Abstract
In response to cellular stress signals, the tumour suppressor p53 accumulates and triggers a host of antineoplastic responses. For instance, DNA damage activates two main p53-dependent responses: cell cycle arrest and attendant DNA repair or apoptosis (cell death). It is broadly accepted that, in response to DNA damage, the function of p53 as a sequence-specific transcription factor is crucial for tumour suppression. The molecular determinants, however, that favour the initiation of either a p53-dependent cell cycle arrest (life) or apoptotic (death) transcriptional programme remain elusive. Gaining a clear understanding of the mechanisms controlling cell fate determination by p53 could lead to the identification of molecular targets for therapy, which could selectively sensitize cancer cells to apoptosis. This review summarizes the literature addressing this important question in the field. Special emphasis is given to the role of the p53 response element, post-translational modifications and protein–protein interactions on cell fate decisions made by p53 in response to DNA damage.
Keywords: p53, apoptosis, cell cycle arrest, transcription, cellular stress
See the Glossary for abbreviations used in this article.
Glossary.
- APO1
apoptosis antigen 1
- ASPP
apoptosis-stimulating of p53 protein
- ATM
ataxia telangiectasia mutated
- Bak
Bcl-2 homologous antagonist/killer
- Bax
Bcl2-associated X
- Bcl2
B-cell lymphoma 2
- BcL-XL
B-cell lymphoma extra large
- CBP
CREB-binding protein
- CHK2
checkpoint homologue kinase 2
- DNA-PK
DNA-dependent serine/threonine protein kinase
- DYRK2
dual-specificity tyrosine-phosphorylation-regulated kinase 2
- E4F1
putative E3 ubiquitin-protein ligase
- Fas
death receptor
- hMOF
human males absent on the first (histone acetyltransferase)
- K120R
Lysine 120 to arginine
- Mdm2
murine double-minute
- MYST
monocytic leukaemia zinc finger protein, yeast Ybf2, yeast Sas2, Tip60 (histone acetyltransferase)
- p53AIP
p53-regulated apoptosis-inducing protein
- p38
mitogen-activating protein kinase
- PIG3
p53-inducible gene 3
- PTM
post-translational modifications
- PUMA
p53-upregulated modulator of apoptosis
- RITA
reactivation of p53 and induction of tumour cell apoptosis
- TAD
transactivation domain
- TIP60
TAT-interacting proteins 60 (histone acetyltransferase)
Introduction
The transcription factor p53 is a tumour suppressor that is often referred to as ‘the cellular gatekeeper’, as it is an essential regulator of cellular stress responses [1]. Under normal physiological conditions, the p53 protein is maintained at low intracellular levels by its negative regulator, the E3 ubiquitin ligase Mdm2, which targets p53 for ubiquitin-dependent degradation through the proteasome [2]. In response to a variety of cellular stresses including DNA damage, the p53–Mdm2 interaction is disrupted and p53 is rapidly stabilized [3]. The accumulated p53 protein is subject to extensive post-translational modifications including phosphorylation, acetylation, methylation, ubiquitination, sumoylation, neddylation and glycosylation (reviewed in references [4,5]). It is broadly accepted that these modifications contribute to the increase in p53 protein stability and modulate its function as a transcription factor [4,5]. Consequently, stabilized p53 is localized to the nucleus in which it can activate or repress the transcription of many genes involved in regulating the main cellular responses to stress, such as cell cycle arrest, DNA repair, senescence and apoptosis, among others (Fig 1; ([3,6,7,8,9,10]). It has, therefore, been proposed that p53 suppresses tumorigenesis by preventing replication of damaged DNA and transmission of potentially harmful mutations [6]. Not surprisingly, mutations in the TP53 gene are common in a large spectrum of human cancers, providing a selective advantage for neoplastic growth [11].
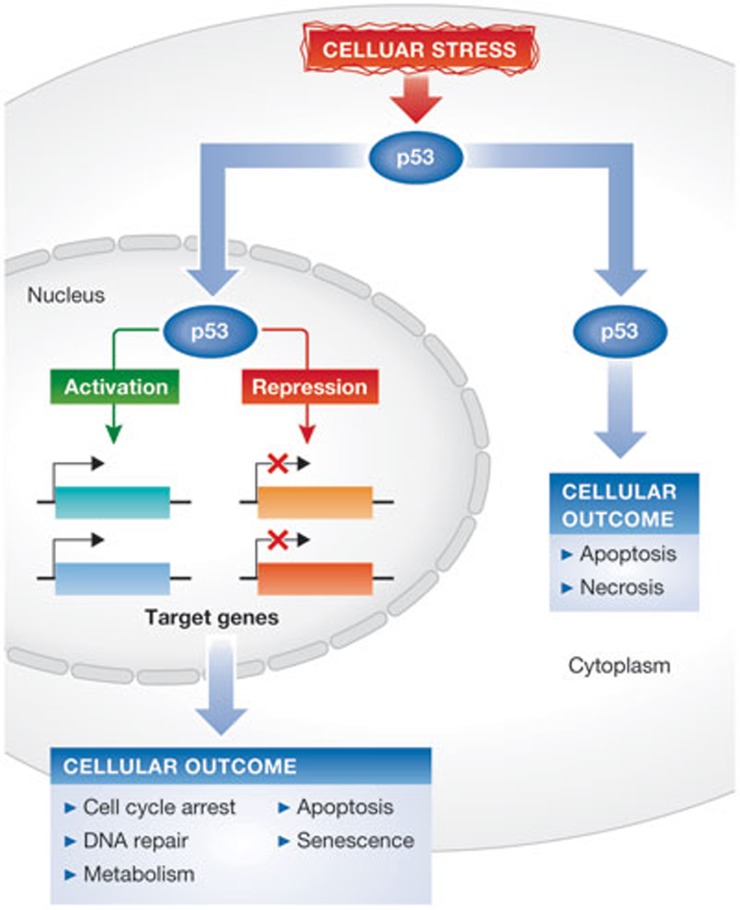
Figure 1.
Cellular stress induces p53-dependent responses. In response to cellular stress such as DNA damage, the p53 protein becomes stabilized. In the nucleus, p53 binds to specific promoters and activates transcription of its target genes. Additionally, p53 can also repress transcription of some of its targets. Depending on which genes are activated or repressed, p53 can induce cell cycle arrest, DNA repair, apoptosis and senescence, as well as regulate other processes such as energy metabolism. Independently of its effects on transcription, cytoplasmic p53 has been reported to trigger apoptosis, as well as necrosis. These responses ensure that damaged DNA is not replicated or transmitted to daughter cells, thus contributing to tumour suppression.
The p53 protein has been well characterized as a sequence-specific transcription factor [12]. It binds to its target gene promoters through a consensus p53 response element (p53RE), consisting of a ten-base-pair palindromic sequence made up of two ‘half sites’—RRRC (A/T)(T/A) GYYY (n) RRRC (A/T)(T/A) GYYY separated by a spacer of 0–13 bases (n) within which R and Y represent a purine and a pyrimidine, respectively [12,13,14]. Depending on cell type, stimulus, or both, p53 has been shown to activate transcriptional programmes and cellular responses [15]. Thus, one of the biggest challenges in the field has been to understand fully how p53 determines cellular fate in response to DNA damage—to let live (arrest) or to kill (apoptosis). Whilst sequence-specific recognition and binding of DNA is crucial to the biological response to p53 activation, a clear understanding of how p53 chooses its target genes remains elusive. To make sense of an enormous body of literature on this topic, this review focuses on three aspects of p53 regulation known to have a role in cell fate determination (life or death)—the p53RE and binding affinity, post-translational modifications and p53 protein–protein interactions.
Role of the p53RE and promoter affinity on gene selectivity
It is well accepted that induction of p53 in cells triggers either cell cycle arrest or apoptosis [1]. Furthermore, transcriptional activation of its cellular targets is thought to be essential for tumour suppression [1,12]. A complete understanding of how p53 chooses its targets to induce specific biological outcomes, such as cell cycle arrest and apoptosis has yet to be elucidated. One of the first mechanisms proposed in the literature suggested that the level of induced p53 in cells can determine cellular outcome [16]. It was reported that a low level of p53 correlates with cell cycle arrest and a higher level with apoptosis [16]. Later, it was shown that p53 binds to distinct classes of p53-binding sites with varying affinities [17]. Taken together, these observations suggest a model in which p53 protein levels and the DNA-binding affinity of p53 have a role in target gene selectivity and in determining cellular outcome (Fig 2). One hypothesis is that high-affinity target gene promoters (those that promote cell cycle arrest) are activated by p53 under low levels of cellular stress, and low-affinity target genes (those that promote apoptosis) require a higher degree of p53 activation or stress. Consistent with this concept, it was later demonstrated that on p53 activation, genes involved in cell cycle arrest were induced at earlier time points than apoptotic targets [7]. Activation of the apoptotic p53 targets, however, correlated with higher levels of p53 and DNA damage. Yet, a different report argued that the central sequence element in a p53RE greatly affects the p53-binding affinity and transactivation activity of its target genes [18]. In that study, the authors concluded that the promoters of apoptotic target genes had lower affinities to p53. Nevertheless, Weinberg et al reported that whilst most high-affinity targets were associated with cell cycle arrest and DNA repair, a fraction of the pro-apoptotic genes also contained a high-affinity p53RE [19]. For example, p53 bound with high affinity to the promoters of the pro-apoptotic genes, PUMA, p53AIP1 and Noxa [19]. Therefore, cell fate choice cannot be explained exclusively on the basis of p53 protein levels or target gene promoter affinity. Perhaps additional factors are required that can prime certain target genes for rapid or selective activation by p53.
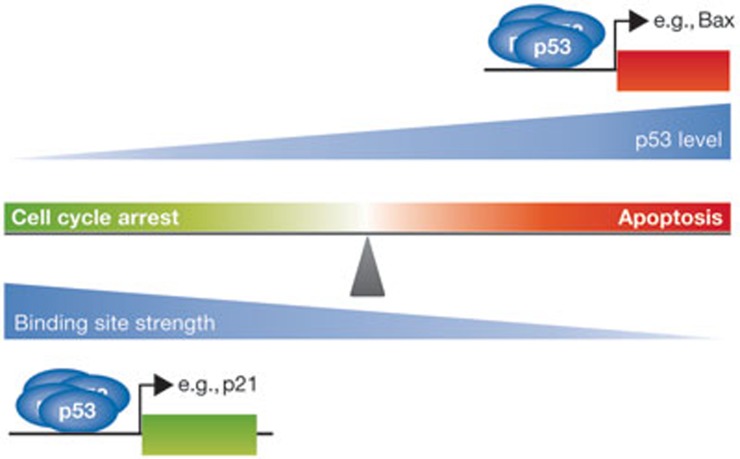
Figure 2.
p53-binding site affinity and cell fate choice. DNA damage results in intracellular accumulation of p53 protein. It has been proposed that at lower levels of damage, p53 preferentially binds to high-affinity target genes, which tend to be involved in mediating cell cycle arrest. Prolonged exposure to DNA damage results in higher levels of p53, which ensures that more p53 is available to bind to weaker sites such as those found in many pro-apoptotic target genes. This view, however, seems to be oversimplified, as the affinity of p53 for some response elements is comparable regardless of whether they are located in genes mediating cell cycle arrest or apoptosis. Nevertheless, the model retains some traction. It remains reasonable that p53 target genes involved in cell cycle arrest are inherently programmed for rapid and short-lived responses. By contrast, pro-apoptotic targets would have a delayed and more sustained response to genotoxic stress. Bax, Bcl2-associated X.
Additional reports suggest that the core promoter composition of p53 targets can have a crucial role in target gene selectivity and, therefore, in dictating the biological outcome [20,21]. The core promoter is a region of DNA required for transcriptional initiation by the RNA polymerase II complex (RNAPII; [22]). In a study by Espinosa et al, the authors reported that basal levels of RNAPII bound to the core promoter of cell cycle arrest genes were constitutively higher compared with those of pro-apoptotic target genes [20]. This observation suggests that cell cycle arrest genes are primed for rapid activation by p53. Consistent with this observation, it was later shown that the affinity of the p21 promoter for RNAPII is intrinsically higher and that preinitiation complexes (PICs) were readily assembled [21]. Interestingly, whilst the affinity of the Fas/APO1 core promoter for RNAPII was determined to be low compared with p21, once the PIC was formed, reinitiation of transcription was more efficient than that of p21 [21]. The authors concluded that the core promoter composition, of p53 target genes, has an essential role in cell fate decisions by regulating the kinetics of gene activation and duration of expression through RNAPII reinitiation [21]. Taken together, these observations support the hypothesis that p53 target genes involved in cell cycle arrest are inherently pre-programmed for a rapid and short-lived response, whilst pro-apoptotic genes have a delayed but sustained response to stress signals. Other evidence supports this model, suggesting that in response to low levels of DNA damage, short pulses of p53 promote cell cycle arrest [23]. By contrast, a high level of DNA damage was shown to induce sustained pulses of p53 and subsequent apoptosis [23]. More recently, it was reported that such p53 pulses have a crucial role in selective activation of genes involved in transient responses to DNA damage, whilst sustained p53 signalling was associated with terminal responses such as cellular senescence and apoptosis [24].
PTM regulate promoter selectivity
In response to DNA damage, p53 undergoes a series of post-translational modifications thought to regulate its stability and biological functions (reviewed in references [4,5]). Although we do not completely understand how p53 determines cell fate (life or death), a few of these modifications are thought to have a significant role in cell fate determination by p53 (Fig 3). For example, ultraviolet (UV) radiation has been reported to induce Ser 33 and Ser 46 phosphorylation on the amino-terminus of p53 by the p38 kinase [25]. Inhibition of p38-mediated phosphorylation of p53, correlated with a decrease in p53-dependent and UV-induced apoptosis [25]. Ser 46 phosphorylation is required for p53-dependent transcriptional activation of the pro-apoptotic factor p53AIP1 in response to high doses of DNA damage [26]. It was shown that p53AIP1 localizes to the mitochondria and facilitates the release of cytochrome c during apoptosis [26]. Additionally, mutation of Ser 46 to alanine prevented p53 from binding to the p53AIP1 promoter but not to the p21 promoter. Therefore, the authors concluded that Ser 46 phosphorylation has a crucial role in p53 target gene selectivity and, thus, in cell fate decision. In support of this model, additional kinases have been shown to phosphorylate p53 on Ser 46 and to have a role in p53-mediated apoptosis. For example, the homeodomain-interacting protein kinase 2 (HIPK2), a nuclear serine/threonine kinase, phosphorylates p53 on Ser 46 in response to DNA damage [27,28]. HIPK2 cooperated with p53 through Ser 46 phosphorylation to trigger UV-induced transcriptional upregulation of pro-apoptotic genes and apoptosis [27,28]. Furthermore, the importance of HIPK2 in cell fate determination by p53 is emphasized by evidence of a tight regulatory loop involving Mdm2-mediated degradation of HIPK2. Under conditions of low DNA damage, p53 induces the degradation of HIPK2 through Mdm2, thus inhibiting its own phosphorylation on Ser 46 [29]. In response to severe DNA damage, reduced Mdm2 levels result in the accumulation of HIPK2 and the induction of p53-dependent apoptosis [29]. In addition to p38 and HIPK2, DYRK2 is another kinase reported to phosphorylate p53 on Ser 46, to have a role in p53 target gene selectivity and p53-dependent apoptosis [25,30]. Studies addressing the relevance of Ser 46 in vivo were conducted by using a p53 knock-in mouse expressing a chimeric protein containing the human p53 DNA-binding domain [31]. Consistent with the in vitro studies, UV-irradiated mouse embryonic fibroblasts expressing the p53HupKIS46A/HupKIS46A humanized protein showed defects in target gene transactivation and apoptosis [31]. Taken together, these studies suggest that Ser 46 phosphorylation on p53 is an important cell fate determinant favouring p53-dependent transcriptional activation of pro-apoptotic target genes.
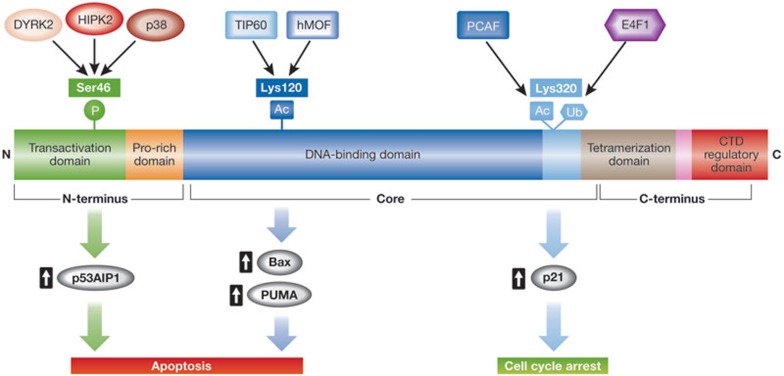
Figure 3.
Post-translational modifications on p53 dictate cell fate decisions. In response to DNA damage, p53 undergoes a series of post-translational modifications thought to be important in regulating p53 activity. Summarized here is a cartoon depicting the most relevant p53 modifications implicated in target gene selectivity and cell fate choice. For example, phosphorylation on Ser 46 and acetylation on Lys 120 enhances the preferential ability of p53 to transactivate genes that induce apoptosis. By contrast, acetylation and ubiquitination of Lys 320 have been implicated in mediating p53-dependent transcriptional activation of pro-arrest genes. Bax, Bcl2-associated X; DYRK2, dual-specificity tyrosine-phosphorylation-regulated kinase 2; E4F1, putative E3 ubiquitin-protein ligase; HIPK2, homeodomain-interacting protein kinase 2; hMOF, human males absent on the first (histone acetyltransferase); p53AIP, p53-regulated apoptosis-inducing protein 1; PCAF, p300/CBP-associated factor; PUMA, p53-upregulated modulator of apoptosis.
Mentioned earlier in the text, p53 is heavily modified in response to DNA damage and additional residues on the N terminus of p53 are also subject to phosphorylation. For example, Ser 15 was reported to be a phosphorylation target of the protein kinases DNA-PK and ATM [32,33]. The role of Ser 15 phosphorylation on cell fate determination, however, is less clear. Conflicting results have argued that Ser 15 phosphorylation on p53 contributes to cell cycle inhibition and apoptosis, as well as p53 stability [34,35,36,37,38,39]. Additionally, it has been suggested that phosphorylation of p53 on Ser 15 by ATM promotes phosphorylation at Ser 6, Ser 9, Thre 18, Ser 20 and Ser 46 [40]. Interestingly, phosphorylation of p53 on Ser 20 by the ATM target CHK2 was reported to induce a cell cycle arrest in G1 [41,42]. In vivo studies that use transgenic mice expressing p53S18A,S23A/S18A,S23A, in which the corresponding Ser 15 and Ser 20 residues were mutated, revealed a decrease in p53 stability and abrogated apoptosis in irradiated thymocytes [43]. These findings suggest that Ser 18 and Ser 23 in the mouse, however, are required for p53 stabilization and efficient induction of apoptosis.
In addition to phosphorylation, acetylation of p53 on Lys 120, which is located in its DNA-binding domain, was shown to enhance p53-dependent apoptosis [44,45,46]. It was reported that on DNA damage, the MYST family of acetyltransferases hMOF and TIP60 rapidly carry out acetylation at Lys 120. Interestingly, K120R mutations are common in human cancers and this single amino acid change abrogates p53-dependent apoptosis, but not cell cycle arrest [45,46]. Consistent with this observation, a K120R p53 mutant retained the ability to transactivate p21 and MDM2, but not the pro-apoptotic genes BAX and PUMA [45,46]. These studies argue that Lys 120 acetylation has a significant role in DNA-damage-induced, p53-mediated apoptosis and support the idea that specific p53 modifications can dictate cell fate.
Lys 320 has also been uniquely implicated in target gene selection by p53. Lys 320 is a substrate for both the atypical ubiquitin ligase E4F1 and the acetyltransferase p300/CBP-associated factor (PCAF; [47,48]). Acetylation of this lysine is important for transcriptional activation of pro-arrest genes such as p21, but not pro-apoptotic genes [49]. This finding was corroborated by using a transgenic mouse model in which the corresponding lysine Lys 317 had been mutated to arginine [50]. The authors reported that cells of several tissues in the L317R mouse responded rapidly to DNA damage by undergoing apoptosis, which also correlated with the induction of pro-apoptotic genes. Therefore, acetylation of Lys 320 (Lys 317 in the mouse) negatively regulates the p53 apoptotic response to DNA damage. Interestingly, monoubiquitination of p53 at Lys 320 by E4F1 also resulted in higher transcriptional output and promoter occupancy of pro-arrest genes such as p21, whilst pro-apoptotic gene expression was unchanged. Although PCAF and E4F1 modify the same amino-acid residue on p53, their activity is mutually exclusive and seems to be competitive [47]. Thus, acetylation and monoubiquitination of Lys 320 on p53 are important in the decision to let live or kill.
The examples presented above represent only a fraction of the p53 post-translational modifications reported in the literature. Additional modifications on several lysine residues on the carboxyl terminus of p53 have also been reported. Whilst a significant body of work has been dedicated to studying the role of the C-terminal lysines, it has been demonstrated that these lysines are not required for p53 transactivation or protein stability [51].
Proteins regulate p53 target gene selectivity
The activity of p53 on specific target gene promoters can also be regulated through interaction with specific transcriptional co-factors during the DNA damage response (Fig 4A). These interacting proteins cooperate with p53 to induce transcriptional activation of specific targets involved in determining cellular fate. In general, such proteins enhance the affinity of p53 binding to a subset of p53REs. In turn, this interaction tips the balance in favour of either life or death. For example, the ASPP protein family, composed of ASPP1, ASPP2 and iASPP, interacts directly with the central core domain of p53 and regulates the apoptotic response [52,53]. Whilst ASPP1 and ASPP2 enhance p53-dependent apoptosis by increasing the affinity of p53 for pro-apoptotic genes, iASPP counteracts this effect [52,53]. Consistent with these observations, in vivo studies suggest that ASSP2 is a haploinsufficient tumour suppressor in mice and cooperates with p53 to suppress tumour growth [54]. ASPP1 and ASPP2, however, have also been shown to enhance apoptosis in a p53-independent manner, by interacting with p53 family members p63 and p73 [55]. This suggests that the role of ASSP proteins in cell fate determination by p53 might involve additional factors that are beyond the scope of this review. Another p53-interacting protein, the prolyl isomerase Pin1 was shown to recognize and bind to phosphorylated p53 on Ser 46, in response to DNA damage, resulting in the dissociation of p53 from iASPP [56]—thus, promoting apoptosis. Furthermore, it has been suggested that Pin1 contributes to p53 stability in response to DNA damage and that it is required for efficient transactivation of its target gene promoters [57,58]. This example, illustrates the cross-talk between p53 modifications and its interacting proteins in determining cell fate. More recently, p90, also known as coiled-coil domain-containing 8, was identified as a p53 co-factor that enhances Tip60-mediated acetylation of p53 on Lys 120 [59]. As mentioned earlier, Tip60-mediated acetylation of p53 on Lys 120 has been reported to promote apoptosis.
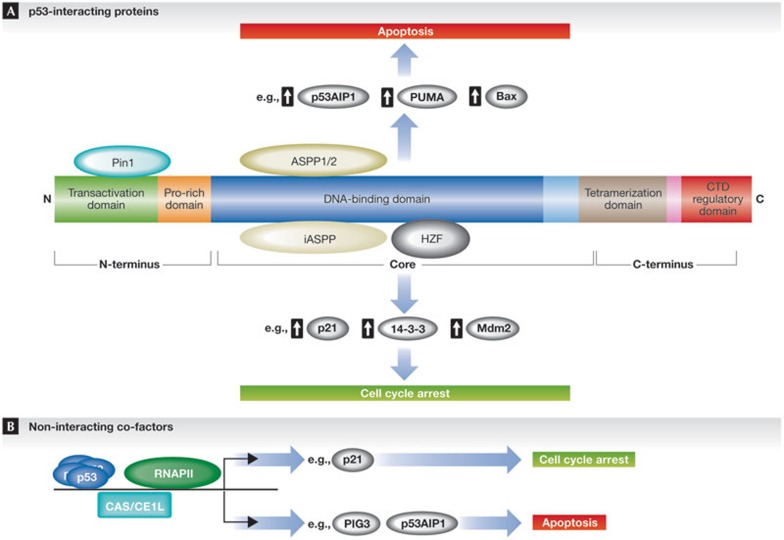
Figure 4.
Interacting proteins contribute to target gene selectivity and cell fate choice by p53. The transcriptional activity of p53 on specific target genes can be influenced by protein–protein interactions of p53 and its co-factors. Interacting proteins or co-factor recruitment to p53 target gene promoters cooperate with p53 to determine which genes are activated and which cellular response is induced. (A) For example, p53 interactions with either Pin1 or ASPP1 and ASPP2 result in preferential activation of pro-apoptotic genes. By contrast, when bound to iASPP or Hzf, p53 preferentially activates genes that mediate cell cycle arrest. (B) Non-interacting co-factors might also have a role in determining which target genes are preferentially activated by p53. For example, RNAPII is found preloaded and enriched on pro-arrest p53 target genes. By contrast, CAS/CSE1L bound to pro-apoptotic p53 target gene promoters cooperates with p53 to induce an apoptotic response. ASPP, apoptosis-stimulating of p53 protein; CAS/CSE1L, cellular apoptosis susceptibility protein; HZF, haematopoietic zinc finger; iASPP, inhibitor of ASPP protein; Pin1, peptidyl-prolyl cis-trans isomerase; RNAPII, RNA polymerase II.
In addition to the p53 co-factors that promote apoptosis, other co-factors can promote cell cycle arrest. For example, the haematopoietic zinc finger protein Hzf has been shown to enhance the p53 affinity and transactivation of pro-arrest genes [60]. The HZF gene was identified as a p53 target gene and reported to engage in a positive feedback loop with p53 that enhances the cell cycle arrest response. DNA damage was shown to induce a p53-dependent apoptotic response in HZF−/− mouse embryonic fibroblasts, in contrast to a cell cycle arrest response observed in wild-type cells [60]. Additionally, p53 bound to Hzf preferentially bound to pro-arrest promoters such as p21 and 14-3-3 [60]. The level of Hzf protein was also shown to correlate inversely with the intensity of genotoxic stress. Under conditions of sustained p53 expression or extensive genotoxic stress, Hzf is degraded through the ubiquitin proteasome pathway, and its degradation induces the expression of pro-apoptotic genes and apoptosis [60]. The authors proposed a model wherein p53 activates transcription of HZF, which on binding to p53, forms a complex that preferentially activates cell cycle arrest genes and results in cell cycle arrest. On the other hand, extensive DNA damage or prolonged p53 expression induces the degradation of the Hzf protein, favouring the induction of pro-apoptotic genes and apoptosis [60].
Some proteins can be recruited to p53 target gene promoters without directly interacting with p53 and can regulate p53 target gene selectivity and its biological functions (Fig 4B). The human cellular apoptosis susceptibility protein (hCAS/CSE1L) was identified as part of a chromatin-bound complex together with p53 at p53 target gene promoters [61]. The authors reported that the hCAS/CSE1 protein selectively regulates the induction of pro-apoptotic genes such as PIG3 and apoptosis. Knockdown of hCAS/CSE1L by using RNA interference resulted in suppression of p53-dependent induction of the pro-apoptotic genes PIG3 and P53AIP1, whilst only a minimal effect was observed on p21 messenger RNA transcription [61]. Furthermore, downregulation of hCAS/CSE1L impaired the apoptotic response to cellular stress. hCAS/CSE1L can, thus, bind to p53 target genes independently of p53 and direct selective, p53-mediated transcription towards cell fate determination after genotoxic stress.
Concluding remarks and perspectives
An essential property of p53 as a tumour suppressor is its ability to induce cell cycle arrest and DNA repair, or apoptotic cell death in response to DNA damage [1]. Whilst it is clear that transcriptional upregulation of specific genes by p53 is crucial in determining cellular outcome, the molecular basis for target gene selectivity remains elusive. To shed light on the considerable amount of data available on this subject, this review summarizes three basic models supported by published literature. Target gene selectivity and cell fate decisions by p53 can be explained, at least in part, by differences in p53 affinity to specific target gene promoters, the effect of post-translational modifications on p53 and the influence of interacting proteins on target gene selectivity. The underlying mechanisms are perhaps more complicated on the basis of the heterogeneity of p53 responses in vivo, which can be both tissue- and stimulus-specific (reviewed in reference [15]).
Other evidence suggests that, in addition to its nuclear function as a transcription factor, p53 might have additional functions outside the nucleus that also have a role in dictating cell fate. For instance, cytosolic p53 was reported to localize to the mitochondria and to interact directly with anti-apoptotic proteins Bcl-XL and Bcl2 [62]. The authors propose that this interaction sequesters Bcl-XL and Bcl2, thereby blocking their ability to bind to and inhibit the pro-apoptic proteins Bak and Bax [62]. Later, Chipuk and colleagues provided additional mechanistic insight into the role of cytosolic p53 in apoptosis [63]. In that study, it was reported that p53 interacts with and activates the pro-apoptotic protein Bax, which triggers mitochondrial permeabilization and activation of the apoptotic cell death programme [63]. Consequently to their initial study, Chipuk et al also showed that p53-mediated transactivation of the pro-apoptotic protein PUMA couples its transcription-dependent and -independent effects on apoptosis [64]. Indeed, the data argue that nuclear p53-mediated expression of PUMA displaces cytosolic p53 from Bcl-XL, which then allows cytosolic p53 to interact with Bax and to induce apoptosis [64]. More recently, Pin1 was shown to enhance stress-induced p53 translocation to the mitochondria and to induce p53-dependent apoptosis [65]. As discussed earlier, Pin1 binds to phosphorylated p53 on Ser 46, has a role in target gene selectivity and promotes apoptosis. Thus, Pin1 is a p53-interacting protein that can influence both the nuclear and cytosolic activity of p53. Interestingly, p53 localized to the mitochondria was linked to oxidative stress-induced necrosis—a form of cell death, which is mechanistically different from apoptosis [66]. In this context, it is also important to mention that a mutant p53 that cannot induce cell cycle arrest, apoptosis or senescence, was reported to retain its tumour-suppression activity by regulating specific genes involved in cellular metabolism [10]. Thus, in addition to its well-characterized functions as a tumour suppressor, p53 can regulate a diverse set of cellular processes involving both transcription-dependent and -independent effects.
Whilst important strides have been made towards understanding fully the molecular basis for cell fate decision-making by p53, there are still many unanswered questions that need to be addressed (see Sidebar A). How are different stimuli, for example, integrated by p53 and translated into a particular cellular outcome? Future studies should aim to define a molecular ‘signature’ for each stimulus and p53 response. One study has already discriminated between p53 transcriptional programmes involved in acute DNA damage response and tumour suppression [67]. In this study, the first transcriptional domain (TAD1) of p53 was reported to have a crucial role in transactivation of genes involved in cell cycle arrest and apoptosis [67]. The TAD1 mutants analysed, however, retained their tumour suppression activity and ability to induce senescence [67]. This study put forth the idea that administering a selective p53 TAD1 inhibitor in combination with chemotherapy could reduce the side effects associated with conventional therapies. Pharmacological compounds such as RITA, which was shown to induce p53-dependent apoptosis by blocking p21 expression through an MDM2-dependent mechanism, are already being used in experimental settings [68,69]. Such analyses are of crucial importance, as different ‘signatures’ might derive from not only different stimuli, but also tissue type and cellular context, and might portend different cell fate outcomes in response to p53 activation. Therefore, elucidating the mechanisms that regulate the ability of p53 to determine cell fate decisions (life or death) could facilitate the discovery of new therapies, which could preferentially sensitize cancer cells to apoptosis.
Sidebar A | In need of answers.
How are stimuli integrated by p53? How does p53 translate stimuli into a biological response?
Is there a unique ‘bar code’ or molecular ‘signature’ required to induce a cell cycle arrest or apoptosis?
What is the ‘bar code’ under mild DNA damage conditions? What is the ‘bar code’ under severe DNA damage conditions?
How do specific post-translational modifications enhance the p53-binding affinity to promoters of either cell cycle arrest or apoptotic genes? Which combinations of stimulus and post-translational modifications are important for target gene selectivity and cell fate determination?
Which p53-binding partners are important for target gene selectivity and cell fate decisions? Which post-translational modifications are important for co-factor recruitment?
How do we manipulate the p53 response to DNA damage using new therapies specifically to sensitize cancer cells to apoptotic cell death?
Footnotes
The authors declare that they have no conflict of interest.
References
- Levine AJ (1997) p53, the cellular gatekeeper for growth and division. Cell 88: 323–331 [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH (1997) Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- Horn HF, Vousden KH (2007) Coping with stress: multiple ways to activate p53. Oncogene 26: 1306–1316 [DOI] [PubMed] [Google Scholar]
- Meek DW, Anderson CW (2009) Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol 1: a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM (2006) Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 6: 909–923 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C (2009) Blinded by the light: the growing complexity of p53. Cell 137: 413–431 [DOI] [PubMed] [Google Scholar]
- Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ (2000) Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev 14: 981–993 [PMC free article] [PubMed] [Google Scholar]
- Mirza A et al. (2003) Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene 22: 3645–3654 [DOI] [PubMed] [Google Scholar]
- Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B (1997) A model for p53-induced apoptosis. Nature 389: 300–305 [DOI] [PubMed] [Google Scholar]
- Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W (2012) Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149: 1269–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC (1991) p53 mutations in human cancers. Science 253: 49–53 [DOI] [PubMed] [Google Scholar]
- Beckerman R, Prives C (2010) Transcriptional regulation by p53. Cold Spring Harbor Perspect Biol 2: a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A (2008) Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412 [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B (1992) Definition of a consensus binding site for p53. Nat Genet 1: 45–49 [DOI] [PubMed] [Google Scholar]
- Jackson JG, Post SM, Lozano G (2011) Regulation of tissue- and stimulus-specific cell fate decisions by p53 in vivo. J Pathol 223: 127–136 [DOI] [PubMed] [Google Scholar]
- Chen X, Ko LJ, Jayaraman L, Prives C (1996) p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev 10: 2438–2451 [DOI] [PubMed] [Google Scholar]
- Resnick-Silverman L, St Clair S, Maurer M, Zhao K, Manfredi JJ (1998) Identification of a novel class of genomic DNA-binding sites suggests a mechanism for selectivity in target gene activation by the tumor suppressor protein p53. Genes Dev 12: 2102–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inga A, Storici F, Darden TA, Resnick MA (2002) Differential transactivation by the p53 transcription factor is highly dependent on p53 level and promoter target sequence. Mol Cell Biol 22: 8612–8625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RL, Veprintsev DB, Bycroft M, Fersht AR (2005) Comparative binding of p53 to its promoter and DNA recognition elements. J Mol Biol 348: 589–596 [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Verdun RE, Emerson BM (2003) p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell 12: 1015–1027 [DOI] [PubMed] [Google Scholar]
- Morachis JM, Murawsky CM, Emerson BM (2010) Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes Dev 24: 135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT (2002) The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev 16: 2583–2592 [DOI] [PubMed] [Google Scholar]
- Zhang XP, Liu F, Cheng Z, Wang W (2009) Cell fate decision mediated by p53 pulses. Proc Natl Acad Sci USA 106: 12245–12250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G (2012) p53 dynamics control cell fate. Science 336: 1440–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulavin DV, Saito S, Hollander MC, Sakaguchi K, Anderson CW, Appella E, Fornace AJ Jr (1999) Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J 18: 6845–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K et al. (2000) p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102: 849–862 [DOI] [PubMed] [Google Scholar]
- D'Orazi G et al. (2002) Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 4: 11–19 [DOI] [PubMed] [Google Scholar]
- Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, Will H, Schmitz ML (2002) Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol 4: 1–10 [DOI] [PubMed] [Google Scholar]
- Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F, Soddu S (2007) MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol Cell 25: 739–750 [DOI] [PubMed] [Google Scholar]
- Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K (2007) DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol Cell 25: 725–738 [DOI] [PubMed] [Google Scholar]
- Feng L, Hollstein M, Xu Y (2006) Ser46 phosphorylation regulates p53-dependent apoptosis and replicative senescence. Cell Cycle 5: 2812–2819 [DOI] [PubMed] [Google Scholar]
- Lees-Miller SP, Sakaguchi K, Ullrich SJ, Appella E, Anderson CW (1992) Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol 12: 5041–5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB (1997) DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev 11: 3471–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiscella M, Ullrich SJ, Zambrano N, Shields MT, Lin D, Lees-Miller SP, Anderson CW, Mercer WE, Appella E (1993) Mutation of the serine 15 phosphorylation site of human p53 reduces the ability of p53 to inhibit cell cycle progression. Oncogene 8: 1519–1528 [PubMed] [Google Scholar]
- Huang LC, Clarkin KC, Wahl GM (1996) p53-dependent cell cycle arrests are preserved in DNA-activated protein kinase-deficient mouse fibroblasts. Cancer Res 56: 2940–2944 [PubMed] [Google Scholar]
- Rathmell WK, Kaufmann WK, Hurt JC, Byrd LL, Chu G (1997) DNA-dependent protein kinase is not required for accumulation of p53 or cell cycle arrest after DNA damage. Cancer Res 57: 68–74 [PubMed] [Google Scholar]
- Shieh SY, Ikeda M, Taya Y, Prives C (1997) DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91: 325–334 [DOI] [PubMed] [Google Scholar]
- Banin S et al. (1998) Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281: 1674–1677 [DOI] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD (1998) Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281: 1677–1679 [DOI] [PubMed] [Google Scholar]
- Saito S, Goodarzi AA, Higashimoto Y, Noda Y, Lees-Miller SP, Appella E, Anderson CW (2002) ATM mediates phosphorylation at multiple p53 sites, including Ser(46), in response to ionizing radiation. J Biol Chem 277: 12491–12494 [DOI] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Appel M, Halazonetis TD (2000) Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev 14: 278–288 [PMC free article] [PubMed] [Google Scholar]
- Shieh SY, Ahn J, Tamai K, Taya Y, Prives C (2000) The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev 14: 289–300 [PMC free article] [PubMed] [Google Scholar]
- Chao C, Herr D, Chun J, Xu Y (2006) Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO J 25: 2615–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet C et al. (2011) Phosphorylation of Tip60 by GSK-3 determines the induction of PUMA and apoptosis by p53. Mol Cell 42: 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W (2006) Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 24: 827–839 [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB (2006) Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 24: 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cam L et al. (2006) E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell 127: 775–788 [DOI] [PubMed] [Google Scholar]
- Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL (1999) p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol 19: 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights CD et al. (2006) Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol 173: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C et al. (2006) Acetylation of mouse p53 at lysine 317 negatively regulates p53 apoptotic activities after DNA damage. Mol Cell Biol 26: 6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummel KA, Lee CJ, Toledo F, Wahl GM (2005) The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci USA 102: 10188–10193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi D et al. (2006) iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat Genet 38: 1133–1141 [DOI] [PubMed] [Google Scholar]
- Samuels-Lev Y et al. (2001) ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell 8: 781–794 [DOI] [PubMed] [Google Scholar]
- Vives V, Su J, Zhong S, Ratnayaka I, Slee E, Goldin R, Lu X (2006) ASPP2 is a haploinsufficient tumor suppressor that cooperates with p53 to suppress tumor growth. Genes Dev 20: 1262–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, Jin B, Duraisingham S, Crook T, Lu X (2004) ASPP1 and ASPP2: common activators of p53 family members. Mol Cell Biol 24: 1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani F, Tocco F, Girardini J, Smith P, Gasco M, Lu X, Crook T, Del Sal G (2007) The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. NatStruct Mol Biol 14: 912–920 [DOI] [PubMed] [Google Scholar]
- Zacchi P et al. (2002) The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 419: 853–857 [DOI] [PubMed] [Google Scholar]
- Zheng H et al. (2002) The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature 419: 849–853 [DOI] [PubMed] [Google Scholar]
- Dai C, Tang Y, Jung SY, Qin J, Aaronson SA, Gu W (2011) Differential effects on p53-mediated cell cycle arrest vs. apoptosis by p90. Proc Natl Acad Sci USA 108: 18937–18942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW (2007) Hzf determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell 130: 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ohkubo S, Tatsuno I, Prives C (2007) hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell 130: 638–650 [DOI] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM (2003) p53 has a direct apoptogenic role at the mitochondria. Mol Cell 11: 577–590 [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR (2004) Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303: 1010–1014 [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR (2005) PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 309: 1732–1735 [DOI] [PubMed] [Google Scholar]
- Sorrentino G, Mioni M, Giorgi C, Ruggeri N, Pinton P, Moll U, Mantovani F, Del Sal G (2013) The prolyl-isomerase Pin1 activates the mitochondrial death program of p53. Cell Death Differ 20: 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM (2012) p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 149: 1536–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady CA et al. (2011) Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinkevich VV et al. (2009) Ablation of key oncogenic pathways by RITA-reactivated p53 is required for efficient apoptosis. Cancer Cell 15: 441–453 [DOI] [PubMed] [Google Scholar]
- Enge M, Bao W, Hedström E, Jackson SP, Moumen A, Selivanova G (2009) MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell 15: 171–183 [DOI] [PubMed] [Google Scholar]