Background: Lytic polysaccharide monooxygenases (LPMOs) represent a recently discovered enzymatic route to cleave carbohydrates.
Results: We report the first basidiomycete LPMO structure and describe enzyme-cellulose interactions with simulation.
Conclusion: We characterize the copper-containing active site and identify loops important for substrate recognition and binding.
Significance: This structure is the first LPMO from a model basidiomycete fungus that contains many LPMO genes.
Keywords: Biofuel, Carbohydrate-binding Protein, Glycoside Hydrolases, Molecular Dynamics, Structural Biology, CBM33, GH61, Lytic Polysaccharide Monooxygenase, Phanerochaete chrysosporium, Copper Monooxygenase
Abstract
Carbohydrate structures are modified and degraded in the biosphere by a myriad of mostly hydrolytic enzymes. Recently, lytic polysaccharide mono-oxygenases (LPMOs) were discovered as a new class of enzymes for cleavage of recalcitrant polysaccharides that instead employ an oxidative mechanism. LPMOs employ copper as the catalytic metal and are dependent on oxygen and reducing agents for activity. LPMOs are found in many fungi and bacteria, but to date no basidiomycete LPMO has been structurally characterized. Here we present the three-dimensional crystal structure of the basidiomycete Phanerochaete chrysosporium GH61D LPMO, and, for the first time, measure the product distribution of LPMO action on a lignocellulosic substrate. The structure reveals a copper-bound active site common to LPMOs, a collection of aromatic and polar residues near the binding surface that may be responsible for regio-selectivity, and substantial differences in loop structures near the binding face compared with other LPMO structures. The activity assays indicate that this LPMO primarily produces aldonic acids. Last, molecular simulations reveal conformational changes, including the binding of several regions to the cellulose surface, leading to alignment of three tyrosine residues on the binding face of the enzyme with individual cellulose chains, similar to what has been observed for family 1 carbohydrate-binding modules. A calculated potential energy surface for surface translation indicates that P. chrysosporium GH61D exhibits energy wells whose spacing seems adapted to the spacing of cellobiose units along a cellulose chain.
Introduction
Nature employs mixtures of glycoside hydrolases (GHs)7 to convert carbohydrate polymers found in plant, fungal, and algal cell walls to soluble sugars (1). Recently, a new class of enzymes was discovered that uses copper-dependent oxidative pathways for the cleavage of glycosidic linkages (2–5). These oxidative enzymes, referred to here as lytic polysaccharide monooxygenases (LPMOs), have garnered significant interest because they enhance degradation of recalcitrant polysaccharides, such as chitin and cellulose, when added to GH mixtures (6, 7). Vaaje-Kolstad et al. (2) first observed lytic activity of chitin-binding protein 21 (CBP21) from the bacterium Serratia marcescens on β-chitin, which produced soluble C1-oxidized chito-oligosaccharides (aldonic acids) in the presence of reductants. CBP21 was originally classified as a family 33 carbohydrate-binding module (CBM33), which are prevalent proteins in biomass-degrading bacteria (8). Soon after, a CBM33 enzyme was characterized that also produces soluble aldonic acids from cellulose (3). Similarities in the structures of CBM33s and family 61 GHs (GH61s) were noted when the structure of Hypocrea jecorina GH61B was determined (9), including a conserved surface-located metal coordination site (2, 9). Shortly after the initial report on CBP21 (2), it was shown by several groups that GH61s also employ a metal-dependent oxidative pathway to cleave glycosidic bonds in cellulose (4, 5, 10–12). The consensus between CBM33 and GH61 activity to date is that enzymes from both families utilize copper as the catalytic metal (5, 13) and that reducing agents, including cellobiose dehydrogenases (4, 11), ascorbate, reduced glutathione, gallate (2, 3, 5, 10, 12), or non-carbohydrate species present in biomass (6, 14), are required for activity.
To date, there are five LPMO structures from five different fungal GH61s available: H. jecorina GH61B (Protein Data Bank (PDB) code 2VTC) (9), Thielavia terrestris GH61E (PDB codes 3EII and 3EJA) (6), Thermoascus aurantiacus GH61A (PDB codes 2YET and 3ZUD) (5), and Neurospora crassa PMO-2 and PMO-3 (PDB codes 4EIR and 4EIS, respectively) (15). Additionally, there are structures of four different CBM33 enzymes available: S. marcescens CBP21 (PDB codes 2LHS, 2BEM, and 2BEN) (7, 16), Vibrio cholerae CBM33 (PDB code 2XWX) (17), Enterococcus faecalis CBM33 (PDB code 4A02) (18), and Burkholderia pseudomallei CBM33 (PDB code 3UAM). The GH61 structures all contain a metal ion in the putative catalytic center of the enzyme, whereas not all CBM33 structures solved to date contain a metal ion. The catalytic centers in all of these enzymes are embedded in a flat protein face containing aromatic and polar residues for putative binding to the surfaces of cellulose and chitin. Aromatic residues are not usually dominating, but some GH61s show arrangements similar to what is found on the binding faces of family 1 CBMs (19–21). The flat catalytic binding surfaces of LPMOs are putatively suited to cleave glycosidic linkages without decrystallizing polymer chains (13, 22, 23), whereas endoglucanases, with a catalytic cleft, are thought to mainly act on more accessible, amorphous regions (24). This may explain why these two enzyme classes are synergistic (2).
To date, most LPMOs have been found to oxidize the C1 position (2–5, 11, 12). However, oxidation of the C4 carbon in the scissile bond to form a 4-keto-aldose moiety has been described for an LPMO from N. crassa (10). Oxidation has also been suggested at the C6 carbon (5, 25). There is no general consensus yet on the spectrum of oxidative chemistry potentially employed by LPMOs, let alone the structural basis of the selectivity of oxidation. There is significant incentive to understand the structural basis of LPMO action because of their observed activity improvements to industrial mixtures (6). Because biomass-degrading enzyme mixtures remain a major cost driver in production of biofuels (26, 27), including LPMOs in the industrial enzyme mixtures offers the potential for significant cost reductions for enzymatic hydrolysis of biomass. Thus, determining the LPMO mechanism of action, screening LPMO activities from natural diversity, and enzyme engineering for higher activity and stability are now under way (8).
It is noteworthy that the known GH61 structures and most of the recent progress on mechanism elucidation are with enzymes from ascomycete fungi. However, wood decomposition in nature is predominantly conducted by basidiomycete fungi (28, 29), which are broadly divided into brown rot and white rot fungi (28). Multiple putative and identified GH61 genes have been found in genomes of both types (28–31), with the number of genes appearing to be larger in white rot than in brown rot fungi (28). It is thus of significant interest to study LPMO structures from basidiomycete fungi. Phanerochaete chrysosporium, in particular, is one of the most extensively studied white rot fungi, and as such, its genome was the first basidomycete sequenced (31). Up to 17 putative P. chrysosporium genes encoding GH61 enzymes (PchGH61s) were initially identified (31).
We previously cloned the P. chrysosporium GH61D gene and expressed the protein, referred to here as PchGH61D (Joint Genome Institute Protein ID: 4691 in Pichia pastoris) (12). We showed that PchGH61D is a copper-dependent LPMO with activity on Avicel, filter paper, and phosphoric acid-swollen cellulose, which oxidizes at the C1 carbon. No soluble sugars oxidized at C4 or C6 were detected (12). In the present study, we present the crystal structure of PchGH61D, the first LPMO structure from a basidiomycete fungus, and we use x-ray absorption fine structure scanning to analyze metal binding. We conduct a structure-based alignment of the PchGH61D structure with other LPMOs to examine the conservation of surface and active site residues. Additionally, we show for the first time the profile of released products when an LPMO enzyme acts on a real biomass substrate, namely pretreated spruce. Last, we use MD simulation to study aspects of the interaction of PchGH61D with the cellulose surface. Overall, this study contributes to the expanding repertoire of LPMO structures and identifies key interactions with the hydrophobic face of cellulose, which will aid in describing the mechanism and specificity of these important enzymes.
EXPERIMENTAL PROCEDURES
Protein Preparation and Crystallization
Recombinant PchGH61D was expressed in P. pastoris and purified using hydrophobic interaction and ion exchange chromatography after endoglycosidase H treatment, as described previously (12). The purified protein solution was incubated with 10 mm EDTA for 3 h and then diluted into 10 mm sodium acetate buffer, pH 5.0, with 1 mm CuSO4 for 30 min. A PD-10 column (GE Healthcare) was used for buffer exchange to 10 mm sodium acetate buffer, pH 5.0. After buffer exchange, the protein was concentrated to 12 mg/ml using a VIVASPIN-6 centrifugal concentrator (10,000 molecular weight cut-off polyethersulfone membrane; Sigma-Aldrich).
The initial search for crystallization conditions for PchGH61D was done with sitting drop vapor diffusion techniques at 20 °C in a MRC2 well crystallization plate (Hampton Research) using the JCSG+ Suite sparse matrix screen (Qiagen). Crystals for structure determination were obtained at 20 °C with 2.1 m dl-malic acid, pH 7.0, as precipitant, mixed 1:1 (v/v) with 12 mg/ml PchGH61D in 10 mm sodium acetate, pH 5.0. Prior to data collection, crystals were soaked briefly in crystallization solution mixed with glycerol at 20% (v/v) final concentration as cryoprotectant and then flash-frozen in liquid N2.
Data Collection and Structure Determination
The x-ray absorption spectrum scan of a PchGH61D crystal was recorded by measuring the fluorescence signal during energy scan near the copper absorption edge, using the PyMCA program at the ID23-1 beamline at the European Synchrotron Radiation Facility (ESRF) (Grenoble, France). X-ray diffraction data were collected at beamline ID14-1 (ESRF) using a single PchGH61D crystal. The diffraction data set was reduced and scaled using the XDS program (32, 33) and the CCP4 program suite (33). Diffraction data to 1.75 Å resolution were used in the scaling and throughout structure refinement.
The PchGH61D structure was solved by molecular replacement using Phaser (34). The search model was a homology model of PchGH61D (12), based on the Thielavia terrestris GH61E (PDB code 3EII) structure, built by the SWISS-MODEL Server (35). REFMAC5 (36) was used for structure model refinements, and manual model rebuilding was performed with Coot (37), using maximum likelihood (σA) weighted 2Fo − Fc electron density maps (38). For cross-validation and R and Rfree calculations, 5% of the data was excluded from the structure refinement (39). Solvent molecules were automatically added using the automatic water picking function in the ARP/wARP package (40). Picked water molecules were selected or discarded manually by visual inspection of the 2Fo − Fc electron density map. The copper ion bound in the active site was introduced at a final stage of the structure refinement. The coordinates for the final structure model and the structure factors have been deposited in the PDB (41) with accession code 4B5Q.
The search for similar structures was carried out using the Dali server (42). The Lsqman program (43) in the Uppsala Software Factory suite was used to provide root mean square deviation (RMSD) values and structure comparison statistics (44). Coot was used for structural analysis (37), and MacPyMOL (Schrödinger, LLC) was used for the preparation of structural figures.
Structure-based Sequence Alignment
Sequences of GH61 enzymes Phchr1|4691 (also known as PchGH61D), 41563, 41650, 41123, 31049, 129325, 121193, 122129, and 10320 (the numbers indicate Protein ID) were retrieved from the P. chrysosporium version 2.0 genome database at the Department of Energy Joint Genome Institute (45). A structure-based sequence alignment of the catalytic domains of GH61s with known crystal structure (PchGH61D, PDB code 4B5Q; TteGH61E, 3EJA; NcrPMO-2, 4EIR; NcrPMO-3, 4EIS; HjeGH61B, 2VTC; TauGH61A, 3ZUD) was made with the help of Dali server constraints (42), to which the other PchGH61 sequences were aligned using the MAFFT program (46) (supplemental Fig. S1). The secondary structure elements of PchGH61D were assigned using the program STRIDE (47). The sequence alignment table was edited in ESPript version 2.2 (48).
PchGH61D Activity Assay
Degradation experiments with PchGH61D were conducted using 0.1% phosphoric acid-swollen cellulose (PASC), prepared as described (49), or 0.5% steam-exploded (225 °C, 10 min) and washed spruce wood chips (50) in 25 mm sodium acetate, pH 5.3, as substrate. The enzyme and ascorbic acid concentrations were 34 μg/ml and 1.5 mm, respectively. The reactions were incubated for 20 h at 50 °C with 900 rpm vertical shaking in an Eppendorf Thermo mixer and then centrifuged at 21,000 × g for 3 min. The content of soluble oxidized oligosaccharides in the supernatants was analyzed by high performance anion exchange chromatography, as described previously (3, 51).
Computational Study of PchGH61D-Cellulose Interactions
To conduct classical MD simulations of PchGH61D with a copper ion bound in the enzyme, the charge redistribution in the active center upon copper binding was examined with electronic structure calculations, as described in the supplemental material and shown in supplemental Fig. S2. CHARMM (52) was used for all simulations. PchGH61D was placed on the hydrophobic face of cellulose 1β with the active site facing the cellulose surface, as shown in supplemental Fig. S2. The cellulose model was taken from a 10-ns equilibrated structure for cellulose 1β from previous work (23). We note that 10 ns was previously demonstrated to be a sufficient equilibration time for studying cellulose surface behavior (53). The hydrophobic face of cellulose on which the protein was placed contains three cellodextrin chains, and the copper atom in the PchGH61D active site was placed directly above a glycosidic linkage on the middle chain. In this orientation, Tyr-28 and Tyr-198 align over the middle chain on the crystal surface, and Tyr-75 aligns over the edge chain. The orientation of the enzyme with respect to cellulose was chosen to be similar to that of the family 1 CBM from H. jecorina Cel7A in that the enzyme was placed such that Tyr-28 and Tyr-198 align along a single chain. Shorter simulations were also conducted with PchGH61D rotated 180° in the opposite direction, which yielded similar results in terms of how the enzyme active site stabilized over the active site (data not shown). Additional details related to the simulation setup and methods can be found in the supplemental material. Last, a potential energy surface (PES) for the PchGH61D-cellulose interaction was also constructed to examine the location of stable energetic wells. This closely follows previous work conducted on the family 1 CBM (19, 54). Details of the PES construction can be found in the supplemental material.
RESULTS AND DISCUSSION
Overall Structure of PchGH61D
PchGH61D crystallized in space group C2 with unit-cell parameters of a = 149.3 Å, b = 37.5 Å, c = 79.8 Å and with a β angle of 117.4°. The asymmetric unit of the crystal contains two non-crystallographic symmetry-related molecules (A and B) related by a 2-fold rotation axis, giving a Matthews coefficient of 2.0 (55). The structure was solved by molecular replacement using a homology model of PchGH61D (12) and was refined at 1.75 Å resolution. The final PchGH61D structure model exhibits crystallographic R and Rfree values of 18.6 and 22.3% and contains a total of 3,781 non-hydrogen atoms, including all 434 amino acid residues, two copper atoms, one mannose residue (in chain A), two glycerol molecules, and 366 water molecules. The amino acid residues are numbered according to the mature protein after signal peptide cleavage, starting with His-1. Statistics of diffraction data and structure refinement are summarized in Table 1. The RMSD values between all Cα atom pairs of the two molecules in the asymmetric unit is 0.11 Å.
TABLE 1.
Diffraction data and refinement statistics for the PchGH61D structure (PDB code 4B5Q)
| Data collection | |
| Beamlinea | ID14:EH1 |
| Wavelength (Å) | 0.933 |
| Space group | C2 |
| Unit cell dimensions | |
| a, b, c (Å) | 149.3, 37.5, 79.8 |
| α, β, γ (°) | 90.0, 117.4, 90.0 |
| Rmerge (%)b,c | 8.6 (56.3) |
| I/σ(I)b | 10.9 (2.1) |
| Completeness (%)b | 98.5 (97.5) |
| Multiplicityb | 3.4 (3.3) |
| Refinement | |
| Resolution(Å) | 40.7–1.75 (1.84–1.75) |
| Rwork/Rfree (%) | 18.6/22.3 |
| RMSD, bonds (Å)d | 0.007 |
| RMSD, angles (degrees)d | 1.165 |
| No. of protein residues | 434 |
| No. of water molecules | 366 |
| No. of metal atoms | 2 |
| Average B factor | |
| Overall (Å2)e | 20.0 |
| Protein (Å2)e | 16.7 |
| Metals (Å2)e | 15.5 |
| Organic ligands (Å2)e | 34.1 |
| Waters (Å2)e | 26.3 |
| Ramachandran outliersf (%) | 0.8 |
a Beamlines at the European Synchrotron Radiation Facility (Grenoble, France).
b Values in parentheses are those for the highest resolution shell.
c Rmerge = ΣhklΣi|I − 〈I〉|ΣhklΣi|I|.
d Data from Engh and Huber (71).
e Calculated using MOLEMAN2 (72).
f Calculated using a strict boundary Ramachandran definition given by Kleywegt and Jones (73).
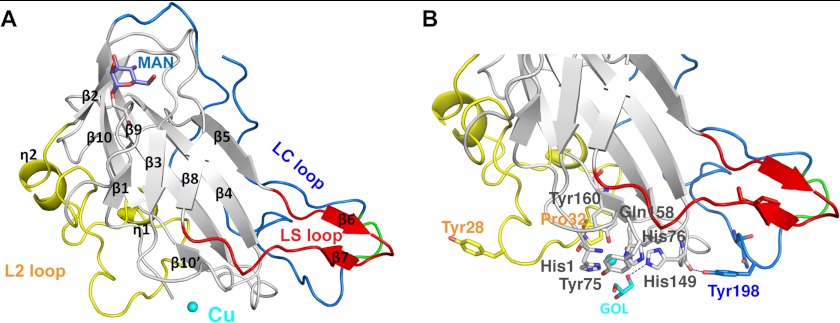
The overall fold of PchGH61D (Fig. 1A) is a β-sandwich fold consisting of two β-sheets, formed by in total eight β-strands. One β-sheet, the front sheet in Fig. 1, includes the β1, β3, and β8 strands, whereas the other includes strands β4, β5, β9, and β10. Strand β2 is involved in forming both β-sheets, which pack onto each other to form the core of the protein. The proposed catalytic center of the enzyme is positioned on a flat surface on one side of the β-sandwich fold. The PchGH61D structure shows three extended loops, which are all involved in shaping the potential substrate-binding surface. The L2 loop region (residues 17–57) includes two short η-helices. The long C-terminal loop (LC loop, residues 170–217) contains no secondary structure elements. In the tip of this loop, the backbone atoms of residues 201–204 (Pro-201, Lys-202, Asn-203, and Phe-204) display much higher B factors (34.8, 40.0, 39.7, and 26.9 Å2, respectively) than the average B factor for the protein (16.7 Å2), indicating high flexibility in this region (Fig. 1, green). The third, shorter LS loop (residues 109–124) forms hydrophobic interactions with the C-terminal LC loop and contains a β-hairpin motif (residues 110–117; β-strands β6 and β7). Residues 109–124 exhibit elevated B factors (average 24.6 Å2), indicating that also the LS loop is quite flexible. Cys-43 and Cys-163 form a disulfide bridge between the L2 loop and strand β10. PchGH61D contains two potential N-linked glycosylation sites, Asn-173 and Asn-203. Both are located on the long C-terminal LC loop (shown in supplemental Fig. S1). Asn-203 is positioned in the more flexible region of the loop, close to the potential substrate-binding surface. Previous results indicated that the protein is indeed N-glycosylated, because the apparent size was reduced upon treatment with endoglycosidase H (12). However, there is no clear electron density at either site for the GlcNAc residue that should remain after deglycosylation with endoglycosidase H. On the other hand, one mannose residue O-linked to Ser-11 is visible and included in molecule A but not in molecule B, where the density is too weak at the corresponding position (Fig. 1A). The electron density map of the PchGH61D structure does not show any indication of methylation of the Nϵ2 of His-1. Such post-translational modification is visible in the electron densities of some GH61s that have been structurally described so far (5, 9, 15), all of which are from filamentous fungi. It may be possible that PchGH61D is not methylated at His-1 because it has been expressed in yeast (P. pastoris). At this stage, we do not know whether methylation occurs when the enzyme is produced by P. chrysosporium itself. Nevertheless, it is notable that the non-methylated PchGH61D enzyme is active (see Ref. 12; see below). It thus seems that methylation of His-1 is not strictly necessary for GH61 activity. Notably highly active CBM33-type LPMOs are not methylated (16, 18).
FIGURE 1.
Features of the PchGH61D crystal structure. A, schematic representation of the PchGH61D structure with the bound copper atom, depicted as a sphere in cyan. The L2 loop (residues 17–57) is colored in yellow, the short LS loop containing a β-hairpin in red (residues 109–124), and the C-terminal LC loop in blue (residues 170–217). All secondary structure elements of the enzyme are labeled according to their position in the protein sequence. The glycosylated residue Ser-11 and the attached O-linked mannose residue are shown in a stick representation in gray and slate blue, respectively. B, close up view of the structure showing potentially important residues at the proposed substrate-binding surface and the metal binding site in a stick representation with the same color coding as in A. The flexible portion of the LC loop (residues 201–204) is colored in green in both panels.
Like other known LPMO structures, PchGH61D exhibits a flat putative binding surface in which the proposed catalytic center is embedded, as shown in detail in Fig. 1B. It is likely that the details of this binding face dictate catalytic specificity to given carbon atoms in cellulose substrates as well as specificity to other polysaccharide substrates (2, 5, 12, 13, 18). Fig. 1B highlights several residues of potential interest, which are discussed further below.
Copper Binding and the Structure of the Catalytic Center in PchGH61D
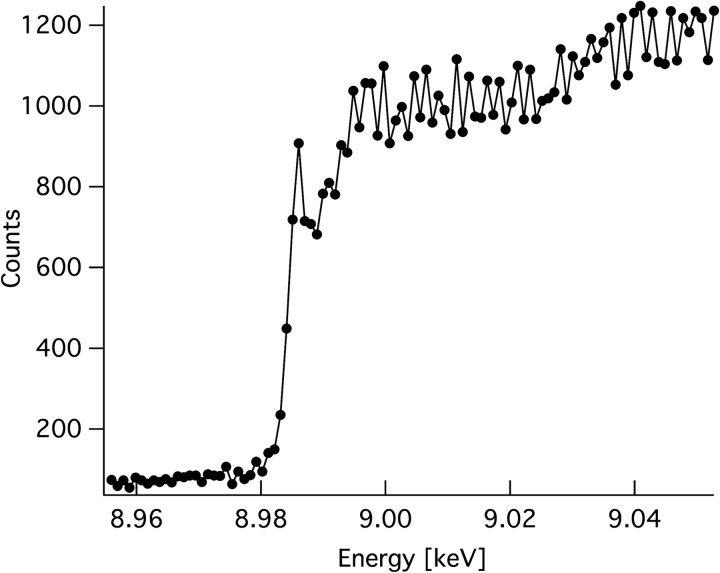
The x-ray absorption spectrum of the PchGH61D crystal shows a characteristic absorption edge at 8.9841 keV (Fig. 2), which indicates that the protein binds copper. Consequently, copper atoms were modeled in the catalytic centers of both protein molecules with full occupancy, based on the strong positive peak in the Fo − Fc map (Fig. 3A). The B factors of the copper atoms in the final model are 14.8 and 16.2 Å2 for molecule A and B, respectively. These low B factors, which are in the same range as the B factors for the protein backbone, indicate that copper is strongly bound.
FIGURE 2.
Copper K-edge fluorescence scan of the PchGH61D crystal. The scan demonstrates that copper is the bound metal.
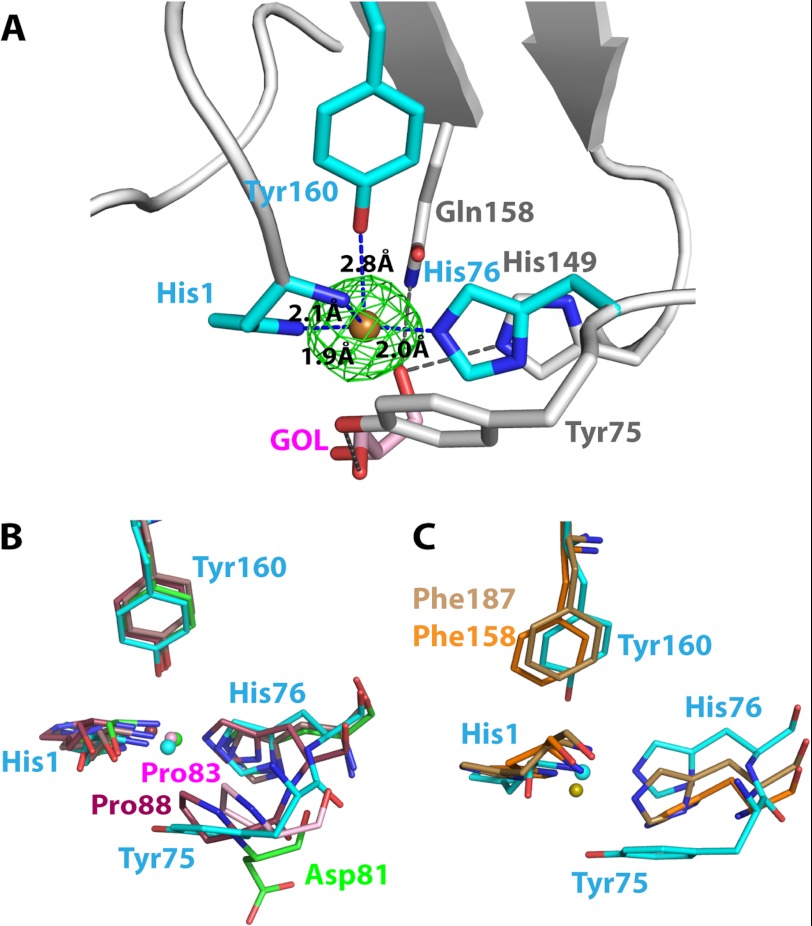
FIGURE 3.
The metal binding site of LPMOs. A, close up view of the PchGH61D in the vicinity of the copper binding site (PDB code 4B5Q). The green Fo − Fc map of the copper atom is contoured at 0.41 e/Å3 (3σ). Cyan-colored residues are coordinated to the copper atom. A glycerol molecule was modeled below the copper atom at the active site, colored in pink (denoted GOL). The bound glycerol molecule is stabilized by His-149, Gln-158, and Tyr-75 (in gray) by hydrogen bonds. B, superposition of the metal binding sites of PchGH61D (PDB code 4B5Q; cyan) with the metal binding sites of NcrPMO-2 (4EIR; green), TauGH61A (3ZUD; pink), and HjeGH61B (2VTC; maroon). The metal ions were modeled as Cu2+ in the first three structures and as Ni2+ in the HjeGH61B structure. C, comparison of the metal binding site of PchGH61D (cyan) with the corresponding non-occupied metal binding sites of SmaCBP21 (2BEM; brown) and EfaCBM33 (4A02; orange).
In the final structure, there were unmodeled Fo − Fc electron densities within 2.0 Å from the copper ion in both the A and B molecule, which may reflect movement of the metal ion between different putative reaction states, similar to what has been suggested for TauGH61A (5). Published GH61 structures show water molecules, a peroxide ion, oxygen molecules, or a sulfate ion in this position (5, 6, 9, 15). The corresponding space in PchGH61D is occupied by contiguous electron density that was interpreted as a glycerol molecule. The glycerol molecule might be stabilized in this position by hydrogen bonds to the side chains of Gln-158, His-149, and Tyr-75 (Fig. 3A).
The copper-binding site in PchGH61D is a type II copper center, which exhibits a hexacoordination. In the geometry, a square planar coordination was created by nitrogen or nitrogen/oxygen atoms (56, 57). In the PchGH61D structure, square coordination is provided by the main-chain amide group (2.1 Å), Nδ (1.9 Å) of His-1, and Nϵ of His-76 (2.0 Å), whereas there is no ligand at the fourth coordination position. In HjeGH61B (PDB code 2VTC (9)), the corresponding position is occupied by a water molecule. The hydroxyl group of Tyr-160 (2.8 Å) occupies one of the axial positions, whereas the other axial position is empty in the hexacoordination geometry. Protein atoms thus occupy four coordination positions, leaving two sites available for ligand binding. In the PchGH61D structure, both available sites are blocked by the bound glycerol molecule (Fig. 3A).
Several differences occur in the catalytic center of PchGH61D compared to other known GH61 and CBM33 structures, as shown in Fig. 3, B and C. One noteworthy variation involves Tyr-75 in PchGH61D, which is positioned on one side of the copper atom (Fig. 3B). The corresponding amino acid is aspartate in NcrPMO-2 and NcrPMO-3 and proline in HjeGH61B and TauGH61A, which suggests variations in exposure of the copper binding site in different GH61s. Interestingly, metal binding sites of CBM33s differ distinctly from those of GH61s. The conserved tyrosine in the GH61s (Tyr-160 in PchGH61D) is a phenylalanine in the CBM33 enzymes (Fig. 3C), lacking the hydroxyl group that participates in metal coordination in GH61s. Thus, in CBM33s, the copper atom is coordinated by three planar interactions only. Interestingly, currently available limited data indicate that GH61s (5) bind copper more strongly than CBM33s (13).
Overall Comparison of LPMO Structures
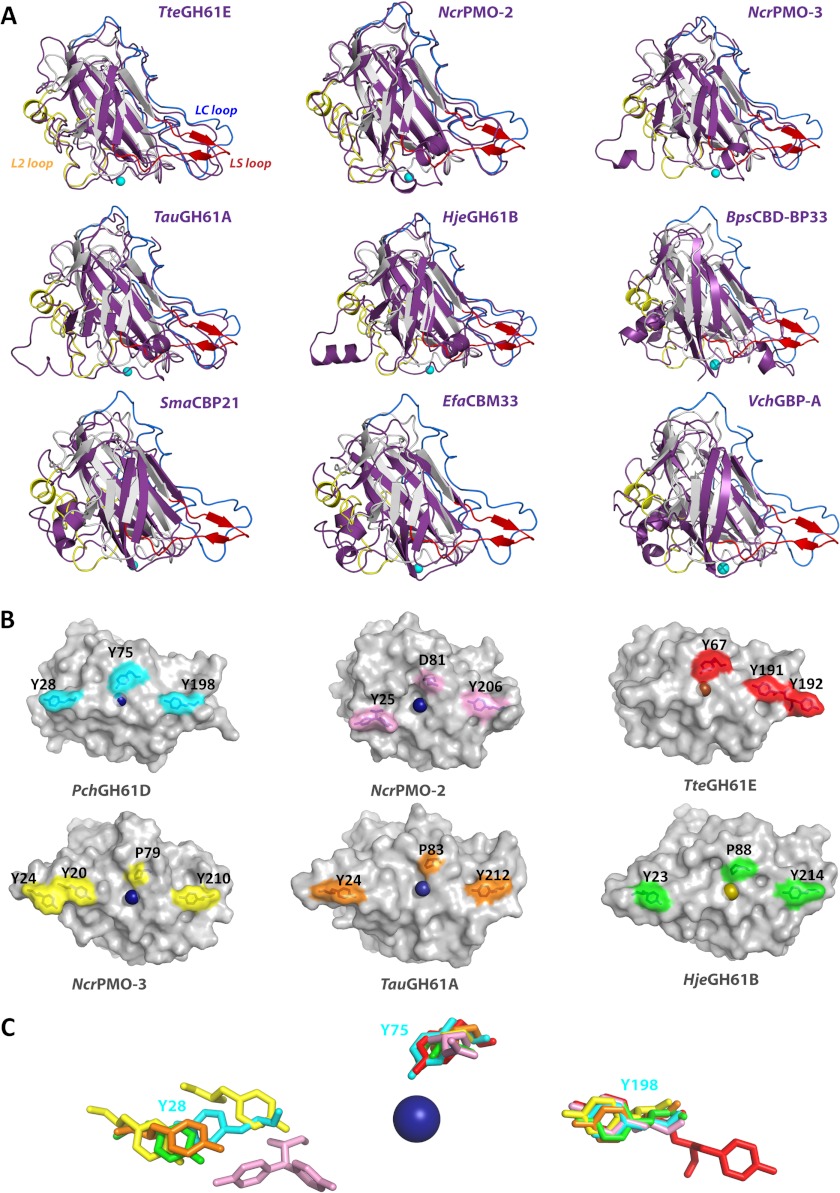
Molecule A of the PchGH61D structure was used to search for similar structures in the PDB (41) using the Dali server (42) (see supplemental Table S1 for listing and quantitative information). The most similar structure is TteGH61E (46% sequence identity with PchGH61D), followed by four other fungal LPMOs: two N. crassa LPMOs, NcrPMO-2 and NcrPMO-3 (38 and 34% identity); TauGH61A (28% identity); and HjeGH61B (30% identity). Lower, but significant, Z-scores were seen for the bacterial LPMOs, earlier classified as CBM33s, namely SmaCBM33 (known as CBP21), BpsCBD-BP33 from B. pseudomallei, EfaCBM33 from E. faecalis, and the N-terminal domain of Gbp-A from V. cholera. For example, the structural comparison of PchGH61D with EfaCBM33 included only 95 Cα atoms, with an RMSD value of 1.82 Å (Z-value = 9.3). Note that to date, there is no structural information for CBM33 domains known to act on cellulose. Fig. 4 shows structural superpositions of PchGH61D with the aforementioned nine different fungal and bacterial LPMOs. The most prominent structural differences are shown and indicate potential determinants of binding affinity and substrate specificity, as discussed below.
FIGURE 4.
Structural comparison of LPMOs. A, superimposed structures of PchGH61D (gray) with other LPMOs (purple): NcrPMO2 (PDB code 4EIR); TteGH61E (3EJA); NcrPMO-3 (4EIS); TauGH61A (3ZUD); HjeGH61B (2VTC); BpsCBD-BP33 (3UAM); SmaCBP21 (2BEM); EfaCBD-CBM33 (4A02); and VchGlc-binding protein A (2XWM). Yellow, blue, and red regions correspond to the L2 loop, LC loop, and LS loop, respectively, in the PchGH61D structure. B, aromatic residues (Tyr-28, Tyr-75, and Tyr-198) on the flat substrate binding surface of PchGH61D are shown on the molecular surface in cyan. The corresponding residues or additional aromatic residues on the surface of other GH61s are colored as follows. Pink, NcrPMO2 (PDB code 4EIR); red, TteGH61E (3EJA); yellow, NcrPMO-3 (4EIS); orange, TauGH61A (3ZUD); green, HjeGH61B (2VTC). The residue numbers are indicated beside the depicted residues. C, superposition of the residues shown in B with the corresponding color, in a stick representation. Tyr-25 in NcrPMO-2 occurs in two conformations in pink.
At the metal binding site, His-149 and Gln-158 are highly conserved in all fungal LPMOs. Their side chains point toward the copper atom, but they are too far away to form coordination interactions (4.7 and 3.9 Å, respectively). His-149 is suitably positioned to provide a hydrogen bond to any ligand binding to one of the two copper coordination positions that are available for substrate binding. In PchGH61D, the His-149 imidazole ring is rotated 180° compared with its counterpart in other structures, probably because it makes a hydrogen bond to one hydroxyl of the glycerol ligand (Figs. 1B and 3A). One imidazole nitrogen is close to the side chain oxygen of Gln-158 (3.4 Å), and if rotated 180°, the distance would be even shorter (3.1 Å). This suggests an interaction between His-149 and Gln-158, although the geometry is far from ideal for a hydrogen bond. Gln-158 in turn interacts with the hydroxyl group of the conserved Tyr-160 that is axially coordinated to the copper atom. It remains to be seen if these residues are conserved because they participate directly in the catalytic mechanism or if their primary role is to maintain the shape and electrostatic properties of the metal binding site.
Structure and sequence comparisons (Fig. 4 and supplemental Fig. S1) show variations in the three loop regions that comprise the putative substrate-binding surfaces in LPMOs. Generally, the LC and LS loop regions are more extended in fungal LPMOs compared with the bacterial LPMOs (Fig. 4A). The L2 loop varies within fungal LPMOs as well; PchGH61D, TteGH61E, and NcrPMO-2 have shorter L2 loops compared with HjeGH61B, TauGH61A, and NcrPMO-3 (Fig. 4A). The only conserved amino acid present in the L2 region is a cysteine, Cys-43 in PchGH61D, that forms a disulfide bond to Cys-163 in the β10 strand. Despite the overall structural diversity shown in Fig. 4A, there are similarities in the exposure of aromatic residues that may impact binding, as highlighted in Fig. 4B. The extended L2 loops in three of the GH61 structures contain tyrosines (Tyr-23 in HjeGH61B; Tyr-24 in TauGH61A; Tyr-20/Tyr-24 in NcrPMO-3) that occupy spatially similar locations on the surface as Tyr-28 in PchGH61D and Tyr-25 in NcrPMO-2 (Fig. 4, B and C). It is noted that the L2 loop in NcrPMO-3 actually contains two tyrosine residues side-by-side at this location. In PchGH61D, weak electron density and high B factors indicate that the Tyr-28 side chain is flexible. Similar flexibility is not seen for the tyrosines in the NcrPMO structures, possibly because they are involved in the crystal packing (15). In TteGH61E, Glu-23 replaces Tyr-28 of PchGH61D. Instead, TteGH61E has an additional exposed tyrosine, Tyr-192, next in sequence to the highly conserved tyrosine in the LC loop, which the other enzymes do not have (Fig. 4B). Tyr-191 and Tyr-192 form a similar substrate-binding motif in TteGH61E as present in CBM1s (20).
P. chrysosporium LPMO Comparison
Nine PchGH61s, including PchGH61D, were chosen for inclusion in the sequence alignment (supplemental Fig. S1) because previous studies had indicated that they may be important for growth on lignocellulosic substrates (58–60). Six of the enzymes contain a C-terminal family 1 CBM (Phchr1|41563, 41650, 31049, 129325, 121193, and 10320), whereas three do not (Phchr1|41123 and 122129 and PchGH61D). Only the predicted catalytic domains were included in the sequence alignment of supplemental Fig. S1. As with LPMO sequences across species, PchGH61s exhibit significant sequence variability. Three PchGH61s exhibit longer L2 loops (Phchr1|129325, 121193, and 10320) similar to HjeGH61B, TauGH61A, and NcrPMO-3, whereas there is no major length variation in the LS or LC loop regions.
With respect to the active site residues, the histidine residues around the copper atom are conserved with the exception of Phchr1|122129, which contains an arginine residue instead of His-76 (PchGH61D numbering). The axial tyrosine residue in PchGH61D, Tyr-160, is conserved with the exception of Phchr1|41123, which contains a gap at this position. As in the known LPMO structures, Gln-158 is completely conserved in all PchGH61s.
In terms of conspicuous residues on the putative PchGH61D binding surface (Fig. 4, B and C), five PchGH61s display aromatic residues in sequence positions similar to Tyr-28, whereas Phchr1|41123, 31049, and 122129 do not. Residues corresponding to Tyr-75 vary considerably within PchGH61s as well and can be aspartate, proline, glycine, alanine, or asparagines. Notably, as discussed above (Fig. 3), Tyr-75 may indirectly affect copper binding, so variation at this position may result in variation in the active sites of PchGH61s (Fig. 3). The third surface-located aromatic residue, Tyr-198, is completely conserved among PchGH61s.
Degradation of PASC and Steam-exploded Spruce by PchGH61D
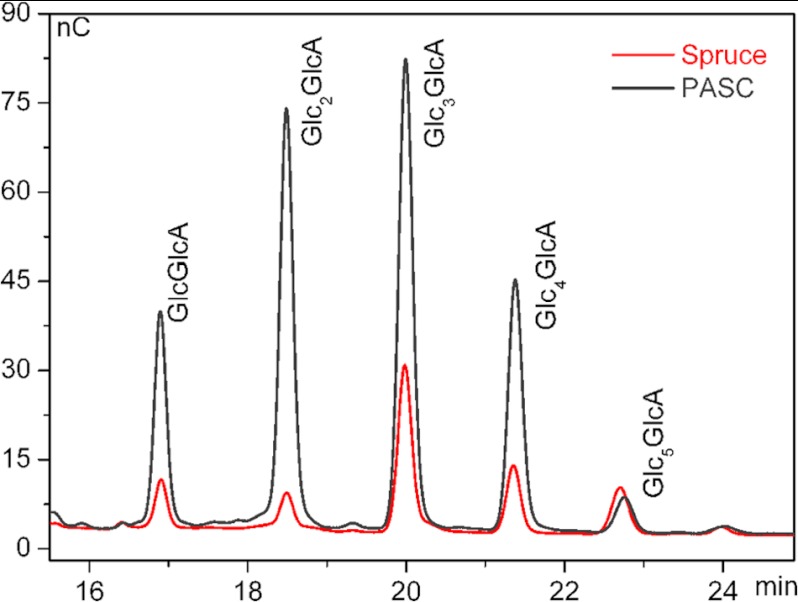
To date, LPMOs, including PchGH61D, have been shown to conduct oxidative cleavage of cellulose. However, the production of oxidized oligomeric products from a heterogeneous biomass substrate, rather than a model cellulose substrate, has not been thoroughly addressed (61). To that end, Fig. 5 demonstrates that PchGH61D produces C1-oxidized cello-oligosaccharides not only from PASC, a model substrate, but also from steam-exploded spruce. The same oligosaccharides are found for both substrates but, as expected, at different amounts. It has previously been claimed (2) that such differences could indicate differences in substrate accessibility. Action on a well ordered hydrophobic surface of cellulose crystal where only every second glycosidic bond would be accessible for the enzyme is likely to yield predominantly even-numbered soluble products, a tendency also observed for the spruce incubation (Fig. 5). Such a periodicity would be less visible for less crystalline substrates where chains are accessible from “any” side, as for PASC.
FIGURE 5.
High performance anion exchange chromatography; chromatogram showing soluble aldonic acids (degree of polymerization 2–6) obtained upon incubation of 0.1% (w/v) PASC or 0.5% (w/v) steam-exploded spruce with 34 μg/ml PchGH61D in 25 μm sodium acetate, pH 5.3, 1.5 mm ascorbic acid for 20 h at 50 °C. Note the difference between the relative amounts of products released from the two substrates. Control reactions without enzyme yielded no detectable aldonic acids (data not shown).
PchGH61D-Cellulose Interactions Studied with MD Simulation
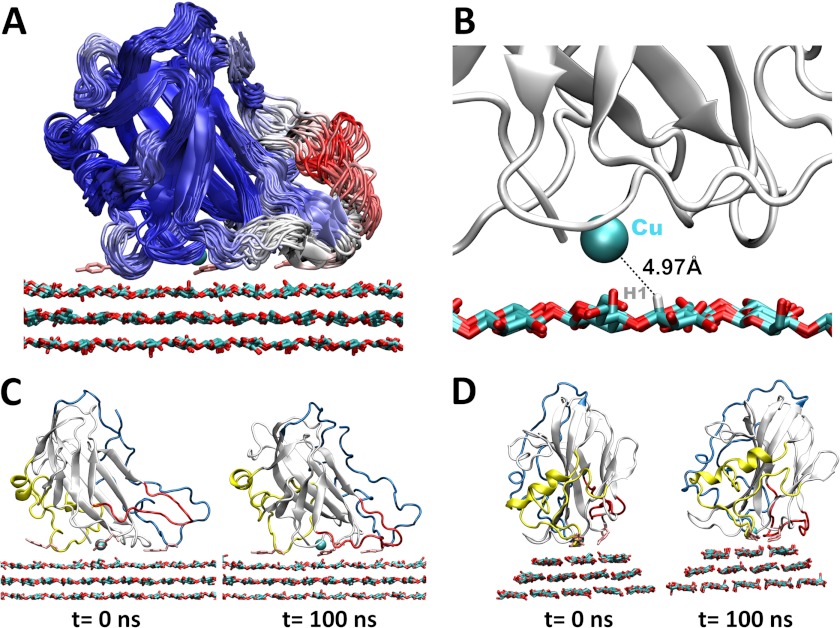
For the MD simulations, PchGH61D was placed on the hydrophobic face of cellulose Iβ to examine the enzyme-substrate interactions, to identify residues that are potentially important for binding, and to examine any conformational changes that occur upon interaction with cellulose. The initial system geometry is shown in supplemental Fig. S3. It has not been determined if LPMOs bind to and perform oxidation on the hydrophobic face of cellulose or chitin microfibrils. However, it has been hypothesized (6, 9) that the aromatic and polar residues on the flat surfaces exhibit structural similarities to Type A CBMs, several of which are known to bind to the hydrophobic face of cellulose I (62, 63). Additionally, the orientation of LPMOs relative to the surface of cellulose is also currently unknown (15). Given the similarity of aromatic residues lining the putative binding face, we aligned PchGH61D in an orientation similar to the orientation of the family 1 CBM from H. jecorina (21, 64). An MD simulation was conducted for 100 ns (supplemental Movie S1). Supplemental Fig. S4 shows the RMSD of the protein relative to the crystal structure, the root mean square fluctuations (RMSF) per residue, and for comparison, the B factors of the PchGH61D crystal structure. Supplemental Fig. S5 shows schematic representations of PchGH61D colored by B factor and RMSF as well for further comparison. The RMSD values indicate that after the initial equilibration of cellulose, conformational changes are minor, and the RMSF results indicate that the primary fluctuations arose almost completely from the LC and LS loops. The B factor results shown in supplemental Fig. S5, although not strictly comparable because the chemical environments of the crystal and simulated PchGH61D enzymes are different, suggest that the LC and LS loops are the most flexible in both cases.
Fig. 6A shows a cluster diagram of PchGH61D on the cellulose surface with the protein backbone colored by RMSF, which shows that there is significant flexibility in the LC and the LS loops. Fig. 6B shows the distance from the active site copper to the hydrogen atom on the C1 carbon, which fluctuates near 5.0 Å during the MD simulation. Although the binding pose of molecular oxygen to copper is not yet known definitively for LPMO enzymes, a distance of 5 Å could most likely bring the superoxo intermediate that is hypothesized to be generated on the copper (13) sufficiently close to abstract a hydrogen atom or conduct nucleophilic attack of the C1 carbon. Fig. 6, C and D, shows the initial and final states of PchGH61D on the cellulose surface from two views. PchGH61D is apparently a C1 oxidizer, but further studies on the catalytic mechanism and on LPMOs with other oxidation preferences are needed to understand the specificity for C1, C4, or possible C6 oxidation.
FIGURE 6.
Simulation results for PchGH61D on the hydrophobic surface of cellulose. A, cluster view of PchGH61D with snapshots taken every 5 ns, colored by RMSF from blue (low) to red (high). The tyrosine side chains (Tyr-28, Tyr-75, and Tyr-198) are shown in pink stick format in the conformation obtained after 100 ns. B, the copper (shown as a cyan sphere) fluctuates at ∼5 Å from the hydrogen atom on the C1 carbon during the MD simulation. C, side view of PchGH61D on the cellulose surface at t = 0 ns and t = 100 ns. The loops are colored as in Fig. 1. D, back view of PchGH61D on the cellulose surface at t = 0 ns and t = 100 ns.
During the simulation, the LC loop becomes quite mobile at ∼30 ns (supplemental Fig. S4A and Fig. 6A). This conformational flexibility is related to a conformational change in the side chains of Phe-112 (in another relatively flexible region) and Phe-204 relative to one another during the MD simulation. This observation agrees with the structural data presented above, showing that part of the LC loop exhibits significantly higher B factors than the rest of the protein (supplemental Figs. S4C and S5). Additionally, concomitantly with the Phe-112 conformational change, the LS loop undergoes a significant translational motion toward the cellulose surface, where it forms hydrogen bonds to an edge cellodextrin chain via both backbone and side chain atoms.
From the MD simulation, we examined the role of the three tyrosine residues (Tyr-28, Tyr-75, and Tyr-198) that may be important for binding, we looked for other conspicuous residues in the putative binding face, and we studied the active site position over cellulose. Table 2 lists the average interaction energy of relevant protein residues with the cellulose surface. The energetic cut-off for examining interactions was 3 kcal/mol on average over the 100-ns MD simulation. As shown, many of the residues that form the putative cellulose binding face in the crystal structure are present in the interaction energy analysis. However, some residues that are not initially bound to the cellulose also appear. Notably, the LS loop that contains Asn-114, Gly-115, and Gln-116 forms long lived contacts with the cellulose surface.
TABLE 2.
Residues that interact with cellulose in the MD simulations
| Residue | Average interaction energy |
|---|---|
| kcal/mol | |
| His-1 | −5.42 |
| Tyr-28 | −10.86 |
| Ser-29 | −6.79 |
| Tyr-75 | −10.17 |
| Asn-114 | −3.81 |
| Gly-115 | −3.21 |
| Gln-116 | −7.40 |
| His-149 | −4.56 |
| Val-150 | −5.23 |
| Tyr-198 | −9.50 |
| Asn-199 | −5.03 |
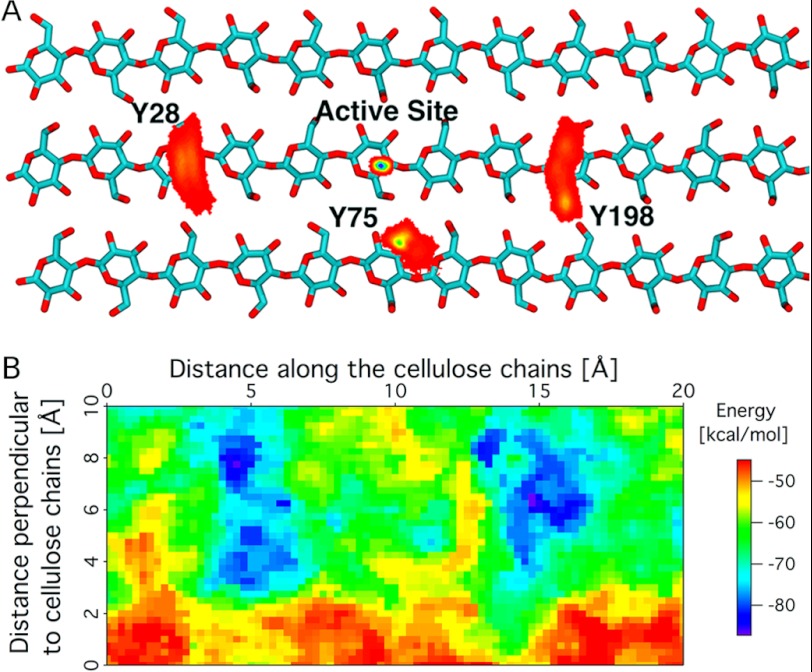
Fig. 7A shows the positions of the three tyrosine residues and the PchGH61D active site over the cellulose surface. Upon docking of PchGH61D on the cellulose surface, Tyr-28 and Tyr-198 align over the same chain, and their position is quite stable during the simulation. Tyr-198 hydrogen-bonds with the adjacent chain as well during the simulation. Additionally, Tyr-75 is bound to the adjacent cellulose chain on the edge of the crystal upon docking, and it retains this conformation over 100 ns. The active site position (defined as the center of mass of His-1, His-76, Tyr-160, and the copper atom) remains in the same position over the cellulose surface during the MD simulation directly above a glycosidic bond in the middle chain. The results obtained here suggest that, overall, the active site is stable near the proposed site of attack.
FIGURE 7.
A, histogram of the tyrosine residues (Tyr-28, Tyr-75, and Tyr-198) and the PchGH61D active site positions on the cellulose surface. The bottom two layers of cellulose are not shown, and the cellulose chains are truncated, both for visual clarity. The color code denotes the position on a 0.1 × 0.1-Å grid on the cellulose surface, with red being low density and blue being the highest density. B, PES of PchGH61D on cellulose. The x direction is along the chains of cellulose, and the y direction is perpendicular to the cellulose chains. Energy minima are found over the putative site of attack, with ∼10-Å separation (i.e. a distance corresponding to a cellobiose unit).
Also, a PES was constructed for the PchGH61D-cellulose interaction, as described in the supplemental material. The PES was constructed with explicit solvation using methods similar to those used in previous work conducted on a family 1 CBM (19, 54). The PES in Fig. 7B suggests that the enzyme is enthalpically stable above glycosidic linkages separated by ∼10 Å, where the enzyme can abstract accessible hydrogen atoms. The stabilization every 10 Å, which is approximately the length of a cellobiose unit, is similar to that observed for a family 1 CBM (19, 54).
We note that this study is limited to MD simulations of the PchGH61D over the putative site of attack, which does not account for diffusion on the surface or the chemical reaction. In a previous study to examine the diffusion and orientational preferences of a family 1 CBM on cellulose, diffusion and orientation on cellulose required 43 μs to reach convergence (21). Because LPMOs are substantially larger, understanding their orientational preferences and studying their diffusion along the surface would probably require simulation times on the order of hundreds of μs without the use of enhanced sampling methods. Thus, the questions of PchGH61D diffusion and orientation on the cellulose surface are outside the scope of the present study. Additionally, we note that LPMOs generally present a challenge to typical protein simulations because of the active site. The approach used here wherein an ad hoc potential was developed may be generalized to other copper monooxygenases, but we stress that the potential developed here is only appropriate for PchGH61D and only then for examining questions for which copper ion diffusion out of the active site is not relevant. Quantum mechanics/molecular mechanics approaches will be necessary to study the reaction mechanism and questions related to binding of other metals in the active site.
CONCLUSIONS
Here, we have solved the first LPMO structure from a basidiomycete fungus, the wood-degrading model organism, P. chrysosporium. This fungus contains up to nine LPMOs known to be expressed when grown on lignocellulosic substrates (12, 59). Because extensive work has been done on other model glycoside hydrolase (65–68) and dehydrogenase enzymes (69, 70) from P. chrysosporium, the P. chrysosporium LPMOs offer an excellent model system to understand the need for multiple oxidative activities to degrade biomass. The PchGH61D structure revealed potentially important residues around the active site and putative binding surface that may impart differences in LPMO specificity and activity. With simulation, we demonstrated that several conformational changes occur upon PchGH61D binding to cellulose, which suggest roles of conserved loops in substrate binding. Going forward in the burgeoning field of LPMO biochemistry, it is likely that a combination of structural, biophysical, and computational studies, such as that presented here, will be necessary to fully understand the diversity of LPMO structures as well as its functional consequences.
Acknowledgments
Computer time for this research was provided by the National Renewable Energy Laboratory Computational Sciences Center, supported by the Department of Energy Office of Energy Efficiency and Renewable Energy under Contract DE-AC36-08GO28308, and by the National Institute of Computational Science Kraken cluster under National Science Foundation XSEDE Grant MCB090159.
This work was supported in part by the Faculty for Natural Resources and Agriculture at the Swedish University of Agricultural Sciences through the research program MicroDrivE and by the Japan Society for the Promotion of Science (JSPS) through a fellowship (to T. I.).

This article contains supplemental Table S1, Figs. S1–S5, and Movie S1.
The atomic coordinates and structure factors (code 4B5Q) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GH
- glycoside hydrolase
- LPMO
- lytic polysaccharide monooxygenase
- PDB
- Protein Data Bank
- CBM
- carbohydrate-binding module
- RMSD
- root mean square deviation
- RMSF
- root mean square fluctuations
- PASC
- phosphoric acid-swollen cellulose
- MD
- molecular dynamics
- PES
- potential energy surface.
REFERENCES
- 1. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy). An expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vaaje-Kolstad G., Westereng B., Horn S. J., Liu Z., Zhai H., Sørlie M., Eijsink V. G. (2010) An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222 [DOI] [PubMed] [Google Scholar]
- 3. Forsberg Z., Vaaje-Kolstad G., Westereng B., Bunæs A. C., Stenstrøm Y., MacKenzie A., Sørlie M., Horn S. J., Eijsink V. G. (2011) Cleavage of cellulose by a CBM33 protein. Protein Sci. 20, 1479–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Phillips C. M., Beeson W. T., Cate J. H., Marletta M. A. (2011) Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 6, 1399–1406 [DOI] [PubMed] [Google Scholar]
- 5. Quinlan R. J., Sweeney M. D., Lo Leggio L., Otten H., Poulsen J.-C., Johansen K. S., Krogh K. B., Jørgensen C. I., Tovborg M., Anthonsen A., Tryfona T., Walter C. P., Dupree P., Xu F., Davies G. J., Walton P. H. (2011) Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. 108, 15079–15084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harris P. V., Welner D., McFarland K. C., Re E., Navarro Poulsen J. C., Brown K., Salbo R., Ding H., Vlasenko E., Merino S., Xu F., Cherry J., Larsen S., Lo Leggio L. (2010) Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61. Structure and function of a large, enigmatic family. Biochemistry 49, 3305–3316 [DOI] [PubMed] [Google Scholar]
- 7. Vaaje-Kolstad G., Horn S. J., van Aalten D. M., Synstad B., Eijsink V. G. (2005) The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 280, 28492–28497 [DOI] [PubMed] [Google Scholar]
- 8. Horn S. J., Vaaje-Kolstad G., Westereng B., Eijsink V. G. (2012) Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 5, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karkehabadi S., Hansson H., Kim S., Piens K., Mitchinson C., Sandgren M. (2008) The structure of a glycoside hydrolase family 61 member, Cel61B from the Hypocrea jecorina. J. Mol. Biol. 383, 144–154 [DOI] [PubMed] [Google Scholar]
- 10. Beeson W. T., Phillips C. M., Cate J. H., Marletta M. A. (2012) Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J. Am. Chem. Soc. 134, 890–892 [DOI] [PubMed] [Google Scholar]
- 11. Langston J. A., Shaghasi T., Abbate E., Xu F., Vlasenko E., Sweeney M. D. (2011) Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl. Environ. Microbiol. 77, 7007–7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Westereng B., Ishida T., Vaaje-Kolstad G., Wu M., Eijsink V. G., Igarashi K., Samejima M., Ståhlberg J., Horn S. J., Sandgren M. (2011) The putative endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative ezyme that cleaves cellulose. PLoS ONE 6, e27807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aachmann F. L., Sørlie M., Skjåk-Bræk G., Eijsink V. G., Vaaje-Kolstad G. (2012) NMR structure of a lytic polysaccharide monooxygenase provides insight into copper binding, protein dynamics, and substrate interactions. Proc. Natl. Acad. Sci. U.S.A. 109, 18779–18784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dimarogona M., Topakas E., Olsson L., Christakopoulos P. (2012) Lignin boosts the cellulase performance of a GH61–61 enzyme from Sporotrichum thermophile. Bioresour. Technol. 110, 480–487 [DOI] [PubMed] [Google Scholar]
- 15. Li X., Beeson W. T., 4th, Phillips C. M., Marletta M. A., Cate J. H. (2012) Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure 20, 1051–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vaaje-Kolstad G., Houston D. R., Riemen A. H., Eijsink V. G., van Aalten D. M. (2005) Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J. Biol. Chem. 280, 11313–11319 [DOI] [PubMed] [Google Scholar]
- 17. Wong E., Vaaje-Kolstad G., Ghosh A., Hurtado-Guerrero R., Konarev P. V., Ibrahim A. F., Svergun D. I., Eijsink V. G., Chatterjee N. S., van Aalten D. M. (2012) The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces. PLoS Pathog. 8, e1002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vaaje-Kolstad G., Bøhle L. A., Gåseidnes S., Dalhus B., Bjørås M., Mathiesen G., Eijsink V. G. (2012) Characterization of the chitinolytic machinery of Enterococcus faecalis V583 and high-resolution structure of its oxidative CBM33 enzyme. J. Mol. Biol. 416, 239–254 [DOI] [PubMed] [Google Scholar]
- 19. Beckham G. T., Matthews J. F., Bomble Y. J., Bu L., Adney W. S., Himmel M. E., Nimlos M. R., Crowley M. F. (2010) Identification of amino acids responsible for processivity in a family 1 carbohydrate-binding module from a fungal cellulase. J. Phys. Chem. B 114, 1447–1453 [DOI] [PubMed] [Google Scholar]
- 20. Kraulis J., Clore G. M., Nilges M., Jones T. A., Pettersson G., Knowles J., Gronenborn A. M. (1989) Determinationn of the 3-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei-A study using nuclear magnetic-resonance and hybrid distance geometry dynamical simulated annealing. Biochemistry 28, 7241–7257 [DOI] [PubMed] [Google Scholar]
- 21. Nimlos M. R., Beckham G. T., Matthews J. F., Bu L., Himmel M. E., Crowley M. F. (2012) Binding preferences, surface attachment, diffusivity, and orientation of a family 1 carbohydrate-binding module on cellulose. J. Biol. Chem. 287, 20603–20612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beckham G. T., Crowley M. F. (2011) Examination of the α-chitin structure and decrystallization thermodynamics at the nanoscale. J. Phys. Chem. B 115, 4516–4522 [DOI] [PubMed] [Google Scholar]
- 23. Beckham G. T., Matthews J. F., Peters B., Bomble Y. J., Himmel M. E., Crowley M. F. (2011) Molecular-level origins of biomass recalcitrance. Decrystallization free energies for four common cellulose polymorphs. J. Phys. Chem. B 115, 4118–4127 [DOI] [PubMed] [Google Scholar]
- 24. Kleywegt G. J., Zou J. Y., Divne C., Davies G. J., Sinning I., Stâhlberg J., Reinikainen T., Srisodsuk M., Teeri T. T., Jones T. A. (1997) The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 Å resolution, and a comparison with related enzymes. J. Mol. Biol. 272, 383–397 [DOI] [PubMed] [Google Scholar]
- 25. Bey M., Zhou S., Poidevin L., Henrissat B., Coutinho P. M., Berrin J. G., Sigoillot J. C. (2012) Comparison of two lytic polysaccharide monooxygenases (GH61) from Podospora anserina reveals differences upon cello-oligosaccharides oxidation. Appl. Environ. Microbiol. 287, 3147–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chundawat S. P., Beckham G. T., Himmel M. E., Dale B. E. (2011) Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu. Rev. Chem. Biomol. Eng. 2, 121–145 [DOI] [PubMed] [Google Scholar]
- 27. Himmel M. E., Ding S. Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., Foust T. D. (2007) Biomass recalcitrance. Engineering plants and enzymes for biofuels production. Science 315, 804–807 [DOI] [PubMed] [Google Scholar]
- 28. Eastwood D. C., Floudas D., Binder M., Majcherczyk A., Schneider P., Aerts A., Asiegbu F. O., Baker S. E., Barry K., Bendiksby M., Blumentritt M., Coutinho P. M., Cullen D., de Vries R. P., Gathman A., Goodell B., Henrissat B., Ihrmark K., Kauserud H., Kohler A., LaButti K., Lapidus A., Lavin J. L., Lee Y. H., Lindquist E., Lilly W., Lucas S., Morin E., Murat C., Oguiza J. A., Park J., Pisabarro A. G., Riley R., Rosling A., Salamov A., Schmidt O., Schmutz J., Skrede I., Stenlid J., Wiebenga A., Xie X., Kües U., Hibbett D. S., Hoffmeister D., Högberg N., Martin F., Grigoriev I. V., Watkinson S. C. (2011) The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 333, 762–765 [DOI] [PubMed] [Google Scholar]
- 29. Fernandez-Fueyo E., Ruiz-Dueñas F. J., Ferreira P., Floudas D., Hibbett D. S., Canessa P., Larrondo L. F., James T. Y., Seelenfreund D., Lobos S., Polanco R., Tello M., Honda Y., Watanabe T., Watanabe T., Ryu J. S., San R. J., Kubicek C. P., Schmoll M., Gaskell J., Hammel K. E., St John F. J., Vanden Wymelenberg A., Sabat G., Splinter BonDurant. S., Syed K., Yadav J. S., Doddapaneni H., Subramanian V., Lavín J. L., Oguiza J. A., Perez G., Pisabarro A. G., Ramirez L., Santoyo F., Master E., Coutinho P. M., Henrissat B., Lombard V., Magnuson J. K., Kües U., Hori C., Igarashi K., Samejima M., Held B. W., Barry K. W., LaButti K. M., Lapidus A., Lindquist E. A., Lucas S. M., Riley R., Salamov A. A., Hoffmeister D., Schwenk D., Hadar Y., Yarden O., de Vries R. P., Wiebenga A., Stenlid J., Eastwood D., Grigoriev I. V., Berka R. M., Blanchette R. A., Kersten P., Martinez A. T., Vicuna R., Cullen D. (2012) Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc. Natl. Acad. Sci. U.S.A. 109, 5458–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Olson A., Aerts A., Asiegbu F., Belbahri L., Bouzid O., Broberg A., Canbäck B., Coutinho P. M., Cullen D., Dalman K., Deflorio G., van Diepen L. T., Dunand C., Duplessis S., Durling M., Gonthier P., Grimwood J., Fossdal C. G., Hansson D., Henrissat B., Hietala A., Himmelstrand K., Hoffmeister D., Högberg N., James T. Y., Karlsson M., Kohler A., Kües U., Lee Y. H., Lin Y. C., Lind M., Lindquist E., Lombard V., Lucas S., Lundén K., Morin E., Murat C., Park J., Raffaello T., Rouzé P., Salamov A., Schmutz J., Solheim H., Ståhlberg J., Vélëz H., de Vries R. P., Wiebenga A., Woodward S., Yakovlev I., Garbelotto M., Martin F., Grigoriev I. V., Stenlid J. (2012) Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol. 194, 1001–1013 [DOI] [PubMed] [Google Scholar]
- 31. Martinez D., Larrondo L. F., Putnam N., Gelpke M. D., Huang K., Chapman J., Helfenbein K. G., Ramaiya P., Detter J. C., Larimer F., Coutinho P. M., Henrissat B., Berka R., Cullen D., Rokhsar D. (2004) Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22, 695–700 [DOI] [PubMed] [Google Scholar]
- 32. Kabsch W. (1993) Automatic Processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 33. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 34. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL Workspace. A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 36. Murshudov G., Vagin A., Dodson E. (1996) Application of maximum likelihood refinement. in The Refinement of Protein Structures: Proceedings of the Daresbury Study Weekend, Science and Engineering Research Council, Daresbury, UK [Google Scholar]
- 37. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pannu N. S., Read R. J. (1996) Improved structure refinement through maximum likelihood. Acta Crystallogr. A 52, 659–668 [Google Scholar]
- 39. Brünger A. T. (1992) Free R value. A novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 40. Evrard G. X., Langer G. G., Perrakis A., Lamzin V. S. (2007) Assessment of automatic ligand building in ARP/wARP. Acta Crystallogr. D Biol. Crystallogr. 63, 108–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr., Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. (1977) The Protein Data Bank. A computer-based archival file for macromolecular structures. J. Mol. Biol. 112, 535–542 [DOI] [PubMed] [Google Scholar]
- 42. Holm L., Rosenström P. (2010) Dali server. Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kleywegt G. J. (1996) Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D Biol. Crystallogr. 52, 842–857 [DOI] [PubMed] [Google Scholar]
- 44. Kleywegt G. J., Jones T. A. (1997) Detecting folding motifs and similarities in protein structures. Methods Enzymol. 277, 525–545 [DOI] [PubMed] [Google Scholar]
- 45. Vanden Wymelenberg A., Minges P., Sabat G., Martinez D., Aerts A., Salamov A., Grigoriev I., Shapiro H., Putnam N., Belinky P., Dosoretz C., Gaskell J., Kersten P., Cullen D. (2006) Computational analysis of the Phanerochaete chrysosporium v2.0 genome database and mass spectrometry identification of peptides in ligninolytic cultures reveal complex mixtures of secreted proteins. Fungal Genet. Biol. 43, 343–356 [DOI] [PubMed] [Google Scholar]
- 46. Katoh K., Misawa K., Kuma K., Miyata T. (2002) MAFFT. A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heinig M., Frishman D. (2004) STRIDE. A web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 32, W500–W502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript. Analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 49. Wood T. M. (1971) The cellulase of Fusarium solani. Purification and specificity of the β-(1–4)-glucanase and the β-d-glucosidase components. Biochem. J. 121, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Horn S. J., Nguyen Q. D., Westereng B., Nilsen P. J., Eijsink V. G. (2011) Screening of steam explosion conditions for glucose production from non-impregnated wheat straw. Biomass Bioenergy 35, 4879–4886 [Google Scholar]
- 51. Westereng B., Agger J. W., Horn S. J., Vaaje-Kolstad G., Aachmann F. L., Stenstrøm Y. H., Eijsink V. G. (2013) Efficient separation of oxidized cello-oligosaccharides generated by cellulose degrading lytic polysaccharide monooxygenases. J. Chromatogr. A 1271, 144–152 [DOI] [PubMed] [Google Scholar]
- 52. Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) CHARMM. The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Matthews J. F., Beckham G. T., Bergenstrahle-Wohlert M., Brady J. W., Himmel M. E., Crowley M. F. (2012) Comparison of cellulose I β simulations with three carbohydrate force fields. J. Chem. Theory Comput. 8, 735–748 [DOI] [PubMed] [Google Scholar]
- 54. Bu L., Beckham G. T., Crowley M. F., Chang C. H., Matthews J. F., Bomble Y. J., Adney W. S., Himmel M. E., Nimlos M. R. (2009) The energy landscape for the interaction of the family 1 carbohydrate-binding module and the cellulose surface is altered by hydrolyzed glycosidic bonds. J. Phys. Chem. B 113, 10994–11002 [DOI] [PubMed] [Google Scholar]
- 55. Matthews B. W. (1968) Solvent content of protein crystals. J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 56. Mccracken J., Peisach J., Dooley D. M. (1987) Cu(II) coordination chemistry of amine oxidases. Pulsed EPR studies of histidine imidazole, water, and exogenous ligand coordination. J. Am. Chem. Soc. 109, 4064–4072 [Google Scholar]
- 57. Klinman J. P. (1996) Mechanisms whereby mononuclear copper proteins functionalize organic substrates. Chem. Rev. 96, 2541–2562 [DOI] [PubMed] [Google Scholar]
- 58. Vanden Wymelenberg A., Gaskell J., Mozuch M., Kersten P., Sabat G., Martinez D., Cullen D. (2009) Transcriptome and secretome analyses of Phanerochaete chrysosporium reveal complex patterns of gene Expression. Appl. Environ. Microbiol. 75, 4058–4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hori C., Igarashi K., Katayama A., Samejima M. (2011) Effects of xylan and starch on secretome of the basidiomycete Phanerochaete chrysosporium grown on cellulose. FEMS Microbiol. Lett. 321, 14–23 [DOI] [PubMed] [Google Scholar]
- 60. Adav S. S., Ravindran A., Sze S. K. (2012) Quantitative proteomic analysis of lignocellulolytic enzymes by Phanerochaete chrysosporium on different lignocellulosic biomass. J. Proteomics 75, 1493–1504 [DOI] [PubMed] [Google Scholar]
- 61. Cannella D., Hsieh C.-W., Felby C., Jørgensen H. (2012) Production and effect of aldonic acids during enzymatic hydrolysis of lignocellulose at high dry matter content. Biotechnol. Biofuels 5, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lehtiö J., Sugiyama J., Gustavsson M., Fransson L., Linder M., Teeri T. T. (2003) The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc. Natl. Acad. Sci. U.S.A. 100, 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Carbohydrate-binding modules. Fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Divne C., Ståhlberg J., Teeri T. T., Jones T. A. (1998) High-resolution crystal structures reveal how a cellulose chain is bound in the 50 angstrom long tunnel of cellobiohydrolase I from Trichoderma reesei. J. Mol. Biol. 275, 309–325 [DOI] [PubMed] [Google Scholar]
- 65. Tsukada T., Igarashi K., Yoshida M., Samejima M. (2006) Molecular cloning and characterization of two intracellular β-glucosidases belonging to glycoside hydrolase family 1 from the basidiomycete Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 73, 807–814 [DOI] [PubMed] [Google Scholar]
- 66. Kawai R., Igarashi K., Samejima M. (2006) Gene cloning and heterologous expression of glycoside hydrolase family 55 β-1,3-glucanase from the basidiomycete Phanerochaete chrysosporium. Biotechnol. Lett. 28, 365–371 [DOI] [PubMed] [Google Scholar]
- 67. Igarashi K., Ishida T., Hori C., Samejima M. (2008) Characterization of an endoglucanase belonging to a new subfamily of glycoside hydrolase family 45 of the basidiomycete Phanerochaete chrysosporium. Appl. Environ. Microbiol. 74, 5628–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Henriksson G., Nutt A., Henriksson H., Pettersson B., Ståhlberg J., Johansson G., Pettersson G. (1999) Endoglucanase 28 (cel12A), a new Phanerochaete chrysosporium cellulase. Eur. J. Biochem. 259, 88–95 [DOI] [PubMed] [Google Scholar]
- 69. Uzcategui E., Ruiz A., Montesino R., Johansson G., Pettersson G. (1991) The 1,4-β-d-glucan cellobiohydrolases from Phanerochaete chrysosporium. 1. A system of synergistically acting enzymes gomologous to Trichoderma reesei. J. Biotechnol. 19, 271–285 [DOI] [PubMed] [Google Scholar]
- 70. Habu N., Igarashi K., Samejima M., Pettersson B., Eriksson K. E. (1997) Enhanced production of cellobiose dehydrogenase in cultures of Phanerochaete chrysosporium supplemented with bovine calf serum. Biotechnol. Appl. Biochem. 26, 97–102 [PubMed] [Google Scholar]
- 71. Engh R. A., Huber R. (1991) Accurate bond and angle parameters for x-ray protein-structure refinement. Acta Crystallogr. A 47, 392–400 [Google Scholar]
- 72. Kleywegt G. J. (1997) Validation of protein models from C-α coordinates alone. J. Mol. Biol. 273, 371–376 [DOI] [PubMed] [Google Scholar]
- 73. Kleywegt G. J., Jones T. A. (1996) Phi/psi-chology. Ramachandran revisited. Structure 4, 1395–1400 [DOI] [PubMed] [Google Scholar]