Abstract
Autophagy is an evolutionarily conserved catabolic mechanism that targets intracellular molecules and damaged organelles to lysosomes. Autophagy is achieved by a series of membrane trafficking events, but their regulatory mechanisms are poorly understood. Here, we report small GTPase Rab12 as a new type of autophagic regulator that controls the degradation of an amino-acid transporter. Knockdown of Rab12 results in inhibition of autophagy and in increased activity of mTORC1 (mammalian/mechanistic target of rapamycin complex 1), an upstream regulator of autophagy. We also found that Rab12 promotes constitutive degradation of PAT4 (proton-coupled amino-acid transporter 4), whose accumulation in Rab12-knockdown cells modulates mTORC1 activity and autophagy. Our findings reveal a new mechanism of regulation of mTORC1 signalling and autophagy, that is, quality control of PAT4 by Rab12.
Keywords: amino-acid transporter, autophagy, mTORC1, recycling endosome, Rab12
Introduction
Macroautophagy (referred to here as autophagy) is a catabolic process conserved among all eukaryotes. Autophagy not only functions as a nutrient supplier in response to the cellular energy concentration but is also involved in quality control of cellular molecules and organelles, cell defence against infection, embryonic development and various diseases [1]. Autophagy is achieved by a series of dynamic membrane trafficking events, including isolation membrane/phagophore elongation, autophagosome formation, and fusion between autophagosomes and lysosomes [1, 2]. However, even though many genes that are essential for autophagy have been identified over the past decade, little is known about the mechanisms that regulate the membrane trafficking involved in autophagy. Small GTPase Rab is a common regulator of membrane traffic in all eukaryotes [3], and several members of the Rab family have been reported to be involved in autophagy, for example, involvement of Rab11 (or Rab11-positive recycling endosomes) in autophagosome formation and maturation [4, 5]. However, the functional relationships between individual members of the mammalian Rab family and autophagy remain largely unknown.
In this study, we comprehensively screened for Rabs that are involved in autophagy by using RNA-mediated interference technologies and found that Rab12 is a new autophagic regulator. We showed that Rab12 regulates activity of mTORC1 (mammalian/mechanistic target of rapamycin complex 1), an upstream autophagic regulator, through controlling the degradation of amino-acid transporter PAT4 (proton-coupled amino-acid transporter 4). Possible mechanisms of Rab12-mediated regulation of mTORC1 activity and autophagy are discussed on the basis of our findings.
Results and Discussion
Screening for Rabs that are involved in autophagy
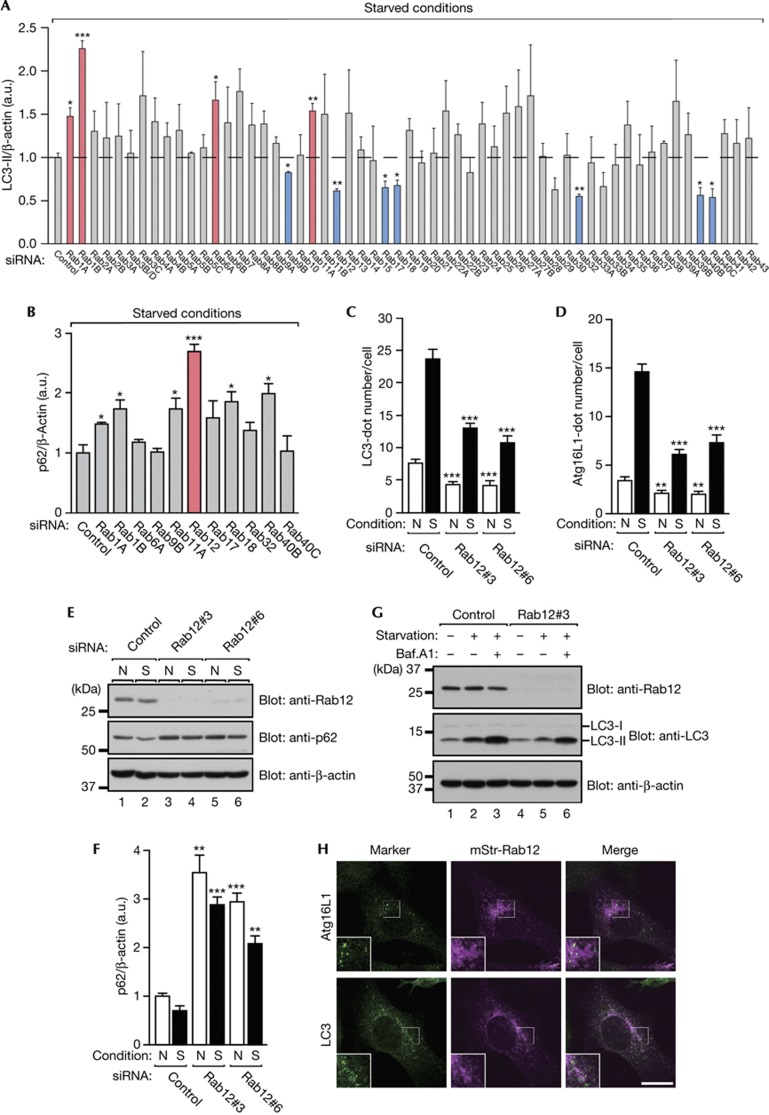
To comprehensively screen for Rabs that regulate mammalian autophagy, we generated effective short interfering RNAs (siRNAs) targeting each mouse Rab isoform (supplementary Fig S1A,B online; supplementary Table S1 online) and proceeded to use these siRNAs to perform screening in two steps. In the first step of our screening procedure, we searched for Rabs, whose knockdown in mouse embryonic fibroblasts (MEFs) altered the amount of LC3-II, a lipidated form of LC3/Atg8 that specifically associates with autophagosomes, under starved conditions (Fig 1A; supplementary Fig S1C online) [6, 7]. Of the 58 Rabs we tested, 4 Rab siRNAs (Rab1A, 1B, 6A and 11A) significantly increased the amount of LC3-II in comparison with the control siRNA, and 7 Rab siRNAs (Rab9B, 12, 17, 18, 32, 40B and 40C) significantly decreased it (Fig 1A). If these candidate Rabs were actually involved in autophagy, the amount of an autophagic substrate should also be affected by the same siRNAs. In the second step of our screening procedure, we therefore investigated whether knockdown of the candidate Rabs identified in the first step influenced the amount of p62 protein, a specific substrate for autophagic degradation, under starved conditions [8]. As shown in Fig 1B and supplementary Fig S1D online, six Rab siRNAs (Rab1A, 1B, 11A, 12, 18 and 40B) significantly increased the amount of p62 protein, but the other five Rab siRNAs had no effect. It should be noted that some of the final candidate Rabs, for example, Rab1 and Rab11, have previously been reported to be involved in autophagy, thereby validating our two-step screening procedure. In the present study, we focused on Rab12, whose knockdown resulted in the greatest increase in the amount of p62 protein among the final candidates identified (Fig 1B).
Figure 1.
Rab12 regulates autophagy. (A) Quantification of the amount of LC3-II protein in mouse Rab-knockdown cells. Representative blots are shown in supplementary Fig S1C online. Error bars represent the means and s.e.m. of three independent experiments. (B) Quantification of the amount of p62 protein in candidates Rab-knockdown cells. Representative blots are shown in supplementary Fig S1D online. Error bars represent the means and s.e.m. of three independent experiments. (C,D) Control and Rab12-knockdown MEFs were cultured under N or S conditions, fixed and then immunostained with the antibodies indicated. Representative images are shown in supplementary Fig S2 online. The mean numbers of LC3-positive (C) or Atg16L1-positive (D) dots per cell are shown. Error bars represent the means and s.e.m. of representative data (n≥80) from three independent experiments. (E) MEFs that had been transfected with the control siRNA or Rab12 siRNA were cultured as in (C,D), and their lysates were analysed by immunoblotting with the antibodies indicated. (F) Quantification of (E). (G) Control and Rab12-knockdown MEFs were cultured under N or S conditions in the absence or presence of 100 μM bafilomycin A1 for 1 h. Cell lysates were analysed by immunoblotting with the antibodies indicated, and the intensity of the LC3-II band was quantified. The normalized amount (arbitrary units) of LC3-II in lanes 1, 2, 3, 4, 5 and 6 is 1.0, 2.3, 4.3, 0.9, 1.6 and 3.8, respectively. (H) MEFs transiently expressing mStr-Rab12 were cultured under starved conditions and immunostained with the antibodies indicated. Scale bar, 20 μm. MEFs, mouse embryonic fibroblasts; N, nutrient-rich; S, starved; siRNA, short interfering RNA. *P<0.05; **P<0.01; ***P<0.005.
Rab12 regulates the efficiency of autophagy
The impact of Rab12 knockdown on autophagy was further evaluated by several independent approaches [9] in which two different Rab12 siRNAs were used. Both siRNAs clearly reduced the number of dots that were positive for LC3 and dots that were positive for Atg16L1 (an isolation membrane marker) under both nutrient-rich conditions and starved conditions (supplementary Fig S2 online; Fig 1C,D). Similarly, the amount of p62 protein was increased by these siRNAs under both conditions (Fig 1E,F). It should be noted, however, that starvation still increased the number of LC3-positive and Atg16L1-positive dots and reduced the amount of p62 protein even in the Rab12-depleted cells in comparison with nutrient-rich conditions, the same as in the control cells (Fig 1C–F). In addition, Rab12 knockdown did not affect autophagic flux as revealed by LC3 turnover assays (Fig 1G) [9], indicating that Rab12 knockdown did not affect lysosomal functions. Moreover, monomeric strawberry (mStr)-tagged Rab12 did not colocalize with Atg16L1 or LC3 at all (Fig 1H). Taken together, these results suggested that Rab12 modulates the signals involved in initiation of autophagy.
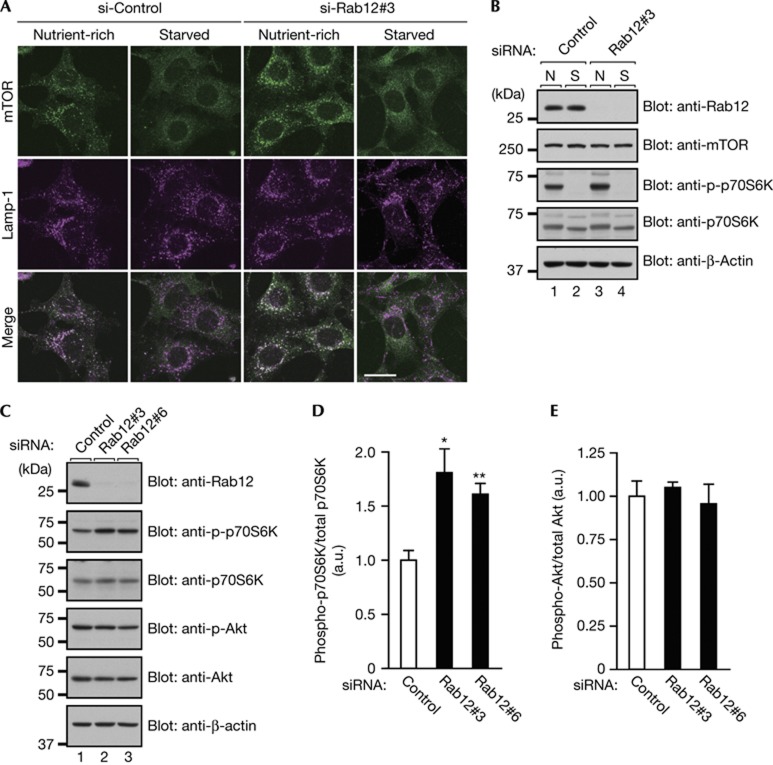
To determine the mechanism by which Rab12 regulates initiation of autophagy, we first focused on mTORC1, a well-known upstream negative regulator of autophagy [10, 11, 12, 13, 14, 15], because several Rab isoforms, including Rab5, have been shown to regulate mTORC1 localization and/or activity [16, 17]. Under nutrient-rich conditions, mTORC1 is targeted to the lysosomal surface by a Ragulator–Rag GTPases complex and activated by Rheb GTPase, and its activation regulates growth and metabolism as well as inhibits autophagy [18, 19, 20, 21, 22, 23, 24]. As we previously found that Rab12 regulates a membrane traffic pathway from recycling endosomes to lysosomes to degrade transferrin receptor (TfR, a recycling endosome marker) [25], we investigated whether the lysosomal targeting of mTORC1 is influenced by Rab12 knockdown (Fig 2A). However, the results showed that under nutrient-rich conditions, Rab12 knockdown did not affect the lysosomal targeting of mTOR, although the signals of mTOR targeting to lysosomes were clearly increased in Rab12-depleted cells in comparison with the control cells under nutrient-rich conditions. This result led us to consider two possibilities: the possibility that Rab12 knockdown leads to increased mTORC1 activity and the possibility that Rab12 knockdown increases the amount of mTOR protein itself, and we performed immunoblotting analyses to determine whether either one of them was correct (Fig 2B). The results showed that Rab12 knockdown increased phosphorylation of ribosomal protein S6 kinase, a readout of mTORC1 activity, without affecting the total amount of mTOR protein. Interestingly, dissociation of mTOR from lysosomes and decreased mTORC1 activity as a result of starvation seemed to occur normally in Rab12-depleted cells (Fig 2A,B), and treatment with rapamaycin, an mTORC1 inhibitor, reduced mTORC1 activity and induced autophagy (that is, p62 degradation and LC3-dot formation) even in the Rab12-depleted cells (supplementary Fig S3 online). By contrast, overexpression of a constitutive active mutant of Rab12 (Rab12-QL) reduced mTORC1 activity (supplementary Fig S4 online). These results taken together indicated that Rab12 functions upstream of mTORC1.
Figure 2.
Effect of Rab12 knockdown on mTORC1 activity. (A) Control and Rab12-knockdown MEFs were cultured under nutrient-rich or starved conditions, fixed and then immunostained with the antibodies indicated. Scale bar, 20 μm. (B) Control and Rab12-knockdown MEFs were cultured as in (A), and their lysates were analysed by immunoblotting with the antibodies indicated. (C) Lysates of MEFs that had been transfected with control or Rab12 siRNA were analysed by immunoblotting with the antibodies indicated. (D,E) Quantification of the phospho-S6K levels and phospho-Akt levels shown in (C). MEFs, mouse embryonic fibroblasts; N, nutrient-rich; S, starved; siRNA, short interfering RNA. *P<0.05; **P<0.01.
We also found that Rab12 knockdown increased mTORC1 activity independent of Akt activity (Fig 2C–E). As mTORC1 is activated by certain amino acids independent of the PI3K-Akt signalling pathway [19, 26, 27, 28], we hypothesized that Rab12 knockdown increases the intracellular amino-acid concentration. To test our hypothesis, we measured the intracellular L-amino-acid concentration in Rab12-depleted cells with an L-Amino-Acid Quantitation Kit (supplementary Fig S5 online). As predicted in our hypothesis, the intracellular L-amino-acid concentration was much higher in the Rab12-depleted cells than in the control cells, suggesting that Rab12 regulates mTORC1 activity through modulation of the intracellular amino-acid concentration.
Rab12 controls the degradation of PAT4
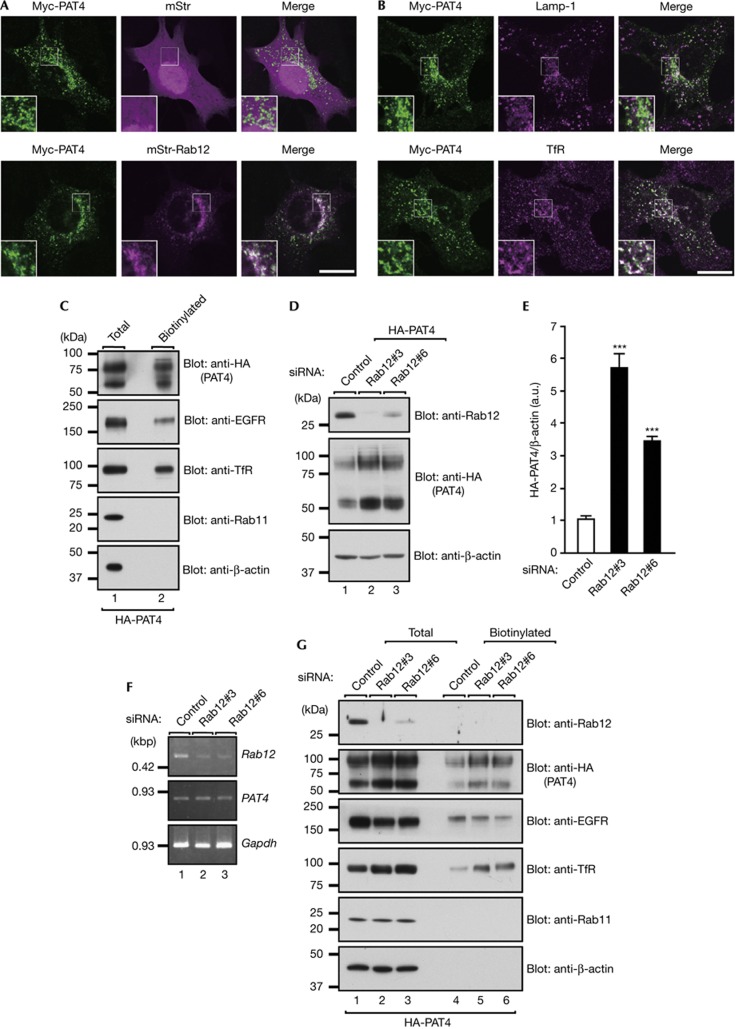
As the intracellular amino-acid concentration is regulated by amino-acid transporters, we hypothesized that Rab12 regulates lysosomal localization or degradation of amino-acid transporters. To test this hypothesis, we focused on the PAT family, because two members of the PAT family that are ubiquitously expressed, PAT1/Slc36a1 and PAT4/Slc36a4, have been shown to affect mTORC1 activity [29, 30, 31, 32]. PAT1, in particular, is specifically localized at lysosomes (supplementary Fig S6A online) and has been reported to regulate mTORC1 activity through export of amino acids from the lysosome lumen into the cytosol [29, 30]. However, detailed colocalization analyses in MEFs indicated that PAT1 only partially colocalized with Rab12 (supplementary Fig S6B online), whereas PAT4 colocalized well with Rab12 and TfR but it did not colocalize with Lamp-1 (Fig 3A,B). We therefore selected PAT4 as the prime candidate for the cargo of Rab12. As both TfR and PAT4 are localized at recycling endosomes, it appeared highly possible that PAT4 cycles between the plasma membrane and recycling endosomes, the same as TfR does. As anticipated, surface biotinylation assays revealed that PAT4 is actually present in the plasma membrane (Fig 3C). However, as Rab11A knockdown, which inhibited the recycling pathway to the plasma membrane, clearly resulted in a reduction in the amount of plasma membrane-localized TfR protein but did not affect the amount of plasma membrane-localized PAT4 protein (supplementary Fig S7 online), unlike TfR protein, PAT4 is not actively recycled back to the plasma membrane by Rab11. Although PAT4 was originally described as a proton-coupled amino-acid transporter, recent evidence indicated that when expressed in Xenopus laevis oocytes [33], PAT4 is most functional at neutral pH, not at low pH, suggesting that PAT4 imports extracellular amino acids into cells through the plasma membrane. As Rab12 regulates lysosomal degradation of TfR through the pathway from recycling endosomes to lysosomes without affecting lysosome function [25], we wondered whether Rab12 also regulates PAT4 degradation. Rab12 knockdown was found to dramatically increase the amount of HA-PAT4 protein in MEFs stably expressing HA-PAT4 (Fig 3D,E) without affecting the PAT4 messenger RNA concentration (Fig 3F). Furthermore, the results of exposure to a lysosomal inhibitor showed that PAT4 is constitutively degraded in lysosomes (supplementary Fig S6C online) and that PAT4 trafficking to lysosomes is inhibited by Rab12 knockdown (supplementary Fig S6D online), indicating that Rab12 regulates constitutive degradation of PAT4 in lysosomes. It was particularly noteworthy that Rab12 knockdown increased the amount of plasma membrane-localized PAT4 (Fig 3G), which is likely to contribute to the rise in the intracellular amino-acid concentration, and thereby result in upregulation of mTORC1 activity and inhibition of autophagy.
Figure 3.
Rab12 regulates constitutive degradation of amino-acid transporter PAT4. (A) MEFs transiently co-expressing Myc-PAT4 and mStr-Rab12 were immunostained with anti-Myc antibody. (B) MEFs transiently expressing Myc-PAT4 were immunostained with the antibodies indicated. Scale bars, 20 μm. (C) Total cell lysates and surface biotinylated proteins from MEFs stably expressing HA-PAT4 were analysed by immunoblotting with the antibodies indicated. (D) Lysates of MEFs stably expressing HA-PAT4 that had been transfected with control or Rab12 siRNAs were analysed by immunoblotting with the antibodies indicated. (E) Quantification of (D). ***P<0.005. (F) PAT4 mRNA levels from MEFs transfected with control or Rab12 siRNA as revealed by reverse-transcription PCR analyses. Gapdh was used as an internal control. (G) Total cell lysates and surface biotinylated proteins from MEFs stably expressing HA-PAT4 that had been transfected with control or Rab12 siRNA were analysed by immunoblotting with the antibodies indicated. Note that the amounts of plasma-membrane-localized PAT4 protein and TfR protein were higher in Rab12-knockdown MEFs, whereas the total amounts of EGFR protein and plasma membrane-localized EGFR protein in these cells were slightly lower, but the mechanisms responsible for these changes are unknown. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MEFs, mouse embryonic fibroblasts; PAT4, proton-coupled amino-acid transporter 4; siRNA, short interfering RNA.
Rab12 regulates mTORC1 activity and autophagy
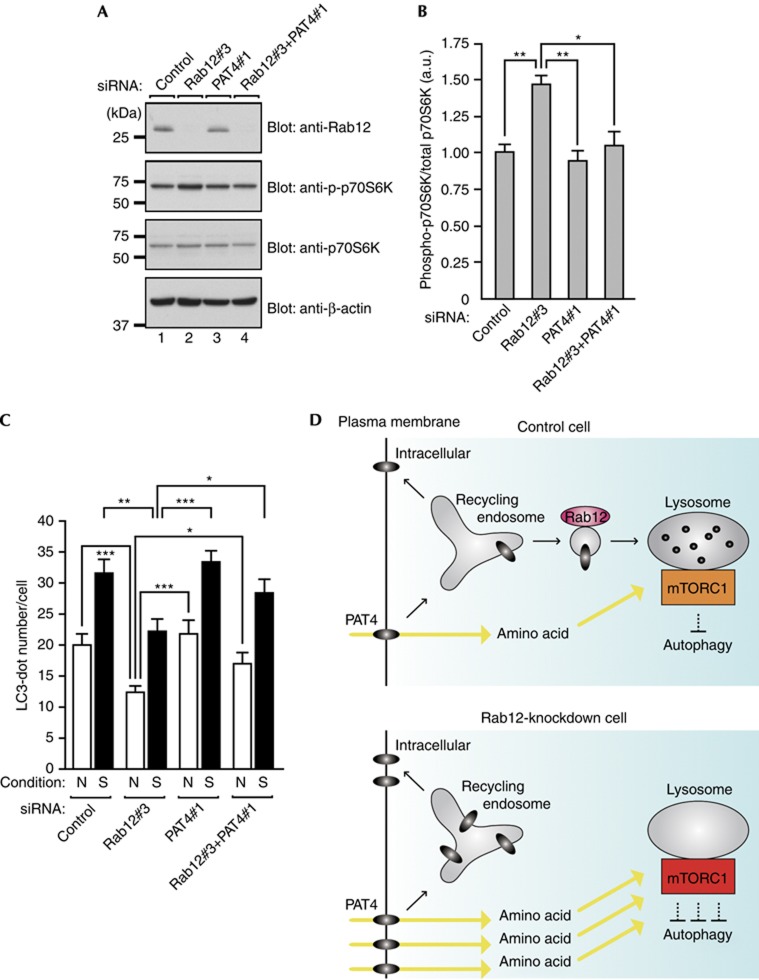
If accumulation of PAT4 in Rab12-depleted cells is the primary cause of the increased mTORC1 activity and decreased autophagic activity, overexpression of PAT4 should mimic Rab12 deficiency. As anticipated, increased phosphorylation of S6K and fewer LC3-positive dots were also observed in HA-PAT4-overexpressing cells (supplementary Fig S8A–D online). Conversely, increased phosphorylation of S6K and a reduced number of autophagosomes, both of which were induced by Rab12 knockdown, were completely rescued by simultaneous knockdown of PAT4 (Fig 4A–C), although PAT4 knockdown alone had little effect on mTORC1 activity or autophagy under our experimental conditions (supplementary Fig S8E–J online). Furthermore, addition of certain L-amino acids, for example, Pro and Trp, both of which have been found to be high-affinity substrates of PAT4 when expressed in Xenopus oocytes [33], to Rab12(QL)-overexpressing MEFs restored phosphorylation of S6K (supplementary Fig S4C,D online). These results allowed us to conclude that Rab12 regulates mTORC1 activity and autophagy through trafficking of PAT4.
Figure 4.
Rab12 regulates mTORC1 activity and autophagy by controlling the PAT4 protein concentration. (A) Lysates of MEFs transfected with the siRNAs indicated were analysed by immunoblotting with the antibodies indicated. (B) Quantification of the phospho-S6K levels shown in (A). (C) MEFs transfected with the siRNAs indicated were cultured under nutrient-rich (N) or starved (S) conditions and were immunostained with anti-LC3 antibody. The mean numbers of LC3-positive dots per cell are shown. (D) A schematic model of inhibition of autophagy followed by increased activation of mTORC1 in Rab12-depleted cells. MEFs, mouse embryonic fibroblasts; N, nutrient-rich; PAT4, proton-coupled amino-acid transporter 4; S, starved; siRNA, short interfering RNA. *P<0.05; **P<0.01; ***P<0.005.
The results of this study revealed an unexpected role of Rab12 in the regulation of mTORC1 activity and autophagy: Rab12 regulates constitutive degradation of amino-acid transporter PAT4, which indirectly modulates mTORC1 activity and autophagy through uptake of amino acids (see the schematic model in Fig 4D). As mTORC1 activity induces translation of cell-division-related genes and inhibits programmed cell death, upregulation of mTORC1 (and/or autophagy dysfunction) is closely associated with cancer/tumour [34, 35, 36]. Intriguingly, PAT4 is broadly expressed in many cancer cell lines [30]. Hence, the Rab12-regulating mechanism that controls mTORC1 activity and autophagy through quality control of PAT4 that we discovered in this study might provide a new target for the treatment of cancer and tumorigenesis.
Methods
Materials. Rab12 rabbit polyclonal antibody, anti-LC3 rabbit polyclonal antibody and anti-Atg16L1 rabbit polyclonal antibody were prepared as described previously [25, 37, 38]. All other commercially available materials are described in the supplementary information online.
siRNA-mediated knockdown. The siRNAs against mouse Rab1∼43, which are summarized in supplementary Table S1 online, and against mouse PAT4#1 (target sequence: 5′-CCAATCAGCCTTGTGTTTA-3′) and PAT4#2 (target sequence: 5′-GCACGCCGATTGTTTCAAA-3′) were chemically synthesized by Nippon EGT (Toyama, Japan). MEFs were cultured for 48 h (to screen for Rabs involved in autophagy) or 72 h (for PAT4 knockdown). For double knockdown of Rab12 and PAT4, MEFs were first transfected with control or PAT4 siRNA and, 24 h later the cells were transfected with control or Rab12 siRNA. After 48 h, the cells were harvested for immunoblotting or starved to induce autophagy for the immunofluorescence analyses.
Immunofluorescence and image analyses. Immunostaining was performed essentially as described previously [39]. In brief, cultured cells were fixed with 4% paraformaldehyde and stained with specific antibodies. The stained cells were examined for fluorescence with a confocal fluorescence microscope (Fluoview 1000; Olympus, Tokyo, Japan) through an objective lens (× 100 magnification, NA 1.45; Olympus) and with Fluoview software (version 2.1c; Olympus). For quantitative analysis, images of the cells were captured at random with the confocal microscope, and the number of the fluorescent dots (for example, LC3 dots and Atg16L1 dots) was counted with the ImageJ software (version 1.42q; NIH, Bethesda, MD).
Quantification of the intracellular L-amino-acid concentration. Control and Rab12-knockdown MEFs grown on 6-cm dishes were rinsed three times with ice-cold PBS and lysed with the lysis buffer. The lysates were analysed by using an L-Amino-Acid Quantitation Kit (BioVision, Milpitas, CA) according to the manufacturer’s note to determine their intracellular L-amino-acid concentration. The values were divided by the number of cells that had been cultured under the same conditions.
Statistical analysis The statistical analyses were performed by using Student’s unpaired t-test, and P-values <0.05 were considered statistically significant.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Megumi Aizawa for technical assistance and members of the Fukuda Laboratory for valuable discussions. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, and Technology (MEXT) of Japan (to M.F.).
Author contributions: T.M. performed experiments and analysed the data. T.M. and M.F. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Mizushima N, Komatsu M (2011) Autophagy: renovation of cells and tissues. Cell 147: 728–741 [DOI] [PubMed] [Google Scholar]
- Mizushima N (2007) Autophagy: process and function. Genes Dev 21: 2861–2873 [DOI] [PubMed] [Google Scholar]
- Stenmark H (2009) Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10: 513–525 [DOI] [PubMed] [Google Scholar]
- Fader CM, Sánchez D, Furlán M, Colombo MI (2008) Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 9: 230–250 [DOI] [PubMed] [Google Scholar]
- Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA (2012) TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol 197: 659–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ et al. (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8: 445–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N et al. (2009) Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol Biol Cell 20: 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20: 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YY, Neufeld TP (2009) An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell 20: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X (2009) ULK1·ATG13·FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284: 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC et al. (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kim E, Yuan H, Inoki K, Goraksha-Hicks P, Schiesher RL, Neufeld TP, Guan KL (2010) Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem 285: 19705–19709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D, Fisher K, Zolov SN, Xiong T, Inoki K, Weisman LS, Saltiel AR (2012) Rab5 proteins regulate the activation and localization of target of rapamycin complex I. J Biol Chem 287: 20913–20921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WS, Adiel C, Melanie D, Mark T, Dietmar EM, Michael NH (2008) TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell 7: 1819–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM (2012) mTOR signaling in growth control and disease. Cell 13: 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL (2008) Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL (2003) Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 17: 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J (2003) Tuberous sclerosis complex gene products, tuberin and hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol 13: 1259–1268 [DOI] [PubMed] [Google Scholar]
- Matsui T, Itoh T, Fukuda M (2011) Small GTPase Rab12 regulates constitutive degradation of transferrin receptor. Traffic 12: 1432–1443 [DOI] [PubMed] [Google Scholar]
- Nicklin P et al. (2009) Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136: 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, Hall MN (2012) Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell 47: 349–358 [DOI] [PubMed] [Google Scholar]
- Ban H, Shigemitsu K, Yamatsuji T, Haisa M, Nakajo T, Takaoka M, Nobuhisa T, Gunduz M, Tanaka N, Naomoto Y (2004) Arginine and leucine regulate p70 S6 kinase and 4E-BP1 in intestinal epithelial cells. Int J Mol Med 13: 537–543 [PubMed] [Google Scholar]
- Sagné C, Agulhon C, Ravassard P, Darmon M, Hamon M, El Mestikawy S, Gasnier B, Giros B (2001) Identification and characterization of a lysosomal transporter for small neutral amino acids. Proc Natl Acad Sci USA 98: 7206–7211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heublein S, Kazi S, Ogmundsdóttir MH, Attwood EV, Kala S, Boyd CAR, Wilson C, Goberdhan DCI (2010) Proton-assisted amino-acid transporters are conserved regulators of proliferation and amino-acid-dependent mTORC1 activation. Oncogene 29: 4068–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll M, Daniel H, Gasnier B (2004) The SLC36 family: proton-coupled transporters for the absorption of selected amino acids from extracellular and intracellular proteolysis. Pflügers Arch Eur J Physiol 447: 776–779 [DOI] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334: 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai SM, Meredith D (2011) SLC36A4 (hPAT4) is a high affinity amino acid transporter when expressed in Xenopus laevis oocytes. J Biol Chem 286: 2455–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW (2004) Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 428: 332–337 [DOI] [PubMed] [Google Scholar]
- Wendel HG et al. (2007) Dissecting eIF4E action in tumorigenesis. Genes Dev 21: 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A et al. (2011) Autophagy-deficient mice develop multiple liver tumors. Genes Dev 25: 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Fujita N, Kanno E, Yamamoto A, Yoshimori T, Fukuda M (2008) Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell 19: 2916–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M (2011) OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J Cell Biol 192: 839–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Fukuda M (2006) Identification of EPI64 as a GTPase-activating protein specific for Rab27A. J Biol Chem 281: 31823–31831 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.