Abstract
The mismatch repair (MMR) system plays a major role in removing DNA polymerization errors, and loss of this pathway results in hereditary cancers characterized by microsatellite instability. We investigated microsatellite stability during DNA replication within human postmeiotic segregation 2 (hPMS2)–deficient and proficient human lymphoblastoid cell lines. Using a shuttle vector assay, we measured mutation rates at reporter cassettes containing defined mononucleotide, dinucleotide, and tetranucleotide microsatellite sequences. A mutator phenotype was observed in the hPMS2-deficient cell line. The mutation rate of vectors containing [G/C]10 or [GT/CA]10 alleles was elevated 20-fold to 40-fold in hPMS2-deficient cells, relative to an hPMS2-expressing cell line. We observed a 6-fold and 12-fold relative increase in mutation rate of [TTTC/AAAG]9 and [TTCC/AAGG]9 sequences, respectively, in hPMS2-deficient cells. Mutational specificity analyses suggested that repair by hPMS2 is biased. In the absence of hPMS2, a greater number of microsatellite expansion versus deletion mutations was observed, and expansion rates of the tetranucleotide alleles were similar. In the presence of hPMS2, we observed a 29-fold decrease in the [TTCC/AAGG]9 expansion rate but only a 6-fold decrease for the [TTTC/AAAG]9 allele. Our data indicate that hPMS2 is more protective of tetranucleotide expansions than deletions and that hPMS2 displays a sequence bias, wherein [TTCC/AAGG] sequences are stabilized to a greater extent than [TTTC/AAAG]. Our results allow for greater accuracy during identification of MMR defects by providing a mutational signature characteristic of hPMS2 defect. This study also provides clues to possible mechanisms of repair by hPMS2 in the context of the MMR system.
Introduction
DNA mismatch repair (MMR) activity is crucial for maintaining genome integrity. The MMR pathway exists as a cooperation of multiple proteins responsible for the recognition of premutational intermediates and initiates repair by recruiting the necessary cellular factors to excise and resynthesize DNA (reviewed in ref. 1). The MutSα (hMSH2 and hMSH6) and MutSβ (hMSH2 and hMSH3) heterodimers have been implicated in mismatch recognition. Recognition is followed by recruitment of a MutL heterodimer, primarily MutLα [hMLH1 and postmeiotic segregation 2 (hPMS2)] and, to a lesser extent, MutLγ (hMLH1 and MLH3; ref. 1). Loss of MMR results in a mutator phenotype and predisposition to cancer (2–5).
Microsatellites, short repetitive sequences of 1 to 6 bp per unit, are prone to strand slippage errors during DNA synthesis (6). Microsatellites are destabilized by loss of MMR, resulting in a diagnostic, genome-wide microsatellite instability (MSI) phenotype. Germline mutations in the hMLH1, hMSH2, hMSH6, or hPMS2 genes are found in patients with hereditary nonpolyposis colorectal cancer (HNPCC; reviewed in ref. 7). The Bethesda panel of microsatellite markers consists of mononucleotide and dinucleotide repeats to detect HNPCC or HNPCC-like phenotypes (8). The classic MSI-high phenotype is defined as alterations at multiple mononucleotide and dinucleotide loci within patient tumor samples and is highly sensitive for identifying patients carrying germline hMLH1 or hMSH2 gene mutations (9). The proportion of germline mutations in hPMS2 is less frequent in HNPCC families due to decreased penetrance of the disease phenotype and the presence of multiple pseudogenes on chromosome 7 that have hindered accurate detection (10–12). Using immunohistochemical staining of tumors together with MSI analyses, Truninger and colleagues were able to show that 1.5% of unselected colorectal carcinomas are due to a PMS2 defect, similar to the incidence of MSH2-deficient tumors (13). Moreover, a comparable number of families with inheritance of biallelic mutations in MMR genes had defects in hPMS2, often with similar severity of phenotype (12). Improving the specificity of MSI marker panels for the detection of mutations within specific MMR genes, such as hPMS2, may aid in the correct identification of germline carriers.
Tetranucleotide microsatellites are highly polymorphic in the human genome, and A/T-rich alleles are among the highest in density of tetramers in the genome (14). A panel of microsatellite markers that include [TTTC/AAAG]n and [TTCC/AAGG]n alleles has been used diagnostically for detection of bladder, head and neck, and lung cancers (15–18). Primary lung cancers displaying MSI at these markers do not display a phenotype consistent with MMR defects (15), and positive primary bladder tumors are not correlated with absence of either MSH2 or MLH1 (16). However, the effect of MMR loss on the stability of these markers has not been examined directly, and tetranucleotide markers have not been evaluated extensively for use in HNPCC testing.
In the current study, we used a forward mutation assay to analyze the effect of hPMS2 loss on the stability of reporter cassettes containing mononucleotide and dinucleotide alleles ([G/C]10 and [GT/CA]10) versus two tetranucleotide alleles ([TTCC/AAGG]9 and [TTTC/AAAG]9) during DNA replication. We show here that the spontaneous mutation rate of the tetranucleotide alleles is increased in hPMS2-deficient cells, relative to proficient cells. Quantitatively, however, the tetranucleotides were destabilized to a lesser extent than either the mononucleotide or dinucleotide microsatellites. We show that the repair capability of hPMS2 varies with respect to tetranucleotide microsatellite motif sequence and that hPMS2-mediated repair is biased toward removal of expansion mutations.
Materials and Methods
Cell lines
LCL1261, an EBV-transformed human lymphoblastoid cell line cultured from a patient with phenotypic evidence of Turcot syndrome (19), was obtained from Dr. Susan Farrington, University of Edinburgh. LCL1261 cells contain R134ter mutation, as well as a 2184delTC mutation, in a second allele (11, 19). Cells were maintained in RPMI 1640 supplemented with 15% fetal bovine serum (Hyclone). LCL721, a nontumorigenic, EBV-transformed human lymphoblastoid cell line established from a clinically normal female donor (20) was cultured, as described previously (21).
hPMS2 complementation of LCL1261 cell line
LCL1261 cells were infected with pseudoviral particles containing an hPMS2 expression vector, which was packaged using the pPACKF1 Lentivector Packaging kit (System Bioscience) for stable gene complementation. hPMS2 cDNA was cloned into the EcoRI site of the pCDF lentiviral expression construct. 293TN cells (60–70% confluent) were transfected with 2 µg of pCDF-hPMS2 or pCDF only, along with 2 µg of pSIF1-H1-siLuc-copGFP as a positive control for infection. Pseudoviral particles containing pCDF-hPMS2 were collected at various time points posttransfection, as per the manufacturer’s protocol. Pseudoviral titers were found to be 5 × 105 infectious particles/mL by flow cytometry. LCL1261 cells were infected with packaged pCDF-hPMS2 or pCDF only at a multiplicity of infection of 1, as per the manufacturer’s protocol. We performed a total of three rounds of infection to ensure complete transduction and integration of the viral expression construct into the genomic DNA. Flow cytometry showed successful infection of pCDFPMS2 (56%) and pCDF only (22%), termed 1261-PMS2 and 1261-vector only, respectively. Expression vector containing cells were selected using 25 µg/mL puromycin (Clontech Laboratories).
Herpes simplex virus type 1 thymidine kinase mutational analysis
Microsatellite sequences were inserted in-frame within the 5′ coding region of the herpes simplex virus type 1 thymidine kinase (HSV-tk) gene. Construction of the [TTCC/AAGG]9, [TTTC/AAAG]9, [G/C]10, and [GT/CA]10 shuttle vectors has been previously described (21–24). Vectors were introduced into lymphoblastoid cells by electroporation. Two to three days after electroporation, plasmid-bearing cells were selected using 50 to 150 µg/mL hygromycin. After an additional 5 to 7 d, cells were cloned in 96-well plates at a density of one to five cells per well. PTB (1 mmol/L sodium pyruvate, 50 mmol/L α-thioglycerol, 20 nmol/L bathocuproindisulfonic acid disodium salt) was added in the 96-well dish to reduce oxidative stress due to cloning for the LCL1261 cells (25). Wells contained 12,000 to 15,000 Balb291 mouse fibroblast cells as a feeder layer for LCL1261. Cells were kept on the feeder layer until they were expanded to p100 plates. Between 5 to 10 plasmid-bearing clones were expanded from 29 to 32 generations in the presence of hygromycin. DNA was isolated using a modified alkaline extraction procedure and processed as described (21, 26). Shuttle vector DNA was electroporated into Escherichia coli strain FT334 and plated on media containing chloramphenicol, with or without 5-fluoro-2′-deoxyuridine (FUdR), for mutational analysis. FUdR selects for cells with an HSV-tk– deficient phenotype, which may occur due to mutations in the HSV-tk promoter or coding sequence or frameshift mutations within the inserted microsatellite allele. The HSV-tk mutation frequency is obtained by dividing the number of FudR + chloramphenicol-resistant colonies by the number of chloramphenicol-resistant colonies. The clonal mutation rate is defined as the mutation frequency per cell generation. For mutational analysis, DNA from 15 to 25 independent mutants per clone, from three clones of each vector, was isolated. Dideoxy sequence analysis was used to determine the type and location of mutations within the HSV-tk gene.
Protein analysis
Nuclear and cytoplasmic fractions were isolated from LCL721 and LCL1261 cells using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce) with addition of a protease inhibitor, according to the manufacturer’s protocol. Protein concentration was determined using the DC protein assay (Bio-Rad Laboratories). Protein lysate (20 µg) was denatured, reduced, and separated by SDS-PAGE using a 10% NuPage bis-Tris gel (Invitrogen), followed by transfer onto polyvinylidene difluoride membrane (Invitrogen). The membrane was blocked with 5% dry milk in TBS containing 0.1% Tween 20 (TBST) for 60 min, then incubated overnight at 400b0C with a rabbit polyclonal antibody (Santa Cruz Biotechnology) to hPMS2 (1:200) or mouse monoclonal antibodies (BD Biosciences) to hMLH1 (1:250), hMSH6 (1:670), MSH2 (1:250), MSH3 (1:250), or TATA binding protein (TBP; 1:2,000). The membrane was rinsed twice with TBST followed by two 15-min washes and incubated with horseradish peroxidase–conjugated goat anti-rabbit (1:2,000) or goat anti-mouse (1:2,000; Jackson ImmunoResearch) for 60 min. Immunoreactive proteins were visualized using ECL+ (Amersham Biosciences) and scanned on a Molecular Dynamics Storm phosphorimager.
Results
The LCL1261 cell line exhibits a mutator phenotype
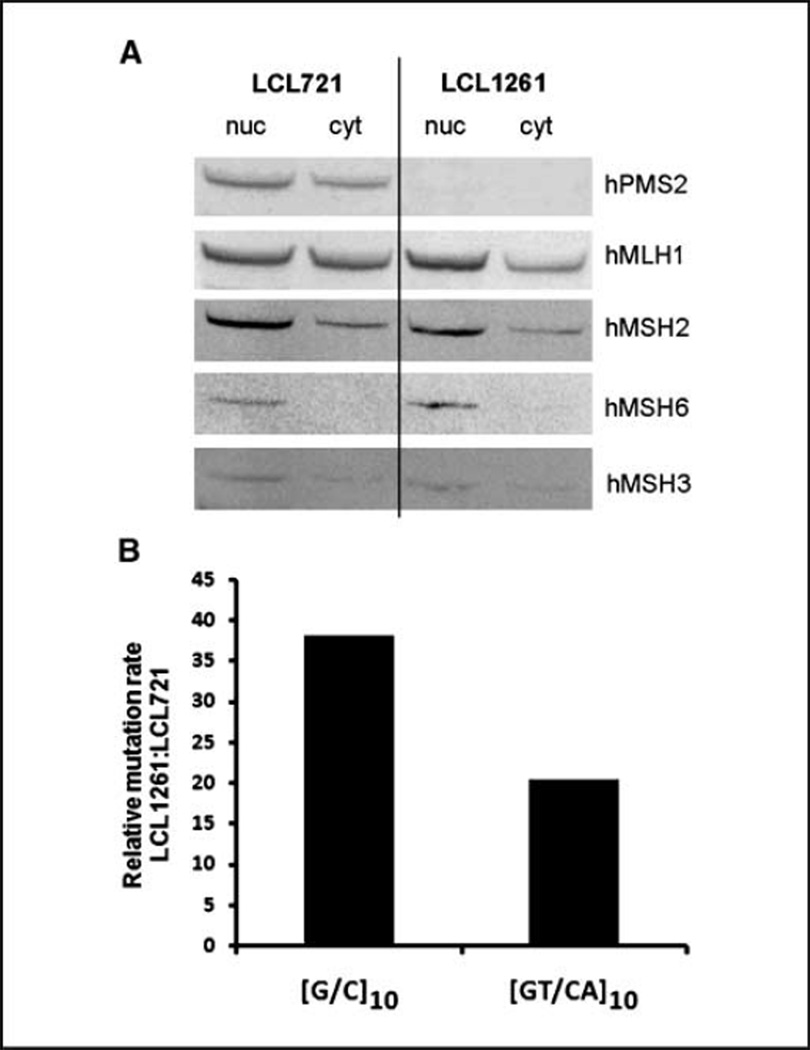
The LCL1261 cell line is a nontumorigenic cell line cultured from a patient with phenotypic evidence of Turcot syndrome (19). The cells have been shown previously to contain a biallelic mutation (R134ter, 2184delTC) in hPMS2, causing the loss of protein production and MMR activity (11, 19). We detected no expression of the hPMS2 protein in the LCL1261 cell line, whereas a prominent hPMS2 protein of 97 kDa was observed in the LCL721 cell line (Fig. 1A). To exclude the possibility of other MMR proteins being misregulated due to hPMS2 loss, we examined the status of hMSH2, hMSH3, hMSH6, and hMLH1 proteins. We found these levels in LCL1261 to be similar to those observed in the MMR proficient LCL721 cell line (Fig. 1A). Full images of all Western analyses can be found in Supplementary Fig. S1. These data show that the MMRdeficient phenotype of LCL1261 cells is likely due to the specific loss of hPMS2 expression. However, we cannot rule out the possibility that the MMR deficiency caused by loss of hPMS2 has subsequently resulted in loss-of-function point mutations within the other MMR genes.
Figure 1.
LCL1261 cells are hPMS2 deficient and display a mutator phenotype. A, nuclear (nuc) and cytoplasmic (cyt) fractions were prepared from LCL721 and LCL1261 cells and analyzed for expression of MMR proteins (20 µg of total protein loaded) by Western blot analysis. The membrane was probed with hPMS2, hMLH1, hMSH2, hMSH6, and hMSH3. B, shuttle vectors containing [G/C]10 and [GT/CA]10 microsatellite alleles were introduced into LCL721 (hPMS2+) and LCL1261 (hPMS2−) cells. Individual plasmid-bearing clones were expanded from 24 to 35 cell generations. After extracting shuttle vector DNA, HSV-tk mutant frequencies were determined for each clone by selection in E. coli. The mutation rate was estimated by calculating the HSV-tk mutation frequency per cell generation. Mutational rate increase in the [G/C]10 and [GT/CA]10 vectors is shown as a proportion of the median mutation rate of LCL1261 clones versus LCL721.
Our ex vivo HSV-tk mutagenesis system uses an oriP-based episomal shuttle vector to study microsatellite mutagenesis in human cells (21). As this is the first report using a shuttle vector system in the LCL1261 cell line, we performed a Dpn1 sensitivity assay to monitor replication of the vector. Nonreplicated DNA from E. coli is methylated at GmATC sequences, and these sites are sensitive to cleavage by the Dpn1 restriction endonuclease. DNA replicated in mammalian cells is not methylated at adenosines and, thus, will be Dpn1 resistant. At 8 days postelectroporation of LCL1261 cells, the oriP-tk shuttle vector was completely Dpn1 resistant, as evidenced by the presence of full-length vector DNA and the disappearance of digestion products (Supplementary Fig. S2). Using copy number standards, we estimate that the vector is present in the population at an average of five copies per cell.
A mutator phenotype for the LCL1261 cell line was previously shown using the genomic BAT40, D2S123, and TGFβR micro-satellite markers (27). We used our previously reported HSV-tk shuttle vector assay to examine MSI within mononucleotide and dinucleotide microsatellites, representative of markers destabilized in HNPCC patient tumors ([G/C]10 and [GT/CA]10). Reporter cassettes containing defined microsatellite sequences in-frame within the coding region of the HSV-tk gene were constructed to monitor spontaneous mutagenesis in human cells (21, 23). The HSV-tk sequences are under no selective pressure during replication in human cells. The episomal nature of our oriP-based shuttle vectors ensures that endogenous repetitive sequences at points of integration do not affect mutagenesis. This allows direct comparative analyses of mutational variation due to the cellular genetic background or the sequence composition of the microsatellite. Shuttle vectors were introduced into LCL721 and LCL1261 lymphoblastoid cells, and the populations were cloned to ensure that the founder cells contained only wild-type vectors. Individual plasmid-bearing clones were expanded 24 to 35 cell generations. After extracting shuttle vector DNA, HSV-tk mutant frequencies were determined for each clone by genetic selection in E. coli. The mutation rate was estimated by calculating the HSV-tk mutation frequency per cell generation.
The median mutation rate of the control shuttle vector containing only the HSV-tk gene was 2.6 × 10−5 (four clones) after replication in LCL1261 cells. For comparison, the corresponding mutation rate for the control vector was 14-fold lower, 1.9 × 10−6, after replication in LCL721 cells (21). For vectors containing the [G/C]10 and [GT/CA]10 alleles, we observed median mutation rates of 2.1 × 10−3 (two clones) and 5.5 × 10−5 (three clones), respectively, after replication in hPMS2-deficient LCL1261 cells. The corresponding median mutation rates in hPMS2-proficient LCL721 cells were 5.5 × 10−5 (10 clones)1 and 2.7 × 10−6 (21), respectively. This corresponds to a 38-fold increase in mutation rate for the [G/C]10-containing vector and a 20-fold increase for the [GT/CA]10-containing vector in an hPMS2-deficient background, relative to an hPMS2-proficient background (Fig. 1B). Overall, these results are consistent with a mutator phenotype of the LCL1261 cell line due to MMR loss.
Mutation rates of [TTCC/AAGG]9 and [TTTC/AAAG]9-containing vectors
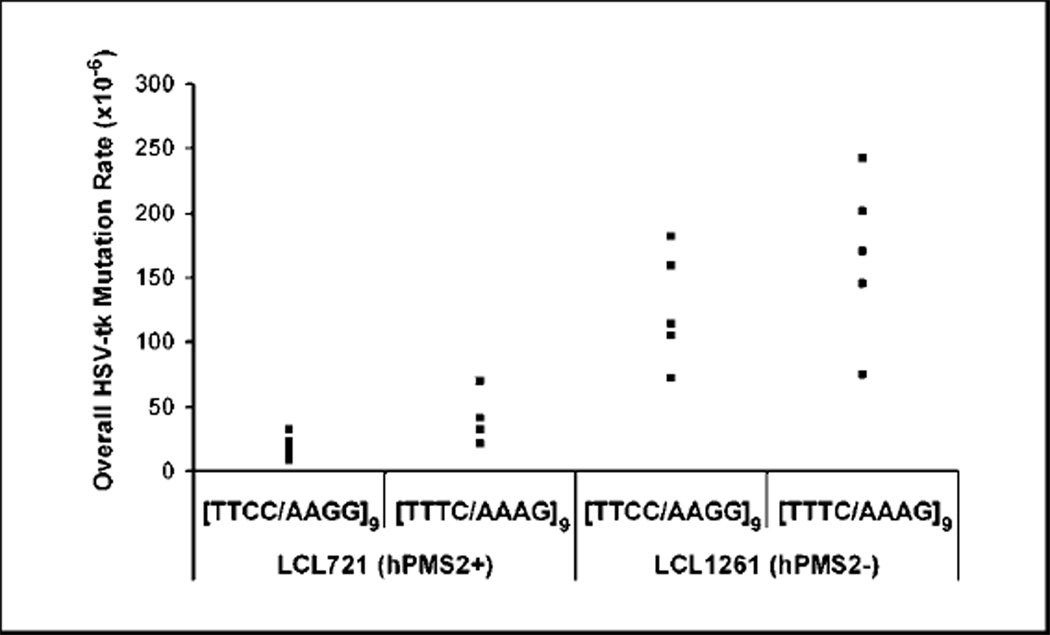
Using the shuttle vector assay, we sought to determine the effect of hPMS2 absence upon the stability of tetranucleotide repeats that have high presence throughout the genome. We determined the median mutation rate of [TTCC/AAGG]9 and [TTTC/AAAG]9-containing vectors after replication in hPMS2-proficient LCL721 cells to be 1.3 × 10−5 and 3.7 × 10−5, respectively (Fig. 2). These values are similar to our previous independent mutational analyses (23). In hPMS2-deficient LCL1261 cells, we observed an 11-fold higher median mutation rate for the [TTCC/AAGG]9 vector (1.4 × 10−4) and a 5-fold increase for the [TTTC/AAAG]9 vector (1.7 × 10−4; Fig. 2). The differences in mutation rates between cell lines are statistically significant (P = 0.0047 and P = 0.0095, respectively, Wilcoxon-Mann-Whitney test).
Figure 2.
PMS2 loss destabilizes tetranucleotide repeats. Points, mutation rate of an independent LCL721 or 1261 clone bearing a tetranucleotide-containing oriP-tk shuttle vector. n = 10, 721/TTCC; n = 4, 721/TTTC; n = 6, 1261/TTCC; n = 6, 1261/TTTC. Outliers not shown on graph: 721/TTCC, 3.16 × 10−3; 1261/TTCC, 3.31 × 10−2. Data were analyzed statistically using the nonparametric Wilcoxon-Mann-Whitney test.
The mutation rates obtained in the shuttle vector assay reflect all types of mutational events occurring in the HSV-tk gene, including base substitutions, frameshifts, and large deletions within the coding sequence, as well as mutations within the microsatellite alleles. To examine the effect of hPMS2 status on the specificity of mutations, we derived mutation spectra for both cell lines by sequencing multiple independent mutants from at least three clones bearing each tetranucleotide vector. We categorized mutations as occurring at the microsatellite or within the HSV-tk coding region (Table 1). For both cell lines, the majority of observed mutants arose within the microsatellite region. The microsatellitespecific mutation rate within the [TTCC/AAGG]9 allele is increased 12-fold, and within the [TTTC/AAAG]9 allele, it is increased 6-fold in hPMS2-deficient cells. As a point of comparison, the HSV-tk gene contains an endogenous [G/C]7 mononucleotide microsatellite at positions 487 to 493 within the coding region, which is highly unstable in MMR-deficient E. coli (22). We calculated the average mutation rate for this [G/C]7 motif after replication in hPMS2-deficient LCL1261 cells to be 1.4 × 10−4 for both vectors, a value that is ~50-fold higher than the average rate observed in LCL721 cells (3.0 × 10−7).2
Table 1.
Mutation rates and spectra of tetranucleotide vectors in human lymphoblastoid cell lines
| Class of mutation | LCL721 (PMS2+) | LCL1261 (PMS2−) | ||
|---|---|---|---|---|
| [TTCC/AAGG]9* | [TTTC/AAAG]9 | [TTCC/AAGG]9* | [TTTC/AAAG]9 | |
| Microsatellite | ||||
| Expansions | 12 (3, 5, 4) | 12 (4, 5, 3) | 42 (14, 16, 12) | 34 (8, 17, 9) |
| Deletions | 14 (1, 8, 5) | 9 (2, 4, 3) | 2 (0, 2, 0) | 8 (2, 1, 5) |
| Other | 1 (1, 0, 0) | 0 | 0 | 0 |
| Coding region† | 6 (2, 0, 4) | 10 (3, 2, 5) | 10 (2, 4, 4) | 11 (5, 5, 1) |
| Total | 33 | 31 | 54 | 53 |
| Median rate | 13 × 10−6 | 37 × 10−6 | 140 × 10−6 | 170 × 10−6 |
NOTE: Mutation spectra of [TTCC/AAGG]9 and [TTTC/AAAG]9 were generated after replication in LCL721 and LCL1261. Numbers represent total number of occurrences, whereas those in parentheses indicate values for individual clones.
The difference in the ratio of expansions to deletions between [TTCC/AAGG]9 and [TTTC/AAAG]9 was statistically significant (P < 0.0001, Fisher’s exact test, 2-sided).
The difference between microsatellite and coding region mutations was not statistically significant between LCL721 and LCL1261 cell lines containing the same vector (P > 0.05, Fisher’s exact text, 2-sided).
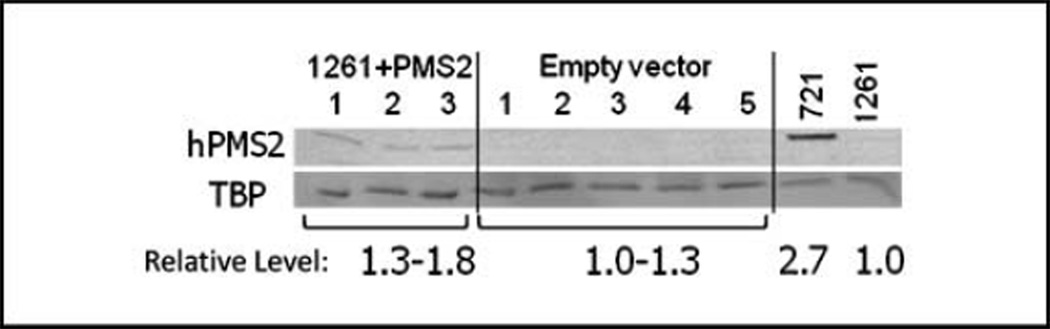
Because the LCL1261 cell line was cultured from a clinically affected individual, the cells may harbor genetic defects in addition to hPMS2, which could affect mutagenesis. To confirm that the mutational difference we observed is, in fact, due to the absence of hPMS2, we stably complemented LCL1261 cells with a lentiviral vector containing either a cDNA for hPMS2 or the vector alone. A previous study has shown that overexpression of hPMS2 results in hypermutability (28). Therefore, our vector design did not include motifs, such as a Kozak sequence, to optimize translation but rather used the native sequence upstream of the start codon found in the hPMS2 cDNA. Protein expression was examined in multiple, clonally derived populations. The cDNA-complemented LCL1261 cells displayed detectable levels of hPMS2 protein, albeit at a relatively lower level than that observed in LCL721 cells. No hPMS2 protein was detectable in either the parental LCL1261 cells or the vector-complemented clones. The cDNA-complemented clonal population with the highest expression, along with a vector-only control clone, was electroporated with the [TTCC/AAGG]9-containing shuttle vector. Clonal populations were once again obtained for mutational rate and specificity analysis. Increased hPMS2 expression (97 kDa) was detected in complemented cell clones containing the [TTCC/AAGG]9 shuttle vector but not in vector-only complemented clones, relative to a TBP loading control (Fig. 3). Levels of hMLH1 protein were unchanged upon lentiviral infection and were similar among cDNA-complemented and vector only–complemented clones (Supplementary Fig. S3). In hPMS2-complemented cells, the median mutation rate of the [TTCC/AAGG]9-containing vector was 2.6 × 10−5 (three clones, range 1.3–4.6 × 10−5), 2-fold lower than that of control vector-complemented cells (5.3 × 10−5, four clones, range 3.7–23 × 10−5). DNA sequence analyses revealed that the microsatellite mutation rate was 2.4-fold lower and the HSV-tk coding region rate was 3.2-fold lower in the hPMS2-complemented cells, relative to the control (Table 2). The mutation rates measured in the hPMS2-complemented cells were not as low as were those measured for the LCL721 cells (Table 2); however, this was expected because the relative level of hPMS2 protein was lower in the complemented cells (Fig. 3). Unexpectedly, the mutation rates measured in the vectorcomplemented control cell line were ~2-fold lower than the parental 1261 cells (Table 2), which may be attributed to attenuation of the mutator phenotype caused by puromycin selection or by lot variation in cell culture media (29). Overall, the mutational results are consistent with complementation of the LCL1261 mutator phenotype by hPMS2.
Figure 3.
hPMS2 expression in complemented 1261 clones. hPMS2 cDNA was delivered into LCL1261 by a Lentivirus-based system for integration into the host genome. A clonal population was obtained and electroporated with [TTCC/AAGG]9 shuttle vector DNA. hPMS2 protein expression of clones containing [TTCC/AAGG]9 shuttle vector DNA was analyzed by Western blot. hPMS2 levels were normalized to TBP loading control and fold change in protein levels relative to LCL1261 parental cells are indicated.
Table 2.
Mutation rates and spectra of [TTCC/AAGG]-containing vectors in patient and cDNA-complemented human lymphoblastoid cell lines
| Mutational event | HSV-tk mutation rate (n × 10−5) | |||
|---|---|---|---|---|
| Complemented clone | Parental cells (median) | |||
| Vector | hPMS2 | LCL1261 | LCL721 | |
| Microsatellite | 5.2 | 2.2 | 11 | 0.95 |
| Expansion | 4.7 | 2.2 | 11 | 0.42 |
| Deletion | 0.43 | nd | 0.51 | 0.49 |
| HSV-tk coding | 1.3 | 0.41 | 2.5 | 0.25 |
| Overall | 6.5 | 2.6 | 14 | 1.3 |
Biased repair of tetranucleotide microsatellites in the presence of hPMS2
In LCL1261 cells, we observed no statistical difference in the mutation rates between the two tetranucleotide vectors (P = 0.25; Fig. 2; Table 1), whereas in LCL721 cells, the mutation rate measured for the [TTCC/AAGG]9 vector was significantly lower than for the [TTTC/AAAG]9 vector (P = 0.011). This comparison suggests that hPMS2 repair is more efficient at the [TTCC/AAGG]9 allele, relative to the [TTTC/AAAG]9 allele. We compared the specificity of mutations arising within the microsatellites between the two cell lines. The microsatellite mutants arising in the hPMS2-deficient cells were highly biased toward the addition of one tetranucleotide unit for both alleles. For the [TTCC/AAGG]9 allele, the observed proportion of expansion to deletion mutations was significantly different in LCL1261 cells compared with LCL721 cells (P < 0.0001, two-sided Fisher’s exact test; Table 1). For the [TTTC/AAAG]9 allele, 81% of mutations arising in LCL1261 cells were expansions compared with 57% in LCL721 cells, although this difference is not statistically significant. Whereas the number of mutants is small, a similar mutation bias was observed at the endogenous [G/C]7 sequence: 90% of the mutants (n = 10) arising within the LCL1261 cells were +1 bp frameshifts compared with 60% (n = 5) in LCL721 cells. Whereas we did not observe any microsatellite changes greater than one unit in LCL1261 cells, changes greater than one unit were present in the LCL721 cell spectra, albeit at a lower frequency than singleunit changes (data not shown).
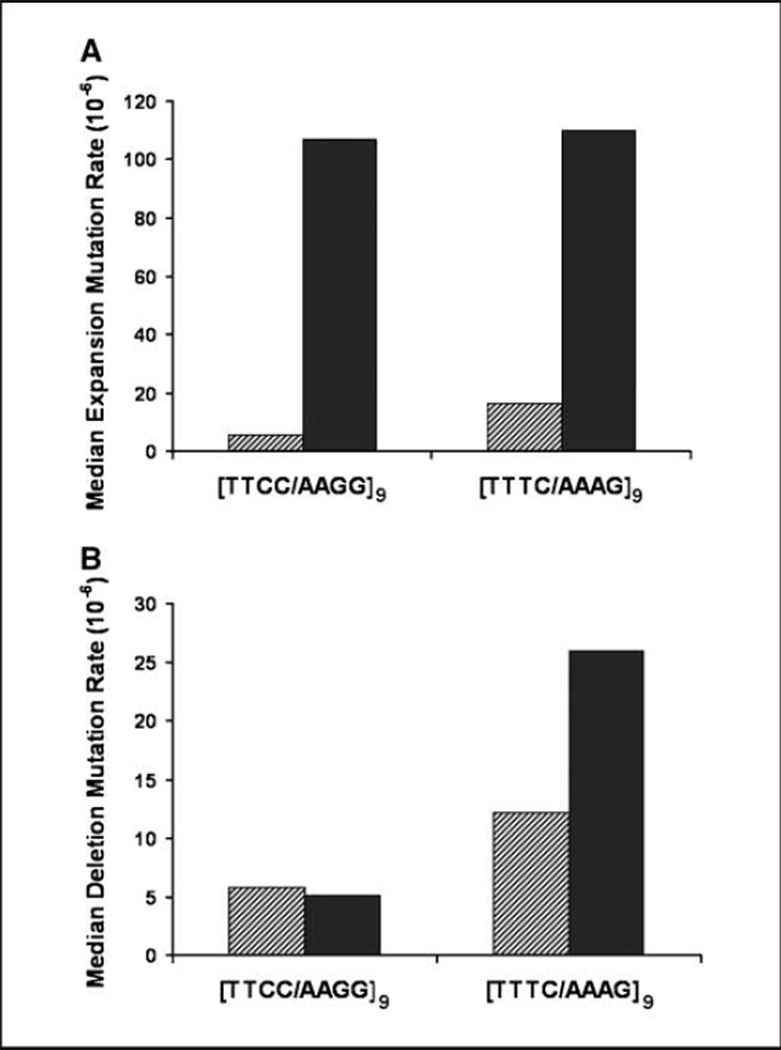
To compare absolute mutation rates between the two alleles, we compared the rate of expansions versus deletions at the microsatellite. In hPMS2-deficient cells, we observed a similar high rate of expansion mutations for both microsatellite alleles (Fig. 4A). In hPMS2-proficient cells, we observed a 25-fold lower expansion rate for the [TTCC/AAGG]9 repeat but only an 8-fold lower expansion rate for the [TTTC/AAGG]9 repeat when compared with the hPMS2-deficient cells. The corresponding microsatellite deletion rates were similar between the two alleles, and a small difference was observed between the two cell types (Fig. 4B). Overall, the mutational specificity data are consistent with strand and sequence-biased repair of tetranucleotide alleles by hPMS2.
Figure 4.
PMS2 is biased toward repair of [TTCC/AAGG] expansions but has little effect on the deletion rate at the microsatellite. A, the proportion of mutants that contained expansions at the microsatellite was multiplied by the median mutation frequency to obtain the expansion mutation rate. B, the proportion of mutants that contained deletions at the microsatellite was multiplied by the median mutation frequency to obtain the deletion mutation rate. Hatched bars, LCL721; solid bars, LCL1261.
Discussion
Tetranucleotide microsatellite repeats are almost as prevalent as dinucleotide repeats and are present throughout every chromosome of the human genome (14). Mechanisms that govern tetranucleotide stability are poorly understood, although instability of this type of microsatellite has been seen in numerous cancers (15–17). Markers containing [TTTC/AAAG]n and [TTCC/AAGG]n repeats are predominantly affected within cancer cell lines displaying tetranucleotide instability (30). In this study, we focus on the MMR repair pathway, specifically hPMS2, and show that it is a critical component for maintaining tetranucleotide repeat stability. In addition, we observed a strong repair bias by hPMS2 favoring the repair of [TTCC/AAGG]9 over [TTTC/AAAG]9 sequences and correction of expansion mutations over deletion mutations.
Somatic frameshifts within a [G/C]8 microsatellite in the BAX gene are observed at a high rate in MMR-deficient tumors and are implicated in the etiology of HNPCC (31). We observed a 50-fold higher mutation rate at the [G/C]7 mononucleotide within the HSV-tk gene after shuttle vector replication in hPMS2-deficient LCL1261 cells, relative to hPMS2-proficient LCL721 cells. The MYCL1 marker is a complex allele containing a [GAAA/CTTT]n tetranucleotide repeat that can show instability in MSI-H tumors (9, 32). Although tetranucleotide repeats have not been routinely used in MSI testing, we observed a 6-fold to 12-fold higher mutation rate at two tetranucleotide sequences in the hPMS2-deficient cell line. A caveat to the general use of tetranucleotide markers for MMR status testing is the elevated mutation rate of some, but not all, tetranucleotide alleles in nontumorigenic MMR-proficient cells. We have previously reported that the spontaneous mutation rate at a [TTTC/AAAG]9 allele in MMR-proficient human cells is 30-fold higher than the HSV-tk gene mutation rate but the rate of the [TTCC/AAGG]9 allele is only 5-fold higher (23). Therefore, MSI at specific tetranucleotide alleles may represent the detection of spontaneous mutations in the population due to the intrinsic mutation rate of the microsatellite locus and genetic selection rather than intrinsic genome instability of the tumor. For example, the ability of the oncogenic EWS/FLI transcription factor to modulate gene expression was recently shown to depend upon the number of consecutive GGAA/TTCC tetranucleotide motifs within a promoter-encoded microsatellite (33). We show here that such an allele, [TTCC/AAGG]9, has a relatively low spontaneous mutation rate in nontumorigenic cells but can be significantly destabilized by the loss of hPMS2 (Fig. 2). Although further experimental studies are clearly required to validate the use of tetranucleotide alleles in testing, our data show that increasing detection sensitivity for rare germline mutations (e.g., hPMS2) or MSI-L tumors may be feasible through use of a specific panel of well-characterized microsatellite markers.
In mouse model systems, microsatellite mutagenesis varies both quantitatively and qualitatively in MMR-deficient cells, depending upon the specific MMR gene that is inactivated (34–36). For example, the frequency of mutations arising within [CA/GT]n markers was shown to be similar in PMS2−/− and MLH1−/− mice, but the types of mutations differed significantly, as PMS2 null mice exhibited a higher proportion of expansion mutations within the microsatellites than did MLH1 null animals (34). Similarly, at the SupFG1 mononucleotide microsatellite reporter, the proportion of +1 bp frameshifts is higher in PMS2−/−, MSH3−/−, and MSH6−/− mice compared with MLH1-null and MSH2-null mice (36). Our data show that a mononucleotide allele is highly mutable and biased toward +1-bp mutations in a nontumorigenic hPMS2-null human cell line. Furthermore, we show a protective role of hPMS2 within tetranucleotide microsatellites that is mostly confined to expansion avoidance. The observed mutational variation may be attributable to differences in the DNA substrate specificity of repair by various MMR subunits or reflect differences in residual MMR activity. For example, inactivation of MLH1 will result in loss of both MutLα and MutLγ activities, whereas PMS2 loss will result only in loss of MutLα. Differential MMR removal of premutational intermediates located on the lagging strand of the DNA replication fork has been reported in Saccharomyces cerevisiae (37, 38). However, the consistent bias toward expansion mutations observed here and in other studies suggests a different type of strand bias that may be specifically due to PMS2 deficiency. In support of this notion, novel alleles of scPMS1 (the S. cerevisiae homologue of hPMS2) that predominantly elevate +1 bp expansions with little or no effect on −1 bp deletions have been described (39). In the slipped-strand mispairing model for microsatellite mutagenesis, premutational looped intermediates are formed by dissociation of the nascent DNA strand from the template strand during DNA synthesis, followed by incorrect reassociation. The Erdeniz and colleagues study suggests that scPMS1 interacts specifically with loops present on the nascent primer strand, regardless of leading or lagging strand orientation during DNA replication (39). Our data indicate that hPMS2 is primarily involved in repair of nascent strand tetranucleotide loops, which are the premutational intermediate for expansion mutations.
Our mutation rate data also indicate that hPMS2 is more protective toward [TTCC/AAGG] repeats than [TTTC/AAAG] repeats. The repair bias we observed in the presence of hPMS2 may result from sequence-specific recognition during initiation of repair. MutSβ may recognize one allele better than the other, thus controlling the recruitment of hPMS2 to the mispair. In yeast, the correction efficiency by MutSβ of expansions within a [G]10 mononucleotide was higher than for a [C]10 sequence, indicating that sequence composition does affect repair (40). In E. coli, the efficiency of MutS repair varies greatly across dinucleotide repeats of different sequence composition (41). Alternatively, the repair bias may be directly dependent on PMS2 activity. The endonuclease activity of PMS2 is required to initiate nick-directed excision of the mispair by the 5′–3′ exonuclease EXO1 (42). PMS2 may be more successful at processing a [TTCC/AAGG] loop than a [TTTC/AAAG] loop, which may explain the different degree of repair. A subset of mutations occurring near the 5′ nick in the lagging strand would be repaired even in the absence of an endonuclease activity (43), further explaining the finding that the [TTTC/AAGG]9 allele is still being repaired, although at a lesser extent.
Finally, the differences between the [TTCC/AAGG]9 and [TTTC/AAAG]9 alleles may reflect biased repair by other MutL complexes. A number of studies have observed that hMLH1 is required for stable expression of hPMS2, possibly due to stability conferred by heterodimerization (13). As shown here and previously reported (13), however, the converse is not true, and stable expression of the hMLH1 protein is observed in the absence of hPMS2. Perhaps, this is due to the fact that other MutL homologues can substitute for hPMS2 and form stable complexes with hMLH1. For example, the alternative MutLγ complex may form to a greater extent in the hPMS2-deficent cell line. MLH3 also contains the conserved metal binding motif DQHA(X)2E(X)4E required for endonuclease activity in PMS2 (42). In the absence of PMS2, MLH3 localizes to the nucleus and contributes to MMR (44). MLH3 has been shown to interact with MutSβ to repair a subset of insertion/deletion loops in S. cerevisiae and mouse cells (45). Although repair activity of MLH3 was not observed for two to four base loops in vitro using human cell extracts (46, 47), the MSI phenotype of mice is enhanced in MLH3−/− PMS2−/− double-null mice, relative to PMS2−/− mice (47). Thus, it can be hypothesized that MLH3 keeps [TTTC/AAAG]9 alterations in check in the absence of hPMS2. Further studies are needed to extend the findings reported here and to better define the specificity of hPMS2 repair.
Supplementary Material
Acknowledgments
Grant Support: NIH grant RO1 CA100060 (K. Eckert) and Jake Gittlen Cancer Research Foundation.
We thank Malcolm G. Dunlop and Susan M. Farrington (University of Edinburgh) for providing us with the LCL1261 cell line; Andrew B. Buermeyer (Oregon State University) for providing us with a hPMS2 expression vector; Maria Baker, Suzanne Hile, Kimberly Jacob, and Kimberly Duncan for their critical reading of the manuscript; and Nate Sheaffer and David Stanford in the Flow Cytometry Core Facility (Pennsylvania State University College of Medicine).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
K.D. Jacob, K.A. Eckert, unpublished data.
K.A. Eckert, unpublished data.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 2.Aaltonen LA, Peltomaki P, Leach FS. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharyya NP, Skandalis A, Ganesh A, Groden J, Meuth M. Mutator phenotype in human colorectal carcinoma cell lines. Proc Natl Acad Sci U S A. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 5.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 6.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 7.Rustgi AK. The genetics of hereditary colon cancer. Genes Dev. 2007;21:2525–2538. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- 8.Boland CR, Thibodeau SN, Hamilton SR, et al. A national cancer institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 9.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendriks YM, Jagmohan-Changur S, van der Klift HM, et al. Heterozygous mutations in PMS2 cause hereditary nonpolyposis colorectal carcinoma (Lynch syndrome) Gastroenterology. 2006;130:312–322. doi: 10.1053/j.gastro.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 11.De Vos M, Hayward BE, Picton S, Sheridan E, Bonthron DT. Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome. Am J Hum Genet. 2004;74:954–964. doi: 10.1086/420796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felton KE, Gilchrist DM, Andrew SE. Constitutive deficiency in DNA mismatch repair. Clin Genet. 2007;71:483–498. doi: 10.1111/j.1399-0004.2007.00803.x. [DOI] [PubMed] [Google Scholar]
- 13.Truninger K, Menigatti M, Luz J, et al. Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer. Gastroenterology. 2005;128:1160–1171. doi: 10.1053/j.gastro.2005.01.056. [DOI] [PubMed] [Google Scholar]
- 14.Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahrendt SA, Decker PA, Doffek K, et al. Microsatellite instability at selected tetranucleotide repeats is associated with p53 mutations in non-small cell lung cancer. Cancer Res. 2000;60:2488–2491. [PubMed] [Google Scholar]
- 16.Catto JW, Azzouzi AR, Amira N, et al. Distinct patterns of microsatellite instability are seen in tumours of the urinary tract. Oncogene. 2003;22:8699–8706. doi: 10.1038/sj.onc.1206964. [DOI] [PubMed] [Google Scholar]
- 17.Danaee H, Nelson HH, Karagas MR, et al. Microsatellite instability at tetranucleotide repeats in skin and bladder cancer. Oncogene. 2002;21:4894–4899. doi: 10.1038/sj.onc.1205619. [DOI] [PubMed] [Google Scholar]
- 18.Singer G, Kallinowski T, Hartmann A, et al. Different types of microsatellite instability in ovarian carcinoma. Int J Cancer. 2004;112:643–646. doi: 10.1002/ijc.20455. [DOI] [PubMed] [Google Scholar]
- 19.Parsons R, Li GM, Longley M, et al. Mismatch repair deficiency in phenotypically normal human cells. Science. 1995;268:738–740. doi: 10.1126/science.7632227. [DOI] [PubMed] [Google Scholar]
- 20.Kavathas P, Bach FH, DeMars R. γ ray-induced loss of expression of HLA and glyoxalase I alleles in lymphoblastoid cells. Proc Natl Acad Sci U S A. 1980;77:4251–4255. doi: 10.1073/pnas.77.7.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hile SE, Yan G, Eckert KA. Somatic mutation rates and specificities at TC/AG and GT/CA microsatellite sequences in nontumorigenic human lymphoblastoid cells. Cancer Res. 2000;60:1698–1703. [PubMed] [Google Scholar]
- 22.Eckert KA, Yan G. Mutational analyses of dinucleotide and tetranucleotide microsatellites in Escherichia coli : influence of sequence on expansion mutagenesis. Nucleic Acids Res. 2000;28:2831–2838. doi: 10.1093/nar/28.14.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckert KA, Yan G, Hile SE. Mutation rate and specificity analysis of tetranucleotide microsatellite DNA alleles in somatic human cells. Mol Carcinog. 2002;34:140–150. doi: 10.1002/mc.10058. [DOI] [PubMed] [Google Scholar]
- 24.Jacob KD, Eckert KA. Escherichia coli DNA polymerase IV contributes to spontaneous mutagenesis at coding sequences but not microsatellite alleles. Mutat Res. 2007;619:93–103. doi: 10.1016/j.mrfmmm.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brielmeier M, Bechet JM, Falk MH, Pawlita M, Polack A, Bornkamm GW. Improving stable transfection efficiency: antioxidants dramatically improve the outgrowth of clones under dominant marker selection. Nucleic Acids Res. 1998;26:2082–2085. doi: 10.1093/nar/26.9.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffin BE, Bjorck E, Bjursell G, Lindahl T. Sequence complexity of circular Epstein-Bar virus DNA in transformed cells. J Virol. 1981;40:11–19. doi: 10.1128/jvi.40.1.11-19.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bacon A, Farrington SM, Dunlop MG. Mutation frequency in coding and non-coding repeat sequences in mismatch repair deficient cells derived from normal human tissue. Oncogene. 2001;20:7464–7471. doi: 10.1038/sj.onc.1204973. [DOI] [PubMed] [Google Scholar]
- 28.Gibson SL, Narayanan L, Hegan DC, Buermeyer AB, Liskay RM, Glazer PM. Overexpression of the DNA mismatch repair factor, PMS2, confers hypermutability and DNA damage tolerance. Cancer Lett. 2006;244:195–202. doi: 10.1016/j.canlet.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 29.Meuth M, Richards B, Schneider B. The conditional mutator phenotype in human tumor cells: correction. Science. 1999;283:641. doi: 10.1126/science.283.5402.639d. [DOI] [PubMed] [Google Scholar]
- 30.Xu L, Chow J, Bonacum J, et al. Microsatellite instability at AAAG repeat sequences in respiratory tract cancers. Int J Cancer. 2001;91:200–204. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1031>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 31.Rampino N, Yamamoto H, Ionov Y, et al. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 32.Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Ruschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 33.Gangwal K, Sankar S, Hollenhorst PC, et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proc Natl Acad Sci U S A. 2008;105:10149–10154. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao X, Buermeyer AB, Narayanan L, et al. Different mutator phenotypes in Mlh1- versus Pms2-deficient mice. Proc Natl Acad Sci U S A. 1999;96:6850–6855. doi: 10.1073/pnas.96.12.6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrew SE, Xu XS, Baross-Francis A, et al. Mutagenesis in PMS2- and MSH2-deficient mice indicates differential protection from transversions and frame-shifts. Carcinogenesis. 2000;21:1291–1295. [PubMed] [Google Scholar]
- 36.Hegan DC, Narayanan L, Jirik FR, Edelmann W, Liskay RM, Glazer PM. Differing patterns of genetic instability in mice deficient in the mismatch repair genes Pms2, Mlh1, Msh2, Msh3 and Msh6. Carcinogenesis. 2006;27:2402–2408. doi: 10.1093/carcin/bgl079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kow YW, Bao G, Reeves JW, Jinks-Robertson S, Crouse GF. Oligonucleotide transformation of yeast reveals mismatch repair complexes to be differentially active on DNA replication strands. Proc Natl Acad Sci U S A. 2007;104:11352–11357. doi: 10.1073/pnas.0704695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavlov YI, Mian IM, Kunkel TA. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr Biol. 2003;13:744–748. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- 39.Erdeniz N, Dudley S, Gealy R, Jinks-Robertson S, Liskay RM. Novel PMS1 alleles preferentially affect the repair of primer strand loops during DNA replication. Mol Cell Biol. 2005;25:9221–9231. doi: 10.1128/MCB.25.21.9221-9231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gragg H, Harfe BD, Jinks-Robertson S. Base composition of mononucleotide runs affects DNA polymerase slippage and removal of frameshift intermediates by mismatch repair in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:8756–8762. doi: 10.1128/MCB.22.24.8756-8762.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bichara M, Pinet I, Schumacher S, Fuchs RP. Mechanisms of dinucleotide repeat instability in Escherichia coli. Genetics. 2000;154:533–542. doi: 10.1093/genetics/154.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLα in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 43.Fang WH, Modrich P. Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J Biol Chem. 1993;268:11838–11844. [PubMed] [Google Scholar]
- 44.Korhonen MK, Raevaara TE, Lohi H, Nystrom M. Conditional nuclear localization of hMLH3 suggests a minor activity in mismatch repair and supports its role as a low-risk gene in HNPCC. Oncol Rep. 2007;17:351–354. [PubMed] [Google Scholar]
- 45.Stone JE, Petes TD. Analysis of the proteins involved in the in vivo repair of base-base mismatches and four-base loops formed during meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics. 2006;173:1223–1239. doi: 10.1534/genetics.106.055616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cannavo E, Marra G, Sabates-Bellver J, et al. Expression of the MutL homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res. 2005;65:10759–10766. doi: 10.1158/0008-5472.CAN-05-2528. [DOI] [PubMed] [Google Scholar]
- 47.Chen PC, Dudley S, Hagen W, et al. Contributions by MutL homologues Mlh3 and Pms2 to DNA mismatch repair and tumor suppression in the mouse. Cancer Res. 2005;65:8662–8670. doi: 10.1158/0008-5472.CAN-05-0742. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.