Abstract
The zebrafish spinal cord primary motor neurons are commonly used as an experimental model to study the molecular mechanisms that regulate axonal pathfinding and neuromuscular junction formation, and for the modeling of human neurodegenerative disorders. This study characterized a 125-bp mnx1 enhancer to direct gene expression in spinal cord motor neurons. A promoter containing three copies of the 125-bp mnx1 enhancer was generated in a Tol2 vector and used to drive EGFP expression either directly or in combination with the Gal4/UAS transcriptional activation system. Both methods induced protein expression for up to five days after fertilization, allowing the observation of the dendritic tree and axonal arborization of single motor neurons within a somitic segment in fixed and live animals. The use of the 125-bp mnx1 promoter for transient transgenic expression or for the generation of stable transgenic fish lines will facilitate the study of motor neuron development and neurodegenerative processes.
Keywords: Danio rerio, motor neuron development, mnx1 enhancer, in vivo imaging, spinal cord
The zebrafish (Danio rerio) motor neurons are accessible for observation from the initial stages of development and are widely used as an experimental model system for the study of the molecular mechanisms involved in axonal pathfinding and neuromuscular junction formation (Eisen et al., 1986; Flanagan-Steet et al., 2005; Jing et al., 2009), and for the analysis of neurodegenerative processes (Boon et al., 2009; Sager et al.). Each somitic hemisegment in the zebrafish embryo is first innervated by three primary motor neurons named rostral (RoP), middle (MiP), and caudal (CaP) (Myers et al., 1986; Westerfield et al., 1986), and by a variable motor neuron (VaP) that develops in half of the somitic segments (Eisen et al., 1990). Cell division of motor neuron precursors ends at approximately 16 hours post fertilization (hpf) (Myers et al., 1986). The CaP motor axon exits the spinal cord at ~18–20 hpf, and a few hour later it is followed by the RoP and MiP axons. Primary motor axons migrate ventrally and establish the first neuromuscular junction with the muscle pioneer cells located at the horizontal myoseptum before choosing cell-type specific pathways to innervate different myotomal territories (Eisen et al., 1986; Eisen et al., 1990; Melancon et al., 1997; Myers et al., 1986; Westerfield et al., 1986). In addition to the primary motor neurons, each somitic hemisegment contains approximately twenty-five secondary motor neurons, which start to exit the spinal cord at 26 hpf, and follow the same axonal trajectories pioneered by the primary motor axons (Myers et al., 1986).
The mnx1 transcription factor is selectively expressed in postmitotic spinal cord motor neurons (Arber et al., 1999; Tanabe et al., 1998). For this reason, the regulatory elements of the mnx1 gene (also known as hb9, hlxb9, scra1, and hoxhb9) can be used as a tool for the temporal and spatial control of protein expression in spinal motor neurons (Flanagan-Steet et al., 2005). However, activation of gene expression in various spinal cord neuron types observed with a 3-kb DNA fragment located upstream of the mnx1 protein coding region (Flanagan-Steet et al., 2005), precludes a widespread use of this promoter region for the manipulation of protein expression in single motor axons beyond the first 24 h of development. For these reason we investigated whether a phylogenetically conserved enhancer sequence of 125 base pairs (bp) identified within the mouse mnx1 promoter region (Nakano et al., 2005) could be used to direct gene expression in zebrafish spinal motor neurons.
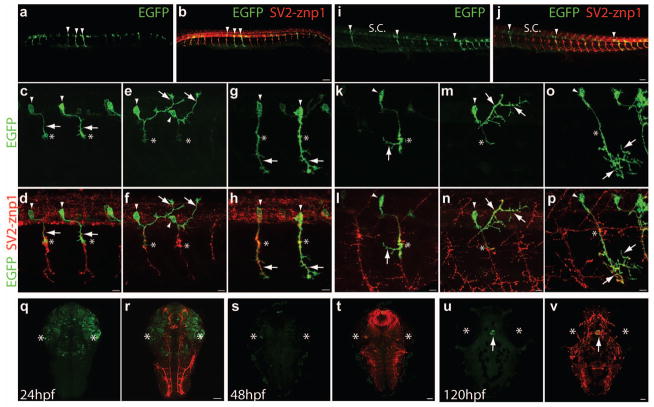
Sequencing of the 3-kb mnx1 zebrafish promoter (a gift from D. Meyer) identified a 125-bp fragment starting at position −1946 upstream of the mnx1 protein coding region with 80% homology to the mouse sequence (Nakano et al., 2005). To determine whether the 125-bp sequence directs gene expression in zebrafish spinal motor neurons, three copies of the fragment were cloned in tandem (mnx1-3x125bp) into the p5E entry vector of the Tol2 Kit (Kwan et al., 2007)(see Table 1 for expression vectors used in this study). The mnx1-3x125bp enhancer was placed upstream of prenylated (pren) enhanced green fluorescent protein (EGFP) into the Tol2 destination vector (pDestTol2pA2) using LR recombination, and the new plasmid (mnx1-3x125bp:prenEGFP) injected into wild type zebrafish embryos. At 24 hpf, from 187 normally developed embryos, 173 (93%) of the embryos contained EGFP expressing cells. Embryos were fixed, immunostained with SV2 and znp1 antibodies to label axons and nerve terminals (Brusés, 2011; Panzer et al., 2005), and observed by confocal microscopy to determine the pattern of EGFP expression (Fig 1). At 24 hpf, EGFP was detected in a few primary motor neurons with an average of 11.4 neurons per embryo (Table 2). Examples of somitic hemisegments with a single labeled primary motor neuron are shown in Figure 1. By 48 hpf, the intensity of EGFP labeling was increased and secondary motor neurons were also observed. However, individual primary motor axons and their entire axonal arborization could be observed in some somitic hemisegments (Fig 1, Table 2), indicating that mnx1-3x125bp was sufficient to direct gene expression specifically in spinal motor neurons. EGFP expression in spinal cord interneurons was rarely observed.
Table 1.
Components of the expression vectors used in the study
| * Expression vector | p5E | pME | p3E |
|---|---|---|---|
| mnx1-3x125bp:prenEGFP | mnx1-3x125-bp enhancer | prenEGFP | SV40 pA |
| mnx1-3x125bp:Gal4-VP16 | mnx1-3x125-bp enhancer | Gal4-VP16 | SV40 pA |
| mnx1-782bp:prenEGFP | mnx1-785-bp | prenEGFP | SV40 pA |
| mnx1-356bp-3x125bp:prenEGFP | mnx1-356bp-3x125bp-enhancer | prenEGFP | SV40 pA |
| 14xUAS:prenEGFP | 14xUAS-E1b | prenEGFP | SV40 pA |
Expressions vectors were generated by combining the elements in p5E, pME, and p3E plasmids into a pDestTol2pA2 vector via LR recombination.
Figure 1.
The mnx1 125-bp enhancer direct gene expression in zebrafish motor neurons. Wild type embryos were injected at the 1-cell stage with a Tol2 plasmid encoding prenEGFP (in green) downstream of mnx1-3x125bp enhancer, fixed at 24 hpf (a – h) and 48 hpf (i – p), and immunostained with SV2 and znp1 antibodies to label all axons (in red). a, b) Low magnification images of a 24 hpf embryo showing a few EGFP labeled spinal cord motor neurons (a) and immunostained with SV2 and znp1 antibodies (arrowheads)(b). c – h) Images from RoP (c, d), MiP (e, f), and CaP (g, h) primary motor neurons. Arrowheads point to the cell soma, arrows point to the motor axons, and the asterisks indicate the horizontal myoseptum. i, j) Low magnification image of a 48 hpf embryo showing EGFP labeled spinal cord motor neurons (i) and immunostained with SV2 and znp1 antibodies (arrowheads)(j). k – p) Images of RoP (k, l), MiP (m, n), and CaP (o, p) primary motor neurons. Arrowheads point to the cell soma of a single motor neuron within a somitic hemisegment, the asterisks indicate the horizontal myoseptum, and the arrows point to the motor axon arborization within the corresponding myotomes. q – v) Images of the head showing transient ectopic EGFP expression. q, r) Head of a 24 hpf embryo in which transient EGFP expression is detected in the eyes and brain. s, t) By 48 hpf EGFP expression is no longer detected in the head. u, v) At 120 hpf EGFP expression is detected only in a group of cells in the epithalamus (arrow). Asterisks indicate the position of the eyes. q, s, and u, EGFP; r, t, and v, EGFP merged with SV2 and znp1 immunostaining. Scale bar in b, j, r, t, and v, 50 μm; scale bar in d, f, h, l, n, and p, 10 μm. a – p) Dorsal is to the top and rostral is to the left. q – v) Rostral is to the top.
Table 2.
Number of primary motor neurons labeled
| Expression vector | Labeled primary motor neurons per embryo | Hemisegments with a single labeled primary motor neurons | ||||
|---|---|---|---|---|---|---|
| 24 hpf | 48 hpf | 120 hpf | 24 hpf | 48 hpf | 120 hpf | |
| mnx1-3x125bp:prenEGFP | 11.4 ± 1.6 n = 22 |
6.5 ± 1.2 n = 19 |
3.1 ± 0.5 | 1.5 ± 0.3 | ||
| mnx1-3x125bp:Gal4-VP16 + 14xUAS:prenEGFP | 6.7 ± 1.4 n = 13 |
7.9 ± 0.5 n = 26 |
3.4 ± 0.7 | 1.0 ± 0.3 | ||
| Tg(mnx1-3x125bp:Gal4-VP16) + 14xUAS:prenEGFP | 9.5 ± 1.2 n = 16 |
10.1± 1.7 n = 12 |
3.5 ± 0.7 n = 10 |
4.1 ± 0.6 | 1.5 ± 0.4 | 0.6 ± 0.3 |
Values are expressed as mean ± SEM; n, number of embryos
However, ectopic EGFP expression was detected in the head of 24 hpf embryos injected with mnx1-3x125bp:prenEGFP including the telencephalon, optic tectum, and eyes (Fig 1). Ectopic EGFP expression levels were lower than the levels detected in motor neurons and became undetectable at 48 hpf (Fig 1). Labeled cells outside the spinal cord could not be associated with any particular cell type. By 120 hpf EGFP expression outside the spinal cord was restricted to a small group of cells within the eipthalamus (Fig 1). Ectopic protein expression in the head has also been observed in transgenic mouse embryos expressing lacZ under a similar 3x-125-bp mnx1 enhancer (Nakano et al., 2005), which was abolished when a 313-bp fragment of the mnx1 promoter region (−9040 to −8727) was combined with the 125-bp enhancer sequence, suggesting that the 313-bp sequence suppresses ectopic gene activation (Nakano et al., 2005). Sequence alignment analysis of the zebrafish 3-kb mnx1 promoter identified a 356-bp sequence located 301 bp upstream of the mnx1 125-bp enhancer with 72% homology to the 313-bp sequence reported in mouse. To test whether the 356-bp fragment affects gene expression in zebrafish, a 782-bp DNA fragment (−2603 to −1821) containing both, the 356-bp conserved region and the 125-bp enhancer was cloned upstream of prenEGFP (mnx1-782bp:prenEGFP). Wild type embryos injected with the mnx1-782bp:prenEGFP vector showed EGFP expression in the spinal cord at 24 hpf while ectopic expression in the head was not detected. However, by 72 hpf EGFP expression in motor neurons was substantially decreased and by 120 hpf EGFP expression was undetectable (data not shown). To determine whether the suppression of ectopic EGFP expression was due to the addition of the 356-bp fragment or due to the use of only 1 copy of the mnx1 125-bp enhancer, the 356-bp DNA fragment was cloned upstream of the mnx1-3x125bp:prenEGFP sequence (mnx1-356bp-3x125bp:prenEGFP) and injected into wild type embryos. Embryos injected with this vector showed ectopic EGFP expression in the head as previously observed with the mnx1-3x125bp:prenEGFP vector, indicating that the transient ectopic EGFP expression outside the spinal cord was likely due to the use of three copies of the mnx1 125-bp.
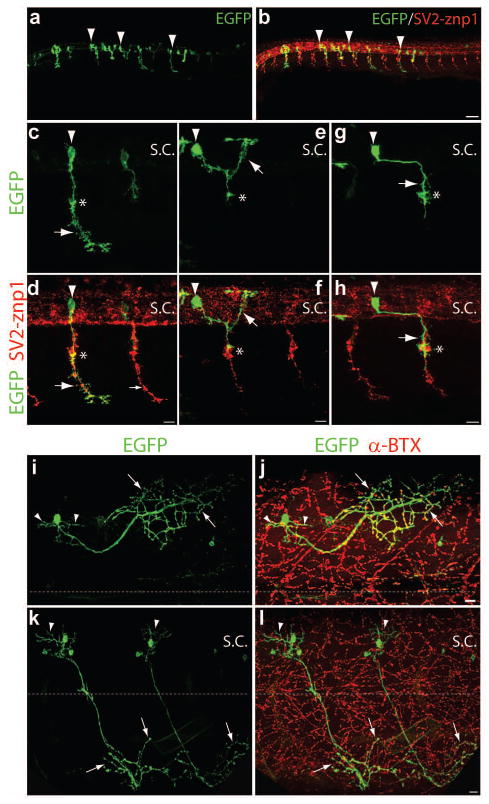
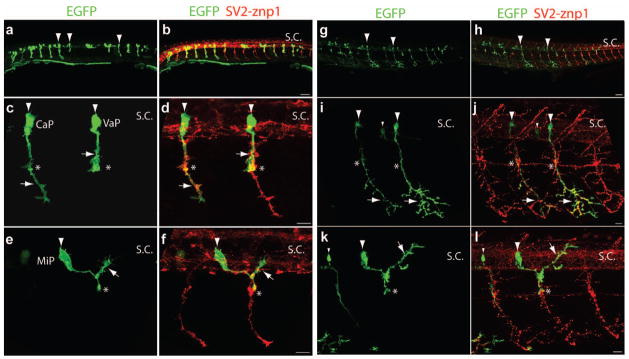
We then examined whether Gal4-VP16 expression under the mnx1-3x125bp enhancer could be used to drive prenEGFP expression. The Gal4-VP16 sequence was cloned downstream of the mnx1-3x125bp enhancer (mnx1-3x125bp:Gal4-VP16), and the plasmid was co-injected with a vector expressing prenEGFP under a 14xUAS element (14xUAS:prenEGFP) into wild type embryos. An average of 6.7 and 7.9 EGFP labeled primary motor neurons per embryo were observed at 24 hpf and 48 hpf respectively (Table 2). Figure 2 shows representative confocal images of different types of primary motor neurons. Although secondary motor neurons were also labeled at 48 hpf, single labeled primary motor neurons within a somitic hemisegment could be identified (Table 2). To facilitate the manipulation of protein expression in spinal motor neurons, stable transgenic fish expressing Gal4-VP16 under the mnx1-3x125bp enhancer (Tg(mnx1-3x125bp:Gal4-VP16)) were generated. F1 Tg(mnx1-3x125bp:Gal4-VP16) embryos were injected with the 14xUAS:prenEGFP vector, fixed at 24 hpf and 120 hpf, immunostained with SV2 and znp1 antibodies, and observed under confocal microscopy (Fig 3). Approximately 10% of the injected embryos contained EGFP expressing motor neurons at 24 hpf with an average of 9.5 primary motor neurons per embryo (Table 2). Primary motor neurons expressing EGFP were also detected in 120 hpf larvae (Fig 3), indicating that mnx1-3x125bp enhancer drives EGFP expression in spinal motor neurons beyond embryonic development.
Figure 2.
Labeling primary motor neurons by transient transgenesis using the mnx1-3x125bp enhancer and Gal4/UAS. Wild type embryos were injected at the 1-cell stage with a mix of mnx1-3x125bp:Gal4-VP16 and 14xUAS:prenEGFP plasmids. Embryos were fixed at 24 hpf (a – f) and 48 hpf (g – l), and immunostained with SV2 and znp1 antibodies (in red). a, b) Low magnification images showing a few motor neurons expressing EGFP in each somitic segment. c – f) Images of primary motor neurons. c, d) CaP and VaP motor neurons in adjacent somitic segments. The arrowheads point to the cell bodies and the arrow to the motor axons. The CaP motor axon extends ventral to the horizontal myoseptum while the VaP axon stalls at the horizontal myoseptum (asterisks). e, f) Images from an MiP neuron (arrowheads) in which the axon extends a dorsal branch (arrows) after reaching the horizontal myoseptum (asterisks). g, h) Low magnification images of a 48 hpf embryo depicting a few motor neurons expressing EGFP (arrowheads). i, j) Two CaP motor neurons (arrowheads) from adjacent somitic hemisegments in which the entire axonal arbor is labeled with EGFP (arrows). One secondary motor neuron is also labeled with EGFP (small arrowheads). k, l) Images from an MiP motor neuron (arrowheads) in which the axon innervates the dorsal myotome after reaching the horizontal myoseptum (asterisks). Small arrowheads point to a secondary motor neuron. Scale bar in b and h, 50 μm; scale bar in d, f, j, and l, 10 μm; S.C., spinal cord. Dorsal is to the top and rostral is to the left.
Figure 3.
Labeling single primary motor neurons in a stable transgenic fish expressing Gal4-VP16 under the mnx1-3x125bp enhancer. F1 Tg(mnx1-3x125bp:Gal4-VP16) embryos were injected at the 1-cell stage with a plasmid encoding 14xUAS:prenEGFP. a – h) 24 hpf embryos immunostained with SV2 and znp1 antibodies. a, b) Low-magnification images showing primary motor neurons labeled with EGFP (arrowheads). c – h) Images from CaP (c, d), MiP (e, f), and RoP (g, h) primary motor neurons. The arrowheads point to the soma, the asterisks indicate the horizontal myoseptum, and the arrows point to the motor axons. Expression of Gal4-VP16 and prenEGFP does not affect motor axons development as compared to the growth of untransfected axons from adjacent somitic segments labeled with SV2 and znp1 antibodies (small arrow in d). i – l) 120 hpf embryos labeled α-bungarotoxin (α-BTX) Alexa-546 conjugated. Examples of MiP (i, j) and CaP (k, l) motor neurons showing axonal arborizations (arrows) within the dorsal and ventral myotomes respectively, and dendritic trees (arrowheads) within the spinal cord labeled with EGFP. Scale bar in B, 50 μm; scale bars in d, f, h, j, and L 10 μm; S.C., spinal cord. Dashed lines in i – l indicate the horizontal myoseptum. Dorsal is to the top and rostral is to the left.
To determine whether the mnx1-3x125bp enhancer directs EGFP expression at sufficient levels to observe motor neurons morphology over time in live embryos and larvae, wild type embryos were injected with the mnx1-3x125bp:prenEGFP vector at the 1-cell stage. At 28 hpf an embryo was embedded in 1% low melting point agarose and observed with a Zeiss LSM510 confocal microscope using an Achroplan 40x/NA0.8 water immersion lens. A z-stack of images was taking at 1 μm intervals every 20 min and each stack was then projected to single plane. Figure 4 shows a CaP motor neuron and magnified images of the growth cone showing extension and retraction of processes over time. Images from the same CaP motor neuron at 48 hpf and 72 hpf show sufficient prenEGFP expression to label the expansion of the axonal arborization within the ventral muscle (Fig 4). Finally, to examine whether stable transgenic expression of Gal4-VP16 under the mnx1-3x125bp enhancer drives protein expression at sufficient levels for in vivo imaging, F1 Tg(mnx1-3x125bp:Gal4-VP16) embryos were injected at the 1-cell stage with the 14xUAS:prenEGFP vector and observed at 120 hpf. Figure 4 (panle j) shows a projection to a single plane of a z-stack from a CaP motor neuron in which the axonal arborization within the ventral muscle and the dendritic extensions from the cell soma within the spinal cord can be observed.
Figure 4.
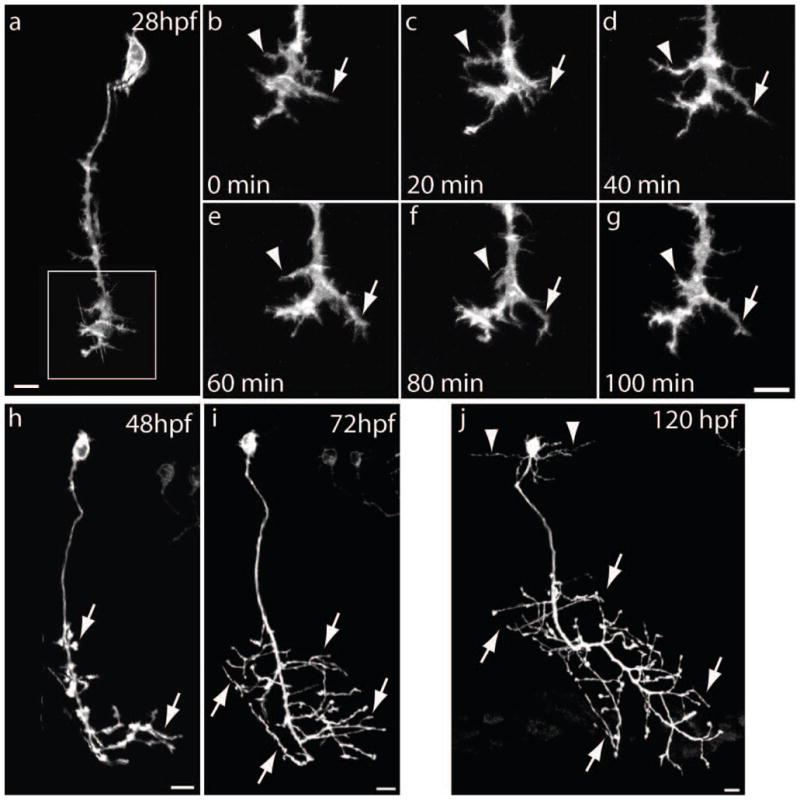
In vivo imaging of primary motor neurons using the mnx1-3x125bp enhancer and prenEGFP. Wild type embryos were injected at the 1-cell stage with the mnx1-3x125bp:prenEGFP plasmid, embedded in agarose at various developmental time-points, and observed with a Zeiss LSM510 confocal microscope using a 40x/0.8NA water immersion lens. a – g) Time-lapse images of a CaP motor neuron from a 28 hpf embryo. b – g) Magnified images of the growth cone framed in panel a. Z-stacks of images were taken every 20 min and show the extension (arrow) and retraction (arrowhead) of growth cone processes. h, i) A CaP motor neuron imaged at 48 hpf and 72 hpf showing a substantial expansion of the axonal arborization (arrows) within the ventral muscle. j) Image of CaP motor neuron at 120 hpf from a Tg(mnx1-3x125bp:Gal4-VP16) live embryo injected at the 1-cell stage with a plasmid encoding 14xUAS:prenEGFP. prenEGFP labels the entire cell including the axonal arborization (arrows) in the ventral muscle and dendritic extensions (arrowheads) within the spinal cord. Scale bars in a, g, h, i, and j, 10 μm. Dorsal is to the top and rostral is to the left.
In summary, the mnx1-3x125bp enhancer directs cell-type specific gene expression that can be used for the analysis of spinal motor neurons. The activity of this promoter beyond embryonic development will facilitate the analysis of gene products involved in neurodegenerative disorders with onsets in postembryonic life. As the mnx1-3x125bp enhancer is only 395-bp long, it can be cloned upstream of a protein coding sequence or it can be combined with other methods using Cre/lox-p recombinase and steroid receptors to provide further temporal control of gene activation (Collins et al.; Matsuda and Cepko, 2007).
Materials and Methods
Fish maintenance and husbandry
Zebrafish were obtained from the Zebrafish International Resource Center (ZIRC, Eugene, OR), raised, maintained as described in The Zebrafish Book (Westerfield, 2000), and staged as previously reported (Kimmel et al., 1995). The zebrafish AB line was used as wild type strain (strain AB-3, stock # 5560). The use and manipulation of animals used in this study has been approved by the Institutional Animal Care and Use Committee from the University of Kansas School of Medicine.
Plasmid Construction
This study used att site-specific recombination-based cloning system (MultiSite Gateway technology, Invitrogen, Carlsbad, CA) from lambda phage to build plasmids. The Tol2 Kit from C–B Chien Laboratory was the main source of plasmids for recombination and backbones for novel entry vector development (Hartley et al., 2000; Kwan et al., 2007). Three kinds of entry vectors were used for LR recombination: 5′ entry vector (p5E), middle entry vector (pME), and 3′ entry vector (p3E). The Tol2 Kit plasmid pDestTol2pA2 (Kwan et al., 2007) was used as a destination vector in all LR recombinations using LR clonase II (Invitrogen), and served as a backbone for the expression plasmids carrying an SV40 polyadenylation (pA) signal. The DNA fragment inserted in this plasmid is flanked by Tol2 transposable elements that facilitate DNA insertion (Suster et al., 2009). The Tol2 is a transposable element identified in medaka that has been successfully used to integrate large pieces of DNA into the zebrafish genome by promoting efficient DNA recombination (Kawakami et al., 1998). All sequences were verified by DNA sequencing.
The following entry vectors were generated:
p5E mnx1-3x125bp enhancer: The mnx1 125-bp enhancer located −1946 to −1821 from the start codon was amplified by PCR using 3-kb mnx1 promoter as template (Flanagan-Steet et al., 2005) and three different sets of primers containing unique restriction sites (underlined): primer set 1: forward XhoI, reverse SphI and HindIII (5′-TCCTCGAGAGTGGTTAGCTGATGA and 5′-ACAAGCTTTTGCATGCCCTCTTATAAGCCTCTTTAA); primer set 2: forward SphI, reverse AvrII and HindIII (5′-TTGCATGCCGAGAGTGGTTAGCTGA and 5′-ACAAGCTTTTCCTAGGCCTCTTATAAGCCTCTTTAA); and primer set 3: forward AvrII, reverse HindIII (5′-TTCCTAGGCGAGAGTGGTTAGCTGA and 5′-TTAAGCTTCCTCTTATAAGCCTCTTTAA). The PCR products were purified, digested with the corresponding restriction enzymes, and sequentially cloned into the p5E-MCS from the Tol2 Kit (Kwan et al., 2007).
p5E mnx1-356bp-3x125bp enhancer: The 356-bp mnx1 sequence, located −2603 to −2247 upstream of the mnx1 protein coding sequence, was obtained by PCR amplification using the 3-kb mnx1 promoter as template (5′-AAAGGTACCCGAAAATATTTATTGCAATAAAAGAG,5′-TAACTCGAGCGCTTTGTCCGCTTCTCA). Cloning was performed via restriction sites KpnI and XhoI (introduced by PCR) upstream of the mnx1 3x125bp enhancer sequence.
p5E mnx1-782bp: The 782-bp mnx1 sequence (−2603 to −1821) which includes the 125-bp enhancer segment and the upstream 356-bp segment was obtained by PCR amplification using the plasmid containing the 3-kb mnx1 promoter as template (5′-AAAATATTTATTGCAATAAAAGAG,5′-TCTTATAAGCCTCTTTAA) and inserted into pENTR™5′-TOPO (Invitrogen) via TOPO-TA cloning reaction.
p5E 14xUAS: A DNA fragment containing 14 UAS elements fused to the fish basal promoter E1b derived from the carp β-actin was PCR amplified using pBluescript 14xUAS-E1B-lyn-EGFP (Koster and Fraser, 2001) as a template and cloned into pENTR™5′-TOPO (Invitrogen) via TOPO-TA cloning reaction.
p3E 14xUAS-prenEGFP: To create a prenylated-EGFP (prenEGFP) expression cassette, the SV40pA and 14xUAS sequences were cloned upstream of EGFPCAAXpA from the Tol2 Kit (Kwan et al., 2007). The resulting cassette was amplified by PCR to introduce attB2 (GGGGACAGCTTTCTTGTACAAAGTGGAATCCTAGGAGATCCAGACATG) and attB3 (GGGGACAACTTTGTATAATAAAGTTGATTGGAAAAAACCTCCCACACCTC) sites (underlined) at the 5′ and 3′ ends respectively, and the PCR product was recombined via Gateway cloning BP reaction using BP clonase II into Tol2Kit pDONR P2R-P3 (p3E) vector (Kwan et al., 2007).
Generation and genotyping of transgenic zebrafish
To generate a stable transgenic fish (Tg(mnx1-3x125bp:Gal4-VP16) expressing the yeast Gal4 transcriptional activator (Brand and Perrimon, 1993; Giniger et al., 1985) fused to a virally derived VP16 activator sequence (Gal4-VP16)(Sadowski et al., 1988) in motor neurons, a Tol2 destination vector (pDestTol2pA2) was created carrying three copies of the 125-bp mnx1 enhancer, followed by the Gal4-VP16 sequence, a SV40 polyadenynlation (pA) signal, and flanked by the Tol2 transposable elements (Kawakami et al., 1998). The plasmid was injected into wild type embryos together with in vitro transcribed transposase mRNA. Messenger RNA was synthesized using the mMESSAGE mMachine kit (Ambion, Austin TX). F0 embryos were raised to adulthood and crossed with wild type animals. DNA was extracted from the eggs with the DNeasy kit (Qiagen, Valencia, CA) and used as template to identify F0 animals with germ-line transmission by PCR amplification with primers annealing to the Gal4 sequence (forward, 5′-ATGAAGCTACTGTCTTCTATCG; and reverse, 5′-TGTCTTTGACCTTTGTTACTAC). F1 Tg (mnx1-3x125bp:Gal4-VP16) embryos were injected with a plasmid encoding carrying 14xUAS:prenEGFP and examined at 24 hpf for EGFP expression in spinal motor neurons.
Embryo microinjection
Plasmid injections were carried out with an air-pressured Picospritzer III microinjector (Parker, Cleveland, OH) using glass micro needles pooled with a two stage Kopf 730 vertical puller (Kopf Instruments, Tujunga, CA). Plasmid DNA was prepared using Endotoxin Free Plasmid kits (Qiagen), diluted in injection solution (0.2M KCl, 0.04% phenol red) at a final concentration of 50 ng/μL, and 1–2 nL were injected into the cell of 1-cell stage embryos.
Immunostaining and confocal microscopy
Embryos were dechorionated, anesthetized and sacrificed in ice-cold E3 embryo medium (NaCl, 5mM; KCl, 0.17mM; CaCl2, 0.33 mM; and MgSO4, 0.33mM) containing 0.4% tricaine (MS222, Ethyl 3-aminobenzoate methanesulfonate salt)(Sigma-Aldrich, St Louis, MO), immersed in ice-cold 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) containing 1% dimethyl sulfoxide (DMSO) in Dulbecco’s phosphate buffer saline (PBS), fixed for 3 h at room temperature (RT) followed by 10 min incubation in methanol at −20°C, washed in PBS, and incubated in blocking solution (2% bovine serum albumin (BSA) in PBS) for 1 h at RT. Embryos were then incubated in primary antibodies diluted in blocking solution overnight at 4°C, washed in PBS, incubated with corresponding secondary antibodies conjugated with Cy3 (Jackson ImmunoResearch, West Grove, PA) washed, deyolked, and mount in Prolong Gold (Invitrogen). The following primary antibodies were used: znp1, mouse monoclonal anti-synaptotagmin (Fox and Sanes, 2007; Trevarrow et al., 1990) (Developmental Studies Hybridoma Bank (DSHB), Iowa city, IA), and SV2, mouse monoclonal anti-synaptic vesicle protein 2 (Buckley and Kelly, 1985) (DSHB). For the labeling of nicotinic acetylcholine receptors, Alexa-546 conjugated α-bungartotoxin (Molecular Probes, Eugine, OR) [10μg/ml] was added to the blocking solution and incubated overnight at 4°C. Embryos were observed and imaged with a Nikon C1Si confocal (Nikon, Tokyo, Japan) mounted on a Eclipse 90i upright microscope using Nikon lenses (10X/NA0.45 Plan-Apo, 20X/NA0.75 Plan-Apo, Pan-Fluor 40X/NA1.3 oil, a Plan-Apo 60X/NA1.4 oil, and a Apo-TIRF 100X/NA1.49 oil).
Live imaging
Zebrafish embryos and larvae were anesthetized in 0.02% tricaine, embedded in 1% low melting point agarose (SeaPlaque, Lonza, Rockland, ME) dissolved in Hank’s solution (NaCl 137 mM, KCl 5.4 mM, Na2HPO4 0.25 mM, KH2PO4 0.44 mM, CaCl2 1.3 mM, MgS04 1.0 mM, NaHCO3 4.2 mM), and placed in a 35 mm tissue culture plastic dish. The dish was filled with Hank’s solution and the specimens observed with a Zeiss LSM510 confocal scanning microscope (Carl Zeiss, Thornwood, New York) mounted on a Zeiss Axioplan up-right microscope using an Achroplan 40x/NA0.8 water immersion lens. The specimens were maintained at 26–28°C, the Hank’s solution was replaced every hour, and confocal z-stacks images obtained every 20 min for up to 6 h. Specimens observed in consecutive days were removed from the agarose, maintain in a 28.5°C incubator in E3 embryo media, and re-embedded for the imaging session. Spontaneous movements and blood cell circulation at the end of the imaging session were used as indicators of embryo viability.
Analysis of DNA sequence homology
The Daio rerio genome (Zv8) was queried with mouse mnx1 313-bp sequence (Nakano et al., 2005), using cross-species megaBLAST algorithm (Altschul SF, 1990). The mnx1 5′ UTR regions of mouse, human, Fugu, and zebrafish were aligned using ClustalX2 to search for phylogenetically conserved sequences. The identified 356-bp and 125-bp fragments in zebrafish were aligned separately with mouse, human, and Fugu mnx1 5′ UTR to determine sequence identity. Identity values were calculated in BioEdit.
Supplementary Material
Acknowledgments
We thank Xia Zhao for her assistance in maintaining the zebrafish colony, embryo injection, and immunostaining. We also thank Chi-Bin Chien from the University of Utah for providing the Tol2 kit plasmids, and Dirk Meyer from the University of Freiburg for providing a plasmid carrying the mnx1 promoter. This work was in part supported by NIH grants P20 RR016475, P20 RR024214, and NICDH HD02528.
References
- Altschul SFGW, Miller W, Myers EW, Lipman DJ. Basic local aligment search tool. Journal of Molecular Bioloy. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arber S, Han B, Mendelsohn M, Smith M, Jessell TM, Sockanathan S. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- Boon KL, Xiao S, McWhorter ML, Donn T, Wolf-Saxon E, Bohnsack MT, Moens CB, Beattie CE. Zebrafish survival motor neuron mutants exhibit presynaptic neuromuscular junction defects. Hum Mol Genet. 2009;18:3615–3625. doi: 10.1093/hmg/ddp310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brusés J. N-cadherin regulates primary motor axons growth and branching during zebrafish embryonic development. J Comp Neurol. 2011 doi: 10.1002/cne.22602. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley K, Kelly RB. Identification of a transmembrane glycoprotein specific for secretory vesicles of neural and endocrine cells. J Cell Biol. 1985;100:1284–1294. doi: 10.1083/jcb.100.4.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RT, Linker C, Lewis J. MAZe: a tool for mosaic analysis of gene function in zebrafish. Nat Methods. 7:219–223. doi: 10.1038/nmeth.1423. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Myers PZ, Westerfield M. Pathway selection by growth cones of identified motoneurones in live zebra fish embryos. Nature. 1986;320:269–271. doi: 10.1038/320269a0. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Pike SH, Romancier B. An identified motoneuron with variable fates in embryonic zebrafish. J Neurosci. 1990;10:34–43. doi: 10.1523/JNEUROSCI.10-01-00034.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan-Steet H, Fox MA, Meyer D, Sanes JR. Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development. 2005;132:4471–4481. doi: 10.1242/dev.02044. [DOI] [PubMed] [Google Scholar]
- Fox MA, Sanes JR. Synaptotagmin I and II are present in distinct subsets of central synapses. J Comp Neurol. 2007;503:280–296. doi: 10.1002/cne.21381. [DOI] [PubMed] [Google Scholar]
- Giniger E, Varnum SM, Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985;40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing L, Lefebvre JL, Gordon LR, Granato M. Wnt signals organize synaptic prepattern and axon guidance through the zebrafish unplugged/MuSK receptor. Neuron. 2009;61:721–733. doi: 10.1016/j.neuron.2008.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Koga A, Hori H, Shima A. Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene. 1998;225:17–22. doi: 10.1016/s0378-1119(98)00537-x. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Koster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–346. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melancon E, Liu DW, Westerfield M, Eisen JS. Pathfinding by identified zebrafish motoneurons in the absence of muscle pioneers. J Neurosci. 1997;17:7796–7804. doi: 10.1523/JNEUROSCI.17-20-07796.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers PZ, Eisen JS, Westerfield M. Development and axonal outgrowth of identified motoneurons in the zebrafish. J Neurosci. 1986;6:2278–2289. doi: 10.1523/JNEUROSCI.06-08-02278.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Windrem M, Zappavigna V, Goldman SA. Identification of a conserved 125 base-pair Hb9 enhancer that specifies gene expression to spinal motor neurons. Dev Biol. 2005;283:474–485. doi: 10.1016/j.ydbio.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Panzer JA, Gibbs SM, Dosch R, Wagner D, Mullins MC, Granato M, Balice-Gordon RJ. Neuromuscular synaptogenesis in wild-type and mutant zebrafish. Dev Biol. 2005;285:340–357. doi: 10.1016/j.ydbio.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sager JJ, Bai Q, Burton EA. Transgenic zebrafish models of neurodegenerative diseases. Brain Struct Funct. 2010;214:285–302. doi: 10.1007/s00429-009-0237-1. [DOI] [PubMed] [Google Scholar]
- Suster ML, Kikuta H, Urasaki A, Asakawa K, Kawakami K. Transgenesis in zebrafish with the tol2 transposon system. Methods Mol Biol. 2009;561:41–63. doi: 10.1007/978-1-60327-019-9_3. [DOI] [PubMed] [Google Scholar]
- Tanabe Y, William C, Jessell TM. Specification of motor neuron identity by the MNR2 homeodomain protein. Cell. 1998;95:67–80. doi: 10.1016/s0092-8674(00)81783-3. [DOI] [PubMed] [Google Scholar]
- Trevarrow B, Marks DL, Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–679. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) 4. Eugene, OR: University of Oregon Press; 2000. [Google Scholar]
- Westerfield M, McMurray JV, Eisen JS. Identified motoneurons and their innervation of axial muscles in the zebrafish. J Neurosci. 1986;6:2267–2277. doi: 10.1523/JNEUROSCI.06-08-02267.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.