Abstract
A central undertaking in synthetic biology (SB) is the quest for the ‘minimal genome’. However, ‘minimal sets’ of essential genes are strongly context-dependent and, in all prokaryotic genomes sequenced to date, not a single protein-coding gene is entirely conserved. Furthermore, a lack of consensus in the field as to what attributes make a gene truly essential adds another aspect of variation. Thus, a universal minimal genome remains elusive. Here, as an alternative to defining a minimal genome, we propose that the concept of gene persistence can be used to classify genes needed for robust long-term survival. Persistent genes, although not ubiquitous, are conserved in a majority of genomes, tend to be expressed at high levels, and are frequently located on the leading DNA strand. These criteria impose constraints on genome organization, and these are important considerations for engineering cells and for creating cellular life-like forms in SB.
Keywords: LUCA, minimal cell, minimal genome, synthetic genomics, chassis, xenobiology
The Holy Grail of SB
The goal of SB is to engineer cells for useful applications, while contributing to our understanding of the origin of life on Earth [1]. A core undertaking in SB has been the quest for the minimal set of genes required to allow cellular life; that is, the ‘minimal genome’ concept (Box 1). The assumption of the minimal genome is to use it as a scaffold onto which genes can be added, followed by its transplantation into a chassis (see Glossary). The final aim is to build upon the chassis through inclusion of specialized genes to create ‘turbo cells’ for applications in the fields of energy production, health, and the environment [2]. These applications require an understanding of minimal genomes onto which these specialized genes can be grafted.
Box 1. From the minimal genome to synthetic genomics.
The quest for the ‘smallest autonomous self-replicating entity’ started in the 1960s when pleuropneumonia-like organisms (Mollicutes) were recognized as the smallest cultivable microorganisms on Earth. With the emergence of molecular biology, the object of the search for the smallest organism shifted towards the organism with the smallest genome [38]. We now know that Mollicutes genomes, with sizes ranging from 600 to 2200 kb, arose from a Streptococcus strain with genome of ∼2000 kb. This gene attrition indicates that the Mollicutes did not arise directly from a ‘founding’ organism [39]. Nevertheless, Mycoplasmas remained model organisms to study the elusive ‘minimal genome’. In 1995, the sequencing of the genome (ca 580 kb) of Mycoplasma genitalium predicted 470 protein-coding genes (CDSs) [40]. Subsequently, transposon mutagenesis in M. genitalium reduced the minimal set to 265 ‘essential’ CDSs [41], but this number later increased to 382 CDSs and 43 structural RNA genes [42]. This discrepancy prompted the synthesis of the ‘minimal genome’ of M. genitalium [43], supported by the idea that it was a crucial prerequisite for the success of SB [44]. Hard work [24] and technological developments [45] enabled the chemical synthesis and transplantation of a minimal genome of Mycoplasma into phylogenetically-related cells [25]. Although this experiment has made SB a priority in biotechnology agendas [46], there are still several issues to be addressed. First, organisms with a modified minimal genome could have impaired reproduction or shortened lifespan. Second, the transplantation method could be valid only for Mycoplasma lacking a cell wall because it uses polyethylene glycol, which is known to fuse cell membranes, and it would therefore be challenging to use this method in other species. Third, the assembly method may be also limited to Mycoplasma because its genetic code is slightly different to that of yeast, the host where the genome was assembled. In Mycoplasma, the UGA triplet codes for tryptophan, but in yeast it determines a stop codon, and therefore the possibility that genes of genomes from other organisms could be toxic for yeast cannot be discarded [47]. Fourth, when the first synthetic cell (amoeba) was reassembled in 1970, the nucleus transplant only produced viable cells when the components (nucleus, cytoplasm, and cell membrane) were taken from the same strain [48]. Thus it remains to be seen whether synthetic genomes can be ‘rebooted’ in phenotypically distant strains. Fifth, even if genome transplantations were successful in distant strains, the expression of many genes could be incompatible, as reported when the genome of Synechocystis was cloned into that of B. subtilis 168 [49]. With the advent of synthetic genomics, other projects have just begun [50].
Attempts to define the minimal genome in microorganisms have employed both random and targeted reductions to strip down nonessential genes as well as comparative genomics. These approaches have yielded valuable information on basic processes required for life, but they have not been entirely successful in their stated goal, and our understanding of minimal genomes still remains limited [3]. Moreover, the lack of consensus in the field as to how to define gene essentiality further complicates the issue.
Here, we suggest using metrics of gene persistence as a constructive way to identify the minimal universal functions that support robust cellular life. We review work combining experiments on gene essentiality with in silico comparative genomics that support our belief that searching for a universal minimal genome is unproductive, highlighting the advantages of this new approach. Assembling rationally designed sets of persistent genes should enable the successful engineering of genomes. A deeper analysis of persistent functions also provides an opportunity to explore the evolution of cells from the origin of life to the extant microbial diversity. This has important implications for designing other evolvable synthetic lifeforms.
The elusive minimal genome
For years, scientists have explored ways to define a universal minimal genome. Some efforts have focused on gene mutagenesis experiments, but ‘minimal gene sets’ have remained problematic because these experiments do not take into account gene–environment interactions. Others have centered on comparative genomics, and this allowed scientists to compare genomes from closely or distantly related microorganisms. But as an ever-increasing number of genome projects were completed, the outcome of the comparisons did not improve. As a consequence, even in combination, these approaches failed to provide a universal minimal genome.
The minimal genome in vivo
The first attempts to delineate minimal gene sets arose from experiments meant to identify novel drug targets by determining which genes were essential for the survival of a pathogen. These studies were carried out on libraries of microorganisms using transposons or antisense RNA expression [4], but the ‘minimal set’ outcomes differed widely in terms of gene number (and often identity), not only in distant organisms but also in the same organism under different (and sometimes similar) conditions (Table 1). This environmental context-dependency reflects the existence of many ‘minimal cells’ with different ‘minimal genome’ versions.
Table 1.
Minimal gene sets obtained by direct and random experimental mutagenesis
| Microorganism | Minimal gene seta | Method | Refs |
|---|---|---|---|
| Cell factories/model organisms | |||
| Acinetobacter baylyi | 205/499 | DGIb | [51] |
| Bacillus subtilis | 217 | DGI | [52] |
| Corynebacterium glutamicum | 658 | RTMc | [53] |
| Caulobacter crescentus | 480 | RTM | [54] |
| Escherichia coli | 620 | RTM | [11] |
| 303 | DGI | [12] | |
| 302 | DGI | [55] | |
| Saccharomyces cerevisiae | 1105 | DGI | [56] |
| Human pathogen | |||
| Bacillus anthracis | 253 | RTM | [57] |
| Francisella tularensis sp. novicida | 396 | RTM | [58] |
| Haemophilus influenzae | 670 | RTM | [59] |
| 136/358 | RTM | [60] | |
| Helicobacter pylori | 255–344 | RTM | [61] |
| Mycobacterium tuberculosis | ∼614 | RTM | [62] |
| Mycoplasma genitalium | 265–350 | RTM | [41] |
| 382d | RTM | [42] | |
| Mycoplasma pulmonis | 321 | RTM | [63] |
| Pseudomonas aeruginosa | 335 | RTM | [64] |
| Salmonella enterica ser. Typhimurium | 257–490 | RTM | [65] |
| Streptococcus pneumoniae | 82 | RTM | [66] |
| Staphylococcus aureus | 71 | RTM | [67] |
| 351 | RTM | [68] | |
| 150/600 | Random antisense RNA | [69] | |
| Vibrio cholerae | 789 | RTM | [70] |
Protein-coding genes.
DGI, direct gene inactivation.
RTM, Random transposon mutagenesis.
The study additionally suggested 43 RNA genes, in other words the first minimal gene set of 405 genes including RNA genes.
Targeted methods were also developed to eliminate one gene at a time to generate collections of knockout strains (Table 1). However, the simultaneous elimination of two individually nonessential genes may lead to a lethal phenotype, an outcome commonly known as ‘synthetic lethality’ due to mutually inclusive mutations [5]. Conversely, some genes may be individually essential but, in combination with a second disruption, the first disruption becomes tolerated. This phenomenon was reported in genes of toxin–antitoxin systems due to mutually exclusive mutations [6]. Therefore, mutually inclusive and exclusive mutations preclude the cumulative elimination of dispensable genes in a single strain. The recent quantitative concept of ‘degree of essentiality’ aims at providing a framework to determine synthetic lethal interactions [7]. This approach could then be combined with advanced genome engineering tools to disrupt dispensable genes rationally in a cumulative manner for applications in SB [8].
The minimal genome in silico
Based on the first two bacterial genomes available, a ‘minimal set’ of 256 genes was identified via comparative genomics, but this number dropped to 63 when 100 genomes were examined [4] and to 0 when 1000 genomes were compared [9]. The number of universally conserved genes, however, often depends on the species of the tree of life chosen. This is illustrated by the fact that two protein-coding (elongation factor and ribosomal protein S12) and two non-coding (16S and 23S rRNAs) genes are conserved in 930 of the available 1000 bacterial genomes, but these genes are not conserved in 70 archaeal genomes [9].
Naturally evolved symbionts with reduced genome sizes were also considered as a way to assess the minimal number of genes required for life, but again this showed little consistency in terms of gene number and identity of essential genes [10]. Indeed, obligatory intracellular symbionts and parasites with host-associated lifestyles have considerably relaxed selection on the maintenance of genes that are not required in their protected environments (e.g., synthesis of essential amino acids or vitamins). Therefore, per definition, ‘minimal genomes’ of symbionts and parasite are ecologically constrained.
Combining computational and experimental approaches to define a minimal genome is also problematic. Comparing minimal gene sets characterized by two studies for Escherichia coli K-12 revealed an overlap of only 205 genes out of 620 genes [11] and 303 genes [12]. This discrepancy is due to differences in the interpretation of essentiality based on bacterial growth (slow versus rapid growth) and the method used to generate the mutant strains (targeted deletion versus random transposon insertion). Furthermore, if the gene set in the latter study is compared with other genomes, the following numbers of conserved genes are found: (i) 282 genes (90%) among three E. coli species, (ii) 147 genes (49%) among 20 different enterobacteria, (iii) 85 genes (28%) among 74 proteobacteria, and (iv) 42 genes (14%) among 171 bacteria [12].
Another example compared the minimal sets obtained in vitro and in silico from symbionts and free-living organisms, and only 206 universal genes were identified [13]. However, when the robustness of the metabolic network derived from this minimal set of 206 genes was explored in silico, the results suggested that this set would make a very fragile network, indicating that more genes would be required for a truly sustainable lifeform [14]. The main issue is that minimal cells endowed with a minimal genome are adapted to ideal environments, typically nutrient-rich media and relatively constant temperature. However, the elimination of stress-response genes, determined as dispensable under ideal conditions, results in cell death upon mild changes of temperature or nutrient availability [15]. In addition, the elimination of genes from the toxin/antitoxin or restriction/methylase systems, which are dispensable for simple growth on solid media, would render cells vulnerable to infections by phages or other microorganisms in a natural environment. Thus, minimal cells are fragile and restricted to various ecological niches.
Genomes as late inventions of cellular life
A major failing of the minimal genome concept is that it assumes a unique origin of cellular life based on the genome of the ‘last universal common ancestor’ (LUCA) [16]. However, recent research based on comparative proteomics hints that LUCA could have given rise to a community of primordial cells, which were in turn the genetic founders of the three domains of life: Archaea, Eubacteria, and Eukarya [17]. Therefore, it is likely that DNA-based genomes may have developed at a late stage of cellular evolution in which the enzymes involved in DNA replication [18], lipid biosynthesis [19], and RNA degradation pathways [20] were invented not once, but multiple times.
Taken together, we believe that these results are indicative of the futility of defining a universal minimal genome, in large part because different essential functions depend on highly diverse environmental constraints, and because life does not appear to have evolved around such a basic unit. Thus, the focus should shift away from the universal minimal genome and towards a more robust and general way to reevaluate the essentiality of a gene. Here, we suggest that the concept of gene persistence can be used to assess the in/dispensability of a gene and provide a list of universal functions shared by living cells that should guide future synthetic biologists as they assemble synthetic constructs.
Gene persistence as a metric of functional essentiality
As discussed above, the number of universally conserved genes, assumed to be essential, drops to 0 as more genomes are compared, especially if many different species from different branches of life are considered. Nevertheless, important genes are preserved and passed on, and this is reflected in gene persistence – in other words, in the fact that some genes, although not ubiquitous, are conserved in the majority of genomes and are distributed throughout the tree of life (Figure 1). This indicates that even if a gene ortholog is not found in the genomes of particular microbial clades, another family of genes might encode the corresponding function.
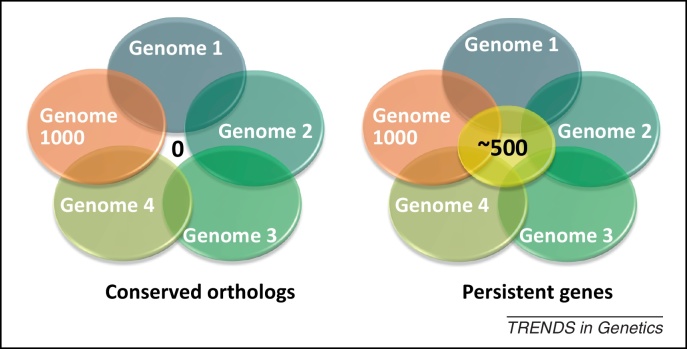
Figure 1.
Different criteria for defining universally conserved genes. When 1000 genomes are compared via comparative genomics, the number of orthologous genes falls to 0 (left), but this number can increase to about 500 persistent genes by comparing orthologs that belong to a quorum of a similar or different genomes from evolutionarily distinct bacteria, above a threshold computed using a measure that retains frequent genes that tend to cluster together (right).
When designing synthetic life, the concept of gene persistence can be used as a metric, replacing gene essentiality. Persistent genes can be identified via gene orthology (Box 2) and are defined by several characteristics: they tend to be expressed at high levels [21], and they are preferentially located on the leading DNA strand [22,23]. This has implications for engineering because replication and transcription occur simultaneously on the same DNA molecule; this biased gene distribution helps in avoiding collisions between the respective machineries. Accordingly, gene persistence suggests strong constraints on genome organization, which should be taken into account by engineers designing robust synthetic cells.
Box 2. Finding persistent genes.
When more than 1000 bacterial genomes were compared with each other no single ortholog was found to be conserved [9]. Nevertheless, major functions allowing cells to make an envelope, develop and maintain energy and intermediary metabolism, and express and replicate genes encoded in DNA must be universally present. Functional ubiquity cannot be equated to sequence/structural ubiquity. Functional constraints, however, are generally important, and once a structure has been found that fulfills a function, it has a tendency (not an absolute need because of possible functional redundancy) to be transmitted to the progeny. As a consequence, orthologs of genes encoding functions essential for life in the short and long term tend to be present in a significant number of genomes. The basis of the idea to identify these ‘persistent’ genes is to look for orthologs that are present in a predefined quorum of genomes. This is of course somewhat arbitrary and, naturally, because the various genomes that have been sequenced have been chosen with biased interests, the absolute number of genomes with the same orthologs cannot be used directly (typically, because dozens of E. coli genomes have been sequenced, genes orthologous to E. coli genes will be found more often than genes for other organisms). Hence one must use a qualifier to compute the cutoff threshold that derives from the quorum, taking into account the phylogenetic distance between organisms: the more distant an organism, the more important its contribution (typically all E. coli strains will count as a single organism) [20,32]. Further refinements must also be included to compute the threshold by taking into account, in particular, the tendency of genes important for life to be located on the DNA-replication leading strand [32] and to cluster together [33]. This heuristic approach allows one to identify some 500 persistent genes in all genomes larger than 1500 kb [31]. Among these about half cannot be inactivated without losing the capacity for model organisms to make a colony on a plate supplemented with rich media under stable conditions (these genes are deemed persistent essential genes). The second half are not essential under such conditions, but become so when the growth medium is metabolically imbalanced (a processed named ‘metabolic frustration’; e.g., excess of serine in the absence of isoleucine [32]), when cells are submitted to random thermal transitions, or if one looks for the ability of a colony to give a long-term progeny (A.D. and A. Sekowska, unpublished).
Engineering constraints for genomes and chasses
One way to assess the persistence of a gene is to conceptually reverse-engineer life. By considering the necessary components of life, one can work backwards to identify the functions that have persisted throughout various organisms that have evolved under many different environmental conditions. Once these functions have been identified, they can be assigned to genes based on the criteria used to define persistence (Box 2).
We know that life requires at least three interrelated components: (i) genetic program, (ii) metabolism, and (iii) compartmentalization. Although we can synthesize a genetic program from scratch relatively easily [24], our ability to design it de novo is still limited [25]. One challenge lies in understanding the physical constraints of the genome such as organization, codon bias, and conformation [26]. Furthermore, we should not forget that any engineered cellular chassis will need safety valves to control osmotic pressure, transporters for discarding useless metabolic products, and the ability to cope with leftovers resulting from macromolecule degradation, all of which are indispensable functions for cellular maintenance and robust growth [27]. The presence of genes involved in these processes must be ubiquitous. However, like chopsticks and forks, things with the same function do not need to resemble each other. As a case in point, an essential function such as degradation of very short RNA leftovers requires nanoRNases that come from a variety of origins (Orn, NrnA, NrnB, NrnC); sometimes these are considered essential because a unique gene exists in a given organism (orn in E. coli), or apparently nonessential because of functional redundancy (nrnA, nrnB) [28]. If this persistent function would be considered nonessential and eliminated in a particular genome, the cells will inevitably age, lose their capacity to generate progeny, and die [29].
An additional important outcome of considering persistent over essential genes in SB is that the former could provide an inventory of essential functions as ‘parts’ to which the corresponding gene sequences could be listed depending on the ‘chassis’. For example, a part could be an essential amino acid, but if a given microorganism lives in a rich environment, the chassis of that organism would have to include a receptor/uptake mechanism. However, if an organism lives in a poor environment, it would have to synthesize the essential amino acid by itself. It is important to mention that the biosynthesis of essential amino acids or coenzymes is highly variable although, as illustrated in the MetaCyc database, for example, that contains multiple pathways for NAD or lysine biosynthesis (respectively, three or six pathways) [30]. Thus, that there are many solutions to the same problem is another aspect that gene persistence considers. Finally, analysis of gene persistence also provides a list of universal functions shared by most bacterial genomes. Accordingly, genomes can be divided in two classes: the ‘paleome’ containing persistent genes and the ‘cenome’ composed of nonpersistent genes [31]. This delineation is important for engineering goals because the paleome corresponds to a universal core, whereas the cenome provides accessory functions for particular niches (vide infra).
Paleome and cenome
The paleome, or old genome, can be defined as an early archive of the origin of life [31]. It contains persistent genes involved in essential functions related to growth, replication, transcription, translation, maintenance, as well as in aging and senescence [31]. The paleome can be further subdivided into two main functionalities: persistent essential genes that allow cells to sustain life, reproduce, and replicate their DNA, and a set of persistent nonessential genes (as determined experimentally in many cases), which are mainly involved in cellular maintenance and stress response [32]. These ‘dispensable’ genes should be particularly important in SB because their elimination can result in cell death upon environmental fluctuations [15]. Structurally, the paleome is composed of about 500 persistent genes (see borderline genes in [31]) distributed in three clusters: (i) core metabolism and synthesis of amino acids, nucleotides, coenzymes, and lipids, (ii) cell division and aminoacyl-tRNA synthetases, and (iii) transcription and translation [31]. A significant proportion of persistent genes allow cells to adapt by evolving while maintaining important functional elements [29].
Although the paleome will allow survival in an optimal environment, many more genes are required for dealing with natural environments. These nonpersistent genes comprise the cenome, or community genome, a set of genes whose functions are necessary for an organism to exploit particular niches by sensing, moving, or scavenging [31]. These genes tend to move from organism to organism by horizontal gene transfer, which accounts for the fact that they tend to cluster together within the genome [33]. The cenome is extremely variable and differs from strain to strain in a given species. Whereas the cenome of a given species is a subset of the corresponding pan-genome of a particular species in a particular niche, the sum of the paleome and all of the cenomes corresponds to the pan-genome of all strains of a given species (Figure 2).
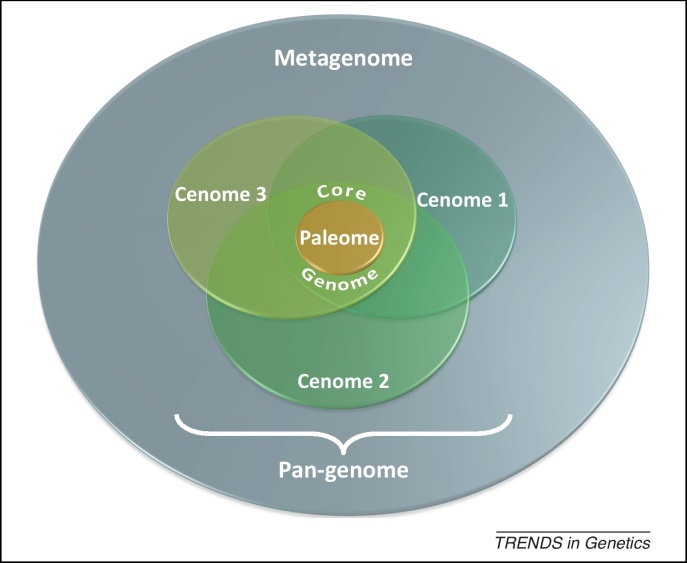
Figure 2.
A universe of gene functions. In a particular environment, the sum of all microbial genes corresponds to the metagenome, which is in turned formed by pan-genomes. A pan-genome is the sum of all genomes of similar strains; each having similar (core genome) or distinct (cenomes) sets of nonpersistent genes. About ∼500 persistent genes form the paleome. As an example, the addition of 1500 nonpersistent genes to the 500 persistent genes of the paleome in E. coli makes a core genome of 2000 genes, whereas the sum of all cenomes of each individual E. coli strain comprises about 18 000 genes [71]. For the time being, the pan-genome of E. coli is composed of roughly 20 000 genes (2000 of the core-genome and 18 000 of the cenomes), the majority of which (80%) is often colocalized on genomic islands [72]. For a particular E. coli strain with a genome of 4500 genes the cenome alone would be about 4000 genes.
Concluding remarks
Like the Holy Grail, a universal DNA ‘minimal genome’ has remained elusive despite efforts to define it. This is partially due to the strong context-dependency of essential genes and the likelihood that DNA-based genomes may have developed at a late stage of cellular evolution. Furthermore, many functions may be fulfilled by a variety of gene products, precluding ubiquitous conservation between species. Therefore, gene essentiality has to be defined within the specific context of the bacterium, growth conditions, and possible environmental fluctuations. This presents a bewildering number of conditions to consider, but gene persistence can be used as an alternative because it provides a more general framework for defining the requirements for long-term survival via identification of universal functions. These functions are contained in the paleome, which provides the core of the cell chassis, whereas the cenome corresponds to nonpersistent genes required to explore a particular niche. These concepts are useful for engineering life for a particular context-dependent application: first, identify a specific chassis (i.e., one suited to the specific environmental conditions), then rationally delete nonpersistent (or truly dispensable) functions [8] to leave behind the paleome and a reduced cenome, and finally add particular sets of functions (extracted from known cenomes or metagenomics projects) helpful for the application in question. It is important to note that it should be easier to design synthetic constructs for scaling-up in a fermenter [27], than for applications in the environment [34], because there are more fluctuations in the latter. In this case, experimentally determining whether a gene is persistent would require evaluating the survival of a mutant in laboratory adaptive-evolution experiments [35], where fluctuation of nutrients or changing environmental conditions could take place, for instance, in a chemostat or turbidostat. This will also enable bottom-up tinkerers [36] and xenobiologists [37] to evolve other similar synthetic lifeforms.
Acknowledgments
The authors are very thankful to the steering committee of the 1st Experimental Round Table Conference (ERTC) on Synthetic Biology, between the Max Planck Society and the Chinese Academy of Sciences, for allowing discussions and the drafting of this manuscript, especially to Nediljko Budisa, Christiane Walch-Solimena, and Gerhard Wegner. We also thank the anonymous referees for their insightful comments and useful suggestions for improving the manuscript. C.G.A.R. is grateful to Manfred T. Reetz for financial support. M.S. acknowledges the financial support provided by the FWF (Austrian Science Fund) projects ‘Investigating the biosafety and risk assessment needs of synthetic biology in Austria (Europe) and China’ (I215-B17) and SYNMOD (I490-B12), as well as the support of FP7 projects METACODE (289572) and ST-FLOW (289326). A.D. acknowledges the support of European Commission FP7 project MICROME (222886-2).
Glossary
- Cenome
nonpersistent genes allowing life in context. It differs in gene number and identity from species to species.
- Chassis
a cellular container, compartment, or envelope containing a metabolic system (together comprising the ‘hardware’), without a genetic program or genome (‘software’).
- Essential gene
a gene necessary for context-dependent growth, mostly involved in basic cellular processes such as translation, transcription, and replication, as well as the synthesis of basic building blocks.
- Metagenome
all microbial genes from a specific environment.
- Minimal cell
the least complex cellular unit supporting life. There are many types of minimal cells depending on the environment.
- Minimal genome
the minimal set of genes required by an organism to support cellular life in an ideal environment. It can be also considered as the minimal species-specific ‘operating system’ or ‘software’ for cellular life.
- Nonessential gene
a gene dispensable for context-dependent growth whose function is generally redundant.
- Paleome
persistent genes that constitute an archive of the origin of life. A closer look at the organization of the paleome allows the exploration of cellular evolution from a basic to a more complex metabolism.
- Pan-genome
the sum of the paleome and the cenome for a particular group of strains.
- Persistent gene
a gene preferentially expressed at higher rates and located on the DNA leading strand, coding for functions either essential for long-term generation of the progeny of a cell (see essential genes) or involved in maintenance and repair. Persistent genes, although not ubiquitous, are conserved in a fair number of bacterial genomes.
- Persistent nonessential gene
a gene showing similar features to persistent genes, and whose elimination is not lethal for short-term growth, but is lethal for long-term growth.
- Synthetic lethality
cellular death due to the mutation of two genes in combination, but where there is no phenotypic effect when the two genes are mutated individually.
References
- 1.de Lorenzo V., Danchin A. Synthetic biology: discovering new worlds and new words. EMBO Rep. 2008;9:822–827. doi: 10.1038/embor.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vickers C.E. Grand challenge commentary: chassis cells for industrial biochemical production. Nat. Chem. Biol. 2010;6:875–877. doi: 10.1038/nchembio.484. [DOI] [PubMed] [Google Scholar]
- 3.Yus E. Impact of genome reduction on bacterial metabolism and its regulation. Science. 2009;326:1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
- 4.Juhas M. Essence of life: essential genes of minimal genomes. Trends Cell Biol. 2011;21:562–568. doi: 10.1016/j.tcb.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Yu B.J. Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat. Biotechnol. 2002;20:1018–1023. doi: 10.1038/nbt740. [DOI] [PubMed] [Google Scholar]
- 6.Smalley D.J. In search of the minimal Escherichia coli genome. Trends Microbiol. 2003;11:6–8. doi: 10.1016/s0966-842x(02)00008-2. [DOI] [PubMed] [Google Scholar]
- 7.Suthers P.F. Genome-scale gene/reaction essentiality and synthetic lethality analysis. Mol. Syst. Biol. 2009;5:301. doi: 10.1038/msb.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feher T. In the fast lane: large-scale bacterial genome engineering. J. Biotechnol. 2012;160:72–79. doi: 10.1016/j.jbiotec.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Lagesen K. Genome update: the 1000th genome – a cautionary tale. Microbiology. 2010;156:603–608. doi: 10.1099/mic.0.038257-0. [DOI] [PubMed] [Google Scholar]
- 10.Klasson L., Andersson S.G. Research on small genomes: implications for synthetic biology. Bioessays. 2010;32:288–295. doi: 10.1002/bies.200900165. [DOI] [PubMed] [Google Scholar]
- 11.Gerdes S.Y. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baba T. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gil R. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 2004;68:518–537. doi: 10.1128/MMBR.68.3.518-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabaldon T. Structural analyses of a hypothetical minimal metabolism. Philos. Trans. R. Soc. Lond. B: Biol. Sci. 2007;362:1751–1762. doi: 10.1098/rstb.2007.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Elia M.A. Are essential genes really essential? Trends Microbiol. 2009;17:433–438. doi: 10.1016/j.tim.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Ouzounis C.A. A minimal estimate for the gene content of the last universal common ancestor – exobiology from a terrestrial perspective. Res. Microbiol. 2006;157:57–68. doi: 10.1016/j.resmic.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 17.Kim K.M., Caetano-Anolles G. The evolutionary history of protein fold families and proteomes confirms that the archaeal ancestor is more ancient than the ancestors of other superkingdoms. BMC Evol. Biol. 2012;12:13. doi: 10.1186/1471-2148-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forterre P., Gadelle D. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereto J. Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem. Sci. 2004;29:469–477. doi: 10.1016/j.tibs.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Engelen S. Distinct co-evolution patterns of genes associated to DNA polymerase III DnaE and PolC. BMC Genomics. 2012;13:69. doi: 10.1186/1471-2164-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rocha E.P., Danchin A. An analysis of determinants of amino acids substitution rates in bacterial proteins. Mol. Biol. Evol. 2004;21:108–116. doi: 10.1093/molbev/msh004. [DOI] [PubMed] [Google Scholar]
- 22.Rocha E.P., Danchin A. Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat. Genet. 2003;34:377–378. doi: 10.1038/ng1209. [DOI] [PubMed] [Google Scholar]
- 23.Rocha E.P., Danchin A. Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res. 2003;31:6570–6577. doi: 10.1093/nar/gkg859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson D.G. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 25.Gibson D.G. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 26.Kepes F. The layout of a bacterial genome. FEBS Lett. 2012;586:2043–2048. doi: 10.1016/j.febslet.2012.03.051. [DOI] [PubMed] [Google Scholar]
- 27.Danchin A. Scaling up synthetic biology: do not forget the chassis. FEBS Lett. 2012;586:2129–2137. doi: 10.1016/j.febslet.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 28.Liu M.F. Identification of a novel nanoRNase in Bartonella. Microbiology. 2012;158:886–895. doi: 10.1099/mic.0.054619-0. [DOI] [PubMed] [Google Scholar]
- 29.Binder P.M., Danchin A. Life's demons: information and order in biology. What subcellular machines gather and process the information necessary to sustain life? EMBO Rep. 2011;12:495–499. doi: 10.1038/embor.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caspi R. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danchin A. The extant core bacterial proteome is an archive of the origin of life. Proteomics. 2007;7:875–889. doi: 10.1002/pmic.200600442. [DOI] [PubMed] [Google Scholar]
- 32.Fang G. How essential are nonessential genes? Mol. Biol. Evol. 2005;22:2147–2156. doi: 10.1093/molbev/msi211. [DOI] [PubMed] [Google Scholar]
- 33.Fang G. Persistence drives gene clustering in bacterial genomes. BMC Genomics. 2008;9:4. doi: 10.1186/1471-2164-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt M., de Lorenzo V. Synthetic constructs in/for the environment: Managing the interplay between natural and engineered Biology. FEBS Lett. 2012;586:2199–2206. doi: 10.1016/j.febslet.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conrad T.M. Microbial laboratory evolution in the era of genome-scale science. Mol. Syst. Biol. 2011;7:509. doi: 10.1038/msb.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwille P. Bottom-up synthetic biology: engineering in a tinkerer's world. Science. 2011;333:1252–1254. doi: 10.1126/science.1211701. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt M. Xenobiology: a new form of life as the ultimate biosafety tool. Bioessays. 2010;32:322–331. doi: 10.1002/bies.200900147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morowitz H.J. The completeness of molecular biology. Isr. J. Med. Sci. 1984;20:750–753. [PubMed] [Google Scholar]
- 39.Maniloff J. The minimal cell genome: ‘on being the right size’. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10004–10006. doi: 10.1073/pnas.93.19.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fraser C.M. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 41.Hutchison C.A. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 42.Glass J.I. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U.S.A. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zimmer C. Genomics. Tinker, tailor: can Venter stitch together a genome from scratch? Science. 2003;299:1006–1007. doi: 10.1126/science.299.5609.1006. [DOI] [PubMed] [Google Scholar]
- 44.Check E. Venter aims for maximum impact with minimal genome. Nature. 2002;420:350. doi: 10.1038/420350b. [DOI] [PubMed] [Google Scholar]
- 45.Lartigue C. Genome transplantation in bacteria: changing one species to another. Science. 2007;317:632–638. doi: 10.1126/science.1144622. [DOI] [PubMed] [Google Scholar]
- 46.Katsnelson A. Synthetic genome resets biotech goals. Nature. 2010;465:406. doi: 10.1038/465406a. [DOI] [PubMed] [Google Scholar]
- 47.Benders G.A. Cloning whole bacterial genomes in yeast. Nucleic Acids Res. 2010;38:2558–2569. doi: 10.1093/nar/gkq119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeon K.W. Reassembly of living cells from dissociated components. Science. 1970;167:1626–1627. doi: 10.1126/science.167.3925.1626. [DOI] [PubMed] [Google Scholar]
- 49.Itaya M. Combining two genomes in one cell: stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15971–15976. doi: 10.1073/pnas.0503868102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dymond J.S. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Berardinis V. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol. Syst. Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi K. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki N. High-throughput transposon mutagenesis of Corynebacterium glutamicum and construction of a single-gene disruptant mutant library. Appl. Environ. Microbiol. 2006;72:3750–3755. doi: 10.1128/AEM.72.5.3750-3755.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Christen B. The essential genome of a bacterium. Mol. Syst. Biol. 2011;7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kato J., Hashimoto M. Construction of consecutive deletions of the Escherichia coli chromosome. Mol. Syst. Biol. 2007;3:132. doi: 10.1038/msb4100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giaever G. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 57.Day W.A., Jr Microarray analysis of transposon insertion mutations in Bacillus anthracis: global identification of genes required for sporulation and germination. J. Bacteriol. 2007;189:3296–3301. doi: 10.1128/JB.01860-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gallagher L.A. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akerley B.J. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. U.S.A. 2002;99:966–971. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gawronski J.D. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salama N.R. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J. Bacteriol. 2004;186:7926–7935. doi: 10.1128/JB.186.23.7926-7935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sassetti C.M. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 63.French C.T. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol. Microbiol. 2008;69:67–76. doi: 10.1111/j.1365-2958.2008.06262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liberati N.T. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Knuth K. Large-scale identification of essential Salmonella genes by trapping lethal insertions. Mol. Microbiol. 2004;51:1729–1744. doi: 10.1046/j.1365-2958.2003.03944.x. [DOI] [PubMed] [Google Scholar]
- 66.Molzen T.E. Genome-wide identification of Streptococcus pneumoniae genes essential for bacterial replication during experimental meningitis. Infect. Immun. 2011;79:288–297. doi: 10.1128/IAI.00631-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bae T. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12312–12317. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chaudhuri R.R. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH) BMC Genomics. 2009;10:291. doi: 10.1186/1471-2164-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ji Y. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]
- 70.Cameron D.E. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8736–8741. doi: 10.1073/pnas.0803281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Touchon M. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lukjancenko O. Comparison of 61 sequenced Escherichia coli genomes. Microb. Ecol. 2010;60:708–720. doi: 10.1007/s00248-010-9717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]