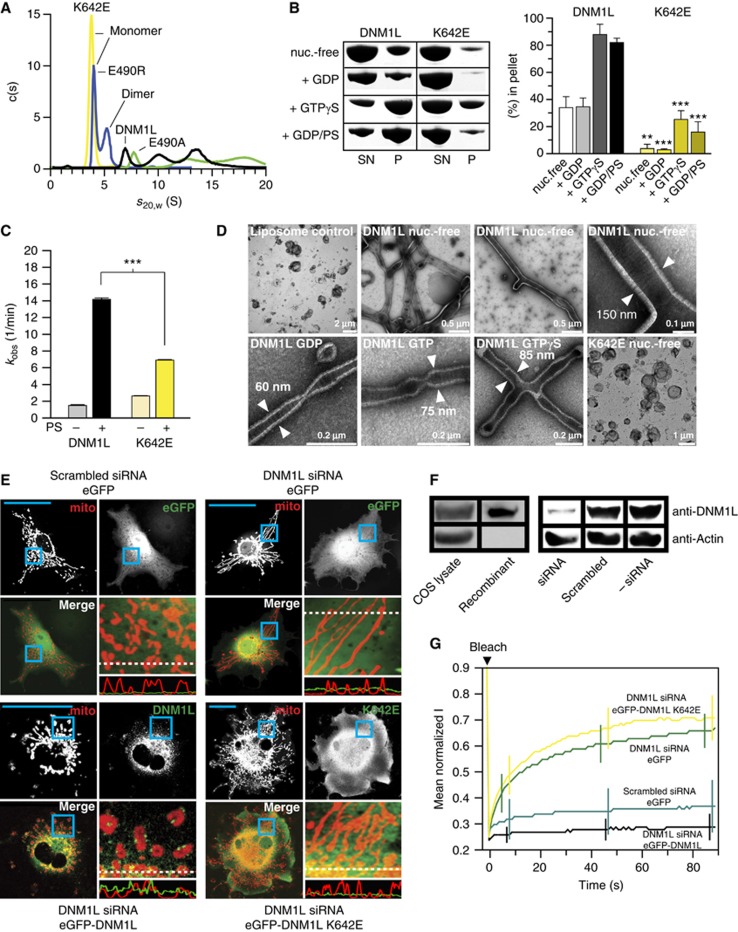
Figure 2.
Dimerization via the stalk mediates oligomerization and mitochondrial remodelling. (A) Analytical ultracentrifugation sedimentation velocity experiments for DNM1L and the indicated interface-2 mutants as described in Figure 1B. Monomer and dimer peaks for E490R and K642E are indicated. As DNM1L, the E490A mutant undergoes rapid exchange reactions between different oligomeric species. (B) Left panel: Sedimentation experiments and liposome binding assays for DNM1L and DNM1L K642E. Sedimentation experiments were performed in the absence and presence of 2 mM GTP-γ-S in the absence of liposomes. Liposome co-sedimentation assays were carried out in the presence of 2 mM GDP, and in the presence or absence of PS liposomes. Lanes from SDS–PAGE are representative for three independent experiments. Boxed lanes are from the same gel. SN, supernatant. P, pellet fraction. Right panel: Quantification of sedimentation and liposome binding assays (error bars represent the s.e.m.). Bars show the percentage of protein found in the pellet with respect of total protein applied on gel. The statistical significance was calculated with respect to the corresponding DNM1L experiments. ***P<0.001; **P<0.01; *P<0.05 (also for all subsequent statistical analyses). (C) Basal and PS liposome-stimulated GTPase activities of DNM1L and the K642E mutant were determined at 37°C (n=2 for each experiment, error bars represent the s.e.m.). The statistical significance was calculated with respect to the corresponding DNM1L experiments. (D) Negative-stain electron microscopic analysis of DNM1L in the presence of PS liposomes and different nucleotides. The K642E mutant did not show any tubulation in the absence and presence of nucleotides. (E) Cellular localization and mitochondrial morphology studies in mito-dsRed expressing COS-7 cells. Cells depleted of DNM1L by siRNA were co-transfected with GFP, siRNA-resistant GFP-DNM1L or GFP-DNM1L K642E, respectively. Scrambled siRNA and co-transfected GFP were used as a control. Magnified boxed regions and a line scan plot with the relative fluorescence of the indicated GFP constructs and mito-dsRed are shown to the lower right of each subpanel. Scale bars: 50 μm. (F) Western blot showing efficient siRNA-mediated knock down of endogenous DNM1L. Scrambled siRNA was used as a control. Actin was stained as a loading control. Antibody efficiency was monitored using a COS cell lysate and recombinant DNM1L in a separate western blot. (G) FRAP assay for mitochondrial network connectivity. Mito-dsRed in an ROI (d=6 μm) containing multiple mitochondria was photobleached and its fluorescence recovery monitored for 90 s. Curves show mean values from 20 independent experiments under the indicated conditions. Prebleach intensities were normalized. For clarity, only three representative error bars are shown for each experiment.
Source data for this figure is available on the online supplementary information page