Abstract
MAP1B, a structural microtubule (MT)-associated protein highly expressed in developing neurons, plays a key role in neurite and axon extension. However, not all molecular mechanisms by which MAP1B controls MT dynamics during these processes have been revealed. Here, we show that MAP1B interacts directly with EB1 and EB3 (EBs), two core ‘microtubule plus-end tracking proteins’ (+TIPs), and sequesters them in the cytosol of developing neuronal cells. MAP1B overexpression reduces EBs binding to plus-ends, whereas MAP1B downregulation increases binding of EBs to MTs. These alterations in EBs behaviour lead to changes in MT dynamics, in particular overstabilization and looping, in growth cones of MAP1B-deficient neurons. This contributes to growth cone remodelling and a delay in axon outgrowth. Together, our findings define a new and crucial role of MAP1B as a direct regulator of EBs function and MT dynamics during neurite and axon extension. Our data provide a new layer of MT regulation: a classical MAP, which binds to the MT lattice and not to the end, controls effective concentration of core +TIPs thereby regulating MTs at their plus-ends.
Keywords: EBs, MAP1B, microtubule dynamics, neuronal development, +TIPs
Introduction
Microtubule (MT) dynamics is crucial during neuronal morphogenesis, allowing rapid morphological changes in neurons in response to extracellular signals. MT dynamics is regulated by different types of MT-associated proteins (MAPs). Among them are classical MAPs, which bind and stabilize MTs along their entire length and MT plus-end tracking proteins (+TIPs) that accumulate specifically at MT growing-ends (reviewed in Amos and Schlieper, 2005).
MT-associated protein 1B (MAP1B) is a classical MAP, highly expressed during neuronal development (Tucker et al, 1988, 1989). MAP1B plays important roles during neurite and axon extension, neuronal migration and growth cone navigation, downstream of different signalling pathways (reviewed in Gonzalez-Billault et al, 2004). MAP1B involvement in these processes partly relies on its function as a regulator of MT stability and dynamics. Although MAP1B promotes MT nucleation, polymerization and stabilization, in vitro and in vivo, different studies indicate that MAP1B is a weaker MT stabilizer than two other classical MAPs, tau and MAP2 (Takemura et al, 1992; Pedrotti and Islam, 1995; Vandecandelaere et al, 1996). MAP1B has been shown to mainly bind to dynamic MTs and its ectopic expression in non-neuronal cells (CHO or COS-7) leads to an increase in the population of dynamic MTs (Goold et al, 1999). In developing neurons, MAP1B is prominently located in growing axons, mainly at their distal part and in growth cones, where the proportion of dynamic MTs is very high (Black et al, 1994). Hippocampal neurons from Map1b hypomorphous mice present a reduced proportion of dynamic MTs in the distal part of the axon that correlates with a delay in axon outgrowth (Gonzalez-Billault et al, 2001). In addition, downregulation of MAP1B by RNA interference in cultured cortical neurons leads to slower growing axons and altered MT growth speed in axons (Tymanskyj et al, 2012). It is therefore likely that MAP1B modulates MT dynamics in neurons, but the molecular mechanisms involved are not clear.
The end-binding (EB) protein family consists of three members (EB1–3) and is viewed as the ‘core’ +TIP family (reviewed in Galjart, 2010), since EB1/3 track MT ends autonomously and hence these proteins mark all growing MTs (Lansbergen and Akhmanova, 2006; Bieling et al, 2007, 2008; Dixit et al, 2009; Komarova et al, 2009; Zimniak et al, 2009). Virtually every known +TIP interacts with EB1/3 and many of them require EB1-like proteins for plus-end tracking. In addition, many +TIPs interact with each other at MT plus-ends (reviewed in Galjart, 2010). During neuronal morphogenesis, EB1/3 (as well as other +TIPs) are present in all neuronal compartments, indicating the existence of local MT polymerization throughout the neuron (Stepanova et al, 2003). In differentiating neuroblastoma cells, EB1 regulates MT growth rate, growth distance and duration and its downregulation leads to a reduction in neurite length (Stepanova et al, 2010). Of the three family members, EB3 is predominantly expressed in brain, in particular in neurons (Nakagawa et al, 2000). EB3 is enriched in growth cones and is involved in the coordination of the interaction between F-actin and dynamic MTs during neuritogenesis (Geraldo et al, 2008). Hence, EBs (EB1/3) function as local regulators of MT dynamics during neuronal development.
We hypothesized that MAP1B and EB1/3 might act in a cooperative manner to regulate MT dynamics during neurite and axon outgrowth. Our results show that overexpression of MAP1B in neuroblastoma cells results in decreased binding of EBs to MT plus-ends. Reciprocally, MAP1B knockdown increases EB1/3 binding to MT growing-ends in correlation with an increase in MT growth speed. Immunofluorescence analyses, co-immunoprecipitation, pull-down and FRAP assays reveal that MAP1B interacts with EBs and sequesters these +TIPs in the cytosol. We provide evidence for an enhanced binding of EB1/3 to MTs and an altered EB3 behaviour in axons and growth cones of MAP1B-deficient neurons. This is reflected in changes in MT growth speed and direction, as well as an increase in MT pausing and looping, which correlate with a delay in axon outgrowth. In summary, we provide molecular insight into how MAP1B regulates locally MT dynamics during neuronal development via its direct interaction with EB1 and EB3 proteins in the cytosol and how this contributes to proper neurite/axon extension.
Results
MAP1B and EB1/3 localize in neurites and growth cones of differentiating neuronal cells
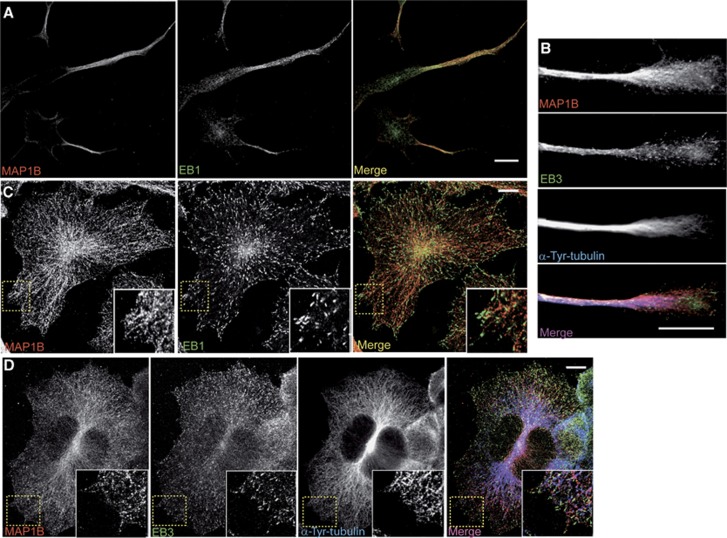
We started analysing the localization of MAP1B and EB1/3 in differentiating mouse neuroblastoma N1E-115 cells, which flatten and elongate neurites upon serum withdrawal. Confocal pictures showed that MAP1B and EB1/3 localized prominently in extending neurites and growth cones (Figures 1A and B). As seen in flat cells, MAP1B localized along the lattice of dynamic (tyrosinated) MTs, whereas EBs accumulated in comet-like dashes at MT plus-ends (Figures 1C and D). These results show that MAP1B and EBs (EB1 and EB3) are enriched in elongating neurites and growth cones and are present at growing (dynamic) MTs in differentiating neuronal cells.
Figure 1.
Localization of MAP1B and EB1/3 in serum-starved N1E-115 neuroblastoma cells. Confocal images of differentiating N1E-115 cells, either bearing neurites (A, B) or flat (C, D). In (A, C), cells were co-stained with anti-MAP1B (N-t, red) and anti-EB1 (green) and in (B, D), cells were triple stained with anti-MAP1B (N-t, red), anti-EB3 (green) and anti-α-Tyrosinated-tubulin (blue). Scale bars=10 μm.
Overexpression of MAP1B results in impaired binding of EBs to MT plus-ends
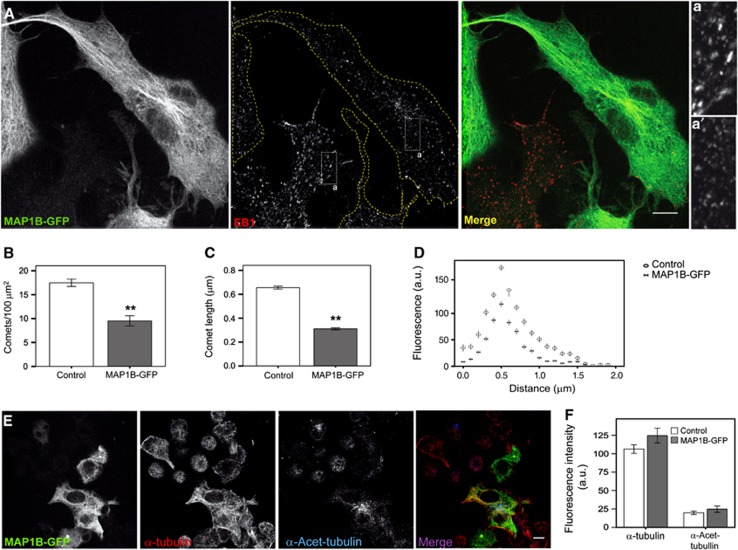
To examine the function of MAP1B on MT dynamics and neurite outgrowth, we first analysed localization of EB proteins in differentiating neuroblastoma cells ectopically expressing GFP-tagged MAP1B. MAP1B-GFP overexpression at low to medium levels led to a significant reduction (∼2-fold) in EB1 comet number (Figures 2A and B). Remaining comets were substantially reduced in length (∼2-fold) (Figures 2A and C) and in overall fluorescence intensity, as compared to comets in control cells (Figure 2D), indicating that EB1 binding to MT plus-ends was impaired by an excess of MAP1B. Remarkably, overexpression of GFP-tagged tau—another classical neuronal MAP—at low to medium levels, did not alter significantly EB1 localization at MT plus-ends (Supplementary Figures S1A–C). Although MAP1B-GFP and GFP-tau localized both in the cytosol and along the MT lattice, cytosolic localization was more prominent for MAP1B-GFP (Figure 2A; Supplementary Figure S1A). Moreover, ectopic expression of moderate levels of MAP1B or tau in N1E-115 cells did not significantly increase MT numbers or bundling (Figures 2A, E and F; Supplementary Figure S1D). Expression of GFP-tau led to a slight rise in MT stabilization (Supplementary Figure S1D). These data show that MAP1B overexpression displaces EB1 from MT plus-ends and indicate that this effect is specific for MAP1B and is not mediated by its actions on MTs. These results suggest a novel phenomenon, namely that crosstalk exists between a classical MAP and a ‘core’ +TIP in neuronal cells.
Figure 2.
MAP1B overexpression displaces EBs from MT plus-ends. (A) Confocal pictures of N1E-115 cells transfected with MAP1B-GFP (green) and stained with an anti-EB1 antibody (red). Scale bar=10 μm. Insets show details of EB1 comets in a control (non-transfected) (a) versus an MAP1B-GFP transfected cell (a′). (B) EB1 comets per 100 μm2 in control cells and cells transfected with MAP1B-GFP. (C) Average length of EB1 comets. Error bars in (B, C) are s.e.m. Number of comets measured was n=419 (control) and n=415 (MAP1B-GFP transfected). (D) Average fluorescence intensity profiles of EB1 dashes in control and MAP1B-GFP-transfected cells. (E) Confocal images of N1E-115 cells transfected with MAP1B-GFP (green). Total MTs were visualized with anti-α-tubulin (red) and stable MTs with anti-α-Acetylated-tubulin (blue). (F) Quantification of the average fluorescence intensity (±s.e.m.) of the whole MT network (α-tubulin) and of stable MTs (α-Acetylated-tubulin) in control and MAP1B-GFP-expressing cells. No significant changes in MT density or stability were observed in transfected cells. **P<0.005.
MAP1B downregulation enhances binding of EBs to MTs
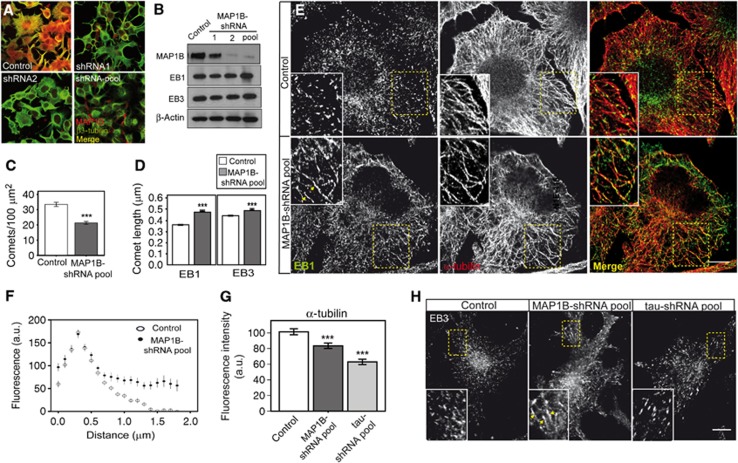
To confirm whether MAP1B interplays with EB1/3 in neuronal cells, we knocked down MAP1B expression and analysed EB1/3 localization in MAP1B-depleted cells. Different shRNA constructs were used to generate N1E-115 cell lines stably depleted of MAP1B. A control cell line was generated by using a scramble shRNA construct. MAP1B levels were reduced to different extents, as verified by immunofluorescence and western blot analysis (Figures 3A and B). EB1 and EB3 levels remained unaffected (Figure 3B). MAP1B-deficient cells presented a significantly reduced amount of EB comets (Figure 3C) in correlation with an enhanced accumulation of EB1/3 at MT plus-ends (Figures 3D–F and H) and a diminished MT density (Figures 3E and G). Moreover, EB1/3 partly localized on MT segments (Figures 3E and H), which, due to their length and the equal fluorescence intensity distribution of EB1/3 along the segments, were clearly distinct from MT comets. A similar increased interaction of EB1 with the MT lattice was found in cells overexpressing medium to high levels of EB1 (Supplementary Figures S2A–C). Thus, MAP1B knockdown leads to a reduction in MT density along with a decrease in EBs comet number and a prominent increase in the binding of EB proteins to MT growing-ends and to the MT lattice. Remarkably, interaction of EB1/3 with MTs was not enhanced in tau-depleted cells (Figure 3H; Supplementary Figures S3A and B), which also presented a reduction in MT density (Figure 3G; Supplementary Figure S3C), indicating that the effect of MAP1B on EBs localization is specific, direct and not due to its action on the MT lattice. Collectively, these results indicate that the interaction of EBs with MTs is—at least partly—dependent on MAP1B levels.
Figure 3.
MAP1B stable downregulation enhances binding of EB1/3 to MTs. N1E-115 stable cell lines generated by lentiviral infection with either scramble shRNA (control) or different specific MAP1B-shRNAs (1, 2, or a pool of 1+2+3). Reduction in MAP1B levels was confirmed by immunocytochemistry (A) (anti-MAP1B (N-t, red) and anti β3-tubulin (green)) and western blot (B), in which MAP1B, EB1 and EB3 levels were analysed and β-actin was used as a loading control. (C) Quantification of average number of EB1 comets/100 μm2 (±s.e.m.) in control and MAP1B-knocked down cells. (D) Average comet length (±s.e.m.) of EB1 or EB3-positive MT tails in control and MAP1B-deficient cells. Number of comets measured in each experimental condition was (a) EB1 staining; n=877 (control), n=1176 (shRNA pool); (b) EB3 staining; n=1497 (control), n=1165 (shRNA pool). Number of cells counted were n=26 (control) and n=55 (shRNA pool). (E) Confocal immunofluorescence pictures of control and MAP1B-silenced cells, co-stained with anti-EB1 (green) and anti-α-tubulin (red). Scale bar=10 μm. Details are presented in insets. (F) Average fluorescence intensity profile of EB1 dashes in control and MAP1B-depleted cells. EB1 interaction is enhanced upon MAP1B knockdown. (G) Quantification of the fluorescence intensity of the MT network (α-tubulin) (±s.e.m.) in control cells and in cells deficient in either MAP1B or tau. (H) Confocal pictures of control (scramble), MAP1B-depleted and tau-depleted cells, stained with anti-EB3. EB3-positive comets and segments are shown at higher magnification in insets. Scale bar=10 μm. Arrowheads in (E, H) point to MT segments highlighted by EB1 (E) or EB3 (H). ***P<0.0005.
Source data for this figure is available on the online supplementary information page.
MAP1B interacts directly with EB1 and EB3
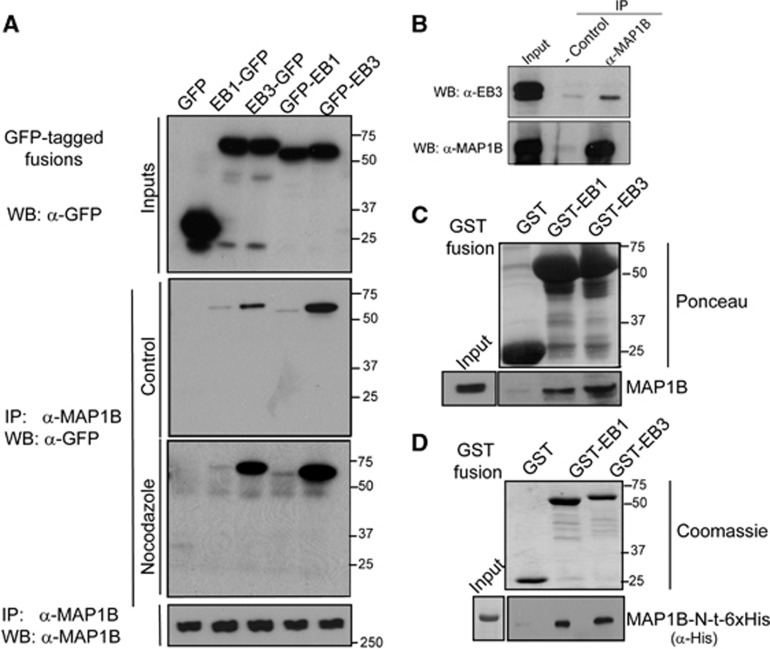
Since our results indicate that the localization of EB1/3 is regulated by MAP1B, we addressed whether MAP1B interacted with these proteins. To test this hypothesis, we first overexpressed EB1 or EB3 fused to GFP (with the tag located either at the N- or at the C-terminus of the proteins) in N1E-115 cells, which were then differentiated by serum starvation, and performed co-immunoprecipitation (co-IP) assays using an antibody against the N-terminal region of MAP1B (Figure 4A). GFP-tagged EB proteins but not GFP, which was used as a negative control, did co-IP with MAP1B. Although expression levels of all GFP-tagged EB constructs were comparable (see input lanes in Figure 4A), clear differences in pull-down efficiencies of EBs with MAP1B were found. In fact, regardless of the position of the GFP tag, EB3 was consistently more abundant in the complex with MAP1B than EB1 (Figure 4A). N-terminal tagging of GFP significantly impairs EBs accumulation at MT plus-ends, whereas the C-terminal tag alters EBs binding to protein partners containing a CAP-GLY domain (such as CLIP-170) (Skube et al, 2010). In our assays, although both GFP-tagged versions of EBs did co-IP with MAP1B, GFP-EBs co-IPed better with MAP1B than EBs-GFP. This suggests that either the interaction is favoured when EBs are not localized at MT plus-ends or that the C-terminal tag partly impairs binding of EB1/3 with MAP1B.
Figure 4.
MAP1B interacts directly with EB1 and EB3. (A) N1E-115 cells were transfected with GFP (control) or GFP-tagged EB1 or EB3, either at their N- or C-terminal regions. Cells were serum starved overnight and co-IP assays were performed with an antibody against MAP1B (N-t). Expression of each construct was confirmed by western blot using anti-GFP (Inputs), and IP of MAP1B was corroborated with anti-MAP1B (lower blot). Co-IP of GFP-tagged EBs with MAP1B in control or Nocodazole-treated cells (10 μM, 20 min) was confirmed by western blot using an anti-GFP antibody. (B) Co-IP of endogenous MAP1B and EB3 proteins from E18 mouse brain lysates. (C) MAP1B from E18 mouse brain lysates was pulled down with GST-EB1 and GST-EB3 but not with GST (control). (D) In vitro pull-down assays of MAP1B-N-t (1–508)-6x-His with either GST or GST-EBs. The MAP1B-N-t (1–508) fragment interacts directly with GST-EBs but not with GST. Expression of each construct was confirmed and shown by Ponceau staining of the nitrocellulose membrane (C) or by Coomassie staining of the acrylamide gel (D) (n=3 in each case).
Source data for this figure is available on the online supplementary information page.
To test whether the interaction between MAP1B and EB1/3 was MT dependent, we treated neuroblastoma cells overexpressing GFP-tagged EBs with Nocodazole (10 μM, 20 min) to induce complete MT depolymerization and release of EBs and MAP1B into the cytosol (Supplementary Figures S4A–C). Co-IP assays (Figure 4A) and confocal microscopy (Supplementary Figure S4C) revealed enhanced interaction between EB3 and MAP1B in Nocodazole-treated cells. Thus, formation of the MAP1B/EB protein complex does not require MT integrity and is favoured when both proteins are present in the cytoplasm. Moreover, endogenous EB3 and MAP1B co-IPed in lysates from E18 mouse brains (Figure 4B), indicating that the complex MAP1B/EB3 occurs in vivo. In addition, MAP1B from E18 mouse brain extracts was pulled-down with both GST-EB1 and GST-EB3 but not with GST alone (Figure 4C).
Interaction of MAP1B with MTs is regulated by phosphorylation by different kinases, such as proline-directed kinases including glycogen synthase kinase-3 (GSK-3) and cyclin-dependent kinase-5 (Cdk5) (Garcia-Perez et al, 1998; Goold et al, 1999), and non proline-directed kinases such as casein kinase 2 (CK2) (Ulloa et al, 1993). We tested whether the formation of the MAP1B/EB3 complex was regulated by phosphorylation by any of these kinases. Co-IP assays showed that binding of EB3-GFP to MAP1B was enhanced in cells treated with inhibitors of GSK-3 (Lithium, 20 mM, 3 h) or Cdk5 (Roscovitine, 20 μM, 3 h) but was not altered in cells treated with a CK2 inhibitor (DMAT, 10 μM, 3 h) (Supplementary Figure S5A). The interaction between endogenous MAP1B and EB3 was also increased upon Cdk5 inhibition (Supplementary Figure S5B). These results indicate that binding of MAP1B to EB3 is regulated by phosphorylation mediated by proline-directed kinases.
We then examined whether the interaction between MAP1B and EBs was direct, by performing in vitro pull-down assays using recombinant GST-EBs and an N-terminal fragment of MAP1B (aa 1–508) with a 6x-His tag. Pull-down assays using glutathione beads showed that (this fragment of) MAP1B interacted directly with both EB1 and EB3 (Figure 4D). Taken together, these results indicate that EB1/3 and MAP1B directly interact and are found in a complex in the cytosol of developing neuronal cells. Although MAP1B interacts with both EB1 and EB3, binding with EB3 is consistently more prominent than with EB1 in cells, in embryonic brain and in vitro.
MAP1B sequesters EBs in the cytosol
Our results suggest that MAP1B might control the localization of EB proteins through their sequestration in the cytosol. To test this hypothesis, we analysed whether overexpression of N-terminal MAP1B (aa 1–508) mimicked the effects of full-length MAP1B on EB1 localization. MAP1B 1–508 does not contain the MT binding domain. (Togel et al, 1998) and as shown above this MAP1B fragment interacts directly with EB proteins (Figure 4D). Ectopically expressed MAP1B 1–508-Myc showed a diffuse cytoplasmic distribution (Supplementary Figures S6A and B) and had no effect on MT density (Supplementary Figures S6B and C). However, cells expressing Myc-tagged MAP1B 1–508 presented a prominent diffuse cytosolic localization of EB1, concomitant with a significant reduction in both the number and length of EB1 comets (Supplementary Figures S6A, D and E), similar to the decrease found in cells overexpressing full-length Myc-tagged MAP1B (Supplementary Figures S6D and E). These results indicate that EB proteins are sequestered by MAP1B in the cytosol.
EB3 cellular mobility is modulated by MAP1B
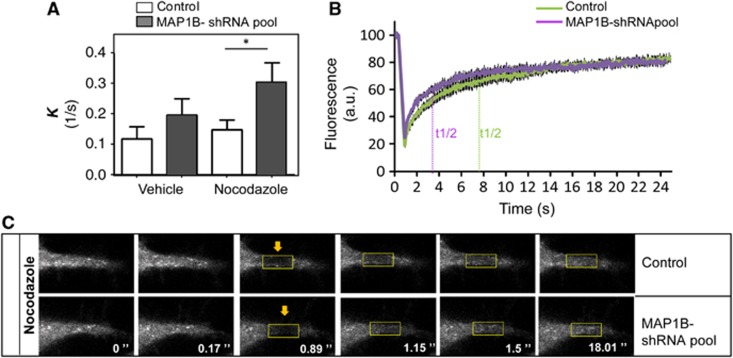
The dynamic behaviour of EB1-like proteins is determined by diffusion and a combination of protein–protein interactions, including fast exchange at MT plus-ends (Dragestein et al, 2008). Since MAP1B preferentially interacts with EB3, we focused on EB3 in the rest of our study. To check whether the direct interaction with MAP1B affects EB3 cellular dynamics during neurite extension, we performed fluorescence recovery after photobleaching (FRAP) assays. Bleaching was done in the distal part of extending neurites of differentiating control or MAP1B-deficient neuroblastoma cells, expressing low levels of EB3-GFP. Since binding reactions with other proteins affect the slow phase of the fluorescence recovery curve, we focused on this region in the FRAP analysis. Interestingly, EB3-GFP fluorescence recovery was faster in MAP1B-depleted cells than in control cells (kcontrol-Vehicle=0.1172±0.0393s−1; kMAP1B-sh-RNA pool-Vehicle=0.1954±0.0531s−1 and t1/2-control-Vehicle=11.70±3.335 s; t1/2-MAP1B-shRNA pool-Vehicle=7.7078±2.079 s) (Figure 5A, Vehicle; t1/2=ln(2)/K). These results indicate that EB3 mobility is increased in the absence of MAP1B. Consistent with these findings, MAP1B-GFP overexpression had opposite effects, with a slight retardation in fluorescence recovery of EB3-mCherry (Supplementary Figure S7). These results indicate that the interaction between EB3 and MAP1B in the distal part of growing neurites results in lowered EB3 mobility.
Figure 5.
EB3 mobility in distal neurites is regulated by MAP1B. Analysis of slow FRAP in control (scramble) and stably MAP1B-depleted (MAP1B-shRNA pool) N1E-115 cells. Cells were transfected with EB3-GFP, serum starved overnight, and either treated with vehicle (DMSO 0.1%, 1 h) or Nocodazole (10 nM, 1 h) and subjected to FRAP in distal neurites. Data collected from three different sets of experiments (∼8–15 cells/experiment) were normalized and fitted with a two-phase association equation. (A) k (1/s) corresponding to the slow phase of fluorescence recovery in vehicle-treated and Nocodazole-treated cells. (B) Curves of averaged actual FRAP data in control and MAP1B-depleted cells, upon Nocodazole treatment. (C) Representative example of time lapses of FRAP in distal neurites of control and MAP1B-silenced cells, treated with Nocodazole. *P<0.05.
Because our biochemical results indicated that the interaction between MAP1B and EBs occurs preferentially in the cytosol, we assessed EB3-GFP dynamics in the presence of Nocodazole. In this experiment, Nocodazole was used at low concentrations (10 nM) to avoid neurite retraction. At this concentration, Nocodazole altered MT dynamics, inducing the removal of most EB3 from MT growing-ends and releasing it in the cytoplasm, as described (Jaworski et al, 2009). Addition of Nocodazole increased EB3-GFP mobility, both in control and in MAP1B-knocked down cells, as shown by an increased k (kControl-Noc.=0.1463±0.0328s−1 and kMAP1B-shRNA pool-Noc.=0.3025±0.0650s−1) and a reduced half-time of fluorescence recovery (t1/2-Control-Noc.=7.7047±1.578 s and t1/2-MAP1B-shRNA pool-Noc.=3.578±0.6378, s) (Figure 5A, Nocodazole, and Figure 5B). Remarkably, Nocodazole enhanced the effect of lack of MAP1B on EB3 dynamics, accelerating fluorescence recovery of EB3-GFP further (Figures 5A–C; Supplementary Figure S8). Taken together, these results confirm that the interaction between MAP1B and EB3 is functional and that MAP1B controls the dynamics/mobility of cytosolic EB3. Moreover, the fact that Nocodazole treatment does not inhibit but exacerbates the effect of MAP1B deficiency on EB3 mobility, supports our biochemical findings showing that complex formation between MAP1B and EB3 occurs in the cytosol.
MAP1B regulates EB3 dynamics at MT plus-ends
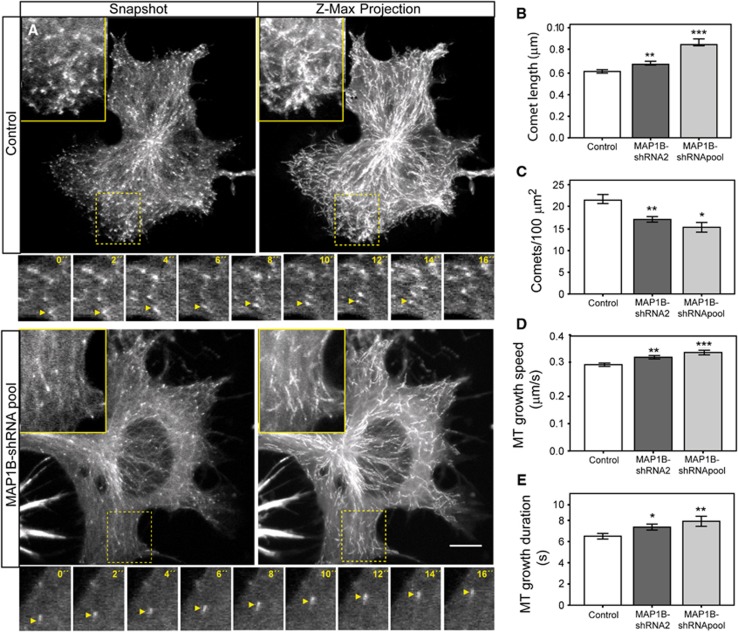
Our results indicate that in differentiating neuronal cells, EB3 mobility and binding to MT plus-ends are negatively regulated by MAP1B. To analyse whether MAP1B controlled MT dynamics at plus-ends, we performed time-lapse confocal experiments in control and MAP1B-depleted N1E-115 cells, transfected with EB3-GFP. EB3-GFP comet displacements (and therefore growing MTs) were manually tracked in living cells. In agreement with the results obtained with endogenous proteins in fixed cells, living cells depleted of MAP1B, presented longer EB3 dashes than control ones (snapshots and insets in Figure 6A and quantification in Figure 6B; Supplementary Movies S1 and S2). MAP1B-silenced cells also displayed a reduction in the density of growing MTs (snapshots and time-lapse insets in Figure 6A and Supplementary Movies S1 and S2), as evidenced by the decrease in the number of EB3-GFP comets per 100 μm2 (Figure 6C). This effect was more prominent close to cell edges, which presented a high concentration of EB3 dashes in control cells but not in depleted cells (snapshots, Z-max projections and insets in Figure 6A).
Figure 6.
MAP1B regulates EB3 dynamics at MT plus-ends. EB3-GFP was transfected into control (scramble) or MAP1B-depleted (with shRNA2 or a pool of shRNAs 1–3) neuroblastoma cells. Cells were serum starved overnight and EB3-GFP displacements were analysed. Examples of still images of control and knocked down cells are shown in (A) (Snapshot). Pictures were taken every 2 s for 30 frames. Z-maximal projections of the whole time-lapse series are shown in (A). Scale bar=10 μm. Details are shown in insets. A fragment of a representative time lapse of EB3-GFP-comet tracking (0–16 s) is shown for control and MAP1B-depleted cells in small insets under the main pictures (A). Arrows follow displacements of individual comets in the selected frames. EB3-GFP comets were manually tracked and different parameters were measured in control and MAP1B-silenced cells. (B) EB3-GFP comet length (μm). (C) Number of comets per 100 μm2. In (B, C), number of comets measured was n=382 (control), n=303 (MAP1B-shRNA2) and n=150 (MAP1B-shRNA pool). (D) MT growth speed (μm/s). Number of EB3-GFP comets tracked in each case was n=187 (control), n=189 (MAP1B-shRNA2) and n=77 comets (MAP1B-shRNApool). (E) Duration of MT growth events (s). Average values±s.e.m. are represented. *P<0.05, **P<0.005, ***P<0.0005.
Moreover, MT plus-end tracking analysis indicated that MAP1B silencing induced a significant increase in MT growth speed (insets with details of time-lapse recordings in Figure 6A; graph in Figure 6D). Furthermore, EB3-GFP comet displacements could be followed during more frames in MAP1B-knocked down cells, indicating that MT growth events persisted longer (Figure 6E). Also, EB3-GFP dashes underwent a reduced number of pauses in depleted cells (Supplementary Figure S9). Notably, as found in immunocytochemical studies, MAP1B-silenced cells frequently showed a faint EB3-GFP localization along segments of the MT lattice even at low expression levels (Figure 6A). In summary, these results show that knocking down MAP1B in differentiating neuroblastoma cells induces an increase in EB3 binding to MT plus-ends that correlates with an increase in MT growth velocity and duration, and a decrease in MT pausing. These data support the novel finding that MAP1B can modulate MT plus-end dynamics by interacting with EB1/3 in the cytosol, thereby negatively regulating binding of EB1/3 to plus-ends.
MAP1B negatively regulates binding of EBs to MT plus-ends during neuronal development
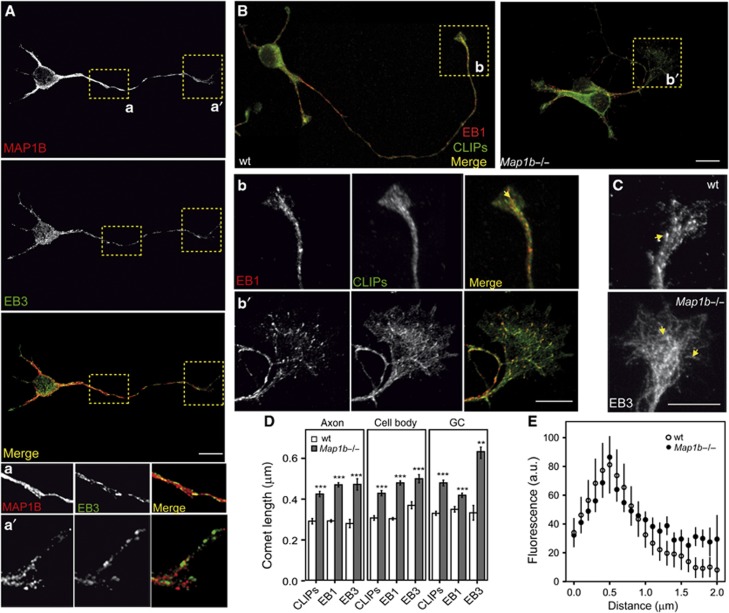
MAP1B has been shown to be involved in axon outgrowth (Gonzalez-Billault et al, 2001, 2002; Montenegro-Venegas et al, 2010; Tymanskyj et al, 2012).We confirmed that our cultures of 1 day in vitro (DIV) primary hippocampal neurons from embryonic E18 MAP1B hypomorphous (Map1b−/−) mice presented shorter axons tipped by enlarged growth cones (Supplementary Figure S10), as described (Gonzalez-Billault et al, 2001; Gonzalez-Billault et al, 2002; Montenegro-Venegas et al, 2010). However, how MAP1B controls MT dynamics in extending axons is still uncertain. Our findings point to MAP1B as a regulator of MT dynamics through its direct interaction with EB proteins in differentiating neuroblastoma cells. For this reason, we checked whether MAP1B also crosstalked with EBs in primary neurons during axon extension. Immunofluorescence images showed that MAP1B and EBs localized to axons and axonal growth cones in 1DIV wild-type (wt) neurons (Figures 7A and B; Supplementary Figure S11A). We then analysed whether lack of MAP1B affected EBs localization in neurons. EB1 comets were present throughout Map1b−/− neurons (Figure 7B), indicating that MT polymerization still occurred in the absence of MAP1B. However, like in MAP1B-deficient neuroblastoma cells, we observed increased EB1 and EB3-comet length in mutant neurons in every cell compartment analysed (axon, cell body and (axonal) growth cones; Figures 7B–D and Supplementary Figure S11B).
Figure 7.
MAP1B negatively regulates binding of EBs to MT plus-ends in developing neurons. Embryonic wild-type (wt) and Map1b−/− primary hippocampal neurons were cultured for 1 day, fixed and stained for MAP1B and/or different +TIPs. (A) Confocal images show wt neurons stained with anti-MAP1B (N-t, red) and anti-EB3 (green). Insets show details of the axon (a) and growth cone (a′). (B) Confocal pictures of wt and Map1b−/− neurons stained with antibodies against EB1 (red) and CLIPs (green). Picture of wt neuron is a composite of two images of the same cell. Insets show details of growth cones of wt (b) and Map1b−/− (b′) neurons. Scale bars=10 μm. (C) Immunofluorescence pictures of growth cones from wt and MAP1B-deficient neurons stained with anti-EB3. Note that EB3 localizes at MT segments in the Map1b−/− growth cone. (D) Average length of MT comets highlighted by CLIPs, EB1 or EB3 in wt versus Map1b−/− neurons. Quantification was performed in different cell compartments (axon, cell body and axonal growth cones). Comets highlighted by different +TIPs were longer in MAP1B-deficient neurons. Number of comets measured per staining and genotype was for CLIPs, n=1043 (wt) and n=1446 (Map1b−/−); for EB1, n=2285 (wt) and n=1999 (Map1b−/−) and for EB3, n=70 (control) and n=144 (Map1b−/− neurons). (E) Average fluorescence intensity patterns of EB3 comets in growth cones of wt and Map1b−/− neurons. **P<0.005, ***P<0.0005.
Cytoplasmic linker proteins (CLIPs) are a family of +TIPs that have recently been shown to regulate neuronal polarization and growth cone dynamics (Neukirchen and Bradke, 2011). Since CLIPs accumulation at MT growing-ends is EB-dependent, we analysed whether binding of CLIPs to MT plus-ends was altered in Map1b−/− neurons. CLIP dashes were longer in every cell compartment analysed in mutant neurons (Figures 7B and D; Supplementary Figure S11B). These data indicate that MAP1B deficiency enhances the binding of both EBs and CLIPs to MT plus-ends throughout neurons. In particular, enlarged growth cones of Map1b−/− neurons displayed longer EBs (and CLIPs) comets than the ones present in wt neurons (insets in Figures 7B–E). Remarkably, EB3, which is more abundantly expressed in brain and enriched in growth cones (Nakagawa et al, 2000; Geraldo et al, 2008), showed an average comet length that was two-fold increased in MAP1B-deficient neurons (Figure 7D), displaying an extended interaction with the MT lattice (Figures 7C and E). On the other hand, EB3 comet pattern was not altered in growth cones of hippocampal tau−/− neurons (Supplementary Figure S11C), suggesting that MAP1B exerts a specific control of EB3 binding to MTs in axonal growth cones during axonogenesis.
Distribution of comets was also different in growth cones of wt and MAP1B mutant neurons. In wt neurons, most comets were distributed throughout the central (C) region of the growth cone in a parallel array, with some dashes invading the most peripheral (P) region (insets in Figures 7B (b and b´) and C). However, in neurons from Map1b hypomorphous mice, most comets spread in a broad array throughout the enlarged C and P regions of growth cones, and the most peripheral ones turn backwards (insets in Figures 7B (b and b´) and C).
Together, these results show that the binding of EBs (and CLIPs) to MT plus-ends in neurons is increased in the absence of MAP1B. In particular, the interaction of EB3 with MTs is especially enhanced in growth cones of MAP1B-deficient neurons, where it can also be found along the MT lattice.
MAP1B controls MT dynamics by regulating EB3 function during axon extension
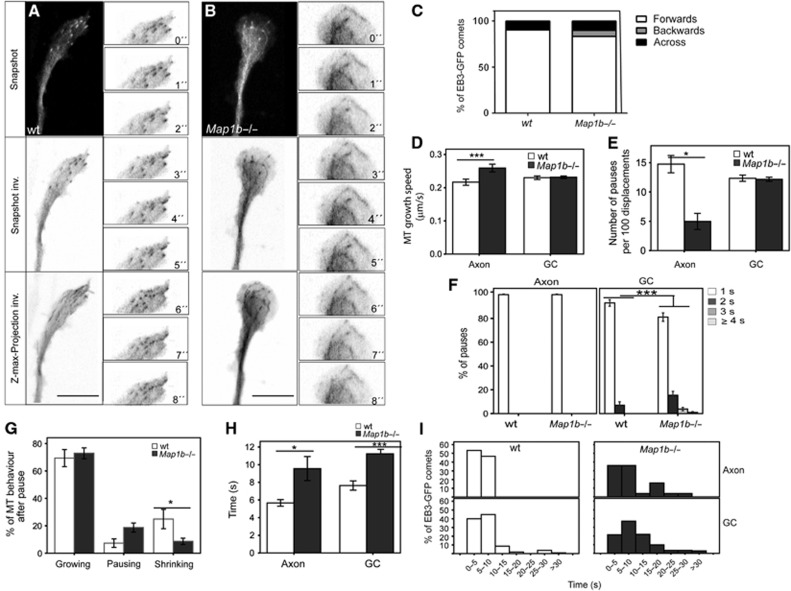
MAP1B has been proposed to control MT dynamics in neurons (Gonzalez-Billault et al, 2001, 2002; Tymanskyj et al, 2012). Our results indicate that MAP1B might locally alter MT dynamics during neuronal development, through its interaction with EB proteins. We first confirmed that the ratio between dynamic and stable MTs was altered in growth cones of cultures of 1DIV hippocampal neurons from Map1b−/− mice, as described (Gonzalez-Billault et al, 2002; Supplementary Figure S10). Moreover, MTs that invaded enlarged growth cones of mutant neurons were looped (Supplementary Figure S10). The presence of looped MTs in growth cones might be either the result of a continued MT polymerization in non-advancing growth cones or of a loss of directionality of MTs as they enter growth cones. To address whether global changes in MT dynamics and looping in growth cones correlate with altered EB3 behaviour at MT plus-ends, wt and Map1b−/− neurons were transfected with EB3-GFP and comet displacements were analysed. We focused on distal axons and growth cones, choosing thinner/longer neurites in which most EB3-GFP dashes moved anterogradely, as described before (Stepanova et al, 2003). In wt neurons, EB3-GFP comets entered in a parallel array through the C and into the P regions of growth cones, with some occasional excursions into filopodia-like extensions (Figure 8A; Supplementary Figure S12A; Supplementary Movies S3 and S5). However, in MAP1B-deficient neurons, most EB3-GFP dashes entered widely spread through the C and the P regions of expanded growth cones, and then a number of EB3 comets moved backwards on curved tracks (Figures 8B and C; Supplementary Figure S12B; Supplementary Movie S4). Moreover, in growth cones from some MAP1B-deficient neurons, EB3-GFP not only localized to MT plus-ends but also bound to stretches of looping MTs (Figure 8B; Supplementary Figure S12B; Supplementary Movies S4 and S6). By tracking of EB3-GFP displacements, we found that MT growth speed was increased in axons but not in growth cones of Map1b−/− neurons (Figure 8D). In addition, we observed that EB3-GFP comets were occasionally pausing. Quantifications showed that the average number of pauses per 100 displacements was reduced in axons of Map1b−/− neurons but did not change in Map1b−/− growth cones (Figure 8E). On the other hand, EB3-GFP comets remained paused longer time in growth cones of MAPB-deficient neurons (Figure 8F). We analysed EB3-GFP comet behaviour after pausing. One second after pausing, EB3-GFP dashes resumed their movement (MT growth), dwelled for a few more seconds (prolonged MT pause) or disappeared (MT shrinking). Map1b−/− neurons presented a significant decrease in the percentage of MTs that shrank immediately after pausing (Figure 8G). Finally, we found that EB3-GFP comets could be followed longer both in distal axons and in growth cones of Map1b−/− neurons (Figures 8H and I). These results indicate that MT growth events persist longer in axons and growth cones of neurons lacking MAP1B.
Figure 8.
MAP1B controls MT dynamics during axon extension by regulating EB3 function in axons and growth cones. Embryonic wt and Map1b−/− primary hippocampal neurons were cultured and transfected with EB3-GFP 4 h after plating. Twenty-four hours later, cells were recorded by time-lapse confocal microscopy and EB3-GFP displacements were followed and analysed. Pictures were taken every second (1 s) for 120 s. (A) Representative examples of growth cones from wt (A) and Map1b−/− neurons (B). Snapshots show still images in each case and Z-Max Projections represent the maximal projection of the 120 images taken. Inverted grey scale is shown to visualize EB3-GFP comets and trajectories more clearly. Insets with details of EB3-GFP comets displacements are shown in (A, B). Quantifications of different parameters related to EB3-GFP displacements were performed in wt and Map1b−/− neurons. We focused on axons and axonal growth cones, with most EB3-GFP comets moving anterogradely. (C) Quantification of the direction of EB3-GFP comet displacements in growth cones. MAP1B-deficient neurons present an increase in the number of comets moving backwards from the leading edge of the growth cone. Number of comets measured per genotype was n=204 (wt) and n=102 (Map1b−/−). Number of neurons taken was n=10 (wt) and n=12 (Map1b−/−). (D) MT growth speed in axons and growth cones (GC). (E) Number of pauses counted per 100 displacements of EB3-GFP comets in axons and GC. (F) Average duration of EB3-GFP comet pauses in axons and GC. (G) Quantification of the different behaviours of comets that undergo pauses in growth cones. After having paused, EB3-GFP comets continued moving (MTs that resumed growing), remained paused (pausing MTs) or disappeared (shrinking MTs). (H) Time EB3-GFP comets were followed in axons and GC (duration of MT growth events). (I) Distribution of duration of MT growth events in axons and GC (from 0–5 to >30 s). Average values±s.e.m. are represented. *P<0.05, ***P<0.0005.
In summary, EB3-GFP behaviour at MT plus-ends is substantially altered in axons and axonal growth cones of MAP1B-deficient neurons. Since EB3 participates in the regulation of MT dynamics at plus-ends, our results indicate that MAP1B locally controls MT dynamics via its regulation of EB3 during axonogenesis.
Discussion
Classical MAPs and +TIPs participate in the regulation of MT dynamics during neuronal development. In this work, we describe a novel interplay between MAP1B, and EB1/EB3, during neurite and axon extension. We show that MAP1B interacts directly with EB1 and EB3 in the cytosol of developing neuronal cells. In this way, MAP1B controls EBs localization, mobility and dynamics in extending neurites/axons and growth cones. Based on our findings, we propose a new function of MAP1B as a direct regulator of the levels of EB proteins that are available for MT end association. Our data provide new insight into how MAP1B controls locally MT dynamics during neurite/axonal outgrowth.
Primary hippocampal neurons from Map1b hypomorphous mice present a delay in axon extension (Gonzalez-Billault et al, 2001, 2002). Downregulation of either MAP1B or EB1 impairs neurite and axon outgrowth and alters MT dynamics in neuronal cells (Gonzalez-Billault et al, 2001; Stepanova et al, 2010; Tymanskyj et al, 2012), suggesting that these proteins regulate neurite and axon elongation in a coordinated manner. We show here that, in developing neuronal cells, MAP1B and EB1/3 are enriched in neurites, axons and growth cones, supporting their roles as modulators of neuritogenesis and axonogenesis.
We furthermore show that MAP1B and EB1/3 interact directly. The fact that EB3 was always more abundantly detected in an MAP1B complex than EB1 indicates that MAP1B interacts preferentially with EB3. While EB1 is ubiquitously expressed, MAP1B and EB3 are predominantly detected in brain, mostly in neurons, where the proteins are enriched in axons and growth cones (Safaei and Fischer, 1989; Black et al, 1994; Nakagawa et al, 2000; Geraldo et al, 2008). This suggests that the interaction between MAP1B and EB3 has a function specifically during axon outgrowth. Of note, EB3 interacts with another neuronal classical MAP, MAP2, in dendrites of mature neurons (Kapitein et al, 2011). Hence, modulation of EB3 localization by classical MAPs might provide an extra mode of local regulation of MT dynamics in neurons.
Proline-directed kinases have been implicated in MAP1B phosphorylation, regulating its binding with MTs (Goold et al, 1999). Remarkably, we found that the interaction between MAP1B and EB3 is regulated by phosphorylation mediated by proline-directed kinases. Thus, it is likely that the formation of the complex MAP1B/EB3 is controlled by multiple signalling pathways that converge at proline-directed kinases during neuronal development. Further studies are required to elucidate the molecular mechanisms involved in the regulation of the interaction between MAP1B and EBs in neuronal cells.
We observed no co-localization of MAP1B and EBs at MTs. Consistently, we found that the interaction between MAP1B and EB1/3 is not mediated by MTs but preferentially occurs in the cytosol. Given the higher intracellular concentration of MAP1B, it might act by sequestering EBs in a complex to control the amount of +TIP that can exchange at MT plus-ends. Indeed, MT distal end localization of EBs in neuronal cells is negatively influenced by MAP1B but not by the neuronal classical MAP tau. MAP1B overexpression displaces endogenous EBs from MT distal ends whereas MAP1B deficiency results in an increased binding of EBs to MT plus-ends. Notably, we found that expression of a cytosolic N-terminal fragment of MAP1B that interacts with EBs but, neither associates with MTs nor alters MT number, impairs binding of EBs to MTs, supporting that EB proteins are sequestered in the cytosol by MAP1B.
EB1/3 do have the capacity to bind along MTs, although their affinity for MT plus-ends is higher, due to structural differences between the MT lattice and plus-ends (Morrison et al, 1998; Maurer et al, 2011, 2012). Indeed, EB1-GFP and EB3-GFP decorate MTs when overexpressed at high levels in cells (Komarova et al, 2005). In our study, upon MAP1B deficiency, an ‘excess’ of EBs would be present in the cytosol to bind MT ends and MT lattices, in some way resembling EBs overexpression (as shown). We did detect binding of EBs to MT segments in the absence of MAP1B but not in the absence of tau indicating a specific and direct role for MAP1B in the control of EBs localization. We propose that in control cells MAP1B prevents binding of EBs to the MT lattice by sequestering EBs in the cytosol, but also by steric hindrance, that is, MAP1B occupies EB binding sites on the lattice. Notably, in mature neurons, in which MAP1B levels are generally low, EB3 has been found along the MT lattice in different locations (e.g., the axon initial segment (Leterrier et al, 2011) and dendrites (Kapitein et al, 2011). Therefore, MAP1B emerges as a controller upstream of EB function in neuronal cells.
EB1/3-comet number and length have been reported to correlate with number of growing MTs and MT growth velocity in the cell, respectively (Bieling et al, 2007, 2008). As mentioned, in MAP1B-depleted neuroblastoma cells, fewer but longer EB comets were observed, in correlation with a reduction in MT density and an increase in MT growth speed. The reduced amount of growing MTs would increase the levels of soluble tubulin, displacing the equilibrium towards MT polymerization and in this way accelerating MT growth. However, if this was the cause for the increased interaction of EBs with MTs found in MAP1B-deficient cells, tau knockdown, which leads to a more marked reduction in the number of MTs, should enhance the binding of EBs to MTs in a similar way. Remarkably, we showed that tau silencing does not enhance the interaction of EBs to MTs. Hence, most of the effects of MAP1B on EBs localization are most likely direct and not mediated by its actions on MT number/density.
An increase in MT growth velocity along with a reduction in pausing was also observed in axons of Map1b−/− neurons, in correlation with the presence of longer EB comets. However, in MAP1B-deficient neurons, MTs that enter growth cones grow as fast as in wt neurons, but undergo an increased proportion of backwards growing events, as well as longer pauses and reduced shrinking. Overall, MAP1B-deficient neurons present longer MT growth events. The differences found between differentiating neuroblastoma cells, axons and growth cones of primary neurons may be due to the different structural and biochemical characteristics of these different cell types and cellular compartments. Moreover, both MAP1B and EB3 are enriched in growth cones and there are many more growing MT ends in the growth cone than in the axon. It is also likely that the presence of cytosolic MAP1B is higher in growth cones than in axons. For these reasons, MAP1B influence on EB3 behaviour might also differ in axons and growth cones.
What is the physiological significance of the regulation of EB1/3 by MAP1B? EB3 dynamics in cells is determined by different factors such as diffusion, exchange on MT plus-ends and interaction with other proteins. Interestingly, our FRAP assays revealed that the interaction between MAP1B and EB3 occurs in vivo, preferentially in the cytosol, in the distal region of extending neurites, in which MAP1B restrains EB3 mobility. MAP1B is a very big protein (>300 kDa) and this might explain why EB3 dynamics is substantially reduced when it is bound to MAP1B. Moreover, since MAP1B is a classical MAP, its concentration is likely to exceed that of a +TIP such as EB3. In non-neuronal cells, in which MAP1B is not expressed, the major mechanism determining EB3 mobility is its rapid binding on/off MT distal ends and the rate limiting step is cytoplasmic diffusion (Dragestein et al, 2008). According to our results, MAP1B thus regulates the ‘effective’ concentration of EB3, the amount that is available for MT plus-end localization. Hence, the rate-limiting step of EB3 mobility in distal neurites/axons and growth cones might be its regulated release from the MAP1B/EB3 complex rather than pure diffusion.
Axon extension and growth cone advance depend on MT organization. EB1/3 proteins participate in MT dynamics at different levels and their functions may differ depending on cell type (Slep, 2010). Our data indicate that, in developing neuronal cells, the effects of MAP1B on EB3 comets, such as alterations in length, number, speed, direction and pausing, correlate with changes in MT dynamics, MT organization and axon extension. Our findings show that MTs grow more persistently in the absence of MAP1B, but cannot push growth cones to advance. This probably induces MT looping, which along with the excessive EB3 binding to MTs, would contribute to MT overstabilization, thereby affecting growth cone advance and axon extension.
EB proteins are essential for the accumulation of other +TIPs, such as CLIPs, at MT plus-ends (reviewed in Akhmanova and Steinmetz, 2008). Indeed, CLIPs binding to MT plus-ends is increased in MAP1B-deficient neurons, probably through the enhanced interaction of EBs to MTs. Since CLIPs are known regulators of MT dynamics during neuronal polarization (Neukirchen and Bradke, 2011), MAP1B may also control axonal extension through an indirect (negative) regulation of the MT plus-end localization of CLIPs, and possibly other +TIPs. Actually, MAP1B has been shown to bind to Lis1 (Jimenez-Mateos et al, 2005), a +TIP that has also been reported to be involved in growth cone remodelling during axon outgrowth (Grabham et al, 2007).
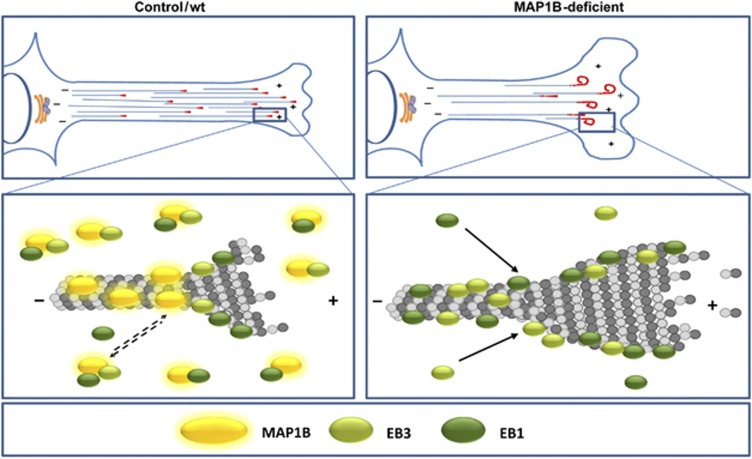
Based on our data, we propose the model shown in Figure 9. In control/wt cells, extending neurites/axons and growth cones have a soluble pool of MAP1B that recruits and partially depletes part of the cytosolic pool of EB1/3 in a complex, reducing the ‘effective concentration’ of EB1/3 available to bind to MT plus-ends. In MAP1B-deficient neuronal cells, more cytoplasmic EB1/3 is free to exchange at MT plus-ends and to interact with low-affinity binding sites along MTs (which are normally hindered by MAP1B). As a result, EB1/3 interaction with MTs would be enhanced, leading to altered MT dynamics, changes in MT directionality, persistent growth and eventually curving and looping. This in turn might induce a delay in growth cone advance and axon extension. In summary, we propose the existence of a new mechanism of EB1/3 regulation by MAP1B that contributes to orchestrate MT dynamics during neuronal differentiation.
Figure 9.
Model of regulation of EBs localization by MAP1B during neurite/axon outgrowth. In control/wt cells, MAP1B negatively controls binding of EB1/3 to MTs in extending neurites/axons and growth cones, thereby regulating MT dynamics and growth cone advance during neuritogenesis and axonogenesis. In MAP1B deficient neuronal cells, interaction of EB1/EB3 to MTs is enhanced, leading to MT overstabilization and looping.
Materials and methods
Cell culture
N1E-115 mouse neuroblastoma cells (ATCC) were cultured in DMEM (Dulbecco’s modified Eagle’s medium) plus 10% fetal bovine serum, 2 mM of L-glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C in a 5% CO2 atmosphere. Cells were induced to differentiate by overnight serum deprivation, Cultures of dissociated hippocampal pyramidal neurons from wt, Map1b hypomorphous (Gonzalez-Billault et al, 2000) and tau knockout mice (Gomez de Barreda et al, 2010) embryonic brain tissue were prepared as described (Banker and Cowan, 1977). Hippocampi were dissected and treated with papain and DNase (Worthington Biochemical Corporation). In all, 10 000 cells were plated on 12 mm glass coverslips coated with 100 μg/ml poly-L-lysine and 10 μg/ml laminin in Neurobasal containing 10% horse serum. After 3 h, plating medium was replaced with Neurobasal medium supplemented with 2 mM L-glutamine, 2 mM D-pyruvate, 1% N2, 2% B27, 100 U/ml penicillin and 100 mg/ml streptomycin. All culturing media and supplements were purchased from Gibco Invitrogen Corporation. Neuronal cultures were maintained for 1DIV in a humidified 37°C incubator with 5% CO2.
Generation of stable cell lines by lentiviral transduction
To produce recombinant lentiviral particles, subconfluent 293T cells were co-transfected with each of the shRNA plasmids (see Supplementary data) along with both pCMVdR8.74 (Addgene) and pMD2G (Addgene) plasmids, using Lipofectamine 2000 (Invitrogen) based on the manufacturer’s protocol. Viruses were collected 48 h post transfection. N1E-115 cells were infected with each virus (or a mix of shRNA viruses) for 24 h and selected with puromycin (Sigma-Aldrich) for 2–3 weeks to generate stable cell lines.
Pull-down assays
GST and His fusion proteins (Supplementary data) were induced in BL21 E. coli, as described (Smith and Johnson, 1988) and purified on glutathione-Sepharose 4B beads (GE Healthcare) and Ni-NTA agarose beads (Qiagen) respectively, according to the instructions of the manufacturer. For GST pull-down assays, individual GST fusion proteins bound to glutathione Sepharose 4B beads were combined with either total mouse brain lysates (E18) or purified His-tagged proteins (in vitro assay). After incubation for 1 h on a rotating wheel at 4°C, beads were separated from the supernatant by centrifugation and washed several times with washing buffer containing 20 mM Tris pH 8, 150 mM NaCl, 1 mM DTT and 0.05% Triton-X100. Beads were resuspended in 2X Laemmli buffer and boiled at 95°C for 5 min. Proteins retained on the beads were analysed by western blotting (Supplementary data).
Immunofluorescence and confocal microscopy
Cells were fixed with 100% methanol-1 mM EGTA for 10 min at −20°C, followed or not by 15 min fixation in 4% paraformaldehyde in PBS at room temperature. Cells were permeabilized/blocked with PBS/0.1% Triton X-100/3% (w/v) bovine serum albumin (BSA) for 1 h. Subsequently, cells were incubated with primary antibodies raised against the indicated proteins, overnight at 4°C. After three washes with PBS, cells were incubated for 1 h at room temperature with secondary antibodies (Invitrogen). Double or triple-stained cells were analysed with a Zeiss LSM510 Meta or Inverted confocal microscope mounted on an Axiovert 200 M or Axioskop2 plus and Axiovert 200 M equipped with a CCD camera.
FRAP experiments
FRAP assays were performed using a LSM510 Multiphoton Laser Scanning and AxioImager M1 (Zeiss) confocal microscope ( × 63 objective, × 1.6 zoom). A selected region of interest (ROI) of 34 × 6 pixels, located at the distal part of extending neurites, was chosen in each experiment. Fluorescence intensity was measured in bleached and non-bleached areas for five scanning cycles before bleaching and then bleaching was performed with scans at 100% laser power (10 cycles iterations, scan velocity=5). Time-lapse images were acquired for 5 s before bleaching and for 50–60 s after bleaching in either vehicle-treated cells (DMSO 0.1%, 1 h) or cells treated with Nocodazole (20 nM, 1 h). Image acquisition was done at 0.084 s intervals for up to 600 cycles.
For FRAP analysis, images of 512 × 512 pixels from the acquired ROI were used. Background values, as well as the bleaching that occurred during image acquisition, were subtracted and corrected. Data were normalized with control fluorescence stated as 100 and average curves were obtained. Average data±s.e.m. were fitted using a two phase association exponential equation (GraphPad): Y=Y0+SpanFast × (1−exp(−k1 × X))+SpanSlow × (1−exp(−k2 × X)) and the following supporting formulas: SpanFast=(Plateau−Y0) × PercentFast × 0.01;SpanSlow=(Plateau−Y0) × (100 × PercentFast) × 0.01. Correlation coefficients obtained in the curve fitting ranged from 0.97 to 0.99. Normalized data from each single cell were also fitted using the mentioned two phase association exponential equation. We focused our study on the slow part of the fluorescence recovery curve, which results from the interaction with other proteins. For that reason, the parameters obtained from these equations and described in Results refer to kslow (1/s) and t1/2 slow (s). t1/2 was calculated as ln(2)/K.
Live-cell imaging
EB3-GFP was transfected into stable N1E-115 cell lines (control or MAP1B-depleted) or into wt or Map1b−/− primary hippocampal neurons (4 h after plating) using Lipofectamine TM 2000 (Invitrogen). Stable cell lines and neurons were filmed 24 h after transfection. Living cells were observed at 36°C with a LSM510 confocal laser-scanning microscope on an Axiovert 200 inverted microscope (Zeiss), as described previously (Stepanova et al, 2003). In most experiments, the optical slice (z-dimension) was set to 1 μm. Other settings that were used (e.g., laser intensity and gain value) differed slightly in the various experiments and were adapted to obtain optimal signal-to-noise ratios. Time-lapse series were acquired every 1 or 2 s for 60 or 30 frames, respectively.
Supplementary Material
Acknowledgments
We are grateful to Drs I Santa-Maria, F Propst and JA Esteban for providing us with constructs, to Dr P Gordon-Weeks for advice and to J Klett (Bioinformatics Department, CBM-SO) for his help with FRAP data fitting. This work was supported by grants from the Spanish Research Ministry (SAF2006-02424 and SAF2011-24841), CIBERNED and Comunidad de Madrid (S2010/BMD2331). ET was supported by the Spanish Research Council (CSIC) and CIBERNED and CLS was funded by CSIC (I3P contract and an intramural project).
Author contributions: ET and CLS performed, analysed and interpreted all the experiments, NG generated most of the reagents, contributed to data interpretation and revised critically the manuscript; JA contributed with critical discussions and revision of the article; CLS conceived the project, designed the research plan and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akhmanova A, Steinmetz MO (2008) Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol 9: 309–322 [DOI] [PubMed] [Google Scholar]
- Amos LA, Schlieper D (2005) Microtubules and maps. Adv Protein Chem 71: 257–298 [DOI] [PubMed] [Google Scholar]
- Banker GA, Cowan WM (1977) Rat hippocampal neurons in dispersed cell culture. Brain Res 126: 397–425 [DOI] [PubMed] [Google Scholar]
- Bieling P, Kandels-Lewis S, Telley IA, van Dijk J, Janke C, Surrey T (2008) CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J Cell Biol 183: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, Dogterom M, Brunner D, Surrey T (2007) Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450: 1100–1105 [DOI] [PubMed] [Google Scholar]
- Black MM, Slaughter T, Fischer I (1994) Microtubule-associated protein 1b (MAP1b) is concentrated in the distal region of growing axons. J Neurosci 14: 857–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Barnett B, Lazarus JE, Tokito M, Goldman YE, Holzbaur EL (2009) Microtubule plus-end tracking by CLIP-170 requires EB1. Proc Natl Acad Sci USA 106: 492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragestein KA, van Cappellen WA, van Haren J, Tsibidis GD, Akhmanova A, Knoch TA, Grosveld F, Galjart N (2008) Dynamic behavior of GFP-CLIP-170 reveals fast protein turnover on microtubule plus ends. J Cell Biol 180: 729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galjart N (2010) Plus-end-tracking proteins and their interactions at microtubule ends. Curr Biol 20: R528–R537 [DOI] [PubMed] [Google Scholar]
- Garcia-Perez J, Avila J, Diaz-Nido J (1998) Implication of cyclin-dependent kinases and glycogen synthase kinase 3 in the phosphorylation of microtubule-associated protein 1B in developing neuronal cells. J Neurosci Res 52: 445–452 [DOI] [PubMed] [Google Scholar]
- Geraldo S, Khanzada UK, Parsons M, Chilton JK, Gordon-Weeks PR (2008) Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat Cell Biol 10: 1181–1189 [DOI] [PubMed] [Google Scholar]
- Gomez de Barreda E, Perez M, Gomez Ramos P, de Cristobal J, Martin-Maestro P, Moran A, Dawson HN, Vitek MP, Lucas JJ, Hernandez F, Avila J (2010) Tau-knockout mice show reduced GSK3-induced hippocampal degeneration and learning deficits. Neurobiol Dis 37: 622–629 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Billault C, Avila J, Caceres A (2001) Evidence for the role of MAP1B in axon formation. Mol Biol Cell 12: 2087–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Billault C, Demandt E, Wandosell F, Torres M, Bonaldo P, Stoykova A, Chowdhury K, Gruss P, Avila J, Sanchez MP (2000) Perinatal lethality of microtubule-associated protein 1B-deficient mice expressing alternative isoforms of the protein at low levels. Mol Cell Neurosci 16: 408–421 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Billault C, Jimenez-Mateos EM, Caceres A, Diaz-Nido J, Wandosell F, Avila J (2004) Microtubule-associated protein 1B function during normal development, regeneration, and pathological conditions in the nervous system. J Neurobiol 58: 48–59 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Billault C, Owen R, Gordon-Weeks PR, Avila J (2002) Microtubule-associated protein 1B is involved in the initial stages of axonogenesis in peripheral nervous system cultured neurons. Brain Res 943: 56–67 [DOI] [PubMed] [Google Scholar]
- Goold RG, Owen R, Gordon-Weeks PR (1999) Glycogen synthase kinase 3beta phosphorylation of microtubule-associated protein 1B regulates the stability of microtubules in growth cones. J Cell Sci 112: Pt 193373–3384 [DOI] [PubMed] [Google Scholar]
- Grabham PW, Seale GE, Bennecib M, Goldberg DJ, Vallee RB (2007) Cytoplasmic dynein and LIS1 are required for microtubule advance during growth cone remodeling and fast axonal outgrowth. J Neurosci 27: 5823–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, Spangler SA, Di Stefano P, Demmers J, Krugers H, Defilippi P, Akhmanova A, Hoogenraad CC (2009) Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron 61: 85–100 [DOI] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Wandosell F, Reiner O, Avila J, Gonzalez-Billault C (2005) Binding of microtubule-associated protein 1B to LIS1 affects the interaction between dynein and LIS1. Biochem J 389: 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Yau KW, Gouveia SM, van der Zwan WA, Wulf PS, Keijzer N, Demmers J, Jaworski J, Akhmanova A, Hoogenraad CC (2011) NMDA receptor activation suppresses microtubule growth and spine entry. J Neurosci 31: 8194–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y, De Groot CO, Grigoriev I, Gouveia SM, Munteanu EL, Schober JM, Honnappa S, Buey RM, Hoogenraad CC, Dogterom M, Borisy GG, Steinmetz MO, Akhmanova A (2009) Mammalian end binding proteins control persistent microtubule growth. J Cell Biol 184: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y, Lansbergen G, Galjart N, Grosveld F, Borisy GG, Akhmanova A (2005) EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol Biol Cell 16: 5334–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G, Akhmanova A (2006) Microtubule plus end: a hub of cellular activities. Traffic 7: 499–507 [DOI] [PubMed] [Google Scholar]
- Leterrier C, Vacher H, Fache MP, d'Ortoli SA, Castets F, Autillo-Touati A, Dargent B (2011) End-binding proteins EB3 and EB1 link microtubules to ankyrin G in the axon initial segment. Proc Natl Acad Sci USA 108: 8826–8831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Bieling P, Cope J, Hoenger A, Surrey T (2011) GTPgammaS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs). Proc Natl Acad Sci USA 108: 3988–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T (2012) EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell 149: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro-Venegas C, Tortosa E, Rosso S, Peretti D, Bollati F, Bisbal M, Jausoro I, Avila J, Caceres A, Gonzalez-Billault C (2010) MAP1B regulates axonal development by modulating Rho-GTPase Rac1 activity. Mol Biol Cell 21: 3518–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison EE, Wardleworth BN, Askham JM, Markham AF, Meredith DM (1998) EB1, a protein which interacts with the APC tumour suppressor, is associated with the microtubule cytoskeleton throughout the cell cycle. Oncogene 17: 3471–3477 [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Koyama K, Murata Y, Morito M, Akiyama T, Nakamura Y (2000) EB3, a novel member of the EB1 family preferentially expressed in the central nervous system, binds to a CNS-specific APC homologue. Oncogene 19: 210–216 [DOI] [PubMed] [Google Scholar]
- Neukirchen D, Bradke F (2011) Cytoplasmic Linker Proteins Regulate Neuronal Polarization through Microtubule and Growth Cone Dynamics. J Neurosci 31: 1528–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrotti B, Islam K (1995) Microtubule associated protein 1B (MAP1B) promotes efficient tubulin polymerisation in vitro. FEBS Lett 371: 29–31 [DOI] [PubMed] [Google Scholar]
- Safaei R, Fischer I (1989) Cloning of a cDNA encoding MAP1B in rat brain: regulation of mRNA levels during development. J Neurochem 52: 1871–1879 [DOI] [PubMed] [Google Scholar]
- Skube SB, Chaverri JM, Goodson HV (2010) Effect of GFP tags on the localization of EB1 and EB1 fragments in vivo. Cytoskeleton (Hoboken) 67: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep KC (2010) Structural and mechanistic insights into microtubule end-binding proteins. Curr Opin Cell Biol 22: 88–95 [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS (1988) Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67: 31–40 [DOI] [PubMed] [Google Scholar]
- Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, Grosveld F, van Cappellen G, Akhmanova A, Galjart N (2003) Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J Neurosci 23: 2655–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T, Smal I, van Haren J, Akinci U, Liu Z, Miedema M, Limpens R, van Ham M, van der Reijden M, Poot R, Grosveld F, Mommaas M, Meijering E, Galjart N (2010) History-dependent catastrophes regulate axonal microtubule behavior. Curr Biol 20: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Takemura R, Okabe S, Umeyama T, Kanai Y, Cowan NJ, Hirokawa N (1992) Increased microtubule stability and alpha tubulin acetylation in cells transfected with microtubule-associated proteins MAP1B, MAP2 or tau. J Cell Sci 103: Pt 4953–964 [DOI] [PubMed] [Google Scholar]
- Togel M, Wiche G, Propst F (1998) Novel features of the light chain of microtubule-associated protein MAP1B: microtubule stabilization, self interaction, actin filament binding, and regulation by the heavy chain. J Cell Biol 143: 695–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP, Binder LI, Matus AI (1988) Neuronal microtubule-associated proteins in the embryonic avian spinal cord. J Comp Neurol 271: 44–55 [DOI] [PubMed] [Google Scholar]
- Tucker RP, Garner CC, Matus A (1989) In situ localization of microtubule-associated protein mRNA in the developing and adult rat brain. Neuron 2: 1245–1256 [DOI] [PubMed] [Google Scholar]
- Tymanskyj SR, Scales TM, Gordon-Weeks PR (2012) MAP1B enhances microtubule assembly rates and axon extension rates in developing neurons. Mol Cell Neurosci 49: 110–119 [DOI] [PubMed] [Google Scholar]
- Ulloa L, Diaz-Nido J, Avila J (1993) Depletion of casein kinase II by antisense oligonucleotide prevents neuritogenesis in neuroblastoma cells. EMBO J 12: 1633–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandecandelaere A, Pedrotti B, Utton MA, Calvert RA, Bayley PM (1996) Differences in the regulation of microtubule dynamics by microtubule-associated proteins MAP1B and MAP2. Cell Motil Cytoskeleton 35: 134–146 [DOI] [PubMed] [Google Scholar]
- Zimniak T, Stengl K, Mechtler K, Westermann S (2009) Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J Cell Biol 186: 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.