Abstract
Cell (2013) 152, 570–583
Dev Cell (2013) 24, 206–214
Recent articles by Klattenhoff et al (2013) and Grote et al (2013) identify long non-coding RNAs, or lncRNAs, important for specifying the cardiac lineage. Depletion of a lncRNA, aptly named Braveheart, resulted in loss of beating cardiomyocytes during embryonic stem (ES) cell differentiation and failure to activate a key network of cardiac transcription factors. Immunoprecipitation of the protein complex associated with Braveheart revealed that the lncRNA physically interacts with epigenetic machinery that regulates cardiac gene expression. Similarly, a second lncRNA, Fendrr, also interacts with epigenetic regulators to promote proper cardiac gene expression and function in vivo in mice. These studies highlight the importance of lncRNAs during lineage commitment and provide a new layer of regulation involved in determining cardiac cell fate.
In the last few years, scientific advances have changed the way we conceptualize the ‘dark matter’ of the genome: it is not junk DNA anymore. This is owing, in part, to deep sequencing technologies that expanded our view of the extent and complexity of the mammalian transcriptome. In 2012, the ENCODE project identified many novel and known non-coding RNAs (ncRNAs) and demonstrated that between 70–90% of the human genome is actively transcribed, although protein-coding genes account for only ∼1% of the genomic sequences (Dunham et al, 2012).
Based on their size and function, ncRNAs are subdivided into classes, including microRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and lncRNAs, among others (Esteller, 2011). miRNAs were initially shown to be important for cardiac differentiation and cardiac development (Zhao et al, 2005), and have subsequently been implicated in myriad events related to cardiovascular development and responses to stress (Cordes and Srivastava, 2009). In many of these cases, miRNAs are embedded in the core transcriptional circuitry, regulated by key transcription factors, and function in positive and negative feedback loops to reinforce cellular decisions. However, the functions of other classes of ncRNAs in cardiovascular biology have been relatively unexplored.
In 1991, Willard and colleagues discovered X-inactive-specific transcript Xist, involved in X inactivation (Brown et al, 1992), representing the first-described mammalian lncRNA. Twenty years of research on Xist is now being revisited as we begin to understand the thousands of newly discovered lncRNA transcripts. LncRNA transcripts are typically greater than ∼200 nucleotides in length and are 5′ capped and polyadenylated like most mRNAs. They have key roles in epigenetics, stem cell biology, cancer, and disease (Lee, 2012). Unlike miRNAs, lncRNAs generally lack strong evolutionary conservation. Like Xist, which is also poorly conserved, lncRNA sequences may be under less evolutionary constraint than protein-coding genes, allowing lncRNAs to evolve rapidly.
In one of the current papers, Klattenhoff et al identified a novel mouse-specific lncRNA transcript, Braveheart (Bvht), which is expressed in early mesodermal progenitors that give rise to the heart. Depletion of Bvht in mouse ES cells resulted in failure to activate key regulators of the cardiovascular programme, including the early mesodermal marker, Mesp1, which is important for emergence of the cardiac lineage (Lindsley et al, 2008). Consequently, mouse ES cells in which Bvht was knocked down did not differentiate into beating cardiomyocytes. The authors further showed that Bvht acts upstream of Mesp1 by interacting with chromatin-modifying complexes that mediate transcriptional repression. Specifically, Bvht interacts directly with the polycomb-repressive complex 2 (PRC2), and this interaction is necessary for PRC2 to dissociate from key developmentally regulated promoters. Removal of PRC2 from promoters of regulators of specific cell fates, such as Mesp1, is important as ES cells adopt a differentiated state. In the absence of Bvht, trimethylation of histone H3 lysine 27 (repressive mark) at the promoters of important cardiac regulatory genes persisted, leading to a failure of cardiac gene expression. In addition, Bvht is abundantly expressed in the mouse neonatal heart, and its depletion in isolated neonatal cardiomyocytes resulted in the failure to maintain cardiac sarcomeric gene expression, such as α- and β-myosin heavy chains. The in vitro data from Klattenhoff et al support an important role for Bvht in determining cardiac cell fate by regulating the epigenetic machinery required to control activation or repression of gene transcription. Whether Bvht is also required for cardiomyocyte differentiation in vivo during cardiac development awaits targeted deletion in mice.
The findings of Boyer and colleagues are similar to other reports of lncRNA interaction with PRC2 that regulate the epigenetic signature and downstream gene transcription in cells (Lee, 2012). Shortly after the Bvht report, Grote et al (2013) described another mesoderm-enriched lncRNA, Fendrr, which is expressed in the lateral plate mesoderm that gives rise to the heart and body wall muscles. Deletion of Fendrr in mice resulted in embryonic lethality owing to impaired heart function and deficits in the body wall. In addition, the authors found that Fendrr interacts with components of PRC and the trithorax group/MLL (TrxG/MLL) to regulate the epigenetic state of mesodermal genes. Unlike Bvht, the interaction of Fendrr with TrxG/MLL promotes trimethylation of histone H3 lysine 4, an activating mark, at promoters of lateral plate mesoderm-specific genes (Grote et al, 2013). These two studies represent the first-described lncRNAs involved in cardiac differentiation and development of the heart, both by epigenetically regulating the cardiac gene network.
Dynamic regulation of cardiac gene expression is important for guiding cells into their proper cardiac lineage, including cardiomyocytes, endothelial cells, smooth muscle cells, and cardiac fibroblasts (Srivastava, 2006). Subtle disruptions in this regulation lead to congenital heart defects and disease (Srivastava, 2006). While many transcription factors are key regulators of the cardiovascular programme, the miRNA class of ncRNAs also are pivotal in cell fate, differentiation, or behaviour of each cell type (reviewed in (Cordes and Srivastava, 2009)) by negatively regulating numerous mRNAs and controlling dosage of cardiac networks. The findings by Klattenhoff et al and Grote et al demonstrate that lncRNAs provide yet another layer of genomic regulation in specific cell types (Figure 1). It will be interesting to determine if there are specific lncRNAs for other cardiovascular cell types that are important for their regulation in an analogous fashion. Furthermore, it will be important to determine if there is a lncRNA counterpart of Bvht in humans, and if not, whether this might help explain species-specific differences in cardiomyocyte biology.
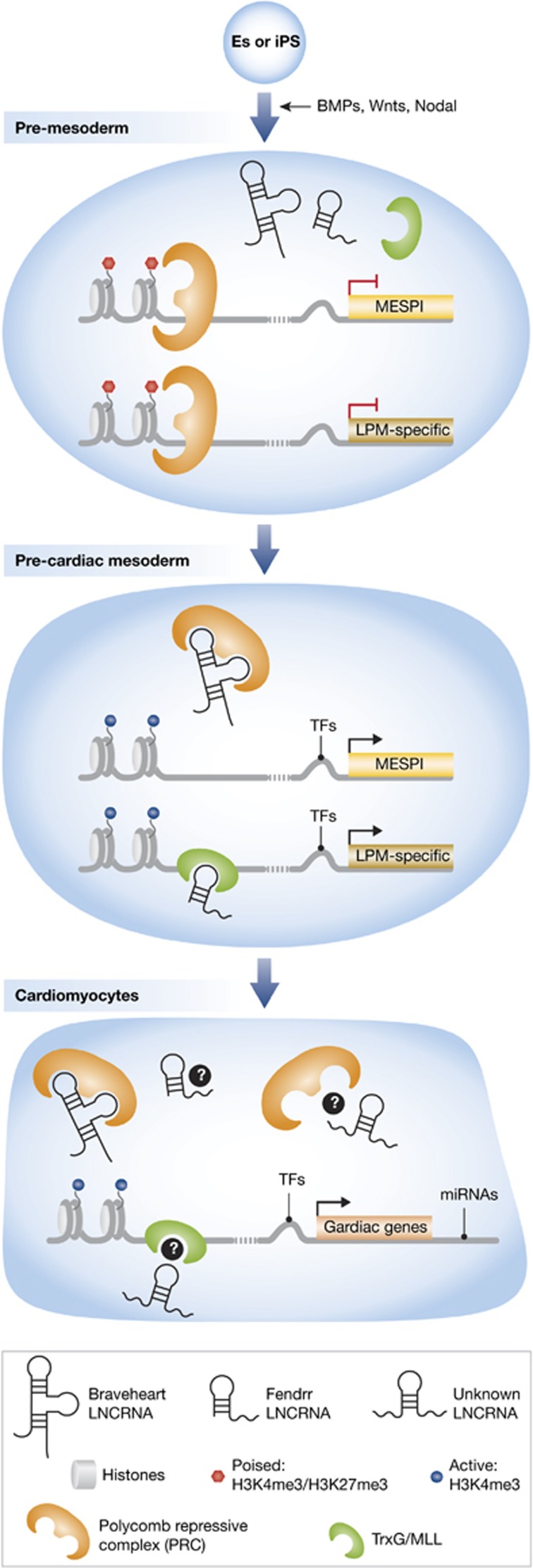
Figure 1.
Epigenetic regulation of cardiac gene expression by lncRNAs. Top, pluripotent cells (ES or iPS) are differentiated by the addition of growth factors and signals, including bone-morphogenetic proteins (BMPs), Wingless proteins (Wnt), and Nodal signals, into pre-mesodermal progenitors; in the pre-mesoderm stage, genes are poised by histone modifications and PRC binding at promoter regions; in the pre-cardiac mesoderm, transcription factors bind upstream of the Mesp1 promoter, and the lncRNA, Braveheart (Bvht), binds to the PRC and removes it from the Mesp1 promoter to activate the pre-cardiac mesoderm programme. Concurrently, the lncRNA, Fendrr, binds the activating complex, TrxG/MLL (shown above), and PRC (not shown) to allow activation of lateral plate mesoderm (LPM)-specific genes; in cardiomyocytes, TFs and miRNAs regulate cardiac-specific gene expression at the 5′ and 3′ ends, respectively. It remains in question whether Bvht, Fendrr, and possibly an unknown lncRNA may bind PRC and TrxG/MLL in cardiomyocytes to add further regulation.
It has become increasingly clear that knowledge of the critical regulators of the genome in individual cell types can be leveraged to control cell fate. Direct reprogramming of skin fibroblasts into induced-pluripotent stem cells by the core transcriptional machinery of ES cells has great promise for modelling human disease and potentially for cell transplantation therapies. To accurately recapitulate cardiac differentiation in a dish, it is important to gain a complete understanding of the layers of transcript regulation.
More recently, direct reprogramming—reprogramming fibroblasts into an alternative mature cell type without first passing through a pluripotent state—represents an exciting new alternative approach for regenerative therapies (Ieda et al, 2010; Vierbuchen et al, 2010). In particular, the use of a combination of developmental transcription factors to reprogram fibroblasts to cardiomyocyte-like cells in vitro was first reported in 2010 (Ieda et al, 2010). The reprogramming process is even more effective in an in vivo setting after injury, harnessing the vast pool of endogenous cardiac fibroblasts in the adult heart (Qian et al, 2012; Song et al, 2012). Applying new knowledge of lncRNAs during differentiation may allow for even greater efficiency of cellular reprogramming for cardiac and other lineages. The pre-requisite for such approaches will be the development of thorough genetic blueprints of the cell types of interest, as was recently reported for the cardiomyocyte lineage (Paige et al, 2012; Wamstad et al, 2012).
As more lncRNAs are investigated at a functional level, many important questions will have to be addressed. Do lncRNAs contribute to the species diversity and complexity of higher organisms? How does lncRNA interaction with PRC or other epigenetic machinery result in specificity of gene regulation? Do lncRNAs positively regulate transcription by interacting with activating complexes? What are the specific functions of sequences throughout the length of the lncRNA? What is the structure–function relationship of the transcript? The coming years will undoubtedly answer these and many other questions, and provide insight into the complex gene networks that control cell fate decisions. As Klattenhoff et al and Grote et al have begun to reveal, the answers are waiting to be discovered for those brave enough to delve into the ‘dark matter’ of our DNA.
Acknowledgments
We are grateful to G Howard and B Taylor for editorial services. DS was supported by grants from NHLBI/NIH (U01 HL100406, U01 HL098179, R01 HL057181, and P01 HL089707), the California Institute for Regenerative Medicine (CIRM), the William Younger Family Foundation, the L.K. Whittier Foundation, and the Eugene Roddenberry Foundation. KRCM is a CIRM Scholar (CIRM Training Grant TG2-01160).
Footnotes
The authors declare that they have no conflict of interest.
References
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF (1992) The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542 [DOI] [PubMed] [Google Scholar]
- Cordes KR, Srivastava D (2009) MicroRNA regulation of cardiovascular development. Circ Res 104: 724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12: 861–874 [DOI] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, Herrmann BG (2013) The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24: 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D (2010) Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA (2013) Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152: 570–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT (2012) Epigenetic regulation by long noncoding RNAs. Science 338: 1435–1439 [DOI] [PubMed] [Google Scholar]
- Lindsley RC, Gill JG, Murphy TL, Langer EM, Cai M, Mashayekhi M, Wang W, Niwa N, Nerbonne JM, Kyba M, Murphy KM (2008) Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem cell 3: 55–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, Moon RT, Stamatoyannopoulos J, Murry CE (2012) A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell 151: 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D (2012) In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485: 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, Hill JA, Bassel-Duby R, Olson EN (2012) Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485: 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D (2006) Making or breaking the heart: from lineage determination to morphogenesis. Cell 126: 1037–1048 [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M (2010) Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463: 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, Erwin G, Kattman SJ, Keller GM, Srivastava D, Levine SS, Pollard KS, Holloway AK, Boyer LA, Bruneau BG (2012) Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 151: 206–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D (2005) Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436: 214–220 [DOI] [PubMed] [Google Scholar]