Abstract
Synthesis of phospholipids, sterols and sphingolipids is thought to occur at contact sites between the endoplasmic reticulum (ER) and other organelles because many lipid-synthesizing enzymes are enriched in these contacts. In only a few cases have the enzymes been localized to contacts in vivo and in no instances have the contacts been demonstrated to be required for enzyme function. Here, we show that plasma membrane (PM)—ER contact sites in yeast are required for phosphatidylcholine synthesis and regulate the activity of the phosphatidylethanolamine N-methyltransferase enzyme, Opi3. Opi3 activity requires Osh3, which localizes to PM–ER contacts where it might facilitate in trans catalysis by Opi3. Thus, membrane contact sites provide a structural mechanism to regulate lipid synthesis.
Keywords: membrane contact sites, phosphatidylcholine synthesis, Opi3, Osh3, Pah1
INTRODUCTION
The structure of the endoplasmic reticulum (ER) of Saccharomyces cerevisiae is somewhat unique among eukaryotes, in that its reticular network, a characteristic of all eukaryotic cells, lies just beneath the plasma membrane (PM). Reconstruction of total yeast ER in individual cells by three-dimensional electron tomography [1] has revealed that PM-associated ER (pmaER) consists of ER tubules and flattened fenestrated ER sheets that are in close apposition to the cytosolic leaflet of the PM. Regions of pmaER that are apposed to the PM are devoid of ribosomes [1], consistent with these being sites of physical contact between ER and PM [2]. PM–ER contacts likely have important roles in all eukaryotic cells [3], and in yeast, pmaER is enriched in lipid-synthesizing enzymes [2], suggesting that PM–ER contacts are important for lipid metabolism. Protein families with lipid-related functions have been found specifically localized at PM–ER contact sites [4,5,6], where they are thought to have roles in non-vesicular lipid transport between PM and ER, although the role for the contacts themselves has not been directly demonstrated. Recently, it has been found that localization of the integral ER phosphatidylinositol phosphate phosphatase, Sac1, to PM–ER contacts regulates phosphatidylinositol 4-phosphate (PI4P) levels in the PM in trans [7]. In trans catalysis by Sac1 requires interaction with Osh3, a soluble lipid-binding protein of the oxysterol-binding protein family that localizes to PM–ER contacts [7]. Consistent with a role for contacts in regulating Sac1, a mutant with reduced pmaER has increased levels of PI4P [8].
We previously found that two genes with roles in ER biogenesis in yeast, SCS2 and ICE2, have an aggravating genetic interaction, and Δscs2Δice2 double mutants have reduced pmaER, suggesting that pmaER performs an essential function required for cell growth [9]. Ice2 is an integral ER protein of unknown function that is required for inheritance of pmaER [10]. Scs2 is a highly conserved tail-anchored protein of the VAP family that localizes proteins containing FFAT motifs to the ER [4] and regulates yeast phospholipid synthesis [11]. Scs2 localizes the oxysterol-binding protein homologues Osh2 and Osh3 to pmaER through interaction with the FFAT motifs in these proteins [4, 6], which is required for regulation of in trans Sac1 activity [7].
RESULTS AND DISCUSSION
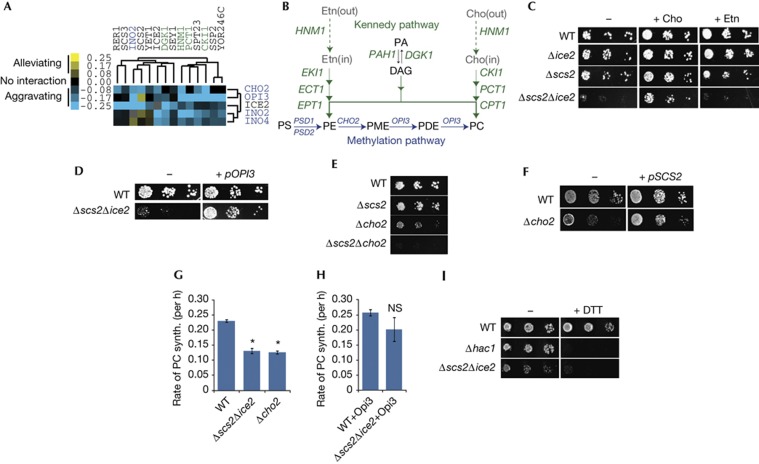
To uncover clues about PM–ER contact site function, we examined the yeast global genetic interaction network [12], which is a comprehensive map of pairwise genetic interactions in which genes with similar functions form coherent clusters. We noticed that SCS2 and ICE2 were in a cluster enriched for genes involved in phospholipid metabolism (Fig 1A). These included genes required to make phosphatidylcholine (PC) by the Kennedy pathway, a salvage pathway that synthesizes PC and PE from the lipid precursors choline, ethanolamine and diacylglycerol (Fig 1B) [13]. This cluster showed predominately aggravating genetic interactions with a cluster containing both PEMT enzymes, encoded by CHO2 and OPI3, which synthesize PC by the methylation pathway (Fig 1A,B). Also in this cluster was the Ino2–Ino4 transcription factor, which activates expression of Kennedy and methylation pathway genes [13], as well as ICE2, further suggesting a function in the methylation pathway.
Figure 1.
Opi3 function is compromised in Δscs2Δice2 cells. (A) Gene clusters containing SCS2 and ICE2 revealed a potential role for PM–ER contacts in phospholipid metabolism. Genes are colour-coded according to pathways in (B). Data were clustered from Costanzo et al [12]. (B) Principal pathways of phospholipid synthesis in S. cerevisiae. Genes encoding enzymes required for each step are shown in italics. (C) Ten-fold serial dilutions of the indicated mutant yeast strains grown on SD media (−) or SD supplemented with either +Cho or +Etn. (D) WT and Δscs2Δice2 yeast grown on SD media transformed with either a plasmid expressing Opi3 (+ pOPI3) or a control plasmid (−). (E) Mutant yeast grown on SD media without choline. (F) Yeast grown on SD media transformed with either a plasmid expressing Scs2 (+ pSCS2) or a control plasmid (−). (G) In vivo PE methylation assay. Log phase yeast were pulse-labelled with [3H]-Etn and the rate of PC synthesis was determined by measuring conversion of PE into PC over time. Error bars, s.e.m. Asterisks, P<0.005 versus WT. (H) PE methylation assay for indicated strains expressing Opi3 from a plasmid (NS versus WT). (I) Yeast grown on SD media containing choline with 1 mM DTT (+DTT) or without (−). Cho, choline; DAG, diacylglycerol; DTT, diothiothreitol; ER, endoplasmic reticulum; Etn, ethanolamine; NS, not significant; PA, phosphatidic acid; PC, phosphatidylcholine; PDE, phosphatidyldimethylethanolamine; PE, phosphatidylethanolamine; PM, plasma membrane; PME, phosphatidylmonomethylethanolamine; PS, phosphatidylserine; SD, synthetic-defined; synth, synthesis; WT, wild-type.
This clustering suggested that PM–ER contacts might regulate PC synthesis. We found that choline, but not ethanolamine, rescued the Δscs2Δice2 slow-growth phenotype (Fig 1C), indicating that the mutant likely had a PC synthesis defect in the methylation pathway. Overexpression of OPI3 in the Δscs2Δice2 mutant rescued its growth defect (Fig 1D), whereas overexpression of CHO2 did not (supplementary Fig S1 online), suggesting that Opi3 function was compromised. Addition of monomethylethanolamine, which is converted into phosphatidylmonomethylethanolamine (PME) by the Kennedy pathway and bypasses the requirement for Cho2, did not rescue growth of Δscs2Δice2 cells (supplementary Fig S1 online) consistent with loss of Opi3 function in the mutant.
Cho2 performs the first PE methylation, whereas Opi3 is primarily responsible for the second and third methylations (Fig 1B), although Opi3 can also methylate first, but at a reduced rate [14]. Because of this redundancy, Δcho2 and Δopi3 cells grow poorly on media lacking choline, but are not obligate choline auxotrophs, whereas Δcho2Δopi3 cells are obligate auxotrophs and are rescued by overexpression of Opi3, but not Cho2 [14]. The aggravating genetic interaction between Scs2 and Cho2 and the alleviating interaction between Scs2 and Opi3 observed in the gene cluster (Fig 1A) suggested that Scs2 might modify Opi3 function directly. Consistent with Scs2 regulating Opi3, we found that Δscs2Δcho2 cells were obligate choline auxotrophs (Fig 1E), and overexpression of Scs2 rescued the choline auxotrophy of the Δcho2 mutant (Fig 1F). The function of ICE2 was less clear; however, we did uncover a genetic interaction with PSD1 that was rescued by ethanolamine (supplementary Fig S1 online), further supporting a role for Ice2 in the methylation pathway as implied by the gene cluster.
To measure Opi3 function in the Δscs2Δice2 mutant, we pulse-labelled cells with [3H]-ethanolamine and monitored conversion of radiolabelled PE into PC. In both wild-type and mutant cells, the rate of incorporation of label into PC was linear for at least the first 3 h of the assay (supplementary Fig S1 online). As expected, we found that the rate of PC synthesis in a Δcho2 mutant control was significantly reduced (Fig 1G). Consistent with decreased Opi3 function, PC synthesis was also reduced in the Δscs2Δice2 mutant (Fig 1G). Overexpression of Opi3 increased the rate of PC synthesis back to levels similar to wild type (Fig 1H; supplementary Fig S1 online). Thus, reduced synthesis of PC in the Δscs2Δice2 mutant likely accounted for its choline auxotrophy. Finally, the Δscs2Δice2 mutant was sensitive to dithiothreitol (Fig 1I), indicating increased ER stress, also consistent with decreased Opi3 function [15].
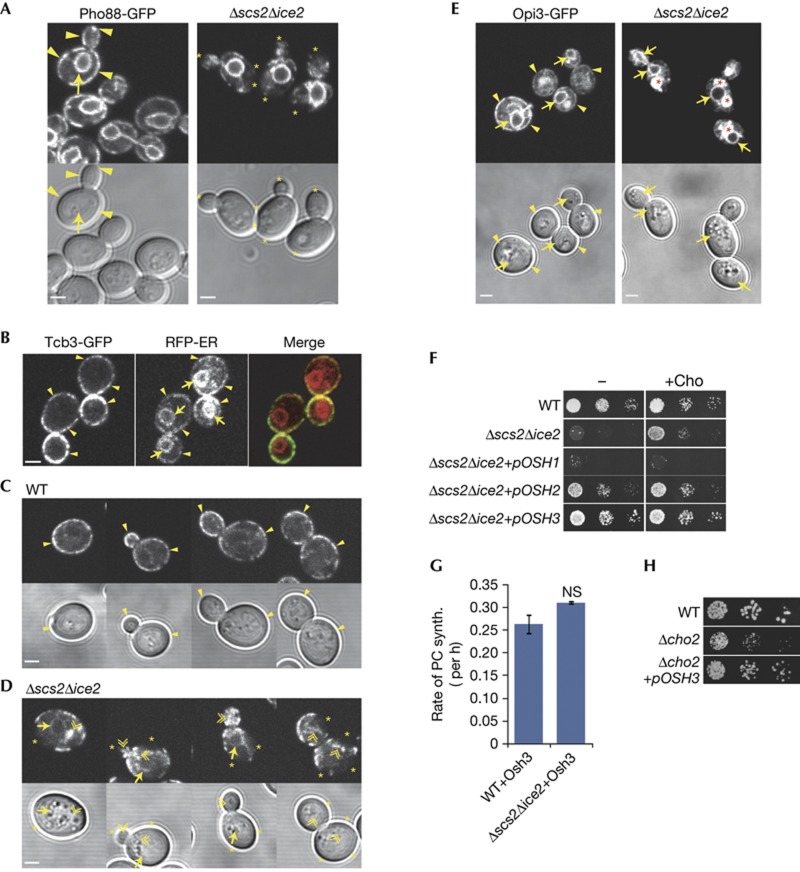
Loss of Opi3 function in the Δscs2Δice2 mutant suggested that Opi3 functioned at PM–ER contacts. Using an endogenous type I integral ER protein Pho88 tagged with green fluorescent protein (GFP), we verified our previous findings that Δscs2Δice2 cells had normal ER tubules and nuclear ER, but greatly diminished ER at the cell cortex (Fig 2A). To characterize PM–ER contacts, we used a specific marker that is an integral ER membrane protein that localizes to PM–ER contacts by interacting directly with the PM [5]. As previously found, Tcb3-GFP localized only to the pmaER domain at the cell cortex, but not the nuclear ER or ER tubules (Fig 2B). Consistent with three-dimensional electron tomography showing that pmaER forms distinct domains in the bud and mother, and is absent from the bud neck [1], Tcb3-GFP was discontinuous through the neck and otherwise localized to pmaER throughout the cell cycle (Fig 2C). In the Δscs2Δice2 mutant, Tcb3-GFP localization was clearly disrupted. pmaER defects were pronounced in small and medium-sized buds, in which Tcb3-GFP often failed to localize to the bud periphery and instead localized to ER tubules near the centre of the bud (Fig 2D). In mothers, Tcb3-GFP also mislocalized to the nuclear ER.
Figure 2.
PM–ER contacts regulate Opi3. (A) Images of yeast cells expressing endogenous Pho88-GFP. Arrows indicate nuclear ER, arrowheads indicate pmaER and asterisks indicate absence of pmaER. (B) Yeast expressing endogenous Tcb3-GFP (green) and RFP-ER (red) expressed from a plasmid. Labels as in (A). (C,D) Yeast expressing Tcb3-GFP staged throughout the cell cycle. Double arrowheads indicate mislocalization of Tcb3-GFP to ER tubules, asterisks indicate absence of pmaER, otherwise labels as in (A). (E) Yeast expressing endogenous Opi3-GFP. Asterisks indicate vacuoles, otherwise labels as in (A). All scale bars, 2 μm. (F) Growth of WT and Δscs2Δice2 yeast overexpressing Osh proteins from plasmids grown on SD media containing galactose in the absence (−) or presence (+Cho) of choline. (G) PE methylation assay for indicated strains expressing Osh3 from a plasmid. Error bars, s.e.m. (H) Growth assays of WT and Δcho2 mutant yeast overexpressing Osh3 from a plasmid grown on SD media containing galactose in the absence of choline. ER, endoplasmic reticulum; GFP, green fluorescent protein; NS, not significant; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PM, plasma membrane; pmaER, PM-associated ER; SD, synthetic-defined; synth, synthesis; WT, wild-type.
Now, we examined the localization of the endogenous Opi3 enzyme tagged with GFP, which appeared functional (supplementary Fig S2 online). In wild-type cells, Opi3-GFP localized throughout the ER (Fig 2E). We observed some extra diffuse staining in the vacuole that was likely a result of turnover of Opi3-GFP. In the Δscs2Δice2 mutant, localization of Opi3-GFP to the nuclear ER remained intact; however, it was almost completely absent from pmaER (Fig 2E). Quantification of Opi3-GFP in the nuclear ER revealed no change in its expression level (supplementary Fig S2 online), indicating that loss of Opi3 function likely resulted from the defect in pmaER in the mutant. Consistent with this, Opi3 overexpression did not seem to rescue pmaER (supplementary Fig S2 online).
Scs2 localizes both Osh2 and Osh3 to PM–ER contacts [4], and we tested for their roles in regulating Opi3. Overexpression of Osh2 partially restored growth of the Δscs2Δice2 mutant, whereas Osh3 fully rescued (Fig 2F). In contrast, Osh1, which is localized to the nucleus–vacuole junction by Scs2 [4], did not rescue, and in fact impeded growth (Fig 2F). Osh3 overexpression restored PC synthesis in the Δscs2Δice2 mutant (Fig 2G), consistent with rescue of growth of the mutant. However, Osh3 did not rescue pmaER in Δscs2Δice2 cells (supplementary Fig S2 online), but it did restore growth of the Δcho2 mutant (Fig 2H), suggesting that Osh3 regulated Opi3 function at contacts.
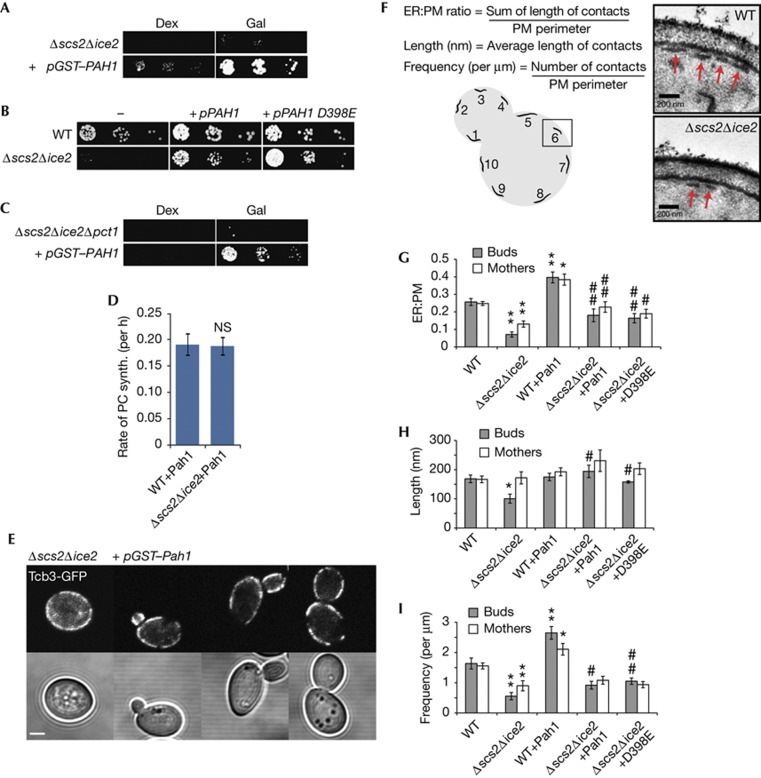
We now exploited the choline auxotrophy phenotype of the Δscs2Δice2 mutant in a screen to identify regulators of PM–ER contact structure. We identified a suppressor plasmid carrying the PAH1 gene (Fig 3A), which encodes a highly conserved phosphatidic acid phosphatase enzyme of the lipin family that catalyses the conversion of phosphatidic acid into diacylglycerol in the Kennedy pathway (Fig 1B) [16]. Surprisingly, a catalytically inactive mutant of Pah1, D398E [17], also rescued the choline auxotrophy of the Δscs2Δice2 mutant (Fig 3B), arguing against rescue via the Kennedy pathway. Consistent with this, Pah1 still rescued a Δscs2Δice2Δpct1 triple mutant with an inactivated Kennedy pathway (Fig 3C). Overexpression of Pah1 rescued PC synthesis in the Δscs2Δice2 mutant (Fig 3D; supplementary Fig S3 online), indicating that Pah1 rescued Opi3 function. However, it did not rescue the Δcho2 mutant (supplementary Fig S3 online), suggesting that Pah1 did not regulate Opi3 directly.
Figure 3.
Pah1 regulates PM–ER contacts. (A,C) Serial dilutions of (A) Δscs2Δice2 and (C) Δscs2Δice2Δpct1 yeast overexpressing GST–Pah1 from a plasmid under control of the galactose promoter (+pGST–PAH1) on SD media containing either glucose (Dex) or galactose (Gal). (B) Yeast expressing either HA–Pah1 or the catalytically inactive D398E mutant from a plasmid grown on SD media lacking choline. (D) Opi3 methylation assay for wild-type and Δscs2Δice2 cells overexpressing GST–Pah1. (E) Images of Tcb3-GFP in Δscs2Δice2 yeast overexpressing GST–Pah1. Scale bar, 2 μm. (F) Schematic of ultrastructural assay and representative transmission electron microscopy images illustrating PM–ER contacts (arrows). (G) Ratio of PM–ER contacts to PM perimeter. **P<10−4 versus WT; *P<0.001 versus WT; ##P<0.01 versus Δscs2Δice2, #P<0.05 versus Δscs2Δice2. (H) PM–ER contact length. *P<0.005 versus WT; #P<0.005 versus Δscs2Δice2. (I) Frequency of PM–ER contacts. **P<0.005 versus WT; *P<0.05 versus WT; ##P<0.005 versus Δscs2Δice2; #P<0.05 versus Δscs2Δice2. Error bars, s.e.m. ER, endoplasmic reticulum; GFP, green fluorescent protein; NS, not significant; PM, plasma membrane; SD, synthetic-defined; synth, synthesis; WT, wild-type.
These results suggested that Pah1 might rescue pmaER in the Δscs2Δice2 mutant. In contrast to Opi3 and Osh3, overexpression of GST–Pah1 appeared to restore Tcb3-GFP localization to pmaER (Fig 3E), and Pah1 also restored Opi3-GFP to pmaER (supplementary Fig S3 online). Therefore, we performed an ultrastructural analysis of PM–ER contacts using transmission electron microscopy. We quantified ER segment length, frequency and the overall ratio of PM–ER contacts to PM perimeter (Fig 3F; supplementary Fig S4 online). In the Δscs2Δice2 mutant, we found that contacts were decreased to ∼7% in buds and ∼13% in mothers, compared with ∼25% of the cell periphery in wild type (Fig 3G). In buds, the decrease resulted from both a decrease in contact site length (Fig 3H) and frequency (Fig 3I). In mothers, the decrease resulted only from reduced frequency as contact site length was unaffected.
Overexpression of both Pah1 and D398E Pah1 in the Δscs2Δice2 mutant restored contacts to near wild-type levels in both buds and mothers (Fig 3G). Rescue resulted from increased contact length and frequency in buds (Fig 3H,I), indicating that Pah1 rescued the defect in formation of pmaER in the bud. In wild-type cells, Pah1 increased contacts to ∼40% of the cell periphery, a result of increased frequency, supporting a physiological role for Pah1 in initiating contacts. Finally, Pah1 rescued the dithiothreitol sensitivity of the Δscs2Δice2 mutant, which did not require activation of the unfolded protein response (supplementary Fig S3 online), further supporting that Pah1 directly rescued the defect in PM–ER contacts by a mechanism that was independent of activation of general stress response pathways.
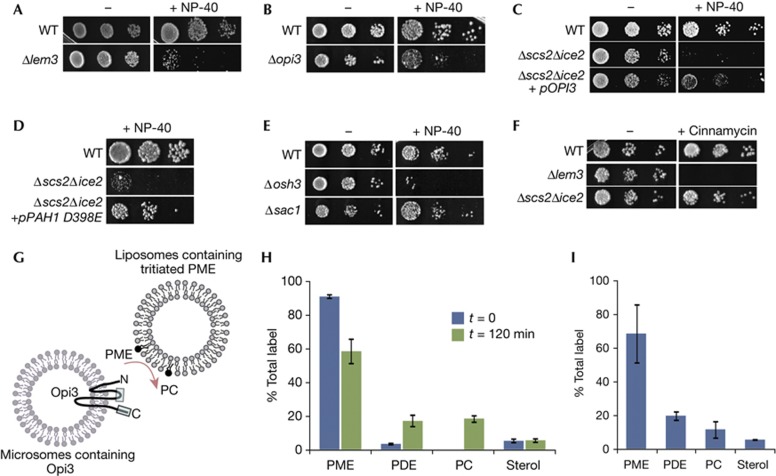
Our results indicate that PM–ER contacts provide a spatial mechanism to regulate synthesis of PC by the yeast PEMT enzyme, Opi3. A consequence of altered Opi3 activity at contacts could be altered PM stability, as has been found in the livers of Pemt−/− mice, which have increased PE:PC in their PM [18]. Yeast mutants with increased PE:PC in the PM are sensitive to non-ionic detergents, which destabilize the bilayer [19]. We found that Δlem3/ros3 cells, which have a buildup of PE in the PM owing to decreased PE flippase activity [20, 21], were sensitive to NP-40 (Fig 4A). We also found that Δopi3 cells, which have increased PME [22], were NP-40 sensitive (Fig 4B), suggesting that PME accumulated in the PM. The Δscs2Δice2 mutant was similarly NP-40 sensitive, which was suppressed by Opi3 (Fig 4C). Importantly, D398E Pah1 also suppressed, consistent with rescue of PM–ER contacts and reconstitution of Opi3 function by Pah1 (Fig 4D). Loss of Osh3 also caused NP-40 sensitivity, further supporting its role in regulating Opi3 at contacts (Fig 4E). This function for Osh3 was independent of its role in regulating PI4P levels in the PM [7], because Δsac1 cells were insensitive to NP-40 (Fig 4E). NP-40 sensitivity was not owing to decreased PE flippase activity, because Δscs2Δice2 cells were insensitive to cinnamycin (Ro09-0198), which binds PE located in the outer leaflet of the PM and lyses cells [20, 21], whereas Δlem3/ros3 cells were highly sensitive (Fig 4F).
Figure 4.
PM–ER contacts affect PM stability in trans. (A–E) Yeast growth assays done in the absence (−) or presence of 0.1% NP-40 (+NP-40) on SD media containing choline. (F) Yeast growth assays done in the absence (−) or presence of cinnaymcin (+) on SD media containing choline. (G) Schematic of in vitro trans-methylation assay. Opi3 topology is on the basis of mammalian PEMT and boxes indicate regions containing catalytic sites [25]. (H) Distribution of label before and 120 min after mixing. (I) Distribution of label in the liposomal fraction 120 min after mixing. Error bars, s.d. ER, endoplasmic reticulum; PC, phosphatidylcholine; PDE, phosphatidyldimethylethanolamine; PM, plasma membrane; PME, phosphatidylmonomethylethanolamine; SD, synthetic-defined; WT, wild-type.
Previous work suggested that Opi3 could act in trans in catalysing the methylation of PME located in a juxtaposed membrane [23]. Hence, the role for PM–ER contacts might be to provide Opi3 access to PE/PME located in the PM for in trans methylation. We tested this possibility using an in vitro assay. We incubated liposomes containing tritiated PME with ER microsomes containing Opi3 and monitored synthesis of PC (Fig 4G). Mixing of liposomes and microsomes resulted in conversion of ∼35% of the PME into phosphatidyldimethylethanolamine (PDE) and PC (Fig 4H). Recovery of the liposomal fraction, which contained the most radiolabel (supplementary Fig S5 online), revealed a similar lipid distribution with ∼30% of the PME converted to PDE and PC (Fig 4I). Thus, accumulation of PDE and PC in the liposomal fraction was consistent with in trans Opi3 activity. We did not detect defects in endocytosis or uptake of NBD–PE/PC in the Δscs2Δice2 mutant (supplementary Fig S5 online), indicating that lipid transport between PM and ER was likely not disrupted, further supporting that Opi3 functioned in trans at contacts.
We propose that PM–ER contacts regulate the activity of Opi3 through both providing its lipid substrate in trans and by restricting access of Opi3 to Osh3 at contacts. Similar to its proposed role in regulation of Sac1 [7], Osh3 might present PME or PE in the PM to Opi3 located at PM–ER contacts. In trans methylation by Opi3 might enable cells to rapidly adjust the PME/PE:PC ratio of the PM, affecting the physical properties of the bilayer. In trans methylation by Opi3 requires that PME is available in the PM. Lipidomic analysis of subcellular fractions of yeast organelles identified PME in the Golgi, but not in any other compartments, including ER microsomes and PM [24]. This suggests that PME is a constituent of the secretory pathway and is trafficked to the PM where it is rapidly converted into PC. Finally, the active site of all eukaryotic PEMTs resides on the cytoplasmic face of the ER membrane, accessible to the PM [25] (Fig 4G), and prokaryotic PEMTs are soluble enzymes that lack transmembrane domains [26] and must, by their nature, associate with the PM in trans. Thus, PM–ER contacts might regulate PC synthesis by providing PEMT enzymes located in the ER access to their lipid substrates located in trans in the PM.
METHODS
Plasmid yeast strains and growth conditions. Details of plasmids and yeast strains used in this study can be found in the supplementary information online. All yeast strains were on the basis of S288C, and growth assays were performed at 30 °C for 24–48 h on synthetic-defined (SD) medium.
Array-based genome-wide suppressor screen. The array-based genome-wide suppressor screen was performed by adapting the synthetic genetic array method to introduce each plasmid from the yeast GST–ORF collection (Open Biosystems) into a Δscs2Δice2 strain and scoring for growth on media lacking choline.
Confocal microscopy. Yeast strains expressing GFP-tagged proteins were grown to log phase and imaged using a Zeiss LSM-5 Pascal confocal microscopy system equipped with a Zeiss × 100 objective (Plan-neofluar, 1.3) by squashing 1–5 μl of live yeast in media between a slide and cover slip.
Transmission electron microscopy of PM–ER contacts. For thin-section electron microscopy, cells were prepared and imaged on a Hitachi H7600 transmission electron microscope. A minimum of 15 budded cells were analysed per condition.
In vivo methylation assay. Briefly, log phase cells were collected and incubated with 24 μCi of [3H]-ethanolamine for 1 h at 37 °C, washed and shaken at 30 °C for the indicated times before total lipids were extracted and analysed by high-performance liquid chromatography.
In vitro Opi3 trans methylation assay. Briefly, microsomes containing Opi3 were isolated from a wild-type strain and liposomes were prepared from lipids extracted from a Δopi3 strain labelled with [3-H] methionine, and microsomes were incubated with [3H]-PME-loaded liposomes at 30 °C for 2 h. Lipids were extracted before or after separating liposomes from microsomes on a 20% (w/v) sucrose cushion by centrifugation, and the conversion of [3H]-methyl-PME was quantitated after thin layer chromatography.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank W. Vogel and also the UBC Bio-imaging Facility for their technical expertise with transmission electron microscopy. This work was supported by the Canadian Institutes of Health Research, the Michael Smith Foundation for Health Research and the Canadian Foundation for Innovation.
Author contributions: S.T. and C.J.R.L. conceived, designed and analysed the experiments. S.T. performed the most experiments. J.T.C constructed yeast strains and performed yeast growth assays. B.P.Y. performed the suppressor screen and with W.A.P developed and performed the in vivo PE methylation assays. A.I.P.M.d.K. and R.C.C. designed and performed the in vitro trans-methylation assays. S.T., B.P.Y. and C.J.R.L wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- West M, Zurek N, Hoenger A, Voeltz GK (2011) A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol 193: 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler H, Gaigg B, Hrastnik C, Achleitner G, Kohlwein SD, Zellnig G, Perktold A, Daum G (2001) A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur J Biochem 268: 2351–2361 [DOI] [PubMed] [Google Scholar]
- Levine T (2004) Short-range intracellular trafficking of small molecules across endoplasmic reticulum junctions. Trends Cell Biol 14: 483–490 [DOI] [PubMed] [Google Scholar]
- Loewen CJ, Roy A, Levine TP (2003) A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J 22: 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A, Prinz WA (2012) A conserved membrane-binding domain targets proteins to organelle contact sites. J Cell Sci 125: Pt 149–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TA, Choi MG, Raychaudhuri S, Mears JA, Ghirlando R, Hinshaw JE, Prinz WA (2009) Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J Cell Biol 187: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD (2011) Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144: 389–401 [DOI] [PubMed] [Google Scholar]
- Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD (2012) ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell 23: 1129–1140 [DOI] [PubMed] [Google Scholar]
- Loewen CJ, Young BP, Tavassoli S, Levine TP (2007) Inheritance of cortical ER in yeast is required for normal septin organization. J Cell Biol 179: 467–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada de Martin P, Du Y, Novick P, Ferro-Novick S (2005) Ice2p is important for the distribution and structure of the cortical ER network in Saccharomyces cerevisiae. J Cell Sci 118: Pt 165–77 [DOI] [PubMed] [Google Scholar]
- Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304: 1644–1647 [DOI] [PubMed] [Google Scholar]
- Costanzo M et al. (2010) The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Han GS (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu Rev Biochem 80: 859–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaki T, Yamashita S (1987) Yeast phosphatidylethanolamine methylation pathway. Cloning and characterization of two distinct methyltransferase genes. J Biol Chem 262: 15428–15435 [PubMed] [Google Scholar]
- Thibault G, Shui G, Kim W, McAlister GC, Ismail N, Gygi SP, Wenk MR, Ng DT (2012) The membrane stress response buffers lethal effects of lipid disequilibrium by reprogramming the protein homeostasis network. Mol Cell 48: 16–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GS, Wu WI, Carman GM (2006) The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem 281: 9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GS, Siniossoglou S, Carman GM (2007) The cellular functions of the yeast lipin homolog PAH1p are dependent on its phosphatidate phosphatase activity. J Biol Chem 282: 37026–37035 [DOI] [PubMed] [Google Scholar]
- Li Z, Agellon LB, Allen TM, Umeda M, Jewell L, Mason A, Vance DE (2006) The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab 3: 321–331 [DOI] [PubMed] [Google Scholar]
- Schüller C, Mamnun YM, Wolfger H, Rockwell N, Thorner J, Kuchler K (2007) Membrane-active compounds activate the transcription factors Pdr1 and Pdr3 connecting pleiotropic drug resistance and membrane lipid homeostasis in Saccharomyces cerevisiae. Mol Biol Cell 18: 4932–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato U, Emoto K, Fredriksson C, Nakamura H, Ohta A, Kobayashi T, Murakami-Murofushi K, Kobayashi T, Umeda M (2002) A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J Biol Chem 277: 37855–37862 [DOI] [PubMed] [Google Scholar]
- Saito K, Fujimura-Kamada K, Hanamatsu H, Kato U, Umeda M, Kozminski KG, Tanaka K (2007) Transbilayer phospholipid flipping regulates Cdc42p signaling during polarized cell growth via Rga GTPase-activating proteins. Dev Cell 13: 743–751 [DOI] [PubMed] [Google Scholar]
- Bilgin M, Markgraf DF, Duchoslav E, Knudsen J, Jensen ON, de Kroon AIPM, Ejsing CS (2011) Quantitative profiling of PE, MMPE, DMPE, and PC lipid species by multiple precursor ion scanning: a tool for monitoring PE metabolism. Biochim Biophys Acta 1811: 1081–1089 [DOI] [PubMed] [Google Scholar]
- Janssen MJ, de Jong HM, de Kruijff B, de Kroon AI (2002) Cooperative activity of phospholipid-N-methyltransferases localized in different membranes. FEBS Lett 513: 197–202 [DOI] [PubMed] [Google Scholar]
- Schneiter R et al. (1999) Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol 146: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields DJ, Altarejos JY, Wang X, Agellon LB, Vance DE (2003) Molecular dissection of the S-adenosylmethionine-binding site of phosphatidylethanolamine N-methyltransferase. J Biol Chem 278: 35826–35836 [DOI] [PubMed] [Google Scholar]
- Klüsener S, Aktas M, Thormann KM, Wessel M, Narberhaus F (2009) Expression and physiological relevance of Agrobacterium tumefaciens phosphatidylcholine biosynthesis genes. J Bacteriol 191: 365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.