Abstract
The intron-encoded proteins bI4 RNA maturase and aI4 DNA endonuclease can be faithfully expressed in yeast cytoplasm from engineered forms of their mitochondrial coding sequences. In this work we studied the relationships between these two activities associated with two homologous intron-encoded proteins: the bI4 RNA maturase encoded in the fourth intron of the cytochrome b gene and the aI4 DNA endonuclease (I-SceII) encoded in the fourth intron of the gene coding for the subunit I of cytochrome oxidase. Taking advantage of both the high recombinogenic properties of yeast and the similarities between the two genes, we constructed in vivo a family of hybrid genes carrying parts of both RNA maturase and DNA endonuclease coding sequences. The presence of a sequence coding for a mitochondrial targeting peptide upstream from these hybrid genes allowed us to study the properties of their translation products within the mitochondria in vivo. We thus could analyze the ability of the recombinant proteins to complement RNA maturase deficiencies in different strains. Many combinations of the two parental intronic sequences were found in the recombinants. Their structural and functional analysis revealed the following features. (i) The N-terminal half of the bI4 RNA maturase could be replaced in total by its equivalent from the aI4 DNA endonuclease without affecting the RNA maturase activity. In contrast, replacing the C-terminal half of the bI4 RNA maturase with its equivalent from the aI4 DNA endonuclease led to a very weak RNA maturase activity, indicating that this region is more differentiated and linked to the maturase activity. (ii) None of the hybrid proteins carrying an RNA maturase activity kept the DNA endonuclease activity, suggesting that the latter requires the integrity of the aI4 protein. These observations are interesting because the aI4 DNA endonuclease is known to promote the propagation, at the DNA level, of the aI4 intron, whereas the bI4 RNA maturase, which is required for the splicing of its coding intron, also controls the splicing process of the aI4 intron. We propose a scenario for the evolution of these intronic proteins that relies on a switch from DNA endonuclease to RNA maturase activity.
Full text
PDF



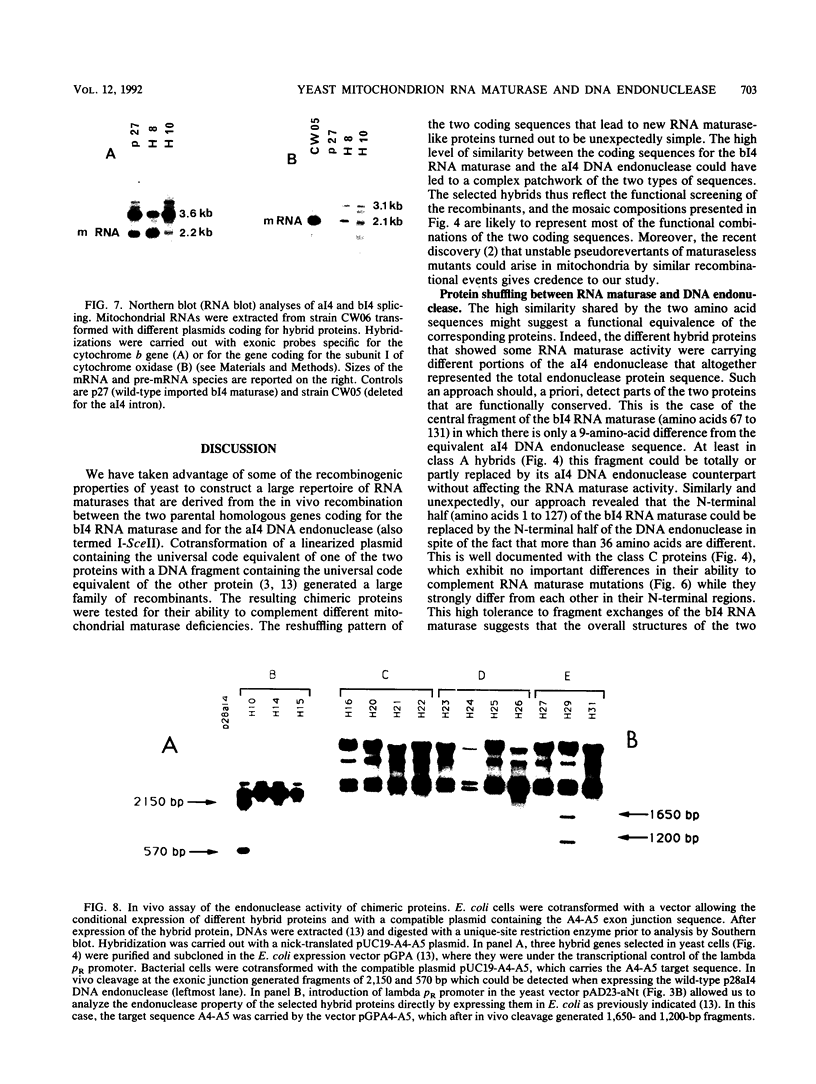


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akins R. A., Lambowitz A. M. A protein required for splicing group I introns in Neurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell. 1987 Jul 31;50(3):331–345. doi: 10.1016/0092-8674(87)90488-0. [DOI] [PubMed] [Google Scholar]
- Anziano P. Q., Moran J. V., Gerber D., Perlman P. S. Novel hybrid maturases in unstable pseudorevertants of maturaseless mutants of yeast mitochondrial DNA. Nucleic Acids Res. 1990 Jun 11;18(11):3233–3239. doi: 10.1093/nar/18.11.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banroques J., Delahodde A., Jacq C. A mitochondrial RNA maturase gene transferred to the yeast nucleus can control mitochondrial mRNA splicing. Cell. 1986 Sep 12;46(6):837–844. doi: 10.1016/0092-8674(86)90065-6. [DOI] [PubMed] [Google Scholar]
- Banroques J., Perea J., Jacq C. Efficient splicing of two yeast mitochondrial introns controlled by a nuclear-encoded maturase. EMBO J. 1987 Apr;6(4):1085–1091. doi: 10.1002/j.1460-2075.1987.tb04862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M. Self-splicing introns in prokaryotes: migrant fossils? Cell. 1991 Jan 11;64(1):9–11. doi: 10.1016/0092-8674(91)90201-9. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D., Quirk S., Clyman J., Belfort M. Intron mobility in phage T4 is dependent upon a distinctive class of endonucleases and independent of DNA sequences encoding the intron core: mechanistic and evolutionary implications. Nucleic Acids Res. 1990 Jul 11;18(13):3763–3770. doi: 10.1093/nar/18.13.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie J. U., Reidhaar-Olson J. F., Lim W. A., Sauer R. T. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science. 1990 Mar 16;247(4948):1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Selfish DNA and the origin of introns. Nature. 1985 May 23;315(6017):283–284. doi: 10.1038/315283b0. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Chapman K. B., Boeke J. D. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell. 1991 May 3;65(3):483–492. doi: 10.1016/0092-8674(91)90466-c. [DOI] [PubMed] [Google Scholar]
- Cherniack A. D., Garriga G., Kittle J. D., Jr, Akins R. A., Lambowitz A. M. Function of Neurospora mitochondrial tyrosyl-tRNA synthetase in RNA splicing requires an idiosyncratic domain not found in other synthetases. Cell. 1990 Aug 24;62(4):745–755. doi: 10.1016/0092-8674(90)90119-y. [DOI] [PubMed] [Google Scholar]
- Colleaux L., D'Auriol L., Galibert F., Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Salle H., Jacq C., Slonimski P. P. Critical sequences within mitochondrial introns: pleiotropic mRNA maturase and cis-dominant signals of the box intron controlling reductase and oxidase. Cell. 1982 Apr;28(4):721–732. doi: 10.1016/0092-8674(82)90051-4. [DOI] [PubMed] [Google Scholar]
- Delahodde A., Goguel V., Becam A. M., Creusot F., Perea J., Banroques J., Jacq C. Site-specific DNA endonuclease and RNA maturase activities of two homologous intron-encoded proteins from yeast mitochondria. Cell. 1989 Feb 10;56(3):431–441. doi: 10.1016/0092-8674(89)90246-8. [DOI] [PubMed] [Google Scholar]
- Dujardin G., Jacq C., Slonimski P. P. Single base substitution in an intron of oxidase gene compensates splicing defects of the cytochrome b gene. Nature. 1982 Aug 12;298(5875):628–632. doi: 10.1038/298628a0. [DOI] [PubMed] [Google Scholar]
- Dujon B. Group I introns as mobile genetic elements: facts and mechanistic speculations--a review. Gene. 1989 Oct 15;82(1):91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- Goguel V., Bailone A., Devoret R., Jacq C. The bI4 RNA mitochondrial maturase of Saccharomyces cerevisiae can stimulate intra-chromosomal recombination in Escherichia coli. Mol Gen Genet. 1989 Mar;216(1):70–74. doi: 10.1007/BF00332232. [DOI] [PubMed] [Google Scholar]
- Hensgens L. A., Bonen L., de Haan M., van der Horst G., Grivell L. A. Two intron sequences in yeast mitochondrial COX1 gene: homology among URF-containing introns and strain-dependent variation in flanking exons. Cell. 1983 Feb;32(2):379–389. doi: 10.1016/0092-8674(83)90457-9. [DOI] [PubMed] [Google Scholar]
- Herbert C. J., Dujardin G., Labouesse M., Slonimski P. P. Divergence of the mitochondrial leucyl tRNA synthetase genes in two closely related yeasts Saccharomyces cerevisiae and Saccharomyces douglasii: a paradigm of incipient evolution. Mol Gen Genet. 1988 Aug;213(2-3):297–309. doi: 10.1007/BF00339595. [DOI] [PubMed] [Google Scholar]
- Hurt E. C., Müller U., Schatz G. The first twelve amino acids of a yeast mitochondrial outer membrane protein can direct a nuclear-coded cytochrome oxidase subunit to the mitochondrial inner membrane. EMBO J. 1985 Dec 16;4(13A):3509–3518. doi: 10.1002/j.1460-2075.1985.tb04110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouesse M., Netter P., Schroeder R. Molecular basis of the 'box effect', A maturase deficiency leading to the absence of splicing of two introns located in two split genes of yeast mitochondrial DNA. Eur J Biochem. 1984 Oct 1;144(1):85–93. doi: 10.1111/j.1432-1033.1984.tb08434.x. [DOI] [PubMed] [Google Scholar]
- Labouesse M., Slonimski P. P. Construction of novel cytochrome b genes in yeast mitochondria by subtraction or addition of introns. EMBO J. 1983;2(2):269–276. doi: 10.1002/j.1460-2075.1983.tb01416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz A. M. Infectious introns. Cell. 1989 Feb 10;56(3):323–326. doi: 10.1016/0092-8674(89)90232-8. [DOI] [PubMed] [Google Scholar]
- Lazowska J., Claisse M., Gargouri A., Kotylak Z., Spyridakis A., Slonimski P. P. Protein encoded by the third intron of cytochrome b gene in Saccharomyces cerevisiae is an mRNA maturase. Analysis of mitochondrial mutants, RNA transcripts proteins and evolutionary relationships. J Mol Biol. 1989 Jan 20;205(2):275–289. doi: 10.1016/0022-2836(89)90341-0. [DOI] [PubMed] [Google Scholar]
- Ma H., Kunes S., Schatz P. J., Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58(2-3):201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Nakagawa K., Morishima N., Shibata T. A maturase-like subunit of the sequence-specific endonuclease endo.SceI from yeast mitochondria. J Biol Chem. 1991 Jan 25;266(3):1977–1984. [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman P. S., Butow R. A. Mobile introns and intron-encoded proteins. Science. 1989 Dec 1;246(4934):1106–1109. doi: 10.1126/science.2479980. [DOI] [PubMed] [Google Scholar]
- Pompon D., Nicolas A. Protein engineering by cDNA recombination in yeasts: shuffling of mammalian cytochrome P-450 functions. Gene. 1989 Nov 15;83(1):15–24. doi: 10.1016/0378-1119(89)90399-5. [DOI] [PubMed] [Google Scholar]
- Raboy V., Kim H. Y., Schiefelbein J. W., Nelson-Jr O. E. Deletions in a dspm insert in a maize bronze-1 allele alter RNA processing and gene expression. Genetics. 1989 Jul;122(3):695–703. doi: 10.1093/genetics/122.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargueil B., Delahodde A., Hatat D., Tian G. L., Lazowska J., Jacq C. A new specific DNA endonuclease activity in yeast mitochondria. Mol Gen Genet. 1991 Feb;225(2):340–341. doi: 10.1007/BF00269867. [DOI] [PubMed] [Google Scholar]
- Sargueil B., Hatat D., Delahodde A., Jacq C. In vivo and in vitro analyses of an intron-encoded DNA endonuclease from yeast mitochondria. Recognition site by site-directed mutagenesis. Nucleic Acids Res. 1990 Oct 11;18(19):5659–5665. doi: 10.1093/nar/18.19.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio C. Group I introns: do they only go home? Trends Genet. 1989 Jun;5(6):168–172. doi: 10.1016/0168-9525(89)90068-1. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. On the origin of RNA splicing and introns. Cell. 1985 Sep;42(2):397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Davies R. W., Scazzocchio C., Brown T. A. Internal structure of a mitochondrial intron of Aspergillus nidulans. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6332–6336. doi: 10.1073/pnas.79.20.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzlau J. M., Saldanha R. J., Butow R. A., Perlman P. S. A latent intron-encoded maturase is also an endonuclease needed for intron mobility. Cell. 1989 Feb 10;56(3):421–430. doi: 10.1016/0092-8674(89)90245-6. [DOI] [PubMed] [Google Scholar]
- Wolf K., Del Giudice L. The variable mitochondrial genome of ascomycetes: organization, mutational alterations, and expression. Adv Genet. 1988;25:185–308. doi: 10.1016/s0065-2660(08)60460-5. [DOI] [PubMed] [Google Scholar]