMed] [Google Scholar]
- 72. Rowland B. D., Peeper D. S. (2006) KLF4, p21, and context-dependent opposing forces in cancer. Nat. Rev. Cancer 6, 11–23 [DOI] [PubMed] [Google Scholar]