Abstract
The selective autophagy receptors Atg19 and Atg32 interact with two proteins of the core autophagic machinery: the scaffold protein Atg11 and the ubiquitin-like protein Atg8. We found that the Pichia pastoris pexophagy receptor, Atg30, also interacts with Atg8. Both Atg30 and Atg32 interactions are regulated by phosphorylation close to Atg8-interaction motifs. Extending this finding to Saccharomyces cerevisiae, we confirmed phosphoregulation for the mitophagy and pexophagy receptors, Atg32 and Atg36. Each Atg30 molecule must interact with both Atg8 and Atg11 for full functionality, and these interactions occur independently and not simultaneously, but rather in random order. We present a common model for the phosphoregulation of selective autophagy receptors.
Keywords: Atg30, Atg32, mitophagy, pexophagy, phosphorylation
INTRODUCTION
Macroautophagy (hereafter called autophagy) is an intracellular bulk degradation system, and is distinct from selective autophagy, which facilitates degradation of specific cargos [1]. Autophagy in yeast is primarily a survival response to nutrient starvation, whereas selective autophagy has a variety of roles, such as cell remodelling to adapt to different environmental conditions and elimination of damaged organelles. Cargo selectivity is mediated via autophagy receptors that simultaneously bind cargos and components of the autophagic machinery [2]. In yeast, four receptors have been described: three in S. cereivisae, Atg19 (cytoplasm-to-vacuole targeting (Cvt) pathway), Atg32 (mitophagy) and Atg36 (pexophagy), and one in P. pastoris, Atg30 (pexophagy) [3,4,5,6,7]. Atg19 interacts directly with the cargo (aminopeptidase I, Ape1) to form the Cvt complex, and subsequently with two autophagy proteins, Atg11 and Atg8 [8]. Atg11 is a required protein for most selective autophagy pathways in yeast and functions as a basic scaffold in assembling the specific phagophore assembly site (PAS) by interacting directly with the receptor, with itself and several other proteins such as Atg1, Atg9 and Atg17 [9] to form the PAS. Selective autophagy receptors interact with Atg8 through WxxL-like sequences, called Atg8-family-interacting motifs (AIMs in yeast) or LC3-interacting regions (LIRs in animals) [10]. Atg19 has such an AIM motif near its carboxy terminus [8]. The hierarchical assembly of Atg8 at the PAS depends on many other autophagy-related (Atg) proteins, suggesting that the binding between Atg19 and Atg8 likely succeeds the Atg11 interaction in growing conditions [11, 12].
Much like Atg19, other autophagy receptors including P. pastoris Atg30 and S. cerevisiae Atg32 and Atg36 (hereafter called Atg30, ScAtg32 and Atg36, respectively) localize with their respective cargos and interact with autophagy proteins. Atg30 and ScAtg32 localize at the cargo surface during organelle biogenesis [4,4,5,6,7]. During pexophagy and mitophagy, these receptors are phosphorylated by an unknown kinase(s), facilitating their interaction with Atg11 and subsequent PAS formation [4, 13]. In addition, Atg30 interacts directly with another scaffold protein, Atg17. Moreover, as a classic autophagy receptor, ScAtg32 interacts with Atg8 through an AIM, but such an interaction is yet to be described for Atg30 and Atg36 [7].
Despite studies involving individual selective autophagy receptors and their interacting partners, little is known about whether and how these interactions are regulated: whether they proceed sequentially or simultaneously; in the same molecule or in two separate molecules; or whether common mechanisms exist for different forms of selective autophagy. We show the existence of a phosphoregulatable AIM on Atg30, Atg32 and Atg36 required for their interactions with Atg8. In addition, we describe putative consensus motifs for Atg8 and Atg11 binding on the receptors. Mutations of these consensus motifs allowed us to study the mechanism of interactions between the receptors and the autophagy proteins. These studies reveal a conserved mode of regulation of selective autophagy pathways, illuminating shared mechanistic principles.
RESULTS AND DISCUSSION
Atg30 interacts with Atg8
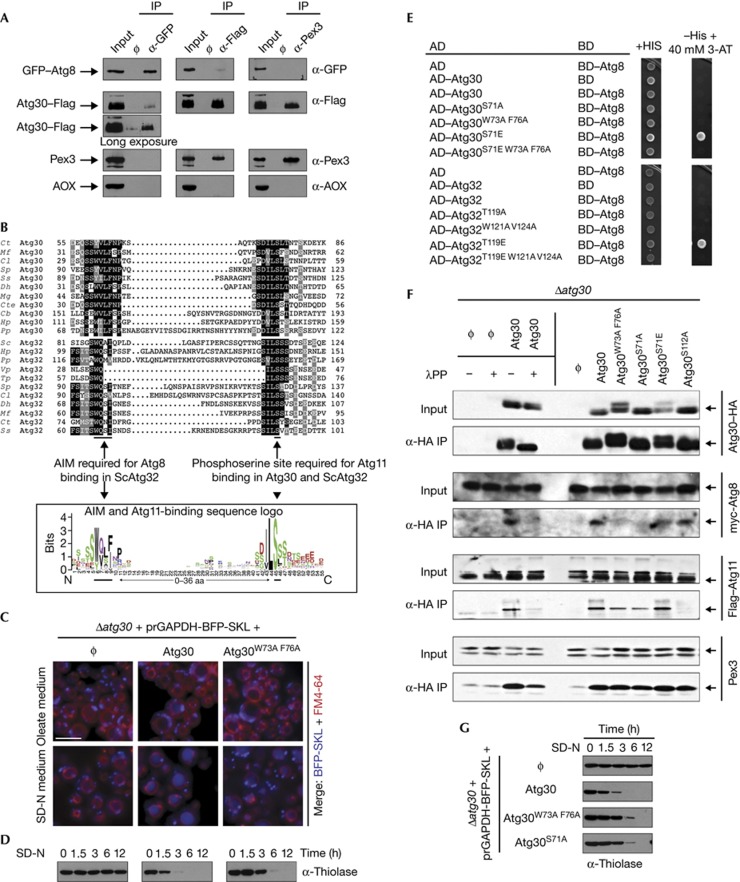
The selective autophagy receptors (Atg19 and ScAtg32) in yeast bind both Atg8 and Atg11 [5, 6, 8]. So far, only Atg11, but not Atg8, is known to interact with Atg30 [4]. By co-immunoprecipitation (IP) experiments, we found that Atg30 also interacts with Atg8 (Fig 1A), suggesting that Atg30 interacts with Atg8 and Atg11 during pexophagy, like the other selective autophagy receptors.
Figure 1.
Atg30 interacts with Atg8 through a cryptic AIM, and phosphorylation upstream of the AIM regulates their interaction. (A) IP of GFP–Atg8 (α-GFP), Atg30–Flag (α-Flag) and Pex3 (α-Pex3) under pexophagy conditions. The abundant peroxisome matrix protein, AOX, was used as a negative control. Input: total lysate; ϕ: IP without antibody. (B) Two multiple sequence alignments, including 11 Atg30 homologues and 11 Atg32 homologues (identical residues are indicated with black boxes, and similar residues with grey boxes), and a sequence logo of the combined multiple sequence alignments from Atg30 and Atg32 homologues listed currently in GenBank. (C,D) Pexophagy experiments of Δatg30 (ϕ), wild-type (Atg30) and Atg30 AIM mutant (Atg30W73A F76A) cells were done by fluorescence microscopy, following the degradation of peroxisomes labelled with BFP fused at its C-terminus to the Ser–Lys–Leu peroxisomal targeting signal 1 (BFP–SKL) and biochemically by monitoring peroxisomal thiolase degradation. Vacuoles were labelled with FM4-64. Scale bar, 5 μm. (E) AH109 cells were transformed with two yeast two-hybrid assay plasmids, AD and BD, which encode the indicated domains fused with Atg30, Atg32 and Atg8 or an empty vector, as negative controls and grown on +His and −His+40 mM 3-AT plates. (F). Δatg30 (ϕ) and Δatg30 cells complemented with Atg30–HA (Atg30) and several Atg30–HA mutants were immunoprecipitated (α-HA IP) under pexophagy conditions. In addition, α-HA IP of Δatg30 cells (ϕ) and Δatg30 cells complemented with Atg30–HA (Atg30) were incubated with (+) and without (−) λPP. Input: total lysate. (G) Pexophagy in atg30 mutants was monitored by following thiolase levels of oleate-induced peroxisomes after shifting cells to SD-N. aa, amino acid; AD, activation domain; AIM, Atg8-family-interacting motif; AOX, alcohol oxidase; BD, binding domain; GFP, green fluorescent protein; HA, haemagglutinin; IP, immunoprecipitation; λPP, λ protein phosphatase.
Atg30 has a cryptic AIM motif
Mutation of a putative AIM (YxxL, amino acids (aas) 330–333 in Atg30), that is not conserved between Atg30 homologues (supplementary Fig S1A online), showed it was unnecessary for pexophagy (supplementary Fig S1B online). The S. cerevisiae mitophagy receptor, ScAtg32, has a phosphorylation-dependent Atg11-binding site with a proximal AIM [13], which led us to another putative AIM-like sequence in Atg30. Atg30 is phosphorylated on Ser112 (S112) and this modification is essential for Atg11 binding and pexophagy [4]. Interestingly, the sequences surrounding the phosphorylation sites, S112 in Atg30 and S114 in ScAtg32, required for Atg11 binding, were similar (Fig 1B). Several sequence alignments of Atg30 and Atg32 homologues highlighted a conserved motif (D/S)ILSSS surrounding the phosphosite required for Atg11 binding (underlined). The AIM in the Atg32 proteins is in close proximity and upstream of the Atg11-binding site, a situation mimicked in Atg30 proteins. The putative AIM in Atg30 (aas 73–76) does not conform to the strict consensus W/F/YxxL/I/V, but has the sequence, W/YxxF [14]. We mutated this cryptic AIM sequence (Atg30W73A F76A) and checked its effect on pexophagy (Figs 1C,D; supplementary Fig S2A–C online). The Δatg30 cells expressing Atg30W73A F76A degraded peroxisomes slower than did wild-type cells, suggesting that this AIM could bind Atg8. The pexophagy defect in this mutant was only partial (Fig 1D), but comparable to the mitophagy defect found in the AIM mutant of ScAtg32 [6].
Phosphoregulatable AIMs in Atg30 and Atg32
In a previous study [4], we had not detected the Atg8–Atg30 interaction by two-hybrid in S. cerevisiae (Y2H), but the recent discovery of the phosphorylation requirement upstream of the AIM/LIR of the OPTN receptor in mammals [15], together with our knowledge that the heterologous P. pastoris Atg30 used in the Y2H was not phosphorylated in S. cerevisiae, suggested that the absence of phosphorylation of Atg30 might have caused the failure of interaction in S. cerevisiae. Interestingly, the hydrophobic core sequences of the AIMs of Atg30 and Atg32 are preceded by several Ser and Thr residues (Fig 1B). We mutated these residues upstream of the putative AIMs, replacing them with a phosphomimic aa (Asp or Glu) or a non-phosphorylatable aa (Ala), and assessed interactions by Y2H (Fig 1E). Atg30 and P. pastoris Atg32 (hereafter called Atg32) with phosphomimic mutations upstream of the AIMs (Atg30S71E and Atg32T119E) did interact with Atg8, but the wild-type proteins and the non-phosphorylatable mutants (Atg30, Atg32, Atg30S71A and Atg32T119A) did not, suggesting that the absence of interaction of wild-type Atg30 with Atg8 was indeed owing to the lack of Atg30 phosphorylation in S. cerevisiae. In addition, we mutated the AIMs of Atg30 to Atg30W73A F76A, Atg30S71E to Atg30S71E W73A F76A, Atg32 to Atg32W121A V124A and Atg32T119E to Atg32T119E W121A V124A. Mutation of the AIMs in Atg30S71E and Atg32T119E abolished the interactions with Atg8, confirming that both Atg30 and Atg32 contain phosphoregulatable AIMs.
We validated independently the phosphorylation requirement for Atg30–Atg8 binding in P. pastoris by Atg30–haemagglutinin (HA) co-IP from different mutants cells (as indicated in the Fig 1F) in the presence (+) or absence (−) of phage λ protein phosphatase. The interactions of both Atg8 and Atg11, but not the control Pex3, with Atg30 were severely affected by the phosphatase treatment. The AIM and S71A mutations in Atg30 also abolished the interaction with Atg8, but not with Atg11 or Pex3. In contrast, and as expected, the phosphomimic mutation (Atg30S71E) did not impair the Atg30–Atg8 interaction.
Definitive evidence of phosphorylation at S71 of Atg30 was obtained by mobility shift detection of phosphorylated proteins and mass spectrometry (MS) of Atg30 purified from P. pastoris cells. First, we confirmed the presence of a phosphorylation site upstream of the AIM of Atg30 (S71) using Phos-Tag acrylamide to improve the separation of phosphoproteins (supplementary Fig S3A online). When S71 was mutated to a non-phosphorylatable S71A (Atg30S71A), some phospho-Atg30 forms shifted to a lower molecular weight, but this protein mobility was rescued by Atg30S71E. In addition, affinity-purified Atg30–HA subjected to MS revealed that Atg30 was phosphorylated at S71 (supplementary Fig S3B,C online).
The physiological relevance of the Atg30 phosphorylation was tested by pexophagy assays (Fig 1G; supplementary Fig S2A,B online) wherein Atg30S71A exhibited delayed pexophagy, similar to the AIM mutant (Atg30W73A F76A). These results indicate that both pexophagy and mitophagy receptors in P. pastoris interact with Atg8 in a phosphorylation-dependent manner and are regulated by an unknown kinase(s). This finding reveals a conserved mode of regulation of Atg8/LC3 binding to autophagy receptors that is likely to be a general and recurring theme across the evolutionary spectrum [15].
Mode of interaction of Atg11 and Atg8 with Atg30
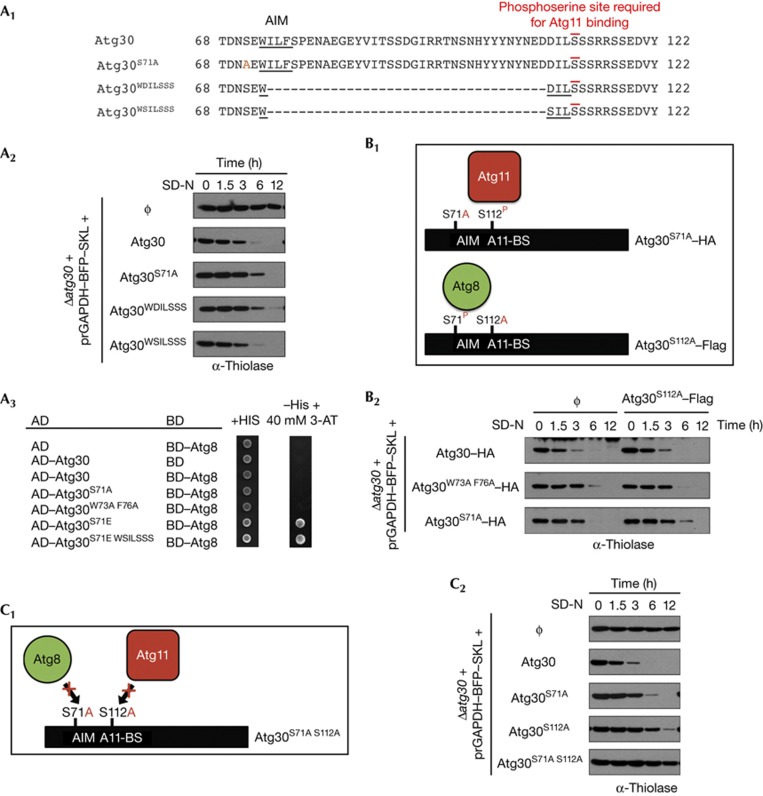
The sequences of Atg32 homologues in yeasts such as Vanderwaltozyma polyspora and Tetrapisispora phaffii contain an Atg11-binding site overlapping the AIM motif, suggesting that Atg8 and Atg11 might interact sequentially on the same receptor molecule (specific or random order) or bind independently to separate receptors (Fig 1B). The overlapping binding domains and steric interference preclude the simultaneous interaction of Atg8 and Atg11 with the receptor(s). We tested this hypothesis by deleting the aas between the AIM and the Atg11-binding motif of Atg30 (Fig 2A1) and testing the mutants in pexophagy assays (Fig 2A2). Two Atg30 truncations (preserving the AIM consensus, underlined in Fig 2A1) analogous to the VpAtg32 and TpAtg32 sequences were tested: (1) Atg30WDILSSS mutant, in which the AIM and Atg11-binding sites overlap and, (2) Atg30WSILSSS mutant, the Asp of the Atg30WDILSSS mutant (underlined) was replaced by Ser to mimic a highly conserved aa near the Atg11-binding site in Atg32 (Fig 1B). The two Atg30 truncations produced distinctive results (Fig 2A2). The Atg30WDILSSS mutant partially complemented the Δatg30 cells, exhibiting a phenotype similar to the Atg8-binding site mutants (Atg30S71A or Atg30W73A F76A), suggesting that this mutant might bind Atg11, but not Atg8. Finally, the Atg30WSILSSS mutant fully complemented Δatg30 cells, showing not only that the overlapping AIM and Atg11-binding sites could function in Atg30, but also that Atg8 and Atg11 cannot bind simultaneously to a single Atg30 molecule. As expected, the mutant Atg30WSILSSS did indeed interact with Atg8 by Y2H (Fig 2A3).
Figure 2.
Interactions between Atg30, Atg8 and Atg11. (A1) Atg30 sequences from aa 68–122 of wild-type (Atg30), the S71A mutant (Atg30S71A) and two different deletions of the sequence between the AIM and phosphosite required for Atg11 binding in Atg30 (Atg30WDILSSS and Atg30WSILSSS). (A2) Pexophagy experiments of Δatg30 cells complemented with appropriate wild-type or mutant Atg30 proteins described in A1. (A3) Two-hybrid assays between Atg30 wild-type or mutants (described in A1) and Atg8. Phosphomimic S71E was included to detect the interaction with Atg8. (B1) Schematic of the two Atg30 molecules, one with an Atg8-binding site mutated and a second with an Atg11-binding site (A11-BS) mutated, used to complement Δatg30 cells by co-expression. P: indicates phosphorylation in vivo. (B2) Pexophagy experiments of Δatg30 cells complemented with the two Atg30 molecules described in B1. (C1) Schematic of the Atg30 mutations, S71A and S112A that impair Atg8 and Atg11 binding, respectively. (C2) Pexophagy assays of Δatg30 cells complemented with Atg30 wild-type and mutants. aa, amino acids; AD, activation domain; AIM, Atg8-family-interacting motif; BD, binding domain; HA, haemagglutinin.
The hypothetical interaction of Atg8 and Atg11 with two different receptor molecules was excluded by an experiment involving the co-expression of two copies of Atg30 in Δatg30 cells, one with the AIM mutation (S71A or W73A F76A) and a second with the Atg11-binding mutation (S112A), followed by pexophagy assays using either endogenous thiolase or thiolase–green fluorescent protein (GFP) (Fig 2B; supplementary Fig S2C online). Atg30S112A did not complement the pexophagy delay of either Atg30S71A or Atg30W73A F76A (Fig 2B2; supplementary Fig S2C online), thereby indicating that Atg8 and Atg11 must interact with the same Atg30 molecule.
On the basis of our findings that the Atg8- and Atg11-binding sites in Atg30 can overlap while maintaining receptor function and the result that both molecules must interact with the same Atg30 for a fully functional receptor, we asked whether there is any obligatory order of binding, using mutants (Atg30S71A S112A and Atg30W73A F76A S112A) that are unable to interact with Atg8 and Atg11 (Fig 2C; supplementary Fig S2A online). We reasoned that an obligatorily sequential binding of the proteins to Atg30, as part of a single pathway, should not have a cumulative effect, but should mimic instead the loss of one or other binding site (Fig 2C1). However, Atg30S71A S112A and Atg30W73A F76A S112A were fully blocked in pexophagy (Fig 2C2; supplementary Fig S2A online), showing that both Atg8 and Atg11 interaction with Atg30 are independently required for optimal pexophagy.
These combined results indicate that Atg8 and Atg11 need to bind to the same Atg30 molecule, neither interaction (Atg8–Atg30 or Atg11–Atg30) is a prerequisite for the other and finally, the two interactions cannot occur simultaneously when their binding sites overlap. We call this mode of interactions independent (on the same molecule) and randomly sequential.
Independence of Atg8 and Atg11 binding to Atg30
Disruption of the Atg30–Atg8 interaction only partially affects selective autophagy ([6], Figs 1D,G, Fig 2; supplementary Fig S2 online). To compare this result in the absence of Atg8, we tested Δatg8 cells for pexophagy and phagophore membrane elongation during pexophagy of both small, oleate-induced and large, methanol-induced peroxisomes (supplementary Fig S4A,B online). In contrast to the delayed pexophagy in the absence of Atg8–Atg30 interaction, Atg8 was indispensable for pexophagy and phagophore membrane elongation (Δatg8, supplementary Fig S4 online). This suggests that Atg8 might interact with other unknown ‘peroxisomal’ proteins or that the interaction between Atg30 and Atg8 is non-essential but a different Atg8 function is crucial for pexophagy.
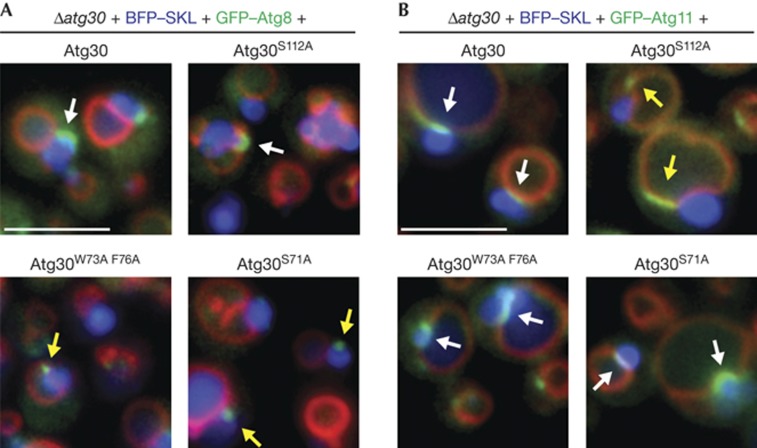
To understand why pexophagy was delayed in the absence of Atg30–Atg8 interaction, we studied Atg8 localization to the phagophore membrane (Fig 3A). No characteristic phagophore membrane was found when the Atg30–Atg8 interaction was abolished (Fig 3A, see S71A and W73A F76A), but instead small Atg8-containing punctae overlapped with peroxisomes. As pexophagy occurs slowly in these mutants, it suggests that Atg8–Atg30 interaction might be required to extend the phagophore membrane, and the Atg8-containing punctae might be phagophore membranes enclosing only small peroxisomes.
Figure 3.
Atg8 and Atg11 localization during pexophagy. (A) Large phagophore membrane formation in WT and the Atg30 mutant cells monitored by GFP–Atg8 during pexophagy conditions. (B) Localization of GFP–Atg11 during pexophagy of methanol-induced peroxisomes in cells expressing WT or mutant Atg30 proteins. White arrows indicate correct localization and yellow arrows indicate mislocalization, or in case of Atg8 localization, indicate absence of phagophore membrane elongation. Peroxisomes were labelled with BFP–SKL and vacuoles with FM4-64. Scale bar, 5 μm. GFP, green fluorescent protein; WT, wild type.
The interaction of Atg11 with the receptor seems to have a more significant role during pexophagy because the disruption of the Atg30–Atg11 interaction (Atg30S112A) strongly delayed peroxisomes degradation [16] (Fig 2C2; supplementary Fig S2A,B online), and this delay was comparable to the absence of Atg11 (Δatg11, supplementary Fig S4A online). Surprisingly, Atg8-labelled phagophore membranes were found in cells expressing Atg30S112A (7.9% of cells) to the same extent as in wild-type (8.1% of cells), despite the severe delay in pexophagy (supplementary Fig S2A,B online).
By analogy to the weak phenotype of the Atg30S71A mutant compared with Δatg8 (strong phenotype), Atg11 has another function in pexophagy beyond its interaction with Atg30, because Δatg11 did not accumulate any phagophore membranes (supplementary Fig S4B online), when compared with the Atg30S112A mutant, which had normal phagophore membranes. This conclusion is not unexpected because Atg11 interacts with and recruits many other proteins.
Finally, we visualized Atg11 in cells impaired in the Atg8–Atg30 and Atg11–Atg30 interactions (Fig 3B). During pexophagy, Atg11 accumulated in the vacuolar membrane region, where the peroxisomes contact the vacuole [17]. Atg11 localization was dependent on Atg11–Atg30 interaction (S112A), but independent of Atg8–Atg30 interaction (S71A or W73A F76A), in agreement with independent binding of Atg11 to Atg30.
In conclusion, the complete loss of Atg8 and Atg11 had a stronger phenotype than just loss of the interaction between Atg8 and Atg11 with the receptors. The likely explanation is that Atg8 and Atg11 interact with several other Atg proteins and therefore are likely to perform other functions in autophagy and selective-autophagy, beyond just the interactions with Atg30. In addition, it is known that Atg17 can substitute for Atg11 to facilitate Ape1 transport to the vacuole during nitrogen starvation [18, 19], and partially for Atg11 during pexophagy [16], which could explain why loss of Atg8 has a more severe effect on pexophagy than the loss of Atg11. The Δatg11 cells are partially complemented by Atg17 and this would also explain why the isolation membrane elongates in the absence of Atg30–Atg11 interaction (Fig 3A). The dependence of the localization of Atg8 and Atg11 exclusively on just their own binding sites in the receptor (Figs 1F, 3) corroborates their independent binding to Atg30. These results are in agreement with the interaction study of ScAtg32 with Atg8 and Atg11 in several autophagy mutants such as Δatg8 and Δatg11, which show that the mitophagy receptor interacts with Atg8 in the absence of Atg11 binding and vice versa [20], and with the finding that Atg19 also mediates an independent dual interaction prApe1-sorting mechanism [21]. An alternative explanation is the possibility of a weak but undetectable interaction under our experimental conditions, between Atg30 mutants and Atg8 or Atg11, which could be enough to support pexophagy and/or phagophore membrane elongation. However, this is very unlikely at least for the Atg11-binding mutant (S112A), because phagophore membrane formation was completely normal (size and number, Fig 3A) but the pexophagy rate was strongly delayed (Fig 2C2).
Common mechanisms for selective autophagy
To extend the model of interaction proposed for Atg30 and the autophagic core machinery proteins, we subjected P. pastoris Atg32 mutants to mitophagy assays. Mitophagy was followed by Tom20 localization (Tom20–mCherry) and Tom20 degradation (free GFP appearance from Tom20–GFP). Atg32 and Tom20 colocalized to mitochondria during growth condition in YPL medium (mid-log growth phase) and were degraded only after cells had reached stationary phase or shifted to SD-N (Figs 4A,B; supplementary Fig S5 online). Tom20 degradation was depended on Ypt7, Atg5 and Atg32 (Figs 4A,B), as expected for mitophagy.
Figure 4.
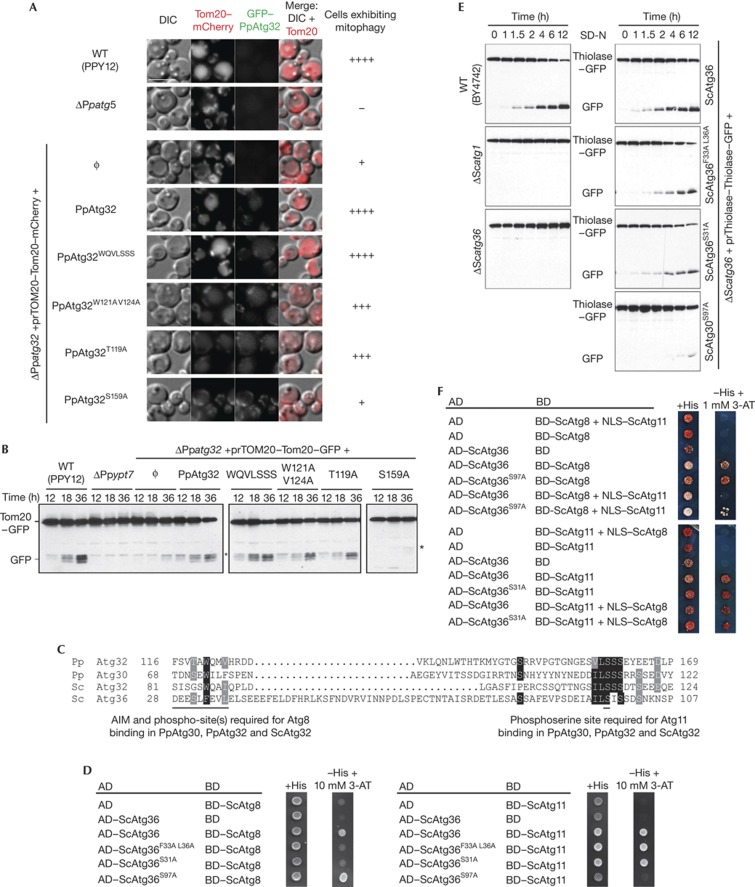
Atg32, ScAtg32 and Atg36 use similar interaction mechanisms as Atg30. (A) Mitophagy experiments of WT (PPY12), Δatg5, Δatg32 (ϕ) and Δatg32 complemented with WT Atg32 (Atg32), Atg32 with a deletion of the sequence between the AIM and phosphosite required for Atg11 binding (Atg32WQVLSSS), Atg32 AIM mutant (Atg32W121A V124A), mutants of the Thr upstream of the Atg32 AIM (Atg32T119A) and mutants altered in the Ser required for Atg11 binding (Atg32S159A). The cells were grown in YPL medium and shifted to SD-N. Mitophagy was followed by the transport of Tom20–mCherry to the vacuole by fluorescence microscopy. Mitophagy was classified as −no mitophagy,+few cells show mitophagy, +++ most cells show mitophagy and ++++ almost all the cells show mitophagy (the intensity and numbers of cells containing Tom20–mCherry in the vacuole was considered). Scale bar, 5 μm. (B) P. pastoris Δatg32 cells (ΔPpatg32) expressing Tom20–GFP and expressing the indicated Atg32 mutants were cultured in YPL medium for 12, 18 and 36 h. Mitophagy was monitored by GFP appearance by immunoblotting with α-GFP antibodies. (C) N-terminal sequence of Atg36 manually aligned against the several sequence alignments of Atg30, Atg32 and ScAtg32. Identical residues are indicated with black boxes, and similar residues with grey boxes. (D) Two-hybrid protein–protein interaction analysis of ScAtg36, ScAtg8 and ScAtg11. The receptors were mutated at the AIM (Atg36F33A L36A), at serine(s) upstream of the AIM (Atg36S31A) and at the Atg11-binding site (Atg36S97A). (E) S. cerevisiae Δatg36 cells (ΔScatg36) expressing thiolase–GFP and expressing the indicated Atg36 mutants were cultured in oleate medium until mid-log growth and then shifted to SD-N. Pexophagy was monitored by GFP appearance by immunoblotting with α-GFP antibodies. (F) Y3H analysis of ScAtg36 with ScAtg8 and ScAtg11. The Y3H technology is on the basis of the yeast two-hybrid system but with the co-expression of third protein as a competitor and indicated in the figure (NLS–ScAtg11 or NLS–ScAtg8). The positive control was the ScAtg36 mutant affected in Atg11 binding (ScAtg36S97A) or in Atg8 binding (ScAtg36S31A), which should be unaffected by the competition of NLS–ScAtg11 or NLS–ScAtg8, respectively. Appropriate auto-activation and interaction controls were also included. AD, activation domain; AIM, Atg8-family-interacting motif; BD, binding domain; GFP, green fluorescent protein; WT, wild type; Y3H, yeast three-hybrid.
First, the aas between the AIM and the Atg11-binding motif of Atg32 (Met123 to Ser156) were deleted, generating a truncated Atg32 (Atg32WQVLSSS) with overlapping binding motifs (supplementary Fig S6A online). Atg32WQVLSSS fully complemented the mitophagy defect of Δatg32 cells (Figs 4A,B) and interacted with Atg8 (supplementary Fig S6B online), suggesting that interactions of the Atg32 with Atg8 and Atg11 might not occur simultaneously. Next, we mutated the AIM on Atg32 (Atg32W121A V124A) and the threonine upstream of the AIM (Atg32T119A), and found slight defects in mitophagy (Figs 4A,B), comparable to the pexophagy defect seen for the equivalent mutation in Atg30 (Figs 1D,G; supplementary Fig S2 online). In contrast, the mutation, S159A, required for Atg11 binding (Atg32S159A) severely impaired mitophagy, comparable to the Δatg32 cells. These results confirmed that Atg32 and Atg30 share the same motif organization for Atg8 and Atg11-binding sites and probably the same molecular mechanisms.
We extended our studies to S. cerevisiae, where Atg36 is the pexophagy receptor [7]. Atg36 does not share sequence homology with Atg30, yet is functionally orthologous, by interacting with the same set of proteins such as Pex3, Atg8 and Atg11. We used our consensus sequence logo (Fig 1B) to screen for a putative AIM followed by an Atg11-binding site in Atg36, and found an N-terminal region with similar sequence organization (Fig 4C). The putative AIM, the Ser (S31) upstream of the AIM and the Ser (S97) in the Atg11-binding site of Atg36 were mutated to Ala and tested by Y2H (Fig 4D). The interaction studies of Atg36 confirmed the presence of a classical AIM (F33-L36), with S31 upstream of the AIM also being required for ScAtg8 binding. Similarly, the Atg11-binding site in Atg36 was confirmed by Y2H with wild-type Atg36 and the mutation, S97A. The physiological roles of these interactions were assayed by free GFP appearance caused by degradation of thiolase–GFP during pexophagy (Fig 4E). Similar to Atg30 and Atg32 point mutations, Atg36 mutants affecting the AIM and upstream Ser (S31) delayed pexophagy to the same extent.
A yeast three-hybrid analysis confirmed the hypothesis that Atg8 and Atg11 cannot interact simultaneously with the receptors, as suggested by the overlapping Atg8 and Atg11-binding motifs found in some receptors in nature (Fig 1B) or recreated by truncation (Fig 2A, Figs 4A,B; supplementary Fig S6 online). We studied this with S. cerevisiae Atg36 because phosphorylation during the Y2H happens normally, making it unnecessary to use phosphimimic mutations of Atg36 to study its interactions with ScAtg8 and ScAtg11. The yeast three-hybrid analysis revealed that ScAtg11 fused to a nuclear localization signal (NLS–ScAtg11) competed with ScAtg8 (binding domain (BD)–ScAtg8) for interaction with ScAtg36 (activation domain (AD)–ScAtg36) and this competition was inhibited by mutation of the Atg11-binding site on ScAtg36 (ScAtg36S97A; Fig 4F). These data confirm that ScAtg11 can displace ScAtg8 for interaction with Atg36. Interestingly, ScAtg8 (NLS–ScAtg8) did not compete with ScAtg11 (BD–ScAtg11) for ScAtg36 (AD–ScAtg36) interaction, in agreement with the pexophagy experiments wherein Atg30 S71E behaved like WT (supplementary Fig S2A,B online). This suggests that Atg11 has a higher affinity for the receptor than Atg8. So, if Atg8 binds first, Atg11 can displace Atg8. However, if Atg11 binds first, modulation of the respective binding affinities, perhaps by phosphorylation of the Atg8 site and/or dephosphorylation at the Atg11 site, would allow Atg8 to displace Atg11.
Finally, we showed that the mitophagy receptor ScAtg32 requires Ser81, 83 and 85 upstream of the published AIM to bind ScAtg8 [6] (supplementary Fig S7 online). Our studies illustrate the evolutionary conservation of selective autophagy receptors in yeast that are characterized by a tripartite interaction with cargo, Atg8 and Atg11, and these interactions are regulated by phosphorylation events.
We summarize the receptor interactions in supplementary Fig S8 online. During organelle biogenesis, the selective autophagy receptors are transported to the target organelle in an inactive form that does not interfere with organelle biogenesis. When selective autophagy is induced, an unknown kinase(s) activates the receptor by phosphorylation of the serine(s)/threonine(s) at the Atg11-binding site (model 1) or upstream of the phosphoregulated AIM (model 2), to allow interactions with Atg11 and Atg8, respectively. Atg11 or Atg8 then dissociates from the receptor, likely via competition on the basis of their intrinsic affinities for the receptor or via the action of an unknown phosphatase(s). A second phosphorylation event happens on the same receptor molecule to facilitate the interaction with the other molecule, Atg8 (model 1) or Atg11 (model 2), as applicable.
Mitophagy and pexophagy display added complexities relative to the Cvt pathway in that their turnover is regulated and their engulfment might be more complex. Mitochondria and peroxisomes must be targeted to the vacuole in a manner that depends on environmental changes, but they must also be degraded in the absence of external cues when they are dysfunctional. The mechanisms proposed here for Atg30, Atg32 and Atg36 outline a general mechanistic framework for most of the selective autophagic receptors in yeast. Whether different types of selective autophagy use the same or different kinases/phosphatases for the regulation of selective autophagy remains to be determined.
METHODS
Strains and plasmids are described in supplementary Tables S1–S4 online. Media and growth conditions are in the supplementary Information online.
In silico analysis. Putative Atg30 and Atg32 homologues were identified using Atg30 (GenBank accession number: AAQ63446) and ScAtg32 (GenBank accession number: DAA08407) protein sequences as described in the supplementary Information online.
Biochemical studies of pexophagy and mitophagy. In P. pastoris, peroxisomes were induced by incubation of cells in oleate medium (starting OD600 of 0.2) for 15 h and transferred to SD-N medium at an OD600 of 2 to induce pexophagy. One millilitre of cells was collected at different times as described in the figures, trichloroacetic acid precipitated and analysed by western blot. In S. cerevisiae, pexophagy was performed as described earlier [22]. Mitophagy in P. pastoris was induced by growth in YPL medium up to stationary phase, as described earlier [23]. Cells were grown in YPL medium starting at 0.1 OD600 and at 12, 18 and 36 h, and 1 ml of cells was trichloroacetic acid precipitated and analysed by western blot.
Yeast two- and three-hybrid analysis. The GAL4-based Matchmaker yeast two-hybrid system (Clontech Laboratories Inc.) was used. Full-length open reading frames were inserted in pGAD–GH (AD) and pGBT9 (BD) plasmids, except for P. pastoris and S. cerevisiae Atg8, where the BD was fused to a truncated Atg8 (Met1 to Phe115). The pBridge plasmid from Clontech Laboratories (for co-expression of a BD fusion protein and a NLS fusion protein) was used exclusively for the three-hybrid assay. The S. cerevisiae strains AH109 and HF7c were used for the two- and three-hybrid assays, respectively. Two transformants from each strain were tested in duplicate in both assays. All strains were plated on SD medium (Leu−, Trp−) as well as SD medium (His−, Leu−, Trp−) containing 3-amino-1,2,4-triazole at the concentration indicated in the figure.
Fluorescence microscopy. Pexophagy assays were performed as described for biochemical studies and pictures were acquired after 3 h in SD-N medium. Large phagophore membrane detection and Atg11 localization: cells were grown in methanol medium (starting OD600 of 0.2) for 15 h and transferred to SD-N medium at an OD600 of 2 for 1 h. The phagophore membrane was labelled with GFP–Atg8 or GFP–Atg26. Mitophagy assays: cells were grown in YPL medium (starting OD600 of 0.2) for 12 h and transferred to SD-N medium at an OD600 of 1, as described in S. cerevisiae [23]. To quantify selective-autophagy defects during experiments, the fluorescence microscopy parameters such as exposition time, gain and binning were kept constant.
Co-IP, protein purification and MS. We used Δypt7 Δatg30 cells, expressing Atg30-Flag or Flag-Atg11 and Atg30–HA, and either GFP-Atg8 or myc-Atg8 expressed from their endogenous promoters. The experiments were performed as described [24], using 1% CHAPS as detergent for Fig 1A or as described in supplementary Methods online for Fig 1F. Protein purification of a mutated Atg30(A81R) to facilitate MS studies was performed as described in supplementary Methods online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank all the lab members, especially Drs Jingjing Liu for Δatg36+thiolase–GFP strain and Taras Nazarko for useful discussion. This work was supported by an NIH grant (GM 069373) to S.S. S.S. also thanks the Chancellor Associates Chair for support of Sarah Burnett and this research.
Author contributions: J.-C.F. designed, performed and analysed the experiments under the supervision of S.S. A.B. and S.F.B. performed experiments. J.-C.F. and S.S. wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Farre JC, Krick R, Subramani S, Thumm M (2009) Turnover of organelles by autophagy in yeast. Curr Opin Cell Biol 21: 522–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ (2001) Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell 7: 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Manjithaya R, Mathewson RD, Subramani S (2008) PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell 14: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ (2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 17: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y (2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell 17: 87–97 [DOI] [PubMed] [Google Scholar]
- Motley AM, Nuttall JM, Hettema EH (2012) Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J 31: 2852–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Huang WP, Stromhaug PE, Klionsky DJ (2002) Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell 3: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ (2005) Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell 16: 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda NN, Ohsumi Y, Inagaki F (2010) Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 584: 1379–1385 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y (2007) Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12: 209–218 [DOI] [PubMed] [Google Scholar]
- Itakura E, Mizushima N (2010) Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6: 764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Kanki T, Hirota Y, Kurihara Y, Saigusa T, Uchiumi T, Kang D (2011) Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol Biol Cell 22: 3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F (2008) Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13: 1211–1218 [DOI] [PubMed] [Google Scholar]
- Wild P et al. (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333: 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarko TY, Farre JC, Subramani S (2009) Peroxisome size provides insights into the function of autophagy-related proteins. Mol Biol Cell 20: 3828–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J et al. (2001) Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol 153: 381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang CW, Klionsky DJ (2005) Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell 16: 3438–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y (2008) Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol Cell 19: 2039–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo-Okamoto N et al. (2012) Autophagy-related protein 32 acts as autophagic degron and directly initiates mitophagy. J Biol Chem 287: 10631–10638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, Huang WP (2007) Atg19 mediates a dual interaction cargo sorting mechanism in selective autophagy. Mol Biol Cell 18: 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R, Jain S, Farre JC, Subramani S (2010) A yeast MAPK cascade regulates pexophagy but not other autophagy pathways. J Cell Biol 189: 303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Kang D, Klionsky DJ (2009) Monitoring mitophagy in yeast: the Om45-GFP processing assay. Autophagy 5: 1186–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Mathewson RD, Manjithaya R, Subramani S (2010) Roles of Pichia pastoris Uvrag in vacuolar protein sorting and the phosphatidylinositol 3-kinase complex in phagophore elongation in autophagy pathways. Autophagy 6: 86–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.