Abstract
Homologous recombination (HR) is essential for genome integrity. Recombination proteins participate in tolerating DNA lesions that interfere with DNA replication, but can also generate toxic recombination intermediates and genetic instability when they are not properly regulated. Here, we have studied the role of the recombination proteins Rad51 and Rad52 at replication forks and replicative DNA lesions. We show that Rad52 loads Rad51 onto unperturbed replication forks, where they facilitate replication of alkylated DNA by non-repair functions. The recruitment of Rad52 and Rad51 to chromatin during DNA replication is a prerequisite for the repair of the non-DSB DNA lesions, presumably single-stranded DNA gaps, which are generated during the replication of alkylated DNA. We also show that the repair of these lesions requires CDK1 and is not coupled to the fork but rather restricted to G2/M by the replicative checkpoint. We propose a new scenario for HR where Rad52 and Rad51 are recruited to the fork to promote DNA damage tolerance by distinct and cell cycle-regulated replicative and repair functions.
Keywords: CDK, DNA damage tolerance, DNA checkpoints, homologous recombination, replication fork dynamics
Introduction
In every cell cycle, DNA accumulates lesions that impair the advance of the replication forks. Eventually, this leads to an accumulation of single-stranded DNA (ssDNA), which activates a number of mechanisms aimed at ensuring that DNA replication passes through DNA lesions and repairing the gaps. Defects in this response cause replication fork stalling and genetic instability in yeast (Vázquez et al, 2008; Putnam et al, 2010) and are associated with cancer in humans (Moynahan and Jasin, 2010). This DNA damage tolerance (DDT) response relies in error-prone translesion synthesis (TLS) and error-free template switch (TS) mechanisms. TLS fills the gap by extending the 3′-end past the damaged template, using specialized DNA polymerases that are able to incorporate a nucleotide opposite the lesion, while TS uses the information of the sister chromatid to bypass damage (Friedberg, 2005). A crucial protein in this decision is the DNA polymerase processivity factor PCNA, which functions as a platform for factors involved in replication, repair, and chromatin assembly (Moldovan et al, 2007). In response to DNA damage, PCNA is ubiquitinated at lysine 164 by the Rad6/Rad18 ubiquitin ligase complex (Hoege et al, 2002), and this modification serves as a target for recruiting TLS polymerases (Lehmann et al, 2007). Alternatively, the ubiquitin residue at lysine 164 can be extended with a K63-linked polyubiquitin chain by the Ubc13/Mms2/Rad5 ubiquitin ligase complex to promote TS (Hoege et al, 2002). Thus, the choice between TLS and TS mechanisms determines whether the repair is mutagenic or not. Consequently, DDT is an essential process for cell-cycle progression, genome integrity, and cancer avoidance.
Homologous recombination (HR) is also necessary for gap filling during DDT (Prakash, 1981; Friedberg, 2005; Heyer et al, 2010). The recombination proteins Rad52 and Rad51 are required for replication fork progression through alkylated DNA (Vázquez et al, 2008; Alabert et al, 2009), and in their absence cells accumulate ssDNA gaps (Lopes et al, 2006; Hashimoto et al, 2010). Since HR uses the genetic information of an intact molecule to repair a DNA break, it might provide the enzymatic activities required for TS. Accordingly, the RAD6 and RAD52 epistasis group of proteins cooperate through a mechanism that forms a sister-chromatid junction (SCJ), which is then further resolved by the helicase/topoisomerase Sgs1/Top3 complex (Liberi et al, 2005; Branzei et al, 2008; Minca and Kowalski, 2010).
The requirement for recombination proteins during DDT is in apparent contradiction with the existence of anti-recombinogenic activities during DNA replication (Fabre et al, 2002). For instance, lysine K164 can also be SUMOylated, even in the absence of DNA damage. This modification is carried out by the Ubc9/Siz1 SUMO ligase complex and recruits the helicase Srs2 to prevent HR (Hoege et al, 2002; Papouli et al, 2005; Pfander et al, 2005). In fact, the activation of the replicative checkpoint by accumulation of ssDNA inhibits HR (Lisby et al, 2004; Meister et al, 2005; Alabert et al, 2009; Barlow and Rothstein, 2009). Thus, HR is often referred to as a double-edged sword; it is necessary for DNA damage repair and tolerance but can also generate genomic rearrangements when it is not properly regulated. However, the molecular scenarios that require or preclude recombination functions are still unknown.
The HR mechanism has been extensively studied in response to DNA double-strand breaks (DSBs) (San Filippo et al, 2008). In contrast, much less is known about how HR is regulated during DDT, despite the fact that ssDNA gaps, and not DSBs, are the major lesions initiating spontaneous recombination (Fabre et al, 2002; Lettier et al, 2006). Indeed, its mode of action is still unclear. An important mechanistic question is whether gap repair by HR is coupled to the replication bypass across the lesion or whether it occurs post-replicatively. The efficiency of the response to replicative DNA damage is not affected when the expression of the group of RAD6 epistasis proteins is restricted to G2/M, suggesting that these mechanisms can operate uncoupled from the replication fork (Daigaku et al, 2010; Karras and Jentsch, 2010). The fact that not only rad18Δ, but also rad52Δ cells, accumulates ssDNA gaps behind the fork under replicative stress (Lopes et al, 2006; Hashimoto et al, 2010) supports the idea that HR can also work uncoupled from the fork. However, the recombination proteins Rad52 and Rad51 are required for replication fork progression through alkylated DNA (Vázquez et al, 2008; Alabert et al, 2009), suggesting that HR has additional, S phase-specific functions that remain to be determined.
A major handicap for studying the role of HR during DDT is the difficulty of discriminating whether a recombinogenic lesion is associated with a ssDNA gap generated by replication fork impairment or with a DSB generated by processing a non-DSB DNA lesion. Furthermore, the few assays able to detect ssDNA gaps infer the role of HR from recombination mutants (Lopes et al, 2006; Gangavarapu et al, 2007), which can sometimes be misleading since the accuracy of HR relies on metastable and reversible intermediates (Heyer et al, 2010). To overcome these problems, we have used the Chromatin Endogenous Cleavage (ChEC) method (Schmid et al, 2004) to follow the binding of recombination proteins to replication forks and to DNA lesions other than DSBs during the cell cycle. We show that Rad52 and Rad51 are recruited to replication forks, where they facilitate DNA synthesis through alkylated DNA by a repair-independent process. Strikingly, the recruitment of Rad52 and Rad51 to chromatin during DNA replication is a prerequisite for the further repair of the lesion by HR, a process that is not coupled to the fork but rather restricted to G2/M by the replicative checkpoint.
Results
Physical evidence for the recruitment of Rad52 and Rad51 to replicative DNA damage other than DSBs
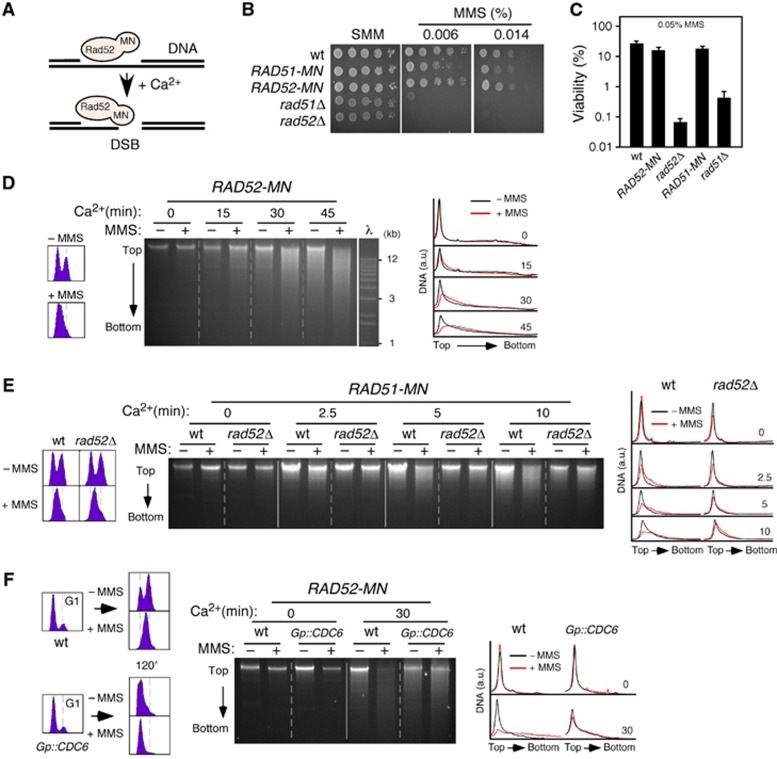
To directly address whether recombination proteins are targeted to DNA lesions other than DSBs, we took advantage of the ChEC method developed by Laemmli and colleagues to map genomic interaction sites of chromatin proteins (Schmid et al, 2004). This method relies on the expression of proteins fused to the micrococcal nuclease (MN); the nuclease domain of these chimeras can be activated with Ca2+ ions and introduce a DNA DSB if they are bound to chromatin. For this, cells are permeabilized with digitonin, which does not affect protein–DNA interactions, and treated with Ca2+ for different times (Schmid et al, 2004). The rationale behind this approach is that a repair protein fused to MN will only generate a detectable cut in the DNA if it is targeted to a lesion that is not a DSB; by contrast, if the lesion is a DSB, the cleavage by the chimera will not enhance the appearance of DSBs (Figure 1A). Therefore, we can infer that recombination proteins bind to DNA if DNA is digested upon exposure to Ca2+.
Figure 1.
Physical evidence for Rad52 and Rad51 binding to non-DSB DNA lesions generated by replication through alkylated DNA. (A) Rationale of the approach. A DNA repair protein fused to MN will induce a detectable cut only if it is bound to a DNA lesion other than a DSB. (B, C) Rad51-MN and Rad52-MN are functional in MMS-induced DDT. (B) Response to chronic MMS treatment; cell growth analysis by 10-fold serial dilutions of the same number of mid-log phase cells. (C) Response to acute high-dose MMS treatment; wild-type, RAD52-MN, RAD51-MN, rad52Δ and rad51Δ cells were grown to mid-log phase, treated or not with 0.05% MMS for 2 h and plated onto YPD medium to determine their viability. The average and s.e.m. of three independent experiments are shown. (D) Rad52 binds to DNA in response to MMS. ChEC analysis of exponentially growing RAD52-MN cells incubated with 0.05% MMS for 2 h. The left and right panels show profiles for DNA content and DNA digestion. (E) Rad51 binds to DNA in response to MMS in a Rad52-dependent manner. ChEC analysis of RAD51-MN and RAD51-MN rad52Δ cells incubated with 0.05% MMS for 2 h. (F) DNA replication is required for Rad52 binding to MMS-induced DNA damage. RAD52-MN and Gp::CDC6 RAD52-MN cells were synchronized in G2/M in YP galactose with nocodazole, incubated for 1 h in YP glucose with nocodazole to remove Cdc6 (Tercero et al, 2003), and released into G1 in YP glucose with α factor. Finally, cells were released for 2 h in YP glucose with or without 0.05% MMS and analysed by ChEC.
We fused MN to the C-terminal ends of Rad52 and Rad51 (Rad52-MN and Rad51-MN), and confirmed that these constructs were as proficient as the wild type in DNA damage repair and tolerance (Figures 1B and C; Supplementary Figure 1A). Rad52, which is essential for most HR events in yeast, binds to the 3′-ended ssDNA molecules generated by resection of a DSB and facilitates the formation of an ssDNA/Rad51 filament that mediates the search and exchange of homologous DNA sequences (San Filippo et al, 2008). To study the role of HR in repairing DNA lesions other than DSBs, cells were treated with methyl-methane sulfonate (MMS), a genotoxic agent that impairs replication fork progression by DNA alkylation (Tercero and Diffley, 2001) and that is highly toxic for cells defective in HR (Prakash and Prakash, 1977). After 2 h either with or without 0.05% MMS, cells were collected, permeabilized, and treated with Ca2+ for different times, and then total DNA was extracted and resolved in agarose gels. In the absence of Ca2+, a single, high molecular DNA band is detected on top of the gel (Figure 1D, time 0). In the absence of MMS, Rad52-MN digested DNA over a time course with Ca2+ (Figure 1D; note the gradual appearance of a smear below the top band in −MMS). Notably, the presence of MMS increased both the kinetics and extent (down to 1 kb) of DNA digestion by Rad52-MN (Figure 1D; compare+relative to −MMS at 30 and 45 min). The top band disappeared leading to a distribution of DNA fragments that peaked at ∼10 kb. This effect was not observed when we expressed only the MN (Supplementary Figure 1B). We also ruled out the possibility that the digestion was due to the fact that most MMS-treated cells remained at S phase (Figure 1D) by showing that DNA digestion by Rad52-MN in cells synchronized in G1, S, or G2/M and released in the absence of DNA damage did not change during the cell cycle (Supplementary Figure 1C). Importantly, and according to our hypothesis, introducing DNA DSBs with zeocin did not affect the kinetics of DNA cleavage by Rad52-MN (Supplementary Figure 1D).
The expression of Rad51-MN also led to a Ca2+-dependent DNA digestion in the presence of MMS, even though less pronounced than that displayed by Rad52-MN. Importantly, this digestion was abolished in rad52Δ (Figure 1E), indicating that Rad52 mediates the binding of Rad51 to chromatin. Taken together, these results provide physical evidence for the recruitment of recombination proteins to non-DSBs DNA lesions.
Next, we investigated whether Rad52 binding to MMS-induced DNA damage is dependent on replication. For this, we used cells expressing CDC6 under the control of the GAL1 promoter (Gp::CDC6). Expression of Cdc6 is essential for replication initiation but not for later cell-cycle events that depend on high levels of CDK activity (Piatti et al, 1995). Wild-type and Gp::CDC6 cells were maintained in glucose for 2 h during the synchronization steps to deplete Cdc6, after which G1 cells were released using medium with or without 0.05% MMS (Figure 1F). As reported, the absence of Cdc6 prevented DNA replication initiation and led to a ‘reductional’ cell division (Piatti et al, 1995). The absence of DNA replication also prevented Rad52-MN from binding to DNA in response to MMS, suggesting that Rad52 is recruited to non-DSBs replicative DNA lesions, presumably ssDNA gaps as suggested from Electron Microscopy studies with recombination mutants (Lopes et al, 2006; Hashimoto et al, 2010).
Recruitment of recombination proteins to MMS-induced DNA damage is coupled to DNA replication
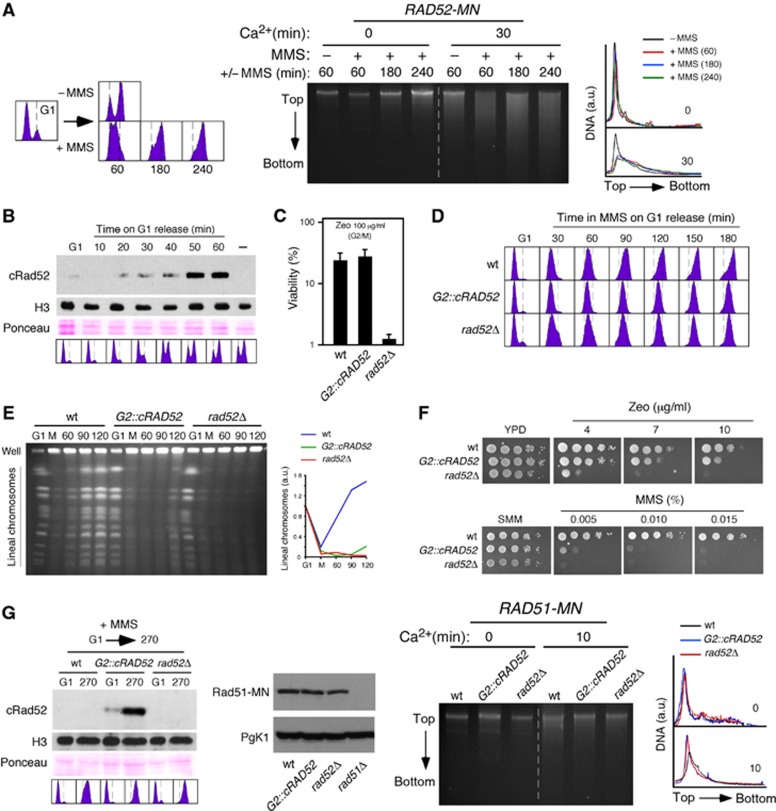
In principle, Rad51 and Rad52 could be targeted to ssDNA gaps at or behind the fork, which are generated either by uncoupling the leading and lagging strands, or by repriming DNA synthesis downstream of damage, respectively. To determine this, cells were synchronized in G1 and released in the presence of 0.033% MMS for different times, and samples were analysed for Rad52-MN DNA cleavage (Figure 2A). Rad52-MN was bound to damaged DNA 1 h upon G1 release, when most cells were at S phase, as inferred from the DNA content profile. However, even at longer time points (3 and 4 h) at which bulk DNA was largely replicated, Rad52-MN was still bound to damaged DNA. This suggests that Rad52 can operate uncoupled from the replication fork at ssDNA gaps left behind the fork.
Figure 2.
The recruitment of Rad52 and Rad51 to MMS-induced DNA damage is coupled to DNA replication. (A) Rad52 can operate uncoupled from the replication fork in response to MMS. ChEC analysis of RAD52-MN cells synchronized in G1 and released in the presence of 0.033% MMS. (B) cRad52 is expressed specifically during G2/M. Western blot analysis of cRad52 in G2::cRAD52 cells synchronized in G1 and released into fresh medium. - indicates an asynchronous culture of a wild-type strain. Ponceau S staining and histone H3 levels were used as loading controls. (C) cRad52 is functional. G2::cRAD52, rad52Δ, and wild-type cells were synchronized in G2/M, incubated with or without 100 μg/ml zeocin for 30 min, and plated onto YPD medium to determine their viability. The average and s.e.m. of four independent experiments are shown. (D, E) Rad52 is required for S-phase progression in the presence of MMS, as determined by Flow cytometry analysis of cells synchronized in G1 and released in the presence of 0.033% MMS (D), and PFGE analysis of cells synchronized in G1, released in the presence of 0.033% MMS for 1 h (M), and then release into fresh media for the indicated times (min) (E). The quantification of lineal chromosomes is shown on the right. (F) Cells cannot tolerate MMS-induced DNA damage when HR is restricted to G2/M. Zeocin and MMS sensitivity of G2::cRAD52, rad52Δ, and wild-type cells. (G) Rad52 and Rad51 recruitment to MMS-induced DNA damage is coupled to DNA replication. ChEC analysis of RAD51-MN, G2::cRAD52 RAD51-MN, and rad52Δ RAD51-MN cells synchronized in G1 and released in the presence of 0.033% MMS for 270 min. The amount of cRad52 during the kinetics and of Rad51-MN 270 min after G1 release on MMS was determined by western blot. An asynchronous culture of rad51Δ was used as a negative control. Histone H3 and Pgk1 were used as loading controls. cRad52 was detected with an antibody against Clb2.
If Rad52 can operate uncoupled from the fork, then one prediction is that HR should be able to repair DNA damage even if HR is restricted to G2/M. To address this point, we limited Rad52 expression to G2/M by inserting the promoter and degron sequences of CLB2 in front of the coding sequence of RAD52 (G2::cRAD52) (Figure 2B). The cRad52 chimera was functional, as determined by analysing the viability of cells arrested in mitosis and then treated for 30 min with zeocin (Figure 2C). As expected for cells lacking Rad52 during S phase, G2::cRAD52 cells were defective in completing DNA replication in the presence of 0.033% MMS as determined by DNA content (Figure 2D) and PFGE (Figure 2E), where only replicated chromosomes enter into the gel. The absence of Rad52 during S phase had little effect on the sensitivity to zeocin-induced DSBs (Figure 2F, top); however, in contrast to our prediction, G2::cRAD52 cells were highly sensitive to MMS (Figure 2F, bottom). This indicates that cells cannot tolerate replicative DNA damage if HR is restricted to G2/M.
The fact that recombination proteins were bound to damaged sites after replication was largely completed but could not tolerate the damage when they were expressed only in G2/M suggests that their recruitment to chromatin is coupled to DNA replication. To assess this possibility, we followed Rad51-MN binding to chromatin in G2::cRAD52 cells by ChEC analysis. We first demonstrated that both cRad52 and Rad51-MN are functional when expressed together (Supplementary Figure 2). Cells were then synchronized in G1, released in the presence of MMS, and analysed after 270 min, at which point most G2::cRAD52 cells remained arrested in mitosis and expressed cRad52 (Figure 2G). As previously shown, Rad52 was required for Rad51-MN binding to chromatin in response to MMS. Importantly, Rad51-MN binding was also prevented when Rad52 expression was restricted to G2/M (Figure 2G). This result, which cannot be explained by reduced levels of Rad51-MN in the mutant relative to the wild type (Figure 2G), demonstrates that the binding of Rad52 and Rad51 to damaged chromatin has to occur during S phase. It is worth noting that this result discards the possibility that the ChEC signal measured the binding of HR factors to ssDNA during the Ca++ treatment, since G2::cRAD52 cells in G2/M have Rad52, Rad51-MN, and ssDNA. In conclusion, our results indicate that the recruitment of recombination proteins is coupled to DNA replication and this step is essential for DDT.
The recruitment of Rad51 to replicating chromatin is required for MMS-induced HR repair
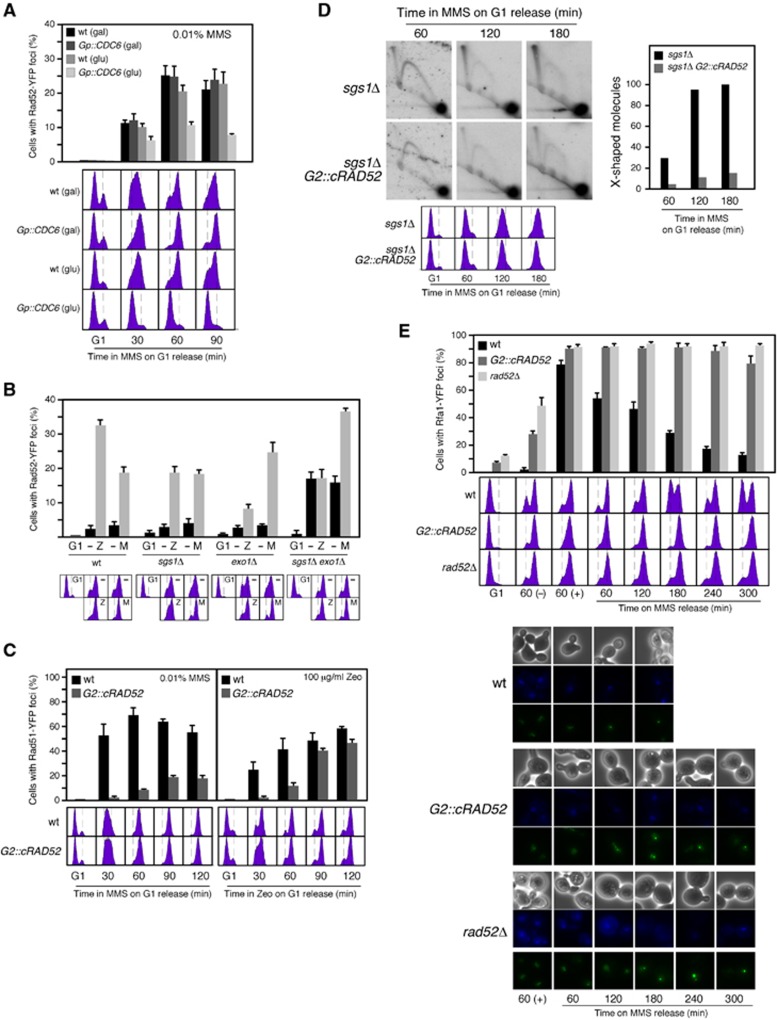
According to our previous results, the recruitment of Rad51 to chromatin during DNA replication should be a prerequisite for the recombinational repair of replicative DNA lesions. DNA damage repair by HR is associated with repair centres that can be detected with the recombination proteins Rad52 and Rad51 fused to the green fluorescence protein (Lisby et al, 2001, 2004). While Rad52-YFP is biologically functional (Lisby et al, 2001), tagging of Rad51 with the fluorescence protein reduces its repair activity (Lisby et al, 2004). We thereby worked with yeast strains expressing both the chimera and the wild-type protein, which are fully functional in the repair of MMS and zeocine-induced DNA damage (Supplementary Figure 3A). Following the aforementioned strategy with Gp::CDC6 cells, we first confirmed that, as expected for replicative DNA damage, replication inhibition in the absence of Cdc6 prevented the assembly of MMS-induced HR foci (Figure 3A).
Figure 3.
Rad51 fork recruitment is a prerequisite for MMS-induced HR repair. (A) DNA replication is required for MMS-induced Rad52-YFP foci. Rad52 foci accumulation in wild-type and Gp::CDC6 cells transformed with pWJ1344 (RAD52-YFP), synchronized in G1 as shown in Figure 1F, and released in YP glucose with 0.01% MMS. (B) Sgs1 and Exo1 are required for zeocin-induced, but not for MMS-induced HR foci. Rad52 foci accumulation in wild-type, sgs1Δ, exo1Δ, and sgs1Δ exo1Δ cells transformed with pWJ1344 (RAD52-YFP), synchronized in G1, and released in the presence of 0.01% MMS (M) or 100 μg/ml zeocin (Z) for 90 and 120 min, respectively. (C) Rad52 expression during S phase is required for MMS-induced, but not for zeocin-induced HR foci. Rad51 foci accumulation in YFP-RAD51 and G2::cRAD52 YFP-RAD51 cells transformed with pWJ1278 (RAD51), synchronized in G1, and released in the presence of 0.01% MMS (left panel) or 100 μg/ml zeocin (right panel). The average and s.e.m. are shown. (D) Rad52 expression during S phase is required for MMS-induced SCJs. 2D gel analysis of X-shaped molecules in sgs1Δ and sgs1Δ G2::cRAD52 cells synchronized in G1 and released in the presence of 0.033% MMS. The amount of X-shaped molecules relative to the total amount of molecules, taken the highest value as 100, is shown. The experiment was repeated with similar results. (E) Rad52 expression during S phase is required for ssDNA gap repair. RPA foci accumulation and FACS analysis in RFA1-YFP1, RFA1-YFP1 G2::cRAD52 and RFA1-YFP1 rad52Δ cells synchronized in G1, released in 0.01% MMS for 1 h, treated with 2.5% sodium thiosulphate to inactivate the MMS, washed and incubated in fresh medium for different times. Bright field, DAPI fluorescence, and Rfa1-YFP foci of selected cells are shown.
Since a single DSB is able to generate a Rad52 focus (Lisby et al, 2003), the foci induced by MMS might reflect the repair of residual DSBs caused by replication through alkylated DNA. To address this point, we analysed the role of Exo1 and Sgs1, required for DNA resection during DSB repair by HR (Gravel et al, 2008; Zhu et al, 2008; Mimitou and Symington, 2008), on MMS-induced HR foci. The double mutant exo1Δ sgs1Δ, and partially the single mutants sgs1Δ and exo1Δ, suppressed zeocin but not MMS-induced recombination foci (Figure 3B), suggesting that MMS-induced foci are indeed associated with non-DSBs DNA lesions. Consistently, exo1Δ sgs1Δ did not prevent the binding of Rad52 to MMS-induced DNA damage as inferred from ChEC analysis (Supplementary Figure 3B). Nevertheless, we do not rule out the possibility that ssDNA gaps may need to be enlarged for efficient DNA repair (e.g., for ssDNA/Rad51 filament formation). According with this idea, exo1Δ sgs1Δ accumulated MMS-induced recombination foci (Figure 3B) and were defective in SCJs formation (Vanoli et al, 2010) as compared to the wild type. In this frame, it is worth noting that this double mutant accumulated high levels of foci in the absence of DNA damage, supporting the idea that spontaneous recombinogenic damage is associated with replicative non-DSB DNA lesions (Fabre et al, 2002; Lettier et al, 2006).
Once established that the repair by HR of MMS-induced non-DSBs replicative lesions is associated with foci, we examined the effect of restricting Rad52 expression to G2/M on MMS-induced YFP-Rad51 foci. G2::cRAD52 and wild-type cells expressing YFP-Rad51 were synchronized in G1 and released in the presence of 0.01% MMS. The lack of Rad52 during S phase in G2::cRAD52 suppressed the formation of Rad51 foci in response to MMS (Figure 3C, left panel). This effect was not due to reduced levels of YFP-Rad51 in the mutant relative to the wild type (Supplementary Figure 3C). Therefore, the recruitment of recombination proteins to chromatin during DNA replication is a prerequisite for the further repair of the lesion. This is an important difference to DSB repair by HR, which is independent of DNA replication (Alabert et al, 2009; Barlow and Rothstein, 2009) and can occur efficiently in G2 (Ira et al, 2004) even if Rad52 is not expressed during S phase (Figures 2C and F). Accordingly, G2::cRAD52 cells synchronized in G1 and released in the presence of zeocin formed YFP-Rad51 foci in G2/M (Figure 3C, right panel; Supplementary Figure 3D).
In cells lacking Sgs1, the repair of MMS-induced DNA lesions leads to an accumulation of SCJs (X-shaped molecules), a process that requires Rad51 and Rad52 (Liberi et al, 2005). Thus, we followed the kinetics of X-shaped molecules in sgs1Δ and sgs1Δ G2::cRAD52 cells synchronized in G1 and released in the presence of 0.033% MMS. As shown in Figure 3D, the absence of Rad52 during S phase led to a dramatic drop in the amount of SCJs, suggesting that Rad52 cannot promote sister-chromatid recombination in response to non-DSB replicative lesions if it is not timely loaded during replication.
An expectation of these results is that G2::cRAD52 cells accumulated unrepaired ssDNA gaps. ssDNA molecules are coated with replication protein A (RPA; formed by Rfa1-3 subunits), and can be detected by using the biologically functional chimera Rfa1-YFP (Lisby et al, 2004). Most wild-type cells released from G1 into medium containing 0.01% MMS for 1 h accumulated Rfa1-YFP foci (∼75%), and this value was even higher in G2::cRAD52 and rad52Δ cells (∼90%) (Figure 3E, top). Inactivation of the MMS and further incubation in fresh medium led to a gradual disappearance of the RPA foci and normal cell-cycle progression in the wild type, whereas both G2::cRAD52 and rad52Δ maintained the initial fraction of cells with foci 4 h after release from MMS and remained arrested in G2/M. Of note, the pattern of RPA foci during the kinetics was independent of Rad52; multiple faint foci after 1 h in the presence of MMS that gradually ended up into 1–2 bright foci after release in fresh medium (Figure 3E, bottom). Faint speckled foci and 1–2 bright foci have been associated with replication and HR, respectively (Lisby et al, 2004; Burgess et al, 2009). These results suggest that the presence of Rad52 during S phase is required for the repair of the replicative ssDNA gaps but not for the recruitment of the DNA lesions to the repair centres.
Rad52 and Rad51 bind to replication forks regardless of the presence of DNA damage
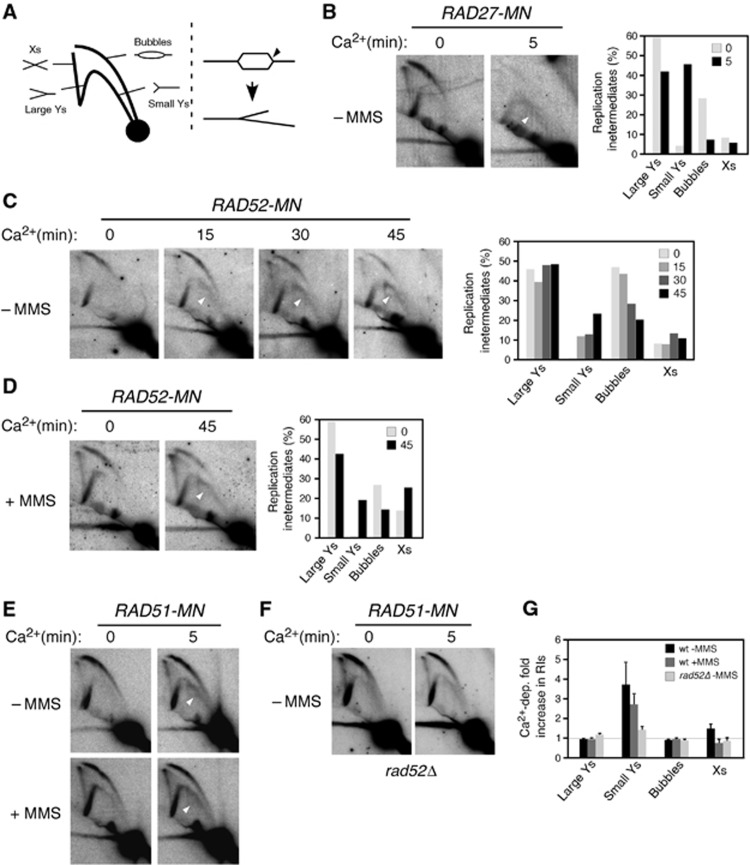
The fact that Rad52 and Rad51 have to be recruited onto chromatin during DNA replication in order to promote DDT suggests that their loading is coupled to the replication fork. To demonstrate at the molecular level that these proteins bind to forks, replication intermediates (RIs) from ChEC-treated cells were analysed by 2D-gel electrophoresis. Here, fork binding of a protein fused to MN and subsequent cleavage would generate a new population of RIs. To validate the assay, and once established that Ca++ treatment did not affect the pattern of RIs (Supplementary Figure 4A), we first analysed Rad27, a nuclease involved in the processing of Okazaki fragments (Zheng and Shen, 2011). We analysed RIs at a fragment that contains the early origin ARS305; RIs initiated from this origin formed a bubble arc that converted to a single-Y arc of large Y-shaped molecules when forks crossed the nearest restriction site (Figures 4A, left and B, −Ca2+). Activation of the nuclease activity of Rad27-MN with Ca2+ led to a huge accumulation (up to 40% of total RIs) of small Y-shaped molecules (Figure 4B, +Ca2+, open arrow). This arc is expected if the bubble is cut at one of the forks (Figure 4A, right) (Martín-Parras et al, 1992). Consistently, a drop in the total amount of bubbles (from 30 to 5%) accompanied the accumulation of small Y molecules (Figure 4B).
Figure 4.
Rad52 and Rad51 bind to replication forks regardless of the presence of MMS. (A) Schematic representation of the migration pattern of RIs by 2D gel electrophoresis (left). The expected molecule resulting from cleavage of a bubble at one of the forks is also shown (right). (B) Rad27 binds to replication forks as determined by ChEC/2D. G1-synchronized RAD27-MN cells were released into S phase for 30 min, permeabilized, and split into two samples that were treated or not with Ca2+; DNA was then extracted, digested with specific restriction enzymes, and analysed by 2D gel electrophoresis. The percentage of RIs at each time point is shown on the right. The experiment was repeated twice with similar results. (C, D) Rad52 binds to replication forks both in the absence and in the presence of MMS. G1-synchronized RAD52-MN cells were released into S phase for 30 min without (C) or with (D) 0.05% MMS, and analysed by ChEC/2D for different times as indicated in (B). The experiments were repeated with similar results (Supplementary Figure 4B). (E–G) Rad51 binds to replication forks in a Rad52-dependent manner. ChEC/2D analysis of RAD51-MN (E) and rad52Δ RAD51-MN (F) cells following the conditions indicated in (C, D). The quantification (G) shows the increase in RIs upon Ca2+ treatment relative to paralleled untreated cultures. The average and s.e.m. from five (wt-MMS), four (rad52Δ-MMS), and three (wt +MMS) independent experiments are shown. (B–F) The arrows show the accumulation of small, Y-shaped molecules upon Ca2+ activation.
Importantly, activation of the nuclease activity of Rad52-MN with Ca2+ led to a substantial accumulation of small Y-shaped molecules (up to 20% of RIs at 45 min in Ca2+) and a concomitant drop in the total amount of bubbles (from 40 to 20%) (Figure 4C; Supplementary Figure 4B, +Ca2+, open arrow). This indicates that Rad52-MN binds to, at least, ∼20% of the forks, a value that might be underestimated if the efficiency of the Rad52-MN nuclease activity at the fork is not maximal.
The cleavage of the fork by Rad52-MN could reflect the recruitment of recombination proteins to forks stalled by spontaneous DNA lesions. Strikingly, however, the presence of 0.05% MMS did not increase the amount of molecules cleaved by Rad52-MN (Figure 4D; Supplementary Figure 4B), despite the fact that this level of alkylation strongly affects replication fork advance at the analysed DNA fragment (compare the arc of large Y-shaped molecules in –MMS versus +MMS in the absence of Ca2+ in Figures 4C–E; DNA alkylation makes the right and left forks advance less symmetrically, resulting in Y-shaped molecules that extend the signal until the inflexion point of the Y-arc). This result suggests that Rad52 is recruited to the fork independently of the presence of DNA lesions.
The activation of the nuclease activity of Rad51-MN also led to an accumulation of small Y-shaped molecules both with and without MMS (3–4% of RIs) (Figures 4E and G). This value was lower than that obtained with Rad52-MN, what is consistent with the bulk DNA digestion analyses (Figure 1), and likely reflects a low Rad51-MN nuclease activity at the fork. Importantly, this accumulation of Y-shaped molecules was prevented in rad52Δ (Figures 4F and G), indicating that it was not the result of an unspecific cleavage by the MN domain, and that, as shown by previous results, Rad52 is required for Rad51 recruitment.
Finally, the fact that MMS increased bulk DNA digestion (Figure 1) but not fork cleavage (Figure 4) by Rad52-MN and Rad51-MN further supports the finding that recombination proteins remain bound to MMS-induced ssDNA gaps left behind the forks (Figure 2A). Thus, our results are consistent with Rad52 and Rad51 interacting both with the fork, likely through transient and DNA damage-independent interactions, and with ssDNA gaps generated by replicative DNA damage and left behind as the fork moves forward.
MMS-induced HR repair occurs after DNA replication
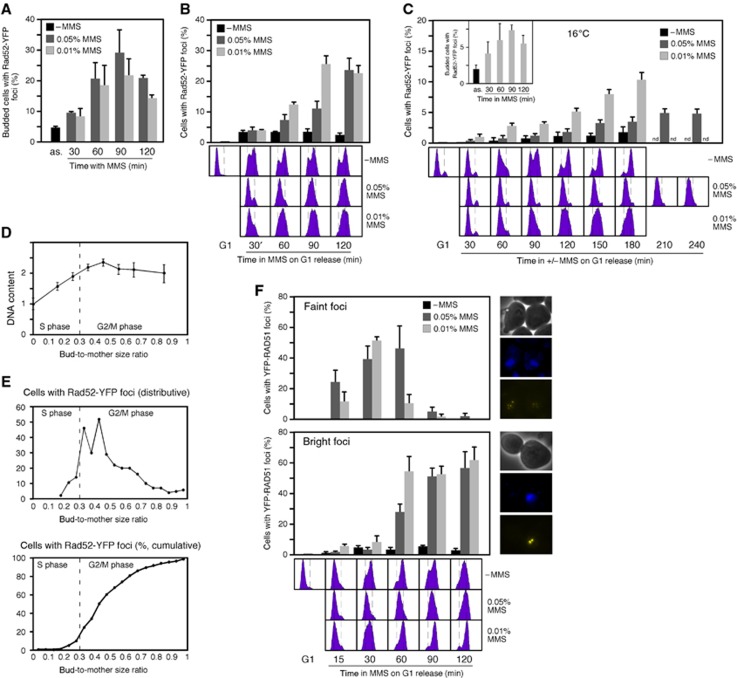
Our analysis of Rad52-YFP foci in response to 0.01% MMS contrasted with a previous study showing that G1 cells released in the presence of 0.033% MMS did not accumulate Rad52 foci (Alabert et al, 2009). However, asynchronous cultures accumulated budded cells with Rad52-YFP foci both at 0.01 and 0.05% MMS (Figure 5A). Given that DNA alkylation slows down replication (Tercero and Diffley, 2001), we hypothesized that HR foci assembly might require completion of DNA replication and therefore be delayed at high MMS concentration. To evaluate this, we analysed the frequency of cells with Rad52-YFP foci in cultures synchronized in G1 and then released in the presence of 0.05% MMS (Figure 5B). These cultures displayed a time-course accumulation of Rad52-YFP foci with a peak at 120 min after G1 release. Notably, the increase in Rad52-YFP foci was small at 60–90 min, at which time most cells were in S phase (Figure 5B), while at those times asynchronous cultures displayed a 4- to 6-fold increase (Figure 5A). In contrast, the same cultures released in the presence of 0.01% MMS progressed faster through S phase and accumulated Rad52-YFP foci earlier (Figure 5B).
Figure 5.
MMS-induced HR foci form after DNA replication. (A) Rad52 foci accumulation in response to different doses of MMS. Asynchronous cultures of wild-type cells transformed with pWJ1344 (RAD52-YFP) were grown in the presence of 0.05 or 0.01% MMS, and the percentage of budded cells with foci was determined by fluorescence microscopy. (B, C) Rad52 foci formation requires completion of DNA replication. Rad52 foci accumulation in wild-type cells transformed with pWJ1344 (RAD52-YFP), synchronized in G1 and released at either 30°C (B) or 16°C (C) into medium with 0.05 or 0.01% MMS. The inset shows the kinetics of Rad52-YFP foci of an asynchronous culture grown in the presence of 0.01% MMS at 16°C. (D) Correlation between DNA content and bud-to-mother size ratio. (E) Rad52 foci form at replication completion. Time lapse of Rad52 foci formation in wild-type cells transformed with pWJ1344 (RAD52-YFP), synchronized in G1 and released in the presence of 0.01% MMS. (F) MMS induces faint (top) and bright (bottom) Rad51 foci during S phase and G2/M, respectively. Rad51 foci accumulation in YFP-RAD51 cells transformed with pWJ1278 (RAD51), synchronized in G1, and released at 30°C into fresh medium with 0.05 or 0.01% MMS. The average and s.e.m. are shown.
To better determine the timing of Rad52-YFP foci accumulation during the cell cycle, the same experiment was conducted at 16°C to prolong the S phase (Figure 5C). At this temperature, the presence of 0.01% MMS led to an accumulation of cells with Rad52-YFP foci over the time course, with a peak at 180 min after G1 release, compared to the peak in asynchronous cultures at 90 min (Figure 5C, inset). This increase over the time course in cells with Rad52-YFP foci occurred concomitantly with an accumulation of cells with a 2C DNA content, suggesting that the assembly of MMS-induced HR repair centres requires completion of DNA replication. Accordingly, most cells synchronized in G1 and released in the presence of 0.05% MMS for 4 h at 16°C neither completed DNA replication nor accumulated Rad52-YFP foci (Figure 5C).
Finally, the timing of the appearance of Rad52-YFP foci in response to 0.01% MMS was followed in individual cells by time-lapse microscopy, using the bud-to-mother size ratio as indicator for cell-cycle progression. For this, we determined the relationship between this ratio and the DNA content, and established a ratio of 0.3 for the completion of DNA replication (Figure 5D), consistent with previous results (Lisby et al, 2004). Rad52-YFP foci appeared in cells with a bud-to-mother size ratio above 0.3 in 95% of cells (Figure 5E), further supporting the notion that HR foci assembly in response to MMS only occurs once DNA replication has concluded.
We also analysed the timing of YFP-Rad51 foci formation in response to MMS. Two types of cells with Rad51 foci were observed: (1) Cells with 1–2 bright foci that accumulated in G2/M and thereby earlier at low than at high MMS concentration (Figure 5F, bottom); these foci are similar to those observed with Rad52-YFP and are also induced by zeocin (Figure 3C). Therefore, they likely represent HR repair centres. (2) Cells with small, faintly speckled foci that accumulated during S phase in a dose-dependent manner (Figure 5F, top), and importantly, did not appear in response to zeocin (Figure 3C). The accumulation of these faint foci correlated with the ChEC signal by HR proteins that occurred in S phase in response to MMS (Figure 1). Therefore, they likely reflect the binding of HR proteins to ssDNA left behind the fork. Both types of MMS-induced YFP-Rad51 foci were suppressed when Rad52 expression was restricted to G2/M (Figure 3C, left panel). The lack of HR foci in unperturbed cells suggests that the amount of Rad52/Rad51 at forks might be insufficient and/or too diffuse to generate a detectable signal.
The replicative checkpoint prevents the assembly of MMS-induced HR repair centres during DNA replication
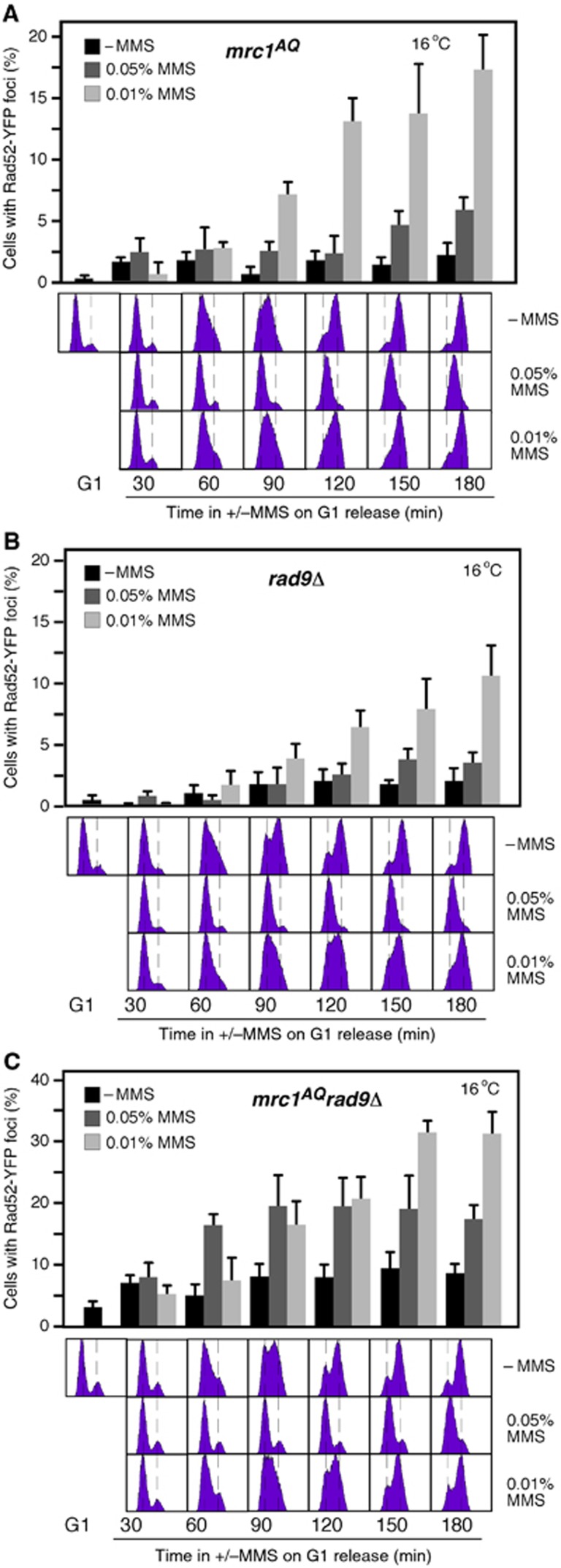
We have shown that cells prevent the assembly of HR repair centres in response to replicative DNA damage until replication is completed, despite having recombination proteins bound to the lesion. Impairment of DNA replication by MMS activates S-phase checkpoints that maintain the integrity and functionality of the advancing forks and inhibit the firing of new origins (Branzei and Foiani, 2010; Berens and Toczyski, 2012). Mrc1 is a specific component of these checkpoints that transduces signals of defective replication to downstream effectors (Alcasabas et al, 2001). We tested whether Mrc1 prevented Rad52 foci formation during S phase in response to MMS. The specific checkpoint defective mutant mrc1AQ (Osborn, 2003) did not release the inhibition on Rad52 foci formation (Figure 6A; compare with the wild type in Figure 5C); however, the DNA damage signalling is partially active in mrc1AQ cells due to the activity of Rad9, the Mrc1 counterpart in the DNA damage checkpoint pathway (Alcasabas et al, 2001). While rad9Δ displayed a similar kinetics as the wild type (Figure 6B), mrc1AQ rad9Δ cells treated with 0.05% MMS accumulated high levels of Rad52-YFP foci at 60–90 min after G1 release, at which point most cells were still in S phase (Figure 6C). Besides, Rad52 foci appeared earlier at 0.05% than at 0.01% MMS. Thus, the cell-cycle regulation of HR foci in response to MMS is lost in mrc1AQ rad9Δ cells. Of note, mrc1AQ rad9Δ cells accumulated more foci at 0.01% than at 0.05% MMS at later times (150–180 min), what might reflect the collapse of the repair centres in the absence of checkpoint activity under conditions of high replicative stress.
Figure 6.
The replicative checkpoint prevents MMS-induced HR foci during S phase. (A–C) Rad52 foci accumulation in mrc1AQ (A), rad9Δ (B), and mrc1AQ rad9Δ (C) cells transformed with pWJ1344 (RAD52-YFP), synchronized in G1, and released at 16°C into fresh medium with 0.05 or 0.01% MMS. The average and s.e.m. are shown.
It is possible that the high levels of Rad52 foci during S phase in mrc1AQ rad9Δ were a consequence of defective replication dynamics by loss of checkpoint functions. Mrc1 and Rad9 are required for the activation of the downstream effectors Rad53 and Chk1 by the checkpoint sensors Mec1 and Tel1 (Alcasabas et al, 2001). While the lack of Tel1 or Chk1 did not affect the kinetics of Rad52 foci, the lack of Mec1 or Rad53 increased the frequency of cells with Rad52 foci (Supplementary Figure 5, note the difference in the scale with the sml1Δ control). However, Rad52 foci appeared earlier at 0.01% than at 0.05% MMS and once cells reached G2/M, despite replication fork stability and dynamic are strongly affected in mec1Δ and rad53Δ (Branzei and Foiani, 2010; Berens and Toczyski, 2012). Therefore, our results suggest a role for the replicative checkpoint in preventing the formation of MMS-induced HR centres during S phase. We analysed the kinetics of Rad52 foci in sml1Δ mec1Δ tel1Δ and sml1Δ rad53Δ chk1Δ to determine whether the sensors Mec1 and Tel1 and the effectors Rad53 and Chk1 have redundant functions in the inhibition of MMS-induced HR foci during S phase. Unfortunately, the high levels of spontaneous Rad52 foci in these mutants did not allow interpreting the results (Supplementary Figure 5).
The cycle-dependent kinase activity of Cdc28 is required for MMS-induced HR repair
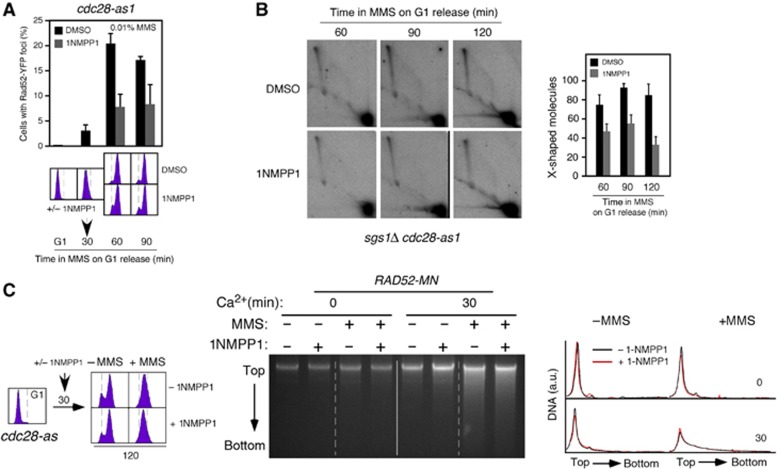
The kinase activity of Cdc28 restricts DSB repair by HR to S/G2 (Aylon et al, 2004; Ira et al, 2004; Barlow and Rothstein, 2009). To examine whether Cdc28 is also required for MMS-induced HR repair, we used cells expressing an allele of CDC28 (cdc28-as1) sensitive to the ATP analogue inhibitor 1NMPP1. G1-synchronized cdc28-as1 cells were released in the presence of 0.01% MMS, and then treated or not with 1NMPP1 after 30 min. Inhibition of the CDK activity of Cdc28 suppressed Rad52-YFP foci formation in response to MMS (Figure 7A).
Figure 7.
The kinase activity of Cdc28 is required for MMS-induced HR repair. (A) The kinase activity of Cdc28 is required for MMS-induced Rad52-YFP foci. Rad52 foci accumulation in cdc28-as1 cells transformed with pWJ1344 (RAD52-YFP), synchronized in G1, and released in the presence of 0.01% MMS. Cells were split into two cultures 30 min after G1 release that were incubated with 1NMPP1 (5 μM) or its vehicle DMSO. The average and s.e.m. are shown. (B) Cdc28 is required for MMS-induced SCJs. 2D gel analysis of X-shaped molecules in sgs1Δ cdc28-as1 cells treated as in (A) but released in the presence of 0.033% MMS. The amount of X-shaped molecules relative to the total amount of molecules, taken the highest value as 100, is shown. The average and s.e.m. of three independent kinetics are shown. (C) Rad52 binding to MMS-induced DNA damage is independent of the Cdc28 kinase activity. cdc28-as1 RAD52-MN cells were synchronized in G1 and released into S phase in the presence of 0.033% MMS for 2 h. Thirty minutes upon G1 release, cells were split into two cultures that were or not incubated with 5 μM 1NMPP1, and the binding of Rad52-MN to DNA followed by ChEC analysis.
Next, we followed the kinetics of recombination foci and X-shaped molecules in sgs1Δ cdc28-as1 cells treated as indicated in Figure 7A but at 0.033% MMS. Inhibition of Cdc28 caused a two-fold reduction in Xs and foci (Figure 7B; Supplementary Figure 6A), but did not affect DNA replication (Supplementary Figure 6B), suggesting that Cdc28 is required for the repair of the ssDNA gaps but not for the replication through alkylated DNA. Of note, the role of Cdc28 may be subsequent to Rad52 loading, because inhibition of the CDK activity of Cdc28 did not suppress Rad52-MN binding to damaged DNA as inferred by ChEC (Figure 7C).
Discussion
The alkylating agent MMS generates replicative, non-DSB recombinogenic DNA lesions
Using chimeras of recombination proteins fused to MN allowed us to show that Rad51 and Rad52 bind to non-DSBs DNA lesions, presumably ssDNA gaps, generated by replication through alkylated DNA (Figure 1). This is further supported at the cytological level by the appearance of small, faintly speckled YFP-Rad51 foci during the S phase of cells treated with MMS but not with zeocin (Figure 5F). These results support conclusions previously inferred from genetic studies with mutants defective in HR (Prakash, 1981; Lopes et al, 2006; Gangavarapu et al, 2007; Hashimoto et al, 2010). We also have shown that recombination foci induced by MMS are not associated with DSBs (Figures 3B and C). Since a single DSB is enough to generate a recombination focus (Lisby et al, 2003), our results strongly suggest that a dose of 0.05% MMS does not lead to DSBs in yeast. These results are consistent with physical analyses of DNA fragmentation showing that very high doses of MMS (0.4%) are required to induce DSBs in yeast (Jachymczyk et al, 1977; Lundin et al, 2005).
Rad52 and Rad51 bind to unperturbed forks and promote replication through alkylated DNA by repair-independent mechanisms
Rad52 and Rad51 are required for replication fork progression through alkylated DNA (Figures 2D and E) (Vázquez et al, 2008; Alabert et al, 2009), a function that is usually attributed to their DNA repair activities. In contrast to this view, we show that Rad52 is bound to MMS-induced non-DSB DNA lesions when replication is largely completed (Figure 2A) and that HR foci do not form until G2 (Figure 5). These results suggest that Rad52 and Rad51 help replication forks bypass DNA lesions by repair-independent mechanisms thus leaving ssDNA gaps behind the fork that are not repaired until S phase is completed. Along this line, Cdc28 lacking CDK activity prevented the repair but not the binding of Rad52 to replicative DNA damage, and accordingly, did not affect replication fork progression (Figures 7A and B; Supplementary Figure 6B).
According to a role assisting stressed replication forks, it was proposed from ChIP assays that Rad51 binds HU-arrested forks (Papouli et al, 2005). By the analysis of RIs from ChEC-treated cells, we present physical evidence that Rad52 and Rad51 bind to unperturbed replication forks and that this binding is not increased in the presence of MMS (Figure 4). Rad51 has been recently shown to be associated with replicating chromatin in human cell lines and X. laevis egg extracts, and in both cases, the presence of genotoxic agents hardly increased the recruitment of Rad51 (Petermann et al, 2010; Hashimoto et al, 2010, 2011). Indeed, the absence of Rad51 in Xenopus extracts and yeast led to an accumulation of ssDNA gaps at the forks that was independent of exogenous DNA damage, which was interpreted to show a replicative role of Rad51 in facilitating fork progression through natural endogenous obstacles, presumably upon recruitment to transiently uncoupled forks (Hashimoto et al, 2010). However, natural impediments are unlikely as frequent and hard to overcome as those generated by 0.05% MMS, as suggested by the impairment of fork progression at that dose (Figure 4; Tercero and Diffley, 2001). Importantly, Rad51 is not required for completion of DNA replication in the absence of genotoxic agents in yeast and Xenopus extracts (Vázquez et al, 2008; Hashimoto et al, 2010). Therefore, we propose that Rad51 is recruited to the fork independently of DNA damage, presumably through transient and highly regulated interactions. In this regard, it is worth noting that the discontinuous synthesis of the lagging strand gives rise to ssDNA/RPA nucleofilaments at the fork, which are the natural substrate for Rad52 to load Rad51 (Sung, 1997).
Our results in yeast demonstrate that Rad52 and Rad51 are required to promote DNA replication through alkylated DNA by a replicative, non-repair function. Recent works in human cell lines have shown that BRCA2 and Rad51 participate in the restart of HU-stalled replication forks by blocking DNA degradation by the nuclease Mre11 (Petermann et al, 2010; Schlacher et al, 2011). Notably, this function does not require their repair activities but rather the formation of stable Rad51/ssDNA filaments (Schlacher et al, 2011). It is therefore possible that a replicative function has been evolutionarily conserved for Rad51 and their mediators Rad52 and BRCA2, aimed at protecting transiently stalled forks. In this regard, the DDT response in bacteria relies on the formation of a RecA/ssDNA nucleofilament by RecFOR, the counterpart of Rad52 (Bichara et al, 2011). Of note, impairment of DNA replication by MMS in cells in which Rad52 expression is restricted to G2/M did not lead to recombination foci (Figure 3C, left), indicating that ‘unprotected’ forks in the presence of MMS do not generate an alternative substrate for HR. Actually, rad52Δ does not lead to the breakage of the ssDNA gaps generated at forks uncoupled by UV light (Lopes et al, 2006). Thus, an alternative but not mutually exclusive possibility is that Rad52 and Rad51 might facilitate the repriming of DNA synthesis downstream of the lesion.
Repair by HR of the DNA lesions generated during DDT is not coupled to the replication fork
An important mechanistic question for elucidating the mode of action of processes that tolerate replicative DNA damage is whether or not they are coupled to the fork. We detected Rad52 binding to MMS-induced replicative non-DSBs lesions not only during S phase but also when the bulk DNA was largely replicated (Figure 2A). In fact, MMS-induced repair foci did not appear until replication was completed (Figure 5). These results are consistent with an accumulation of ssDNA gaps behind the forks of rad52Δ and rad51Δ cells treated either with MMS or with UV light (Lopes et al, 2006; Hashimoto et al, 2010), and support a post-replicative model of DNA repair by HR during DDT. Likewise, the RAD6 DDT pathway can operate behind the fork (Daigaku et al, 2010; Karras and Jentsch, 2010); thus, our results provide a temporal framework for models that propose that the RAD6 and RAD52 pathways cooperate in the repair of replicative DNA damage (Branzei et al, 2008; Minca and Kowalski, 2010).
Rad52 and Rad51 binding to replicating chromatin is a prerequisite for HR repair during DDT
Despite the fact that ssDNA gaps are repaired post-replicatively, cells that express Rad52 only in G2/M were defective in DDT (Figures 2D–F and 3C–E). We show that this defect was due to the fact that Rad51 cannot be loaded into the lesion (Figure 2G). Therefore, Rad52 and Rad51 binding to chromatin during DNA replication is a prerequisite for further repair of the ssDNA gaps. This remarkable finding marks an important mechanistic distinction to DSB-induced HR, during which recombination proteins are recruited to the lesion in a replication-independent manner (Alabert et al, 2009; Barlow and Rothstein, 2009), and establishes a new scenario for studying HR and genome integrity. Though we cannot formally exclude the possibility that the loading of Rad52 and Rad51 onto ssDNA lesions is not coupled to the fork but occurs at a time window and/or cell-cycle stage, the fact that Rad52 loads Rad51 onto the fork regardless of the presence of DNA damage and that these proteins are required for replication fork progression through alkylated DNA led us to propose that the recruitment of Rad52 and Rad51 for DDT is coupled to the fork. We propose a model where the loading of Rad52 and Rad51 onto ssDNA gaps may be a direct consequence of repriming DNA synthesis downstream of the blocking lesion (Figure 8). If the recombination proteins are not loaded in a timely manner, then the gaps left behind the fork cannot act as a substrate for HR and remain unrepaired, although our analysis of RPA1 foci suggests that the lesions are recruited to the repair centres (Figure 3E). An important consequence of the coupling of recombination proteins to the fork is that ensures an error-free repair of the replicative DNA lesions. Furthermore, the fact that Rad52 and Rad51 cannot be directly targeted to ssDNA gaps may preclude recombination proteins from ssDNA intermediates generated during alternative repair processes, such as nucleotide excision repair, base excision repair, or mismatch repair.
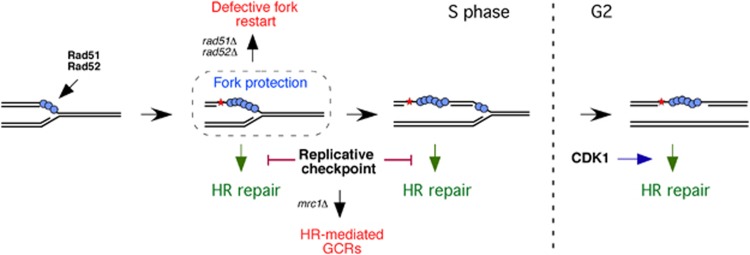
Figure 8.
Model for the roles and cell-cycle regulation of Rad52 and Rad51 in DDT. Rad51 and Rad52 bind to ssDNA/RPA1 filaments at advancing replication forks. When DNA adducts, such as alkylated bases, block the fork, Rad52 and Rad51 facilitate fork bypass by unknown replicative functions. Repriming of DNA synthesis downstream of the DNA lesion leaves Rad52 and Rad51 loaded at the ssDNA left behind the fork, which promote DNA repair in G2/M in a process that requires the CDK activity of Cdc28. In the meantime, the replicative checkpoint prevents gap repair and the accumulation of HR-mediated GCRs. Importantly, Rad52 and Rad51 binding to the forks during DNA replication is a prerequisite for the further repair of the ssDNA gaps.
MMS-induced recombinational repair is cell-cycle regulated by the replicative checkpoint and the CDK activity of Cdc28
The checkpoint-specific functions of Mrc1 and Rad9 prevent the assembly of MMS-induced HR repair centres during S phase (Figure 6). This inhibition is not lost in mec1Δ and rad53Δ (Supplementary Figure 6), which are strongly affected in replication fork stability and dynamics (Branzei and Foiani, 2010; Berens and Toczyski, 2012). This observation, together with the fact that Mrc1 is not required for the maintenance and restart of MMS-stressed forks (Tourrière et al, 2005), makes unlikely that the accumulation of MMS-induced HR foci during S phase was due to defective replication dynamic in mrc1AQ rad9Δ. Instead, we propose that the Mrc1 branch of the S-phase checkpoint restricts HR repair during DDT to G2/M. Likewise, activation of the replicative checkpoint inhibits the repair of DSBs (Alabert et al, 2009; Barlow and Rothstein, 2009) and HU-stalled forks (Lisby et al, 2004; Meister et al, 2005; Barlow and Rothstein, 2009) by HR. We extend this inhibitory role to replicative ssDNA gaps, an unexpected result if we keep in mind that Rad52 and Rad51 are required for DNA replication in the presence of MMS despite the fact that this drug activates the checkpoint (Vázquez et al, 2008; Alabert et al, 2009). Our results provide an explanation to this apparent contradiction by showing that Rad52 and Rad51 have two cell cycle-regulated functions and only the repair one is inhibited by the replicative checkpoints. We therefore propose that a general function of the S-phase checkpoint is to inhibit HR repair, likely because stressed forks are more susceptible to unscheduled recombination and/or because the repair at HR centres might interfere with replication. Besides, restricting HR to G2/M ensures the presence of an identical template to repair the damage. Consistently, the Mrc1-mediated checkpoint prevents HR-dependent gross chromosomal rearrangements (GCRs; Putnam et al, 2009).
We show that Cdc28 regulates ssDNA gap repair (Figures 7A and B). Cdc28 is also required for DSB repair by HR (Aylon et al, 2004; Ira et al, 2004; Barlow and Rothstein, 2009). Therefore, HR is cell-cycle regulated by the replicative checkpoint and Cdc28. However, while Mrc1 and Cdc28 act by preventing or promoting, respectively, Rad52 recruitment at DSBs (Alabert et al, 2009; Barlow and Rothstein, 2009), they may operate at a step subsequent to Rad52 binding to ssDNA gaps (Figure 7C). In the case of DSBs, Mrc1 and Cdc28 act by regulating the resection of the 5′-ended strands (Aylon et al, 2004; Ira et al, 2004; Huertas et al, 2008; Alabert et al, 2009; Chen et al, 2011). Likewise, Cdc28 and Mrc1 might regulate ssDNA gap repair by controlling DNA resection if Rad51-mediated strand invasion requires a minimal amount of ssDNA. An alternative but not mutually exclusive possibility is that the checkpoint promoted the anti-recombinogenic activities of Srs2 and/or Sgs1, which suppress HR at earlier steps and also prevent HR-mediated GCRs (Putnam et al, 2009). Finally, the replicative checkpoint might be required for targeting the lesions to the repair centres. Further experiments will be necessary to elucidate the mechanisms by which CDK and replicative checkpoints coordinate DNA replication and HR.
In conclusion, the cell-cycle regulation of the replicative and repair functions of Rad52 and Rad51 is essential to prevent unscheduled recombination events and may determine the timing and mode of action of other DDT activities. Understanding of these mechanisms should help to clarify the complex and yet unclear genetic interactions between the RAD52 and RAD6 group of proteins.
Materials and methods
Yeast strains, growth conditions, plasmids, and standard methods are included in Supplementary materials and methods and Supplementary Figure 7.
In vivo ChEC analysis
ChEC of native cells was performed as reported (Schmid et al, 2004) from 50 ml cultures grown under different conditions and arrested with sodium azide (0.1% final concentration). Similar results were obtained by directly processing the cultures. For cleavage induction, digitonin-permeabilized cells were incubated with 2 mM CaCl2 at 30°C under gentle agitation. Total DNA was isolated (see Supplementary data) and resolved into 0.8% TAE 1 × agarose gels. Gels were scanned in a Fuji FLA5100, and the signal profile quantified using ImageGauge. The area of the DNA digestion profiles was equalized to eliminate DNA loading differences. Each ChEC experiment was repeated three times (except for Figure 7C, Supplementary Figures 1 and 3, which were repeated twice) with similar results.
Analysis of RIs
To analyse the RIs shown in Figures 3D and 7B, cells from a 100-ml culture were arrested with sodium azide (0.1% final concentration) and cooled down in ice, and total DNA was isolated with the G2/CTAB protocol as previously described (Clemente-Ruiz and Prado, 2009). To analyse the RIs shown in Figure 4; Supplementary Figure 4 (ChEC/2D analysis), total DNA was isolated as detailed in Supplementary data. DNA samples were digested with EcoRV and HindIII, resolved by neutral/neutral two-dimensional gel electrophoresis, blotted onto HybondTM-XL membranes, and analysed by hybridization with the 32P-labelled probes Or (Figure 4; Supplementary Figure 4) or A (Figures 3D and 7B; Clemente-Ruiz and Prado, 2009). All signals were quantified in a Fuji FLA5100 with the ImageGauge analysis program.
Analysis of Rad52-YFP foci
To detect HR foci cells were transformed with plasmids pWJ1344, pWJ1213 (expressing RAD52-YFP), or pWJ1278 (expressing RAD51; for YFP-Rad51 foci analysis), fixed with formaldehyde as described (Monje-Casas et al, 2007) and visualized with a Leica CTR6000 fluorescence microscope. A total number of 300 cells were analysed for each time point. The average and s.e.m. of three independent experiments performed with three different transformants, using two independent transformation of each strain, are shown. The timing of Rad52-YFP foci appearance was followed in individual cells by time-lapse microscopy, using the bud-to-mother size ratio as an indicator of cell-cycle progression (see Supplementary data).
Supplementary Material
Acknowledgments
We thank UK Laemmli, S Jentsch, S J Elledge, F Monje-Casas, JA Tercero, P San-Segundo, and RE Wellinger for various strains and reagents; E Vigueras for helpful discussion about the analysis of RIs; and P San-Segundo, P Huertas, and F Cortés-Ledesma for critical reading of the manuscript. RG-P and MJC-L were recipients of pre-doctoral training grants (FPI) from the Spanish Ministry of Science. AMM-C was a recipient of a contract of the Juan de la Cierva program from the Spanish government. Research was funded by the Spanish Ministry of Science (BFU2006-08336 and BFU2009-09036) and the Junta de Andalucia (P07-CVI-032000).
Author contributions: RG-P performed ChEC, CHEF, 2D gels, and recombination foci and viability analyses, and contributed to planning and interpreting experiments; AM-C performed westerns and recombination foci analyses, and contributed to interpreting experiments; MJC-L performed 2D gels, westerns, and foci and viability analyses; FP coordinated the project, planned and designed experiments, interpreted results, wrote the manuscript and assembled the figures.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alabert C, Bianco JN, Pasero P (2009) Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. EMBO J 28: 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ (2001) Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3: 958–965 [DOI] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M (2004) The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 23: 4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Rothstein R (2009) Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase. EMBO J 28: 1121–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens TJ, Toczyski DP (2012) Keeping it together in times of stress: checkpoint function at stalled replication forks. Mol Cell 45: 585–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichara M, Meier M, Wagner J, Cordonnier A, Lambert IB (2011) Postreplication repair mechanisms in the presence of DNA adducts in Escherichia coli. Mutat Res 727: 104–122 [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2010) Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M (2008) SUMOylation regulates Rad18-mediated template switch. Nature 456: 915–920 [DOI] [PubMed] [Google Scholar]
- Burgess RC, Lisby M, Altmannova V, Krejci L, Sung P, Rothstein R (2009) Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J Cell Biol 185: 969–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Niu H, Chung W-H, Zhu Z, Papusha A, Shim EY, Lee SE, Sung P, Ira G (2011) Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol 18: 1015–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Ruiz M, Prado F (2009) Chromatin assembly controls replication fork stability. EMBO Rep 10: 790–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigaku Y, Davies AA, Ulrich HD (2010) Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 465: 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F, Chan A, Heyer W-D, Gangloff S (2002) Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci USA 99: 16887–16892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC (2005) Suffering in silence: the tolerance of DNA damage. Nat Rev Mol Cell Biol 6: 943–953 [DOI] [PubMed] [Google Scholar]
- Gangavarapu V, Prakash S, Prakash L (2007) Requirement of RAD52 Group Genes for Postreplication Repair of UV-Damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol 27: 7758–7764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V (2010) Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17: 1305–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Puddu F, Costanzo V (2011) RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat Struct Mol Biol 19: 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W-D, Ehmsen KT, Liu J (2010) Regulation of Homologous Recombination in Eukaryotes. Annu Rev Genet 44: 113–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Huertas P, Cortés-Ledesma F, Sartori AA, Aguilera A, Jackson SP (2008) CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachymczyk WJ, Chlebowicz E, Swietlinska Z, Zuk J (1977) Alkaline sucrose sedimentation studies of MMS-induced DNA single-strand breakage and rejoining in the wild type and in UV-sensitive mutants of Saccharomyces cerevisiae. Mutat Res 43: 1–10 [DOI] [PubMed] [Google Scholar]
- Karras GI, Jentsch S (2010) The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 141: 255–267 [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM (2007) Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair 6: 891–899 [DOI] [PubMed] [Google Scholar]
- Lettier G, Feng Q, de Mayolo AA, Erdeniz N, Reid RJD, Lisby M, Mortensen UH, Rothstein R (2006) The role of DNA double-strand breaks in spontaneous homologous recombination in S. cerevisiae. PLoS Genet 2: e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M (2005) Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev 19: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R (2003) Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol 5: 572–577 [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA 98: 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM (2006) Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21: 15–27 [DOI] [PubMed] [Google Scholar]
- Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman ASH, Helleday T (2005) Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res 33: 3799–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Parras L, Hernández P, Martínez-Robles ML, Schvartzman JB (1992) Initiation of DNA replication in ColE1 plasmids containing multiple potential origins of replication. J Biol Chem 267: 22496–22505 [PubMed] [Google Scholar]
- Meister P, Taddei A, Vernis L, Poidevin M, Gasser SM, Baldacci G (2005) Temporal separation of replication and recombination requires the intra-S checkpoint. J Cell Biol 168: 537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minca EC, Kowalski D (2010) Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Mol Cell 38: 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan G-L, Pfander B, Jentsch S (2007) PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A (2007) Kinetochore orientation during meiosis is controlled by aurora B and the monopolin complex. Cell 128: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M (2010) Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature 11: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn AJ (2003) Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev 17: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD (2005) Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell 19: 123–133 [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T (2010) Hydroxyurea-Stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37: 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Moldovan G-L, Sacher M, Hoege C, Jentsch S (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nat Cell Biol 37: 428–433 [DOI] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K (1995) Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J 14: 3788–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash L (1981) Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol Gen Genet 184: 471–478 [DOI] [PubMed] [Google Scholar]
- Prakash L, Prakash S (1977) Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics 86: 33–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hayes TK, Kolodner RD (2009) Specific pathways prevent duplication-mediated genome rearrangements. Nature 460: 984–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hayes TK, Kolodner RD (2010) Post-replication repair suppresses duplication-mediated genome instability. PLoS Genet 6: e1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257 [DOI] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M (2011) Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145: 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Durussel T, Laemmli UK (2004) ChIC and ChEC. Mol Cell 16: 147–157 [DOI] [PubMed] [Google Scholar]
- Sung P (1997) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem 272: 28194–28197 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Longhese MP, Diffley JFX (2003) A central role for DNA replication forks in checkpoint activation and response. Mol Cell 11: 1323–1336 [DOI] [PubMed] [Google Scholar]
- Tourrière H, Versini G, Cordón-Preciado V, Alabert C, Pasero P (2005) Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell 19: 699–706 [DOI] [PubMed] [Google Scholar]
- Vanoli F, Fumasoni M, Szakal B, Maloisel L, Branzei D (2010) Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genet 6: e1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez MV, Rojas V, Tercero JA (2008) Multiple pathways cooperate to facilitate DNA replication fork progression through alkylated DNA. DNA Repair 7: 1693–1704 [DOI] [PubMed] [Google Scholar]
- Zheng L, Shen B (2011) Okazaki fragment maturation: nucleases take centre stage. J Mol Cell Biol 3: 23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung W-H, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.