Background: Rta is a transcription factor encoded by Epstein-Barr virus (EBV) that activates the transcription of vial lytic genes and promotes lytic development.
Results: RNF4 enhances the ubiquitination of Rta, thus decreasing lytic replication and virions production.
Conclusion: RNF4 targets SUMO-2-conjugated Rta and promotes Rta ubiquitination.
Significance: RNF4 is a SUMO-targeted ubiquitin ligase of Rta that modulates the amount of Rta during EBV lytic progression.
Keywords: DNA Viruses, E3 Ubiquitin Ligase, Protein Degradation, Sumoylation, Ubiquitination, Epstein-Barr Virus, Rta, RNF4
Abstract
Epstein-Barr virus (EBV) encodes a transcription factor, Rta, which is required to activate the transcription of EBV lytic genes. This study demonstrates that treating P3HR1 cells with a proteasome inhibitor, MG132, causes the accumulation of SUMO-Rta and promotes the expression of EA-D. GST pulldown and coimmunoprecipitation studies reveal that RNF4, a RING-domain-containing ubiquitin E3 ligase, interacts with Rta. RNF4 also targets SUMO-2-conjugated Rta and promotes its ubiquitination in vitro. Additionally, SUMO interaction motifs in RNF4 are important to the ubiquitination of Rta because the RNF4 mutant with a mutation at the motifs eliminates ubiquitination. The mutation of four lysine residues on Rta that abrogated SUMO-3 conjugation to Rta also decreases the enhancement of the ubiquitination of Rta by RNF4. This finding demonstrates that RNF4 is a SUMO-targeted ubiquitin E3 ligase of Rta. Finally, knockdown of RNF4 enhances the expression of Rta and EA-D, subsequently promoting EBV lytic replication and virions production. Results of this study significantly contribute to efforts to elucidate a SUMO-targeted ubiquitin E3 ligase that regulates Rta ubiquitination to influence the lytic development of EBV.
Introduction
As a reversible posttranslational modification process, ubiquitination influences diverse biological functions (1, 2). Ubiquitin (Ub)3 is covalently attached to protein substrates by a cascade of enzymatic reactions mediated by activating enzymes (E1), conjugating enzymes (E2), and ligases (E3). As is commonly known, ubiquitination leads to degradation of a target protein by 26 S proteasome; the degradation is associated with a polyUb chain that is linked through Lys-48 of Ub (3, 4). After linking to a target protein, the process of polyubiquitination is reversed by deubiquitinases (5). Similar to ubiquitin, a small ubiquitin-like modifier (SUMO) is covalently attached to lysine residues of target proteins via an isopeptide bond by a mechanism resembling that of ubiquitination (6). Eukaryotes express three SUMO isoforms (SUMO-1, SUMO-2, and SUMO-3) of which SUMO-2 and SUMO-3, which have internal consensus modification sites, allow the formation of polySUMO chains on target proteins (7, 8). SUMO-1, rather forming a SUMO chain like SUMO-2 and SUMO-3, modifies a protein by linking a single molecule to lysine residues or serves as a terminator of a SUMO chain (8, 9). Sumoylation often affects protein functions, including transcription, subcellular localization, DNA repair, protein-protein interaction, and protein stability (10–13). In addition to covalent SUMO modification, proteins also interact noncovalently with SUMO via a region called SUMO-interacting motifs (SIMs) (14). SIM is critical to protein sumoylation and mediating interaction with other SUMO-modified proteins (15–19).
Schimmel et al. (20) found that treating cells with a 26 S proteasome inhibitor, MG132, accumulates the proteins conjugated by SUMO-2 but not those conjugated by SUMO-1, suggesting that ubiquitination depends on sumoylation by SUMO-2 (11, 21, 22), and polySUMO chains formed by SUMO-2 serves as an ubiquitination signal. The finding leads to the discovery of a family of SUMO-targeted ubiquitin ligases (STUbLs) that selectively ubiquitinate sumoylated proteins via SIMs (16, 21, 23–25). These STUbLs are important to cellular functions because the dysfunction of the gene in yeast causes the accumulation of SUMO-conjugated proteins and genome instability (23). Meanwhile, Ring-finger protein 4 (RNF4) is a human protein of the STUbL family containing SIM and the RING domain (16, 26) that promotes ubiquitination of SUMO-2-conjugated PML (15, 27). RNF4 also promotes the ubiquitination of kinetochore protein CENP-I, hypoxia-inducible factor 2α (HIF2α), and Sp1 by the same mechanism (28–30). A structural study indicated that dimeric RNF4 RING domains facilitate ubiquitin transfer by binding to the E2-ubiquitin thioester, thus promoting efficient transfer of ubiquitin to its substrates (31). Furthermore, RNF4 interacts with DNA glycosylase and apurinic/apyrimidinic site endonuclease to active demethylation (32) and is important to DNA damage response and promotes DNA double-strand break repair (33–35). Additionally, Tax oncoprotein from HTLV-1 is targeted by RNF4 to relocalize from the nucleus to the cytoplasm (36).
Although normally maintained under latent conditions in B lymphocytes, Epstein-Barr virus (EBV) must enter a lytic cycle to produce infectious virions. During reactivation from latency, EBV expresses two transcription factors, Rta and Zta, that are encoded by BRLF1 and BZLF1, respectively, to activate the transcription of lytic genes (37–43). Posttranslational modifications of Rta, including phosphorylation and sumoylation, have been shown to be critical to regulate Rta functions (44–50). Our previous studies demonstrated that Rta interacts with SUMO E2 and E3 ligases, including PIAS1, PIASxα, and PIASxβ, and is conjugated to SUMO-1 (44–46). Additionally, LF2 enhances Rta conjugation to SUMO-2/3 (47, 48), but the SUMO modification of Rta does not involve LF2-mediated repression of the functions of Rta (49). This study demonstrates that Rta is ubiquitinated via the interaction with RNF4. Moreover, RNF4 targets SUMO-2-conjugated Rta to enhance Rta ubiquitination, thus decreasing the stability of Rta and affecting EBV lytic progression.
EXPERIMENTAL PROCEDURES
Cell Lines and EBV Lytic Induction
P3HR1 cells were cultured in RPMI 1640 medium. 293T cells were cultured in Dulbecco's modified Eagle's medium. The media were supplemented with 10% fetal calf serum. To induce the EBV lytic cycle, P3HR1 cells were treated with 3 ng/ml 12-O-tetradecanoylphobol-13-acetate (TPA) and 3 mm sodium butyrate for 24–48 h (51, 52).
Plasmids
Plasmid pGEX-4T1, which expresses GST, was purchased from Amersham Biosciences. Plasmids pHis-UbcH5a, pGEX-RNF4, and pEGFP-RNF4 encode His-UbcH5a, RNF4, and GFP-RNF4, respectively.4 Plasmids pBMLF1, pCMV-R, pFLAG-Rta, and pET-Rta were described elsewhere (41, 44, 46). Plasmid pFLAG-Ub, which expressed FLAG-tagged ubiquitin, was constructed by inserting a PCR-amplified DNA fragment that encoded ubiquitin into the EcoRI-HindIII sites in pCMV-Tag2B (Stratagene). Plasmid pT-E1E2S2 contains SUMO E1-, SUMO E2-, and SUMO-2-coding sequences cloned downstream of a T7 promoter (53). Plasmids that express FLAG-RNF4, an RNF4 RING finger mutant that contains two point mutations, C136S and C139S (FLAG-RNF4-CS1), and RNF4, in which all the SIMs were mutated (FLAG-RNF4-mtSIM), were provided by Ronald T. Hay (15, 54). Plasmid pEGFP-RNF4-CS1 was constructed by inserting an RNF4-CS1 fragment amplified from FLAG-RNF4-CS1 into XhoI-BamHI sites in pEGFP-C1 (Clontech). Plasmids pCR-SUMO-1 and pCR-SUMO-2 that expressed FLAG-tagged SUMO-1 and FLAG-tagged SUMO-2, respectively, were constructed by inserting a PCR-amplified SUMO-1 or SUMO-2 DNA fragment into pCR3.1 (Invitrogen) (44). Plasmids pHA-Ub, pHA-SUMO-1, and pHA-SUMO-2 were provided by Shih-Chung Chang. Plasmid pHA-Rta(4K-R) that contains lysine-to-alanine substitutions at amino acid positions 426, 446, 517, and 530 in Rta was constructed by a PCR mutagenesis method (55). Plasmids pHA-Rta and pHA-3K-R were described earlier (44). Plasmid pEGFP-PML was generated by inserting a PML cDNA fragment that was isolated from pcDNA3-myc-PML (56) into pEGFP-C1.
Protein Expression and Purification
Escherichia coli BL21(DE3)(pET-Rta, pT-E1E2S2) was treated with isopropyl 1-thio-β-d-galactopyranoside to induce the expression of His-tagged polySUMO-2-Rta according to a method described earlier (53). His-Rta and His-UbcH5a were purified from the E. coli BL21(DE3)(pET-Rta) and E. coli BL21(DE3)(pET-Ubc5a) lysates, respectively, using Ni2+-NTA-Sepharose beads using the method described elsewhere (44).
GST Pulldown Assay
GST, GST-RNF4, and His-Rta were purified from E. coli BL21(DE3)(pGEX-4T1), E. coli BL21(DE3)(pGEX-RNF4), and E. coli BL21(DE3)(pET-Rta), respectively. GST pulldown was performed according to a method described elsewhere (44).
Immunoprecipitation Assay
P3HR1 cells, which were treated with TPA and sodium butyrate for 24 h, and 293T cells, which had been transfected by plasmids for 24 h, were treated with 5 μm MG132 for 12 h. Lysates were prepared using HEPES buffer (12 mm HEPES and 0.5% Nonidet P-40) and centrifuged at 13,800 × g for 5 min. Anti-Rta antibody (Argene) or anti-RNF4 antibody (Sigma) was mixed with the supernatant at 4 °C for 1 h. Protein-A- or protein-G-agarose beads (30 μl) (Oncogene, Boston, MA) were added to the lysate, and the mixture was incubated under shaking for 1 h at 4 °C. The beads were collected by centrifugation and washed three times with HEPES buffer. Proteins binding to the beads were eluted by adding 20 μl of 2× electrophoresis sample buffer and analyzed by immunoblotting using anti-Rta and anti-RNF4 antibodies. To detect sumoylated or ubiquitinated proteins, cells were harvested and washed with PBS containing 10 mm N-ethylmaleimide. Cells were solubilized and sonicated in 100 μl of SUMO protective buffer (44, 57) containing 1% SDS and then incubated at 95 °C for 10 min. The supernatant was then diluted with 900 μl of phosphate-buffered saline (PBS) containing 0.5% Nonidet P-40 and incubated with anti-FLAG antibody that was conjugated to M2-agarose beads (Sigma), anti-HA antibody (Roche Applied Science), or anti-Rta antibody to perform immunoprecipitation. Cells were incubated for 24 h after transfection and then treated with MG132 for an additional 12 h to detect sumoylated or ubiquitinated proteins.
Immunofluorescence Analysis
P3HR1 cells were transfected with pEGFP-C1 or pEGFP-RNF4 and then treated with TPA and sodium butyrate to activate the EBV lytic cycle. After culturing for 24 h, cells were treated with 5 μm MG132 for an additional 12 h. Cells were harvested by centrifugation, plated on poly-l-lysine (Sigma)-coated coverslips, and fixed with 4% paraformaldehyde in PBS for 30 min. Immunostaining was performed using anti-Rta monoclonal antibody and rabbit anti-SUMO-2 monoclonal antibody (Cell Signaling). Cells were then treated with Alexa Fluor 594-conjugated goat anti-mouse IgG polyclonal antibody (Invitrogen), Alexa Fluor 488-conjugated goat anti-mouse IgG polyclonal antibody (Invitrogen), Alexa Fluor 594-conjugated goat anti-rabbit IgG polyclonal antibody (Invitrogen), or Cy5-conjugated goat anti-mouse IgG polyclonal antibody (Invitrogen). Nuclei were visualized by staining using 5 μg/ml 4′-6-diamidino-2-phenylindole (DAPI). Finally, cells were examined under a Zeiss confocal laser-scanning microscope (Model LSM780) (Oberkochen, Germany).
In Vitro Ubiquitination Assay
The reaction mixture (20 μl) contained 4.8 μg of bovine ubiquitin (Sigma), 56 ng of E1 protein (Boston Biochem), 160 ng of purified His-UbcH5a protein, GST-RNF4-glutathione beads, and His-Rta or poly-SUMO-2-conjugated His-Rta in a reaction buffer that contained 5 mm MgCl2, 2 mm ATP, and 0.2% Triton X-100 in PBS. Reaction mixtures were incubated for 1 h at 32 °C with shaking. Proteins were separated by SDS-polyacrylamide gel electrophoresis and detected by immunoblotting. Similar reaction mixtures contained FLAG-ubiquitin (Sigma), E1, UbcH5a, GST-RNF4, and His-Rta, or SUMO-2-Rta was added in 100 μl of SUMO-protective buffer containing 1% SDS and then boiled at 95 °C for 10 min. The mixtures were then diluted with 900 μl PBS containing 0.5% Nonidet P-40. Ub-Rta was immunoprecipitated using anti-FLAG antibody and separated by SDS-polyacrylamide gel electrophoresis and detected by immunoblotting using anti-Rta antibody.
Transient Transfection and Luciferase Assay
P3HR1 cells (5 × 106) were transfected with 5 μg of plasmids under the conditions of 240 V, 975 microfarads by electroporation using a BTX ECM630 electroporator (BTX Instrument). Plasmid DNA was transfected into 293T cells using Turbofect in vitro transfection reagent (Fermentas). To examine promoter activities, cells were harvested and washed with PBS, then lysed in 30 μl lysis buffer containing 25 mm Tris phosphate (pH 7.8), 2 mm dithiothreitol, 2 mm 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol, and 1% Triton X-100. Luciferase activity was measured according to a method described earlier using a luminometer (Orion II; Berthod, Bad Wildbad, Germany) (58). Each transfection experiment was performed at least three times, and each sample in the experiment was prepared in duplicate.
Quantitative Reverse Transcription-PCR
Total RNA was extracted from P3HR1 cells at 0–36 h after treatment with 1.25 μm MG132. cDNA was amplified using random hexamers and Moloney murine leukemia virus reverse transcriptase (Promega). Real-time quantitative PCR was performed according to the method as described (41). Primers used for amplification of BRLF1 and actin were described earlier (59).
Analysis of Protein Stability
293T cells were cotransfected with pCMV-R and RNF4 shRNA or control shRNA and then cultured for 40 h; thereafter, cells were treated with 100 μg/ml cycloheximide (Sigma) to inhibit protein synthesis. Cells were harvested at different time points after the treatment. The amounts of RNF4, Rta, and α-tubulin were determined by immunoblot analysis.
Knockdown of RNF4 Expression Using shRNA
RNF4 shRNAs and plasmids including pVSV-G, pCMVDR8.91, and pLKO-shRNA were purchased from the National RNAi Core Facility, Genomic Research Center, Academia Sinica, Taipei, Taiwan. Two plasmids (300 ng) that express RNF4 shRNAs (target sequences 5′-CGGGCTTCTGACTGCTCCATA and 5′-CATCTGCATGGACGGATACTC) were transfected into 293T cells using Turbofect in vitro transfection reagent (Fermentas). Inhibition of RNF4 expression was examined by immunoblotting using anti-RNF4 antibody at 48 h after transfection. For lentiviral infection, RNF4 shRNAs were cotransfected into HEK-293T cells with pVSV-G and helper plasmids pCMVDR8.91 using Turbofect reagent. Plasmid pLKO-shRNA was used as a negative control. The culture supernatants were harvested and used to transduce P3HR1 cells to knockdown endogenous RNF4 expression according to the method provided by the National RNAi Core Facility. Stable cell lines were selected using 0.5 μg/ml puromycin.
Determination of DNA Replication and the Copy Number of EBV Genome
P3HR1 cells that expressed RNF4-shRNA were treated with TPA and sodium butyrate to induce the lytic cycle. Viral replication was assayed by real-time PCR using primers for the oriLyt DNA region then normalized to cellular actin DNA as described earlier (60). To determine the copy number of EBV, virus particles released into the culture medium were harvested by ultracentrifugation at 25,000 × g for 2 h using P3HR1 cells that had been treated with TPA and sodium butyrate for 5 days. qPCR was conducted according to the method as described (61).
RESULTS
Stabilization of Sumoylated Rta (SUMO-Rta) by MG132
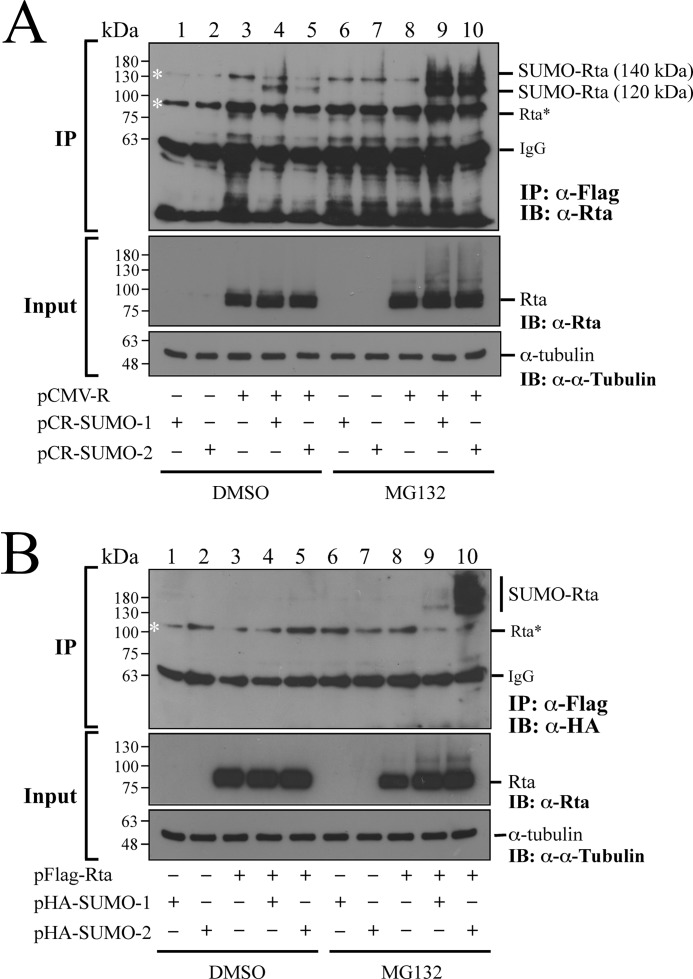
293T cells were cotransfected with plasmids that express Rta and SUMO-1 or Rta and SUMO-2 to investigate how MG132 treatment affected Rta sumoylation. Immunoblot analysis using anti-Rta antibody revealed the presence of Rta in the lysates from the cells transfected with pCMV-R (Fig. 1A, Input, lanes 3–5 and 8–10). Meanwhile, a band that was larger than Rta, which is likely SUMO-Rta, was detected in the lysates from the cells that were cotransfected with pCMV-R or pCMV-R and pCR-SUMO-1 or pCR-SUMO-2 and treated with MG132 (Fig. 1A, Input, lanes 8–10). A control experiment showed that anti-FLAG antibody did not immunoprecipitate Rta that was conjugated by FLAG-SUMO if the cells were not transfected with pCR-SUMO-1 or pCR-SUMO-2 (Fig. 1A, IP, lanes 3 and 8). This study also cotransfected cells with pCMV-R and pCR-SUMO-1 or pCR-SUMO-2. Proteins were then immunoprecipitated using anti-FLAG antibody after denaturing proteins in the lysates at 95 °C. Subsequent immunoblot analysis using anti-Rta antibody revealed a SUMO-Rta band of 120 kDa (Fig. 1A, IP, lanes 4 and 5). Meanwhile, treating the cells with MG132 after transfection caused the accumulation of 120-kDa SUMO-Rta and the appearance of a 140-kDa SUMO-Rta band (Fig. 1A, IP, lanes 9 and 10); the amounts of these two proteins were considerably larger than the 120-kDa SUMO-Rta detected in the cells untreated with MG132 (Fig. 1A, IP, lanes 4 and 5). Notably, the 140-kDa SUMO-Rta co-migrated with a protein that was detected nonspecifically under the experimental conditions (Fig. 1A, IP, lanes 1–8). In fact, bands of 90 and 135 kDa, which are not Rta and sumoylated Rta, respectively, are often nonspecifically detected when an experiment involves immunoprecipitation using anti-FLAG antibody and immunoblot analysis using anti-Rta antibody (46, 62, 63). This study also cotransfected 293T cells with pFLAG-Rta and pHA-SUMO-1 or pFLAG-Rta and pHA-SUMO-2. Experimental results indicated that FLAG-Rta was expressed after cotransfection (Fig. 1B, Input). Furthermore, HA-SUMO-Rta in the lysates was immunoprecipitated using anti-FLAG antibody after the proteins in the lysate were denatured by heat and then immunoblotted using anti-HA antibody to reveal FLAG-Rta that was conjugated by HA-SUMO-1 or HA-SUMO-2. Those results further demonstrated that SUMO-Rta was undetected in the lysates from the cells cotransfected with pFLAG-Rta and pHA-SUMO-1 or pFLAG-Rta and pHA-SUMO-2 (Fig. 1B, IP, lanes 4 and 5). However, SUMO-Rta was detected if the cells were treated with MG132 after cotransfection (Fig. 1B, IP, lanes 9 and 10), suggesting that MG132 treatment stabilizes SUMO-Rta.
FIGURE 1.
Accumulation of SUMO-modified Rta after MG132 treatment. A, 293T cells were transfected with pCR-SUMO-1 (lanes 1 and 6), pCR-SUMO-2 (lanes 2 and 7), or pCMV-R (lanes 3 and 8) or cotransfected with pCMV-R and pCR-SUMO-1 (lanes 4 and 9) or pCR-SUMO-2 (lanes 5 and 10) in the presence of 5 μm MG132 (lanes 6–10) or DMSO (lanes 1–5). At 24 h after transfection, the cells were washed with PBS containing 10 mm N-ethylmaleimide. B, 293T cells were transfected with pHA-SUMO-1 (lanes 1 and 6), pHA-SUMO-2 (lanes 2 and 7), pFLAG-Rta (lanes 3 and 8) or cotransfected with pFLAG-Rta and pHA-SUMO-1 (lanes 4 and 9) or pFLAG-Rta and pHA-SUMO-2 (lanes 5 and 10) and then treated with MG132 (lanes 6–10) or DMSO (lanes 1–5). In A, SUMO-Rta was immunoprecipitated (IP) using anti-FLAG antibody and detected by immunoblotting (IB) using anti-Rta antibody. In B, SUMO-Rta was immunoprecipitated using anti-FLAG antibody and detected using anti-HA antibody. Asterisks indicate bands detected nonspecifically. Rta* indicates where the Rta band is supposed to locate.
MG132 Treatment and EA-D Expression
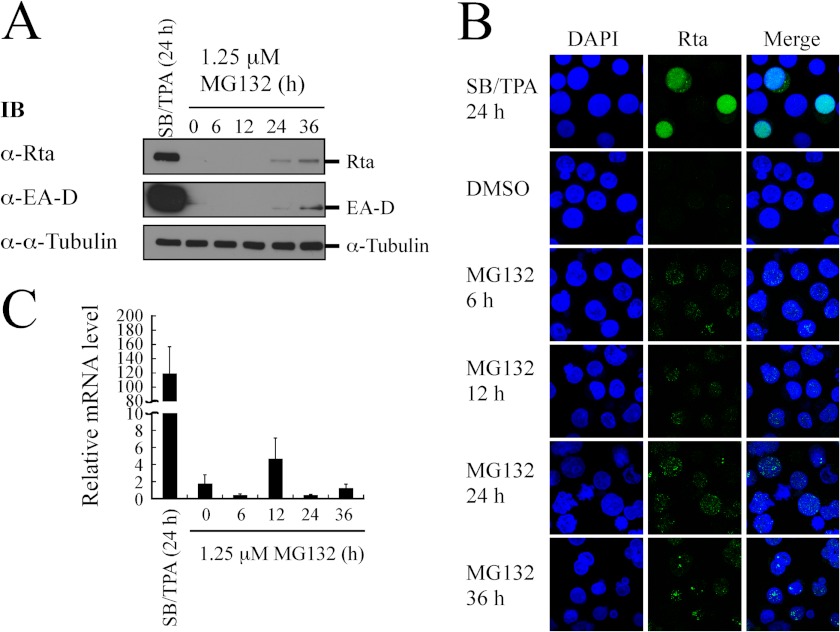
This study investigated whether the stabilization of SUMO-Rta by MG132 caused the expression of an EBV early protein, EA-D. Accordingly, P3HR1 cells were treated with MG132 for 36 h without lytic induction. Immunoblot analysis revealed that the amounts of Rta and EA-D increased substantially at 36 h after the treatment (Fig. 2A), indicating that the treatment with MG132 causes the expression of EA-D (Fig. 2A). Indirect immunofluorescence analysis also revealed the presence of Rta as speckles in the cells after 6 h of MG132 treatment (Fig. 2B). The number and intensity of Rta dots increased over the 36-h incubation period (Fig. 2B), verifying that MG132 stabilizes Rta. Meanwhile, qRT-PCR revealed that MG132 treatment increased only a slight amount of the BRLF1-BZLF1 transcript (Fig. 2C), possibly due to leaky transcription of the genes in P3HR1 cells (64). Additionally, the amount of mRNA detected after MG132 treatment was about 1% or less than that after lytic induction (Fig. 2C), implying that the enhanced EA-D and Rta expression after MG132 treatment is probably not due to transcriptional activation.
FIGURE 2.
Expression of EA-D after MG132 treatment. A, P3HR1 cells were treated with 1.25 μm MG132 for 36 h. Cells treated with sodium butyrate (SB) and TPA (SB/TPA) for 24 h were used as a positive control. Proteins in the lysate were detected by immunoblotting (IB) using anti-Rta, anti-EA-D, and anti-α-tubulin antibodies. B, indirect immunofluorescence analysis was performed using P3HR1 cells that were treated with MG132 for 36 h. Cells treated with SB/TPA or DMSO for 24 h were used as a control. Cells were incubated with anti-Rta monoclonal antibody. DAPI staining revealed the nucleus. Cells were observed under a confocal laser-scanning microscope. C, total RNA from P3HR1 cells that had been treated with MG132 was analyzed by qRT-PCR. The level of BRLF1-BZLF1 expression was normalized relative to those of actin. The data shown are the averages and S.D. of the results from two independent experiments, and each sample in the experiment was prepared in duplicate.
Ubiquitination of Rta in Vivo
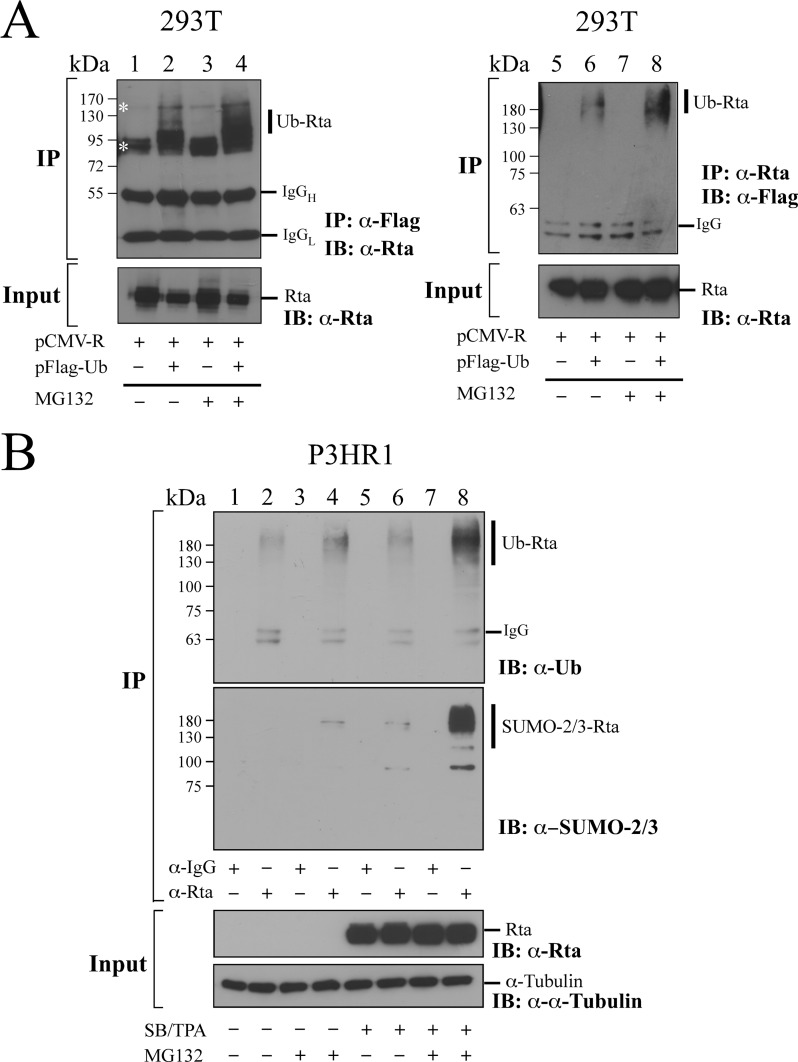
MG132 is a proteasome inhibitor that prevents the degradation of proteins that are conjugated to ubiquitin. The fact that MG132 stabilizes SUMO-conjugated Rta implies that Rta is conjugated to ubiquitin before its degradation. This study attempted to demonstrate that Rta is ubiquitinated by cotransfecting 293T cells with pCMV-R and pFLAG-Ub. At 24 h after transfection, ubiquitinated proteins in the lysate were immunoprecipitated using anti-FLAG antibody. Ubiquitinated Rta (Ub-Rta) was subsequently detected by immunoblotting using anti-Rta antibody. The results indicated that cotransfecting the cells with both plasmids was necessary for detecting Ub-Rta (Fig. 3A, IP, lanes 2 and 4); transfecting the cells with pCMV-R alone was insufficient for such detection (Fig. 3A, IP, lanes 1 and 3). Additionally, the amount of Ub-Rta increased if the cells were treated with MG132 after transfection (Fig. 3A, IP, lane 4). We also found that after the cells were cotransfected with pCMV-R and pFLAG-Ub, Ub-Rta was immunoprecipitated by anti-Rta antibody and detected by immunoblotting using anti-FLAG antibody (Fig. 3A, IP, lane 6); treating the cells with MG132 further increased the amount of Ub-Rta (Fig. 3A, IP, lane 8). Furthermore, this study examined whether Rta was ubiquitinated in P3HR1 cells. If the cells were untreated with TPA and sodium butyrate, a trace amount of Ub-Rta was detected (Fig. 3B, IP, lane 2). Also, treating the cells with MG132 increased the amount of Ub-Rta (Fig. 3B, IP, lane 4). Meanwhile, after lytic induction for 24 h, the presence of Ub-Rta displayed a weak signal (Fig. 3B, IP, lane 6); treating the cells with MG132 after lytic induction resulted in the detection of an abundant amount of Ub-Rta (Fig. 3B, IP, lane 8). A control experiment involving immunoprecipitation in which an anti-IgG antibody was used did not allow for the detection of Ub-Rta (Fig. 3B, IP, lanes 1, 3, 5, and 7), demonstrating that Rta is conjugated to ubiquitin. Furthermore, this study used the same lysate to demonstrate that Rta was conjugated by SUMO-2/3. Accordingly, immunoblot analysis using anti-SUMO-2/3 antibody revealed that SUMO-2/3-Rta was undetected if the cells were untreated with TPA, sodium butyrate, and MG132 (Fig. 3B, IP, lane 2); treating the cells with MG132 or TPA and sodium butyrate resulted in the detection of a weak SUMO-2/3-Rta band (Fig. 3B, IP, lanes 4 and 6). Furthermore, treating the cells with MG132 after lytic induction substantially increased the amount of SUMO-2/3-Rta in the cells (Fig. 3B, IP, lane 8). A negative control showed that SUMO-Rta was undetected if immunoprecipitation involved anti-IgG antibody (Fig. 3B, IP, lanes 1, 3, 5, and 7). These results indicated that Rta was conjugated by SUMO-2/3 and the treatment with MG132 causes the accumulation of SUMO-2/3-Rta.
FIGURE 3.
Ubiquitination of Rta in vivo. A, 293T cells were transfected with pCMV-R (lanes 1, 3, 5, and 7) or cotransfected with pCMV-R and pFLAG-Ub (lanes 2, 4, 6, and 8). Cells were treated (lanes 3, 4, 7, and 8) or untreated (lanes 1, 2, 5, and 6) with MG132 for 12 h after transfection for 24 h. Proteins in the cell lysate was immunoprecipitated (IP) using anti-FLAG antibody (lanes 1–4) or anti-Rta antibody (lanes 5–8) after denaturing the proteins in the lysates at 95 °C. The proteins were then detected by immunoblotting (IB) using anti-Rta antibody (lanes 1–4) or anti-FLAG antibody (lanes 5–8). B, P3HR1 cells were treated (lanes 5, 6, 7, and 8) or untreated (lanes 1, 2, 3, and 4) with sodium butyrate and TPA. After incubation for 24 h, cells were treated with MG132 (lanes 3, 4, 7, and 8) or DMSO (lanes 1, 2, 5, and 6) for 12 h. Proteins in cell lysates were immunoprecipitated using anti-Rta antibody and detected by immunoblot analysis with anti-SUMO-2/3 antibody, anti-ubiquitin (Ub), and anti-Rta antibody. Anti-IgG (lanes 1, 3, 5, and 7) was used in IP as a negative control. Asterisks indicate nonspecific bands. Ub-Rta, ubiquitinated Rta; IgGH, the heavy chain of IgG; IgGL, the light chain of IgG.
Interaction of RNF4 with Rta
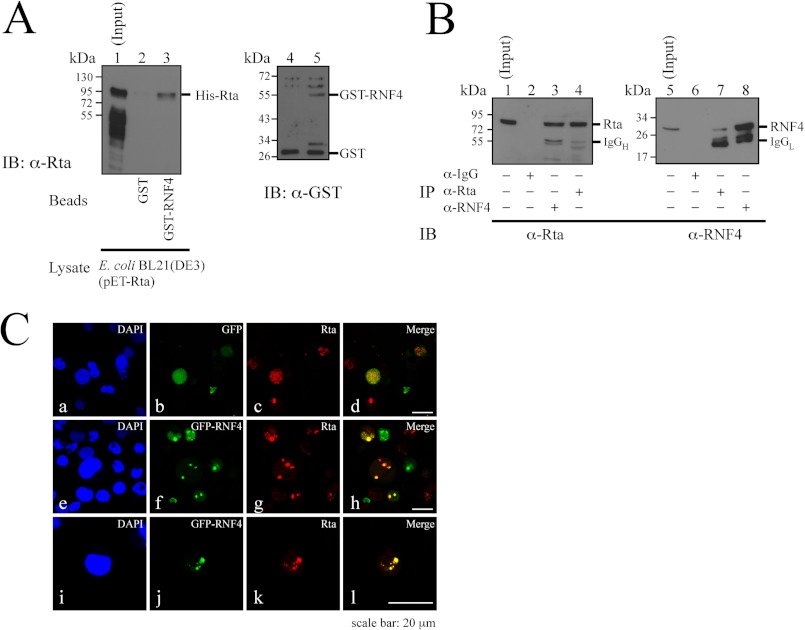
Because RNF4 is known to promote ubiquitination of SUMO-2-conjugated proteins (15, 16, 27), this study investigated whether Rta interacts with RNF4 in a GST pulldown study. Glutathione-Sepharose beads were added to the lysate prepared from E. coli transformed with plasmids that express GST and GST-RNF4 (Fig. 4A, lanes 4 and 5). After washing, the beads were added to a lysate from E. coli BL21(DE3)(pET-Rta), which expresses His-Rta. Proteins that were bound to the beads were eluted and analyzed by immunoblotting using anti-Rta antibody. The result revealed that Rta in the lysate (Fig. 4A, lane 1) was retained by GST-RNF4-glutathione-Sepharose beads (Fig. 4A, lane 3) but not by GST-glutathione-Sepharose beads (Fig. 4A, lane 2), indicating that RNF4 binds to Rta directly. Thereafter, coimmunoprecipitation assay was performed using P3HR1 cells that had been treated with TPA and sodium butyrate for 24 h. Immunoblot analysis revealed that Rta in the lysate (Fig. 4B, lane 1) was immunoprecipitated by anti-Rta antibody (Fig. 4B, lane 4) and coimmunoprecipitated with RNF4 by anti-RNF4 antibody (Fig. 4B, lane 3). However, Rta was not immunoprecipitated by anti-IgG antibody (Fig. 4B, lane 2). A parallel experiment revealed that RNF4 was present in the P3HR1 cell lysate (Fig. 4B, lane 5), immunoprecipitated by anti-RNF4 antibody (Fig. 4B, lane 8), and coimmunoprecipitated with Rta by anti-Rta antibody (Fig. 4B, lane 7). However, anti-IgG antibody did not immunoprecipitate RNF4 (Fig. 4B, lane 6). Indirect immunofluorescence also revealed that GFP-RNF4 colocalized with Rta and displayed a punctate distribution in the nucleus in P3HR1 cells after the cells were transfected with pEGFP-RNF4, treatment with TPA, sodium butyrate, and MG132 (Fig. 4C, h). As is generally known, lytic induction using TPA and sodium butyrate of EBV in B lymphocyte cells is usually inefficient and does not cause the expression of lytic proteins in all the cells (65–68), explaining why only a subset of P3HR1 population was lytically activated and expressed Rta (Fig. 4C, g and h). The study also found that GFP was diffused throughout the cells when the cells were transfected with an empty vector (Fig. 4C, a–d).
FIGURE 4.
Interaction between Rta and RNF4. A, bacterially expressed GST and GST-RNF4 were bound to glutathione-Sepharose beads. The beads were then mixed with a lysate from E. coli BL21(DE3)(pET-Rta). Proteins bound to GST-glutathione-Sepharose (lane 2) and GST-RNF4-glutathione-Sepharose (lane 3) beads were analyzed by immunoblotting (IB) using anti-Rta antibody. Lane 1 was loaded with 1% of the cell lysate. GST proteins that were bound to the beads were eluted and analyzed by immunoblot analysis using anti-GST antibody (lanes 4 and 5). B, P3HR1 cells were treated with TPA and sodium butyrate. Proteins in the lysate were subsequently immunoprecipitated (IP) using anti-IgG (lanes 2 and 6), anti-Rta (lanes 4 and 7), or anti-RNF4 (lanes 3 and 8) antibodies. Immunoblotting was performed using anti-Rta (lanes 1–4) and anti-RNF4 (lanes 5–8) antibodies. Input lanes were loaded with 1% of the lysates. IgGH, heavy chain of IgG; IgGL, light chain of IgG. C, P3HR1 cells were transfected with pEGFP-C1 (a–d) or pEGFP-RNF4 (e–l) and then treated with 5 μm MG132, sodium butyrate, and TPA for 24 h. Cells were incubated with anti-Rta monoclonal antibody. DAPI staining revealed the nucleus. Cells were observed under a confocal laser-scanning microscope. d, h, and l are merged images.
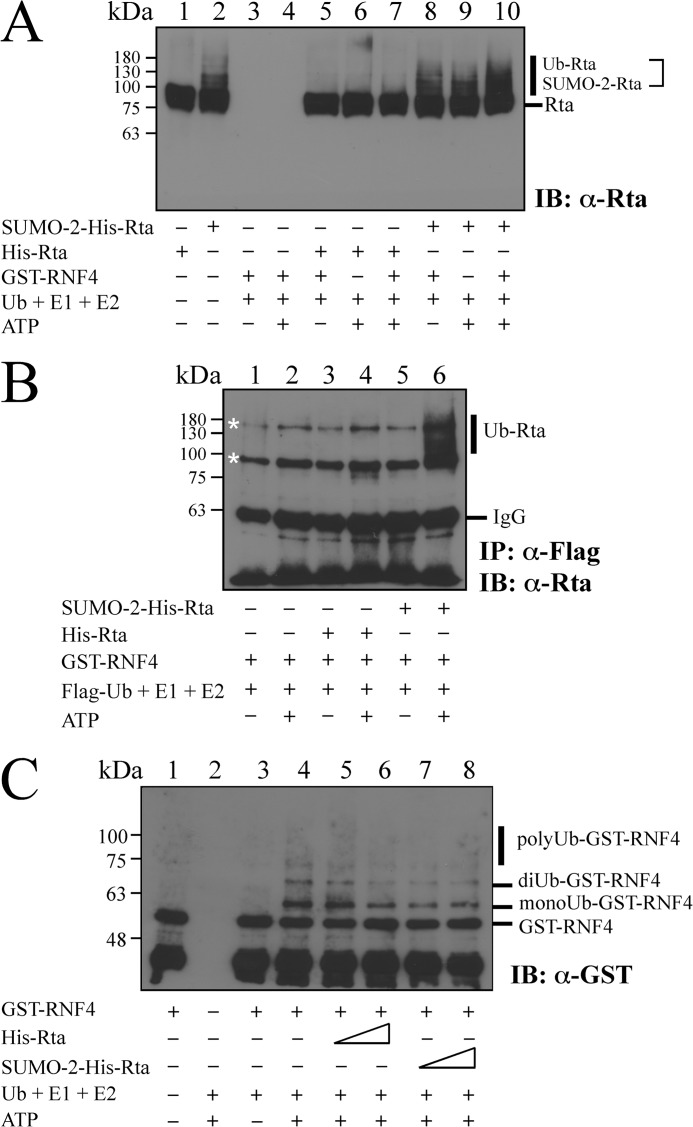
Promotion of Rta Ubiquitination by RNF4 in Vitro
An in vitro ubiquitination assay was performed using purified Rta, ubiquitin E2 enzyme UbcH5a, GST-RNF4, and ubiquitin-activating E1 enzyme. Ub-Rta in the reaction mixture was then detected by immunoblotting using anti-Rta antibody (Fig. 5A). The results showed that His-Rta purified from E. coli BL21(DE3)(pET-Rta) was not sumoylated (Fig. 5A, lane 1). However, His-Rta was sumoylated by SUMO-2 in E. coli BL21(DE3)(pT-E1E2S2) (53) (Fig. 5A, lane 2), although a large amount of Rta was unsumoylated. The assay revealed that neither a blank, which contained only ubiquitin, E1, E2, and ATP (Fig. 5A, lanes 3 and 4), nor adding His-Rta to the blank reaction mixture resulted in protein ubiquitination (Fig. 5A, lanes 5–7). However, adding an Rta mixture that contained SUMO-2-His-Rta to the reaction produced a band that was higher than that of SUMO-2-His-Rta when GST-RNF4 was present, suggesting that SUMO-2-His-Rta is ubiquitinated (Fig. 5A, lanes 9 and 10). The results also demonstrated that ubiquitination of SUMO-2-His-Rta required ATP (Fig. 5A, lanes 8 and 10). An in vitro ubiquitination assay was also conducted using FLAG-ubiquitin, purified His-Rta, an Rta mixture that contained SUMO-2-His-Rta, ubiquitin E2 enzyme UbcH5a, GST-RNF4, and ubiquitin-activating E1 enzyme to verify that RNF4 promotes the ubiquitination of SUMO-2-His-Rta. Ubiquitinated proteins were then immunoprecipitated using anti-FLAG antibody and detected by immunoblot analysis using anti-Rta antibody (Fig. 5B). The results revealed that SUMO-2-Rta was modified by FLAG-ubiquitin only when the reaction included GST-RNF4 (Fig. 5B, lane 6). The results also revealed that the ubiquitination of Rta depended on ATP (Fig. 5B, lanes 5 and 6). The above results indicate that SUMO-2-His-Rta is a substrate for RNF4 in vitro and RNF4 targets SUMO-2-His-Rta to promote Rta ubiquitination. The possibility that Rta influences the ubiquitination of RNF4 was excluded by a similar analysis. The results confirmed that GST-RNF4 autoubiquitinated in the reaction containing E1, E2, Ub, and ATP (Fig. 5C, lane 4), resulting in the detection of monoubiquitinated, diubiquitinated, and polyubiquitinated RNF4 by immunoblotting using anti-GST antibody (Fig. 5C, lane 4). However, adding His-Rta or SUMO-2-His-Rta did not increase the level of GST-RNF4 ubiquitination (Fig. 5C, lanes 5–8), revealing that Rta does not affect the ubiquitination of RNF4.
FIGURE 5.
Enhancement of Rta ubiquitination by RNF4 in vitro. A, conjugation of ubiquitin to Rta was analyzed in vitro using purified Rta, an Rta mixture that contains SUMO-2-conjugated Rta, and GST-RNF4 in the presence of ubiquitin (Ub), ubiquitin E1-activating enzyme, and ubiquitin E2 enzyme (UbcH5a). Ubiquitinated proteins were examined by immunoblotting (IB) using anti-Rta antibody. B, similar in vitro Ub assay was conducted using FLAG-ubiquitin (FLAG-Ub), purified Rta, SUMO-2-conjugated Rta, E1, E2, and GST-RNF4. Thereafter, Ub-Rta was immunoprecipitated (IP) using anti-FLAG antibody and then detected by immunoblotting using anti-Rta antibody. C, additionally, the ubiquitination of RNF4 was examined in vitro with purified GST-RNF4, Rta, or SUMO-2-conjugated Rta in the reaction mixture containing ubiquitin (Ub), ubiquitin E1-activating enzyme, UbcH5a, or ATP. Ubiquitinated proteins including monoubiquitinated, diubiquitinated, and polyubiquitinated GST-RNF4 were detected by immunoblot analysis using anti-GST antibody. Asterisks indicate bands detected nonspecifically.
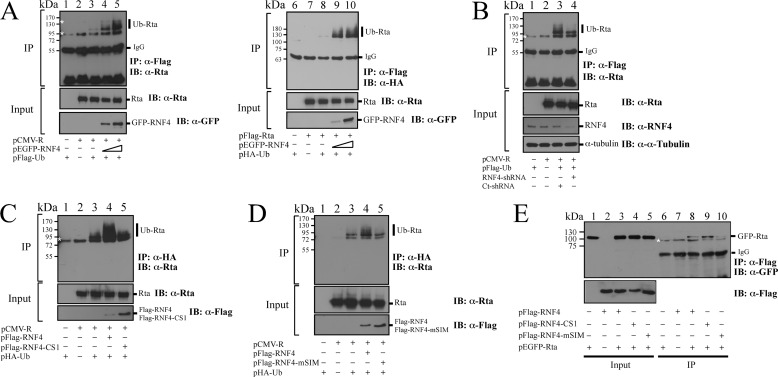
Enhancement of Rta Ubiquitination by RNF4 in Vivo
After demonstrating that RNF4 promotes ubiquitination of Rta in vitro, this study verified the function of RNF4 as an STUbL of Rta in vivo. Accordingly, 293T cells were cotransfected with pCMV-R, pEGFP-RNF4, and pFLAG-Ub, and proteins in the lysate were then immunoprecipitated by anti-FLAG antibody. Immunoblotting using anti-Rta antibody revealed that ubiquitinated Rta was undetected after cells were transfected with pCMV-R or pFLAG-Ub (Fig. 6A, IP, lanes 1 and 2). However, a little Ub-Rta was detected after cells were cotransfected with plasmids that express Rta and FLAG-Ub (Fig. 6A, IP, lane 3). Transfecting the plasmids expressing Rta, FLAG-Ub, and GFP-RNF4 substantially increased the amount of Ub-Rta that was detected by immunoblotting (Fig. 6A, IP, lane 4). Meanwhile, GFP-RNF4 appeared to influence Rta ubiquitination in a dose-dependent manner (Fig. 6A, IP, lanes 4 and 5). 293T cells were also cotransfected with pFLAG-Rta, pEGFP-RNF4, and pHA-Ub, and proteins in the cell lysate were immunoprecipitated using anti-FLAG antibody and examined by immunoblotting using anti-HA antibody. The results revealed that Ub-Rta was detected when cells were cotransfected with pFLAG-Rta, pHA-Ub, and pEGFP-RNF4 in a dose-dependent manner (Fig. 6A, IP, lanes 9 and 10) but not in the cells that were not transfected by pEGFP-RNF4 (Fig. 6A, IP, lanes 6–8). A similar study also attempted to determine how RNF4 shRNA affected Rta ubiquitination. Our control experiment first demonstrated that transfecting RNF4 shRNA caused the accumulation of GFP-PML (data not shown), which is a known substrate of RNF4 (15, 27), to demonstrate that the transfected shRNA is functional. Subsequently, 293T cells were transfected with RNF4 shRNA to determine whether RNF4 reduces Rta ubiquitination. In a control experiment, immunoblotting did not detect Ub-Rta in cells that were transfected with only pCMV-R or pFLAG-Ub (Fig. 6B, IP, lanes 1 and 2). Ub-Rta was detected in cells that were cotransfected with pCMV-R, pFLAG-Ub, and control shRNA (Fig. 6B, IP, lane 3). However, introducing RNF4 shRNA reduced the amount of Ub-Rta (Fig. 6B, IP, lane 4). A similar transfection experiment was performed using the RING mutant of RNF4 (FLAG-RNF4-CS1), which showed that although FLAG-RNF4 promoted the ubiquitination of Rta (Fig. 6C, IP, lanes 3 and 4), FLAG-RNF4-CS1 did not influence the ubiquitination of Rta (Fig. 6C, IP, lane 5). Furthermore, FLAG-RNF4 with mutation at its SIM domains (FLAG-RNF4-mSIM) also failed to promote the ubiquitination of Rta (Fig. 6D, IP, lane 5), suggesting that RNF4-mediated Rta ubiquitination depends on the SIM motifs of RNF4. This study also examined the interaction of Rta with RNF4 mutants, including RNF4-CS1 and RNF4-mSIM. Accordingly, 293T cells were cotransfected with pEGFP-Rta and pFLAG-RNF4, pFLAG-RNF4-CS1, or pFLAG-RNF4-mSIM. Proteins in the lysate were immunoprecipitated by anti-FLAG antibody and detected by immunoblotting using anti-GFP antibody. Analysis results indicated that Rta interacted with FLAG-RNF4 and FLAG-RNF4-CS1 (Fig. 6E, lanes 8 and 9). Mutation of SIM in RNF4 revealed a weak interaction with Rta (Fig. 6E, lane 10) even though the expression level of FLAG-RNF4-mSIM was higher (Fig. 6E, lane 5) than that of FLAG-RNF4-CS1 and FLAG-RNF4 (Fig. 6E, lanes 3 and 4).
FIGURE 6.
RNF4 and the ubiquitination of Rta. A, 293T cells were cotransfected with pCMV-R, pEGFP-RNF4, and pFLAG-Ub (lanes 1–5). At 24 h after transfection cells were treated with 5 μm MG132 for another 12 h. Proteins in the lysate were immunoprecipitated (IP) using anti-FLAG antibody. Proteins bound to FLAG-M2 beads were analyzed by immunoblot analysis (IB) using anti-Rta and anti-GFP antibodies. Similarly, 293T cells were cotransfected with plasmids that express FLAG-Rta, GFP-RNF4, and FLAG-Ub (lanes 6–10). At 24 h after transfection, cells were treated with 5 μm MG132 for another 12 h. Proteins in the lysate were immunoprecipitated using anti-FLAG antibody. Proteins bound to FLAG-M2 beads were analyzed by immunoblot analysis using anti-HA, anti-Rta, and anti-GFP antibodies. B, shown is the effect of knockdown of RNF4 on the ubiquitination of Rta. 293T cells were cotransfected with plasmids that express FLAG-Ub, Rta, and RNF4 shRNA or control shRNA (Ct-shRNA) and then treated with 5 μm MG132 for 12 h. Ubiquitinated Rta was immunoprecipitated using anti-FLAG antibody and detected by immunoblotting using anti-Rta antibody. C, 293T cells were cotransfected with pHA-Ub, pCMV-R, and pFLAG-RNF4 or pFLAG-RNF4-CS1. At 24 h after transfection, cells were treated with 5 μm MG132 for another 12 h. Proteins in the cell lysate were immunoprecipitated using anti-HA antibody. Immunoprecipitated proteins were detected by immunoblotting with anti-Rta antibody. D, a similar experiment as described in C was conducted except that pFLAG-RNF4-CS1 was replaced with pFLAG-RNF4-mSIM. E, interaction of Rta with RNF4 mutants is shown. 293T cells were cotransfected with pEGFP-Rta and plasmids that express FLAG-RNF4, FLAG-RNF4-CS1, or FLAG-RNF4-mSIM. A coimmunoprecipitation assay was performed using anti-FLAG antibody, and immunoprecipitated proteins were detected by immunoblotting with anti-GFP antibody (lanes 6–10). Asterisks indicate bands detected nonspecifically. Ub-Rta, ubiquitinated Rta.
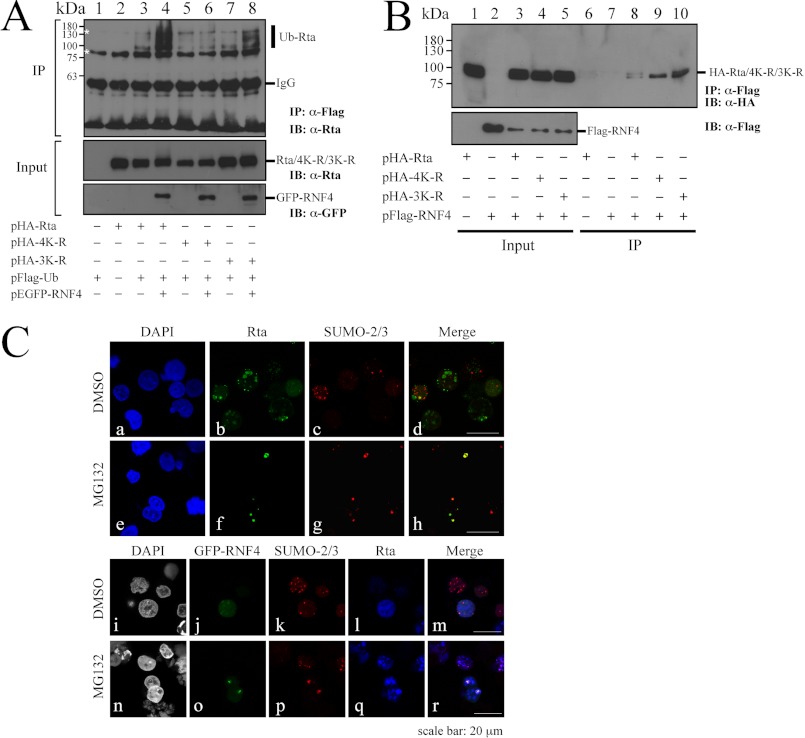
SUMO-dependent Ubiquitination of Rta by RNF4
An earlier study indicated that mutation of Rta at lysine residues 426, 446, 517, and 530 eliminates SUMO-3 conjugation to Rta (49). Our earlier investigation also demonstrated that Rta is conjugated to SUMO-1 via lysine residues 19, 213, and 517 (44). Therefore, in this study, pHA-3K-R and pHA-4K-R, which encodes mutant Rta with mutations of the three and four lysine residues with which SUMO-1 and SUMO-2/3 conjugate, respectively, are generated to examine how these mutations affect Rta ubiquitination. Here, the transfection study revealed that although overexpressing FLAG-Ub and GFP-RNF4 caused the ubiquitination of Rta in 293T cells (Fig. 7A, IP, lane 4), cotransfecting plasmids that encoded FLAG-Ub, HA-3K-R, and GFP-RNF4 did not seem to affect Rta ubiquitination (Fig. 7A, IP, lane 8). However, the degree of Rta ubiquitination was reduced further when pHA-4K-R rather than pHA-3K-R was cotransfected (Fig. 7A, IP, lane 6), suggesting that sumoylation of the 4K-R lysine residues that are conjugated by SUMO-2/3 is important to Rta ubiquitination. Furthermore, this study examined whether Rta mutants interact with RNF4. 293T cells were cotransfected with pFLAG-RNF4 and pHA-Rta, pHA-3K-R, or pHA-4K-R. A coimmunoprecipitation assay revealed that despite the weak interaction of HA-Rta and RNF4 (Fig. 7B, IP, lane 8), mutation of the 3K-R lysine or the 4K-R lysine residues in Rta did not change the interaction with RNF4 (Fig. 7B, IP, lanes 9 and 10), suggesting that the decrease in the amount of Ub-Rta in HA-4K-R is irrelevant to the interaction of RNF4 and HA-4K-R. Moreover, the colocalization among Rta, SUMO-2, and RNF4 was examined using indirect immunofluorescence analysis. Correspondingly, P3HR1 cells were transfected with pEGFP-RNF4 followed by treatment of the cells with TPA and sodium butyrate for 24 h. The results revealed that Rta colocalized with SUMO-2 in the nucleus as dots when cells were treated with MG132 (Fig. 7C, h). Rta was also colocalized with SUMO-2 and RNF4 in the nucleus after MG132 treatment (Fig. 7C, r), suggesting that RNF4 targets SUMO-2-conjugated Rta to enhance its ubiquitination after lytic induction.
FIGURE 7.
SUMO-dependent ubiquitination of Rta by RNF4. A, 293T cells were cotransfected with pFLAG-Ub, pEGFP-RNF4, and pHA-Rta (lanes 2–4), pHA-4K-R (lanes 5 and 6), or pHA-3K-R (lanes 7 and 8). At 24 h after transfection, cells were treated with 5 μm MG132 for another 12 h. Proteins in the lysate were immunoprecipitated (IP) using anti-FLAG antibody. Proteins were then detected by immunoblotting (IB) using anti-Rta and anti-GFP antibodies. B, interaction of RNF4 with Rta is shown. 293T cells were cotransfected with pFLAG-RNF4 and plasmids that express HA-Rta, HA-3K-R, or HA-4K-R. A coimmunoprecipitation assay was performed using anti-FLAG antibody, and immunoprecipitated proteins were detected by immunoblotting using anti-HA antibody. C, P3HR1 cells were transfected with plasmids pEGFP-RNF4 (a–r), and then cells were treated with 5 μm MG132 or DMSO for 12 h after lytic induction by TPA and sodium butyrate. Cells were incubated with anti-Rta monoclonal antibody and anti-SUMO-2 polyclonal antibody. DAPI staining revealed the nucleus. Thereafter, cells were observed under a confocal laser-scanning microscope. d, h, m, and r are merged images.
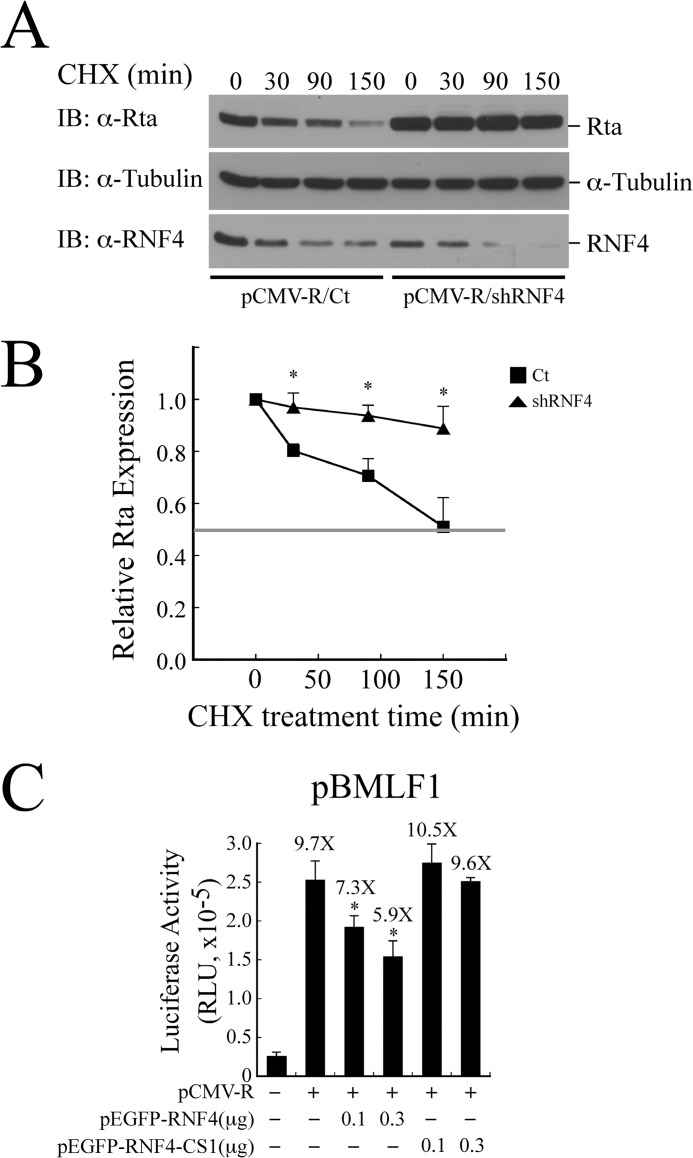
Effect of RNF4 on the Stability and the Transactivation Ability of Rta
Whether the expression of RNF4 shRNA influences the stability of Rta was investigated by cotransfecting 293T cells with plasmids pCMV-Rta and RNF4 shRNA or control shRNA. At 40 h after transfection, cells were treated with cycloheximide (CHX) for 150 min to inhibit protein synthesis. The result showed that the amount of Rta decreased ∼50% at 150 min if cells were transfected with control shRNA (Fig. 8, A and B). Yet the level of Rta was reduced 12% at 150 min in the presence of RNF4 shRNA (Fig. 8, A and B), demonstrating that RNF4 reduces the stability of Rta. This study also examined whether RNF4 affects the transactivation ability of Rta via ubiquitination by undertaking a transient transfection study in which 293T cells were cotransfected with pBMLF1, pCMV-R, and pEGFP-RNF4. According to those results, overexpressing Rta increased the promoter activity by 9.7-fold (Fig. 8C). RNF4 inhibited the transactivation activity of Rta in a dose-dependent manner from 7.3- to 5.9-fold after transfection with 0.1–0.3 μg of pEGFP-RNF4 (Fig. 8C). Notably, the promoter activity activated by Rta was not significantly reduced if cells were cotransfected with the same amounts of an RNF4 mutant, RNF4-CS1 (Fig. 8C), indicating that RNF4 decreases the ability of Rta to transactivate an EBV lytic promoter. Our results further demonstrated using the RNF4-mSIM to compete the function of RNF4 is ineffective (data not shown), likely due to its weak interaction with Rta (Fig. 6E).
FIGURE 8.
Effect of RNF4 on the stability and transactivation activity of Rta. A, 293T cells were transfected with plasmids pCMV-R and control shRNA (Ct-shRNA) or RNF4 shRNA. Cycloheximide (CHX) was added at 40 h after transfection to inhibit protein synthesis. Whole cell lysate was prepared at 0, 30, 90, and 150 min after transfection. Proteins in the lysate were detected by immunoblotting (IB) using anti-Rta, anti-α-tubulin, and anti-RNF4 antibodies. B, a densitometric analysis of Rta level normalized to α-tubulin was plotted using Image J software. Data are presented as the mean with S.D. and represent three independent experiments. The intensity corresponding to 50% of the initial value is indicated by the horizontal line. C, 293T cells were cotransfected with the reporter plasmid pBMLF1 and pCMV-R, pEGFP-RNF4, or pEGFP-RNF4-CS1. Luciferase activities were monitored 24 h post-transfection. Each transfection experiment was performed three times, and each sample in the experiment was prepared in duplicate. The value from each experiment was analyzed statistically with the least square means method. *, p < 0.05.
Role of RNF4 in EBV Lytic Development
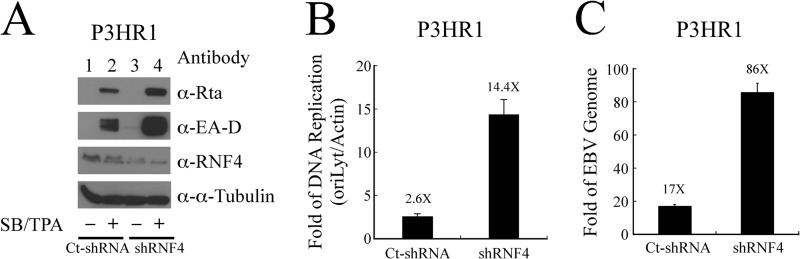
The expression of RNF4 in P3HR1 cells was attenuated using lentiviral-based shRNA (Fig. 9A). Rta and EA-D were expressed when cells were infected with lentiviral control shRNA after lytic induction (Fig. 9A, lanes 1 and 2). Additionally, knockdown of RNF4 expression by RNF4 shRNA increased the expression of Rta and EA-D (Fig. 9A, lanes 2 and 4). Moreover, lytic DNA replication was examined by performing qPCR. The result revealed that the inhibition of RNF4 expression using shRNA increased the level of viral DNA replication by 5.5-fold after the cells were treated with TPA and sodium butyrate (Fig. 9B). Finally, virion production was examined using qPCR. According to those results, knockdown of RNF4 expression also increased the production of virions by 5-fold after lytic induction (Fig. 9C), demonstrating that RNF4 inhibits EBV lytic progression.
FIGURE 9.
Role of RNF4 on EBV lytic development. A, P3HR1 cells were infected by the lentivirus that contains RNF4 shRNA (shRNF4) or control shRNA (Ct-shRNA) under the selection of puromycin. Thereafter, cells were treated with sodium butyrate and TPA (SB/TPA) for 48 h, and proteins in the lysate were examined by immunoblotting using anti-Rta, anti-EA-D, anti-RNF4, and anti-α-tubulin antibodies. B, moreover, cells that were harvested from A were lysed, and EBV lytic DNA replication assay was assayed by qPCR. The amount of EBV DNA was normalized with the amount of actin DNA that was determined in the same assay. C, the lentiviral-transduced P3HR1 cells were treated with TPA and sodium butyrate for 5 days. EBV DNA from viral particles that were released into the culture medium was determined by qPCR after the DNA extraction. The copy number of EBV genome was calculated by using maxi-EBV that had been isolated from E. coli as a standard.
DISCUSSION
Rta is a transcription factor that activates viral early genes, subsequently affecting lytic progression (48, 49, 59, 69–73). Hence, the stability of Rta is essential to its ability to influence the lytic cycle. Our results demonstrate for the first time that Rta is a substrate of RNF4 that targets SUMO-2-conjugated Rta and enhances the ubiquitination of Rta, ultimately inhibiting EBV lytic progression.
Our earlier study established that RanBPM promotes Rta sumoylation and enhances its transactivation activity (46). According to those results, treating cells with 0.5 μm MG132 stabilizes sumoylated Rta in P3HR1 cells (46). Because MG132 inhibits the proteolytic activity of 26 S proteasome complex, preventing the degradation of ubiquitinated proteins (74), the fact that MG132 stabilizes sumoylated Rta (Fig. 1) implies that Rta is conjugated by ubiquitin via polySUMO-2 chains. Based on the result herein, MG132 indeed stabilizes SUMO-1 and SUMO-2-conjugated Rta (Fig. 1, A and B, IP, lanes 9 and 10). When cells were untreated with MG132, the sumoylated Rta was relatively unstable even in the presence of N-ethylmaleimide, which is a SUMO hydrolase inhibitor (Fig. 1, A and B, IP, lanes 4 and 5). Moreover, because SUMO-1 acts as a chain terminator by linking to the ends of polySUMO-2/3 chains (9), distinguishing the SUMO-1 on the polySUMO chain from multiple monosumoylated Rta in vivo may be difficult. Therefore, MG132 may protect Rta that is conjugated by both SUMO-1 or SUMO-2/3 (Fig. 1). This study also finds that MG132 treatment does not substantially activate the transcription of BRLF1-BZLF1 genes in P3HR1 cells (Fig. 2C) but accumulates endogenously expressed Rta, which are verifiable by immunoblot and immunofluorescence analyses (Fig. 2, A and B), suggesting that MG132 affects the stability rather than the transcriptional functions of Rta.
This study demonstrates that Rta is a substrate of RNF4, which is an STUbL that contains SIM and RING domains (15, 54), causing the ubiquitination of SUMO-2-conjugated PML (15). According to our results, RNF4 enhances the ubiquitination of SUMO-2-conjugated Rta in an in vitro ubiquitination assay (Fig. 5B). Earlier studies demonstrated that RNF4 interacts with either the polySUMO-2 chain on PML or Sp1 directly to promote ubiquitination (15, 27, 28), although whether RNF4 preferentially binds to SUMO-2-conjugated substrates remains unknown (22, 75, 76). GST pulldown assays herein reveal that Rta interacts with RNF4 (Fig. 4A), suggesting that the direct interaction between the two proteins or the interaction with the SUMO-2 chain on Rta is important to Rta ubiquitination. This interaction is also verified by immunofluorescence analysis (Fig. 4C). According to earlier studies, GFP-RNF4 is distributed diffusely in the nucleus (36) but has a punctate distribution and colocalizes with PML nuclear bodies after treatment with arsenic (77). The present study reveals that GFP-RNF4 is diffused in cells that are untreated with MG132. However, MG132 treatment causes the punctate distribution (Fig. 4C), possibly because MG132 treatment changes the cellular distributions of the proteasome and the proteins that are associated with the PML nuclear bodies (78).
This study demonstrates that RNF4 enhances Rta ubiquitination (Figs. 5 and 6). The first piece of evidence is that an abundant amount of ubiquitinated proteins is formed in vitro in the reaction involving SUMO-2-Rta, GST-RNF4, ubiquitin, E1, E2, and ATP (Fig. 5A, lane 10). Immunoprecipitation of these ubiquitinated proteins reveals the formation of Ub-Rta, as detected by immunoblotting using anti-Rta antibody (Fig. 5B), suggesting that SUMO-2-conjugated Rta is a preferred substrate for RNF4 in vitro (Fig. 5B, lane 6). However, demonstrating the occurrence of this phenomenon in vivo is relatively difficult. Moreover, although apparently not influencing the ubiquitination of RNF4 (Fig. 5C, lanes 5–8), Rta likely does not function as an E3 ligase despite the fact that the homologs of Rta in KSHV and murine herpesvirus 68 have E3 ligase activity in vitro (79–82). Additionally, introducing RNF4 shRNA to the cell significantly reduced the level of ubiquitinated Rta (Fig. 6B), indicating that RNF4 is critical to Rta ubiquitination. The fact that mutation of SIMs or a RING domain in RNF4 abolishes the ubiquitination of Rta (Fig. 6, C and D) also supports the role of RNF4 as an STUbL to promote Rta ubiquitination. Moreover, mutations in the SIM domain in RNF4 decrease the interaction of the protein with Rta (Fig. 6E) and abolish Rta ubiquitination (Fig. 6D), demonstrating the importance of such an interaction. The phenomenon in which ubiquitination of the 4K-R mutant of Rta significantly reduces in vivo (Fig. 7A) also suggests that SUMO-2 modification at these four lysine residues is important to Rta ubiquitination; RNF4 also functions as an STUbL of Rta that promotes ubiquitination via polySUMO-2 chains. Furthermore, a previous study indicated that Rta is modified by SUMO-3 (49). By using the SUMO-3-defective Rta mutant (HA-4K-R), this study demonstrates that HA-4K-R is ubiquitinated at a level significantly lower than that of HA-Rta (Fig. 7A). HA-3K-R (44) has a similar ubiquitination pattern to that of wild-type Rta (Fig. 7A), verifying that Rta is not ubiquitinated by RNF4 via sumoylation by SUMO-1. This finding is consistent with the fact that SUMO-2-Rta is a preferred substrate for RNF4 in vitro (Fig. 5B). This study also demonstrates that MG132 treatment of P3HR1 cells leads to the accumulation of SUMO-2-Rta and ubiquitinated Rta simultaneously after lytic induction (Fig. 3B). This finding verifies that Rta is ubiquitinated via SUMO-2 chains. Confocal microscopy analysis also reveals that Rta, SUMO-2, and RNF4 colocalize in the nucleus if cells are treated with MG132 (Fig. 7C). This finding demonstrates that RNF4 indeed binds to SUMO-2-Rta in vivo. In sum, results of this demonstrate that RNF4 is an STUbL of Rta.
This study finds that RNF4 is a cellular STUbL that targets Rta for ubiquitination (Figs. 5 and 6) to decrease the transactivation ability and stability of Rta (Fig. 8). We also demonstrate that inhibiting RNF4 expression enhances the expression of EA-D (Fig. 9A) and promotes lytic DNA replication and virions production (Fig. 9, B and C). Although the influence of Rta ubiquitination on the EBV lytic cycle need to be verified further, this study provides insight into the mechanism that modulates the amount of Rta by the cell, which may be important to EBV lytic development.
Acknowledgments
We thank Shih-Tung Liu for criticism and suggestions. We also thank Ronald T. Hay for providing FLAG-RNF4-CS1 and FLAG-RNF4-mtSIM plasmids and Shih-Chung Chang for providing pHA-Ub, pHA-SUMO-1, and pHA-SUMO-2.
This work was supported by National Health Research Institute Grant NHRI-EX102-10030BI, National Science Council NSC-100-2311-B-002-011, and a National Science Council program, Funding for science and technology personnel engaging in short-term foreign research, for a visit to Kumamoto University, Japan.
Y. C. Yang, Y. Yoshikai, S. W. Hsu, H. Saitoh, and L. K. Chang, manuscript in preparation.
- Ub
- ubiquitin
- SUMO
- small ubiquitin-like modifier
- SIM
- SUMO-interacting motif
- STUbL
- SUMO-targeted ubiquitin ligase
- EBV
- Epstein-Barr virus
- TPA
- 12-O-tetradecanoylphobol-13-acetate
- qPCR
- quantitative PCR
- IP
- immunoprecipitate
- RNF4
- Ring finger protein 4
- PML
- promyelocyte leukemia.
REFERENCES
- 1. Pickart C. M., Fushman D. (2004) Polyubiquitin chains. Polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 2. Grabbe C., Husnjak K., Dikic I. (2011) The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 12, 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 [DOI] [PubMed] [Google Scholar]
- 4. Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Komander D. (2009) The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 37, 937–953 [DOI] [PubMed] [Google Scholar]
- 6. Schwartz D. C., Hochstrasser M. (2003) A superfamily of protein tags. Ubiquitin, SUMO, and related modifiers. Trends Biochem. Sci. 28, 321–328 [DOI] [PubMed] [Google Scholar]
- 7. Johnson E. S. (2004) Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 8. Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., Naismith J. H., Hay R. T. (2001) Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35374 [DOI] [PubMed] [Google Scholar]
- 9. Matic I., van Hagen M., Schimmel J., Macek B., Ogg S. C., Tatham M. H., Hay R. T., Lamond A. I., Mann M., Vertegaal A. C. (2008) In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell Proteomics 7, 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilkinson K. A., Henley J. M. (2010) Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Praefcke G. J., Hofmann K., Dohmen R. J. (2012) SUMO playing tag with ubiquitin. Trends Biochem. Sci. 37, 23–31 [DOI] [PubMed] [Google Scholar]
- 12. Bekker-Jensen S., Mailand N. (2011) The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 585, 2914–2919 [DOI] [PubMed] [Google Scholar]
- 13. Lyst M. J., Stancheva I. (2007) A role for SUMO modification in transcriptional repression and activation. Biochem. Soc. Trans. 35, 1389–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hecker C. M., Rabiller M., Haglund K., Bayer P., Dikic I. (2006) Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281, 16117–16127 [DOI] [PubMed] [Google Scholar]
- 15. Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. (2008) RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10, 538–546 [DOI] [PubMed] [Google Scholar]
- 16. Sun H., Leverson J. D., Hunter T. (2007) Conserved function of RNF4 family proteins in eukaryotes. Targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kerscher O. (2007) SUMO junction. What's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8, 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. González-Santamaría J., Campagna M., García M. A., Marcos-Villar L., González D., Gallego P., Lopitz-Otsoa F., Guerra S., Rodríguez M. S., Esteban M., Rivas C. (2011) Regulation of vaccinia virus E3 protein by small ubiquitin-like modifier proteins. J. Virol. 85, 12890–12900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perry J. J., Tainer J. A., Boddy M. N. (2008) A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 33, 201–208 [DOI] [PubMed] [Google Scholar]
- 20. Schimmel J., Larsen K. M., Matic I., van Hagen M., Cox J., Mann M., Andersen J. S., Vertegaal A. C. (2008) The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol. Cell Proteomics 7, 2107–2122 [DOI] [PubMed] [Google Scholar]
- 21. Uzunova K., Göttsche K., Miteva M., Weisshaar S. R., Glanemann C., Schnellhardt M., Niessen M., Scheel H., Hofmann K., Johnson E. S., Praefcke G. J., Dohmen R. J. (2007) Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282, 34167–34175 [DOI] [PubMed] [Google Scholar]
- 22. Geoffroy M. C., Hay R. T. (2009) An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 10, 564–568 [DOI] [PubMed] [Google Scholar]
- 23. Prudden J., Pebernard S., Raffa G., Slavin D. A., Perry J. J., Tainer J. A., McGowan C. H., Boddy M. N. (2007) SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26, 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heideker J., Perry J. J., Boddy M. N. (2009) Genome stability roles of SUMO-targeted ubiquitin ligases. DNA Repair 8, 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mullen J. R., Brill S. J. (2008) Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J. Biol. Chem. 283, 19912–19921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Häkli M., Lorick K. L., Weissman A. M., Jänne O. A., Palvimo J. J. (2004) Transcriptional coregulator SNURF (RNF4) possesses ubiquitin E3 ligase activity. FEBS Lett. 560, 56–62 [DOI] [PubMed] [Google Scholar]
- 27. Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de Thé H. (2008) Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10, 547–555 [DOI] [PubMed] [Google Scholar]
- 28. Wang Y. T., Yang W. B., Chang W. C., Hung J. J. (2011) Interplay of posttranslational modifications in Sp1 mediates Sp1 stability during cell cycle progression. J. Mol. Biol. 414, 1–14 [DOI] [PubMed] [Google Scholar]
- 29. van Hagen M., Overmeer R. M., Abolvardi S. S., Vertegaal A. C. (2010) RNF4 and VHL regulate the proteasomal degradation of SUMO-conjugated hypoxia-inducible factor-2α. Nucleic Acids Res. 38, 1922–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mukhopadhyay D., Arnaoutov A., Dasso M. (2010) The SUMO protease SENP6 is essential for inner kinetochore assembly. J. Cell Biol. 188, 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Plechanovová A., Jaffray E. G., McMahon S. A., Johnson K. A., Navrátilová I., Naismith J. H., Hay R. T. (2011) Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat. Struct. Mol. Biol. 18, 1052–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu X. V., Rodrigues T. M., Tao H., Baker R. K., Miraglia L., Orth A. P., Lyons G. E., Schultz P. G., Wu X. (2010) Identification of RING finger protein 4 (RNF4) as a modulator of DNA demethylation through a functional genomics screen. Proc. Natl. Acad. Sci. U.S.A. 107, 15087–15092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yin Y., Seifert A., Chua J. S., Maure J. F., Golebiowski F., Hay R. T. (2012) SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26, 1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Galanty Y., Belotserkovskaya R., Coates J., Jackson S. P. (2012) RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26, 1179–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luo K., Zhang H., Wang L., Yuan J., Lou Z. (2012) Sumoylation of MDC1 is important for proper DNA damage response. EMBO J. 31, 3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fryrear K. A., Guo X., Kerscher O., Semmes O. J. (2012) The Sumo-targeted ubiquitin ligase RNF4 regulates the localization and function of the HTLV-1 oncoprotein Tax. Blood 119, 1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chevallier-Greco A., Manet E., Chavrier P., Mosnier C., Daillie J., Sergeant A. (1986) Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5, 3243–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kenney S., Holley-Guthrie E., Mar E. C., Smith M. (1989) The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J. Virol. 63, 3878–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Holley-Guthrie E. A., Quinlivan E. B., Mar E. C., Kenney S. (1990) The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J. Virol. 64, 3753–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Giot J. F., Mikaelian I., Buisson M., Manet E., Joab I., Nicolas J. C., Sergeant A. (1991) Transcriptional interference between the EBV transcription factors EB1 and R. both DNA-binding and activation domains of EB1 are required. Nucleic Acids Res. 19, 1251–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chang L. K., Chuang J. Y., Nakao M., Liu S. T. (2010) MCAF1 and synergistic activation of the transcription of Epstein-Barr virus lytic genes by Rta and Zta. Nucleic Acids Res. 38, 4687–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ye J., Gradoville L., Daigle D., Miller G. (2007) De novo protein synthesis is required for lytic cycle reactivation of Epstein-Barr virus, but not Kaposi's sarcoma-associated herpesvirus, in response to histone deacetylase inhibitors and protein kinase C agonists. J. Virol. 81, 9279–9291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ye J., Gradoville L., Miller G. (2010) Cellular immediate-early gene expression occurs kinetically upstream of Epstein-Barr virus bzlf1 and brlf1 following cross-linking of the B cell antigen receptor in the Akata Burkitt lymphoma cell line. J. Virol. 84, 12405–12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chang L. K., Lee Y. H., Cheng T. S., Hong Y. R., Lu P. J., Wang J. J., Wang W. H., Kuo C. W., Li S. S., Liu S. T. (2004) Post-translational modification of Rta of Epstein-Barr virus by SUMO-1. J. Biol. Chem. 279, 38803–38812 [DOI] [PubMed] [Google Scholar]
- 45. Liu S. T., Wang W. H., Hong Y. R., Chuang J. Y., Lu P. J., Chang L. K. (2006) Sumoylation of Rta of Epstein-Barr virus is preferentially enhanced by PIASxβ. Virus. Res. 119, 163–170 [DOI] [PubMed] [Google Scholar]
- 46. Chang L. K., Liu S. T., Kuo C. W., Wang W. H., Chuang J. Y., Bianchi E., Hong Y. R. (2008) Enhancement of transactivation activity of Rta of Epstein-Barr virus by RanBPM. J. Mol. Biol. 379, 231–242 [DOI] [PubMed] [Google Scholar]
- 47. Calderwood M. A., Venkatesan K., Xing L., Chase M. R., Vazquez A., Holthaus A. M., Ewence A. E., Li N., Hirozane-Kishikawa T., Hill D. E., Vidal M., Kieff E., Johannsen E. (2007) Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U.S.A. 104, 7606–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Calderwood M. A., Holthaus A. M., Johannsen E. (2008) The Epstein-Barr virus LF2 protein inhibits viral replication. J. Virol. 82, 8509–8519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heilmann A. M., Calderwood M. A., Johannsen E. (2010) Epstein-Barr virus LF2 protein regulates viral replication by altering Rta subcellular localization. J. Virol. 84, 9920–9931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zacny V. L., Wilson J., Pagano J. S. (1998) The Epstein-Barr virus immediate-early gene product, BRLF1, interacts with the retinoblastoma protein during the viral lytic cycle. J. Virol. 72, 8043–8051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davies A. H., Grand R. J., Evans F. J., Rickinson A. B. (1991) Induction of Epstein-Barr virus lytic cycle by tumor-promoting and non-tumor-promoting phorbol esters requires active protein kinase C. J. Virol. 65, 6838–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luka J., Kallin B., Klein G. (1979) Induction of the Epstein-Barr virus (EBV) cycle in latently infected cells by n-butyrate. Virology 94, 228–231 [DOI] [PubMed] [Google Scholar]
- 53. Uchimura Y., Nakamura M., Sugasawa K., Nakao M., Saitoh H. (2004) Overproduction of eukaryotic SUMO-1- and SUMO-2-conjugated proteins in Escherichia coli. Anal. Biochem. 331, 204–206 [DOI] [PubMed] [Google Scholar]
- 54. Poukka H., Aarnisalo P., Santti H., Jänne O. A., Palvimo J. J. (2000) Coregulator small nuclear RING finger protein (SNURF) enhances Sp1- and steroid receptor-mediated transcription by different mechanisms. J. Biol. Chem. 275, 571–579 [DOI] [PubMed] [Google Scholar]
- 55. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 56. Tojo M., Matsuzaki K., Minami T., Honda Y., Yasuda H., Chiba T., Saya H., Fujii-Kuriyama Y., Nakao M. (2002) The aryl hydrocarbon receptor nuclear transporter is modulated by the SUMO-1 conjugation system. J. Biol. Chem. 277, 46576–46585 [DOI] [PubMed] [Google Scholar]
- 57. Sapetschnig A., Rischitor G., Braun H., Doll A., Schergaut M., Melchior F., Suske G. (2002) Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 21, 5206–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chang P. J., Chang Y. S., Liu S. T. (1998) Role of Rta in the translation of bicistronic BZLF1 of Epstein-Barr virus. J. Virol. 72, 5128–5136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chang F. R., Hsieh Y. C., Chang Y. F., Lee K. H., Wu Y. C., Chang L. K. (2010) Inhibition of the Epstein-Barr virus lytic cycle by moronic acid. Antiviral Res. 85, 490–495 [DOI] [PubMed] [Google Scholar]
- 60. Wiedmer A., Wang P., Zhou J., Rennekamp A. J., Tiranti V., Zeviani M., Lieberman P. M. (2008) Epstein-Barr virus immediate-early protein Zta co-opts mitochondrial single-stranded DNA binding protein to promote viral and inhibit mitochondrial DNA replication. J. Virol. 82, 4647–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chiu Y. F., Tung C. P., Lee Y. H., Wang W. H., Li C., Hung J. Y., Wang C. Y., Kawaguchi Y., Liu S. T. (2007) A comprehensive library of mutations of Epstein Barr virus. J. Gen. Virol. 88, 2463–2472 [DOI] [PubMed] [Google Scholar]
- 62. Sun W., Tan X., Shi Y., Xu G., Mao R., Gu X., Fan Y., Yu Y., Burlingame S., Zhang H., Rednam S. P., Lu X., Zhang T., Fu S., Cao G., Qin J., Yang J. (2010) USP11 negatively regulates TNFα-induced NF-κB activation by targeting on IκBα. Cell. Signal. 22, 386–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Oh Y., Jeon Y. J., Hong G. S., Kim I., Woo H. N., Jung Y. K. (2012) Regulation in the targeting of TRAIL receptor 1 to cell surface via GODZ for TRAIL sensitivity in tumor cells. Cell Death Differ. 19, 1196–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Crawford D. H., Ando I. (1986) EB virus induction is associated with B-cell maturation. Immunology 59, 405–409 [PMC free article] [PubMed] [Google Scholar]
- 65. Countryman J., Gradoville L., Bhaduri-McIntosh S., Ye J., Heston L., Himmelfarb S., Shedd D., Miller G. (2009) Stimulus duration and response time independently influence the kinetics of lytic cycle reactivation of Epstein-Barr virus. J. Virol. 83, 10694–10709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Countryman J. K., Gradoville L., Miller G. (2008) Histone hyperacetylation occurs on promoters of lytic cycle regulatory genes in Epstein-Barr virus-infected cell lines which are refractory to disruption of latency by histone deacetylase inhibitors. J. Virol. 82, 4706–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gradoville L., Kwa D., El-Guindy A., Miller G. (2002) Protein kinase C-independent activation of the Epstein-Barr virus lytic cycle. J. Virol. 76, 5612–5626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Murata T., Kondo Y., Sugimoto A., Kawashima D., Saito S., Isomura H., Kanda T., Tsurumi T. (2012) Epigenetic histone modification of Epstein-Barr virus BZLF1 promoter during latency and reactivation in Raji cells. J. Virol. 86, 4752–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Feederle R., Kost M., Baumann M., Janz A., Drouet E., Hammerschmidt W., Delecluse H. J. (2000) The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19, 3080–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liu C., Sista N. D., Pagano J. S. (1996) Activation of the Epstein-Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J. Virol. 70, 2545–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gruffat H., Manet E., Rigolet A., Sergeant A. (1990) The enhancer factor R of Epstein-Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res. 18, 6835–6843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chang L. K., Chung J. Y., Hong Y. R., Ichimura T., Nakao M., Liu S. T. (2005) Activation of Sp1-mediated transcription by Rta of Epstein-Barr virus via an interaction with MCAF1. Nucleic Acids Res. 33, 6528–6539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Adamson A. L., Darr D., Holley-Guthrie E., Johnson R. A., Mauser A., Swenson J., Kenney S. (2000) Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J. Virol. 74, 1224–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee D. H., Goldberg A. L. (1998) Proteasome inhibitors. valuable new tools for cell biologists. Trends Cell Biol. 8, 397–403 [DOI] [PubMed] [Google Scholar]
- 75. Wang Z., Prelich G. (2009) Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol. Cell Biol. 29, 1694–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Parker J. L., Ulrich H. D. (2012) A SUMO-interacting motif activates budding yeast ubiquitin ligase Rad18 towards SUMO-modified PCNA. Nucleic Acids Res. 40, 11380–11388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Geoffroy M. C., Jaffray E. G., Walker K. J., Hay R. T. (2010) Arsenic-induced SUMO-dependent recruitment of RNF4 into PML nuclear bodies. Mol. Biol. Cell 21, 4227–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mattsson K., Pokrovskaja K., Kiss C., Klein G., Szekely L. (2001) Proteins associated with the promyelocytic leukemia gene product (PML)-containing nuclear body move to the nucleolus upon inhibition of proteasome-dependent protein degradation. Proc. Natl. Acad. Sci. U.S.A. 98, 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yang Z., Yan Z., Wood C. (2008) Kaposi's sarcoma-associated herpesvirus transactivator RTA promotes degradation of the repressors to regulate viral lytic replication. J. Virol. 82, 3590–3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yu Y., Wang S. E., Hayward G. S. (2005) The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 22, 59–70 [DOI] [PubMed] [Google Scholar]
- 81. Gould F., Harrison S. M., Hewitt E. W., Whitehouse A. (2009) Kaposi's sarcoma-associated herpesvirus RTA promotes degradation of the Hey1 repressor protein through the ubiquitin proteasome pathway. J. Virol. 83, 6727–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dong X., He Z., Durakoglugil D., Arneson L., Shen Y., Feng P. (2012) Murine gammaherpesvirus 68 evades host cytokine production via replication transactivator-induced RelA degradation. J. Virol. 86, 1930–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]