Abstract
Matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) has emerged as a novel powerful MS methodology that has the ability to generate both molecular and spatial information within a tissue section. Application of this technology as a new type of biochemical lipid microscopy may lead to new discoveries of the lipid metabolism and biomarkers associated with area-specific alterations or damage under stress/disease conditions such as traumatic brain injury or acute lung injury, among others. However there are limitations in the range of what it can detect as compared with liquid chromatography-MS (LC-MS) of a lipid extract from a tissue section. The goal of the current work was to critically consider remarkable new opportunities along with the limitations and approaches for further improvements of MALDI-MSI. Based on our experimental data and assessments, improvements of the spectral and spatial resolution, sensitivity and specificity towards low abundance species of lipids are proposed. This is followed by a review of the current literature, including methodologies that other laboratories have used to overcome these challenges.
Keywords: Lipidomics, MALDI imaging, Traumatic brain injury, Acute lung injury, Lipid peroxidation, Docosahexaenoic acid
1. INTRODUCTION
“It is always wise to look ahead, but difficult to look further than you can see.” Winston Churchill
1.1 Multiple roles and molecular diversity of phospholipids and their oxidation products
Lipids are fundamental components of tissue architecture and are critical for tissue function. According to current estimates, the mammalian lipidome is comprised of 10,000 to 100,000 individual species of lipid molecules (Schuhmann et al., 2011) making it more diversified than the proteome or genome. This remarkable diversity of tissue-specific lipids originates from a few hundred lipid classes, including phospholipids (PLs), glycolipids, sphingolipids, and neutral lipids (Shevchenko and Simons, 2010). Phospholipids in particular consist of a headgroup, a phosphoester-connected glycerol backbone, and (usually ester-linked) two fatty acid chains. Different headgroups distinguish different phospholipid classes. These headgroups tend to be exposed to the aqueous environment while the hydrophobic fatty acid chains are usually embedded in cellular and subcellular membranes. There are over 15 common fatty acid residues that exist in varying combinations in different phospholipids. One notable exception from this general plan of architecture is cardiolipin (CL), a phospholipid that contains four fatty acid chains and is found exclusively in mitochondria. Its molecular diversity can thus be much larger. Most tissues such as lung, liver, and heart have only a few major species of CL in high abundance (Minkler and Hoppel, 2010). Brain tissue, on the other hand, is extremely diverse with respect to the number of CL species. A typical CL spectrum of murine brain exhibits approximately 10-12 major CL clusters, most of which have multiple individual CL species at several isobaric masses. This gives over eighty major molecular species of brain CL, all at relatively similar abundances (Kiebish et al., 2008) (Bayir et al., 2007) (Tyurin et al., 2008a) (Sparvero et al., 2010) (Samhan-Arias et al., 2012).
In addition to their structural role in membranes, polyunsaturated lipids are precursors of important signaling molecules that are formed by multistage oxygenation of polyunsaturated fatty acid residues (PUFAs) – linoleic (C18:2), arachidonic (C20:4), eicosapentaenoic (C20:5), docosapentaenoic (C22:5) and docosahexaenoic (C22:6) acids (Guichardant et al., 2011). Usually, phospholipase A2—catalyzed hydrolysis precedes the enzymatic oxygenation stages and represents a rate-limiting stage in the overall biosynthetic process. A huge number of different oxygenated fatty acids formed via this general pathway act as potent intra- and extracellular regulators of multiple biological functions (Marcheselli et al., 2003). Alternatively, esterified fatty acid residues of phospholipids that contain multiple double bonds are prone to oxidative modifications yielding predominantly hydroperoxy-, hydroxy-, epoxy- and oxo-functionalities to phospholipids (Domingues et al., 2008) (Fuchs et al., 2011) (Kim et al., 2011). Phospholipids with oxidized fatty acid residues can undergo subsequent hydrolysis by specialized phospholipases – e.g., lipoprotein lipase A2 – to yield oxygenated fatty acids and lyso-phospholipids (Vaishnav et al., 2010). The oxidatively modified phospholipids can also generate a number of secondary degradation products with truncated fatty acid chains (Tyurin et al., 2009) (Yin et al., 2011) (Liu et al., 2011) (Hall et al., 2010). Peroxidation of polyunsaturated phospholipids may occur as a random – likely non-enzymatic - free radical oxidation commonly associated with oxidative stress and injury (Girotti, 1998) (Hall et al., 2004) (Singh et al., 2006). This type of oxidative damage can result from a variety of insults, including overload with catalytic transition metals (Valko et al., 2005), chemical poisoning with carbon-tetrachloride (Weber et al., 2003), and ischemia/reperfusion (Zhang et al., 1996). In many cases, phospholipid peroxidation and tissue damage and cell death can be selective whereby different phospholipids undergo oxidative modification at different rates (Tyurin et al., 2008b).
Two anionic phospholipids - a mitochondria-specific CL and extra-mitochondrial phosphatidylserine (PS) - are the most reactive to oxidative stress during apoptotic cell death (Tyurin et al., 2008a) (Korytowski et al., 2011). Of the many polyunsaturated fatty acid chains that these phospholipids exhibit, only a select few are prone to biological oxidative modification during apoptosis (Sparvero et al., 2010) due to the involvement of a specific catalyst, cytochrome c, in the oxidation process (Kagan et al., 2005) (Tyurin et al., 2010). While these features of random (non-enzymatic) and selective (enzymatic) peroxidation reactions have been known for a long time, their spatial confinements to specific regions and to specific types of cells and subcellular compartments have not been definitively established. This is mostly due to the insufficiency of methodological approaches for the detailed and accurate mapping of lipids and lipid oxidation products in tissues.
1.2 Mass Spectrometry and matrix-assisted laser desorption/ionization (MALDI) imaging in lipidomics
Mass Spectrometry (MS) has emerged as the analytical technology with sufficient sensitivity and specificity to quantitatively analyze the complexities of biological materials at the molecular level (Gross, 2011) (Murphy et al., 2011b). The advent of soft ionization technologies (laser desorption or electrospray ionization) enabled mass spectra of biological molecules (proteins, lipids, etc) to be obtained. Coupling MS to separation techniques (such as liquid chromatography (LC)) and selective fragmentation by tandem mass spectrometry (MS/MS) markedly improves the analysis of complex mixtures. MALDI and electrospray ionization (ESI) are two of the most prominent soft-ionization MS methods for the direct, label-free analysis of intact lipids and each has its own strengths and limitations (Fuchs et al., 2010) (Han et al., 2012). MALDI analysis can be performed on intact tissue sections (MALDI imaging or MALDI-MSI1) to provide spatial localization for various lipid species, while ESI-LC-MS of lipid extracts allows for a global assessment of lipid species (Delvolve et al., 2011) (Goto-Inoue et al., 2011) (Zemski-Berry et al., 2011) (Fernández et al., 2011) (Amstalden van Hove et al., 2010). Although lipid extracts are devoid of spatial information, microdissected regions of interest within a tissue section and their subsequent extraction can determine localization of lipid species (Burnum et al., 2009) (Hankin and Murphy, 2010). Once an array of MALDI mass spectra is obtained using a pulsed laser across a tissue section, it is possible to create a lipid-ion image that is associated with a particular mass-to-charge (m/z) ratio. This image represents the spatial distribution and relative abundance of that particular lipid-ion, and can be correlated with histological features (Burnum et al., 2009) (Delvolve et al., 2011) (Carter et al., 2011). By adding internal standards, absolute abundances of several lipid classes can be measured with high precision using MALDI-MSI (Landgraf et al., 2011). MALDI-MSI does not require specific antibodies such as those used in immunohistochemical methods. This allows for direct analysis and discovery of molecular species. In contrast to radioactive or fluorescent labeling-based techniques, one is not limited to a small number of detectable species. Breakdown products (including lysophospholipids and oxidatively modified species) are also detectable as distinct molecular species (Koizumi et al., 2010) (Wang et al., 2010).
Substantial progress has been made in the field of MALDI-MSI of lipids; however, there are still many unresolved issues that need to be addressed (Lagarrigue et al., 2011). While MALDI-MSI is a unique source of valuable information on spatial distribution of lipids, issues on the completeness of the assessments of the lipidome it yields and the overall quantitative aspects of lipid MSI-maps still await further study. One of the important limitations of MALDI-MSI that is observed but not often discussed is that it detects fewer species of lipids than ESI-LC-MS. Reasons include ion suppression by highly abundant lipid species, uncontrolled in-source decay (ISD), specific matrix requirements for successful MALDI-MSI and the ambiguity of some lipid species with respect to mass. Although highly abundant classes of lipids such as phosphatidylcholine (PC) correlate well between MALDI-MSI and ESI-LC-MS, species from less abundant classes such as phosphatidylethanolamine (PE) do not (Hankin and Murphy, 2010). One of the useful approaches may be based on comparative studies of spectra from MALDI-MSI of lipids in a tissue section to those obtained by traditional LC-MS analyses of lipid extracts from serial sections. By using these comparisons, in the current study we attempted to evaluate the advantages and limitations of MALDI-MSI in a more quantitative way and offer suggestions to potentially overcome certain limitations and ambiguities in applying this powerful technology for the spatial analysis of lipids and their oxidative metabolism. Two types of tissue samples - the brain and the lung – have been employed to illustrate current limitations and possible approaches to circumvent them as well as potential perspectives in the applications of MALDI-MSI of phospholipids. The methodologies covered in the Discussion section are not just the examples from our own laboratory (as illustrated in the Results section) but also include many other techniques developed by other groups. This will be particularly useful for the newcomers since MSI is a relatively new field of both mass spectrometry and lipidomics. Inevitably this resulted in a “hybrid” type of paper that combines features of an original contribution with those of a review.
2. RESULTS
2.1 MALDIMSI in negative and positive modes: regional correlation with phospholipid and sphingolipid abundances
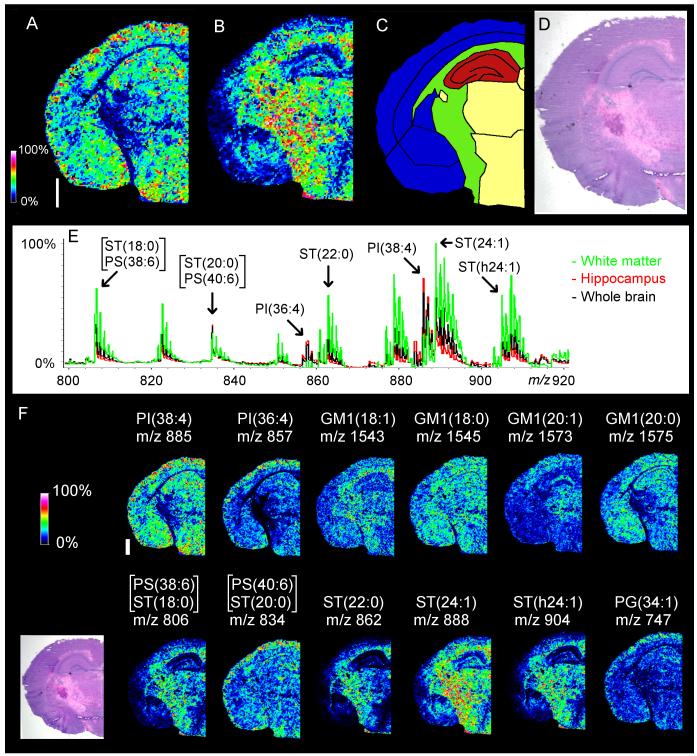
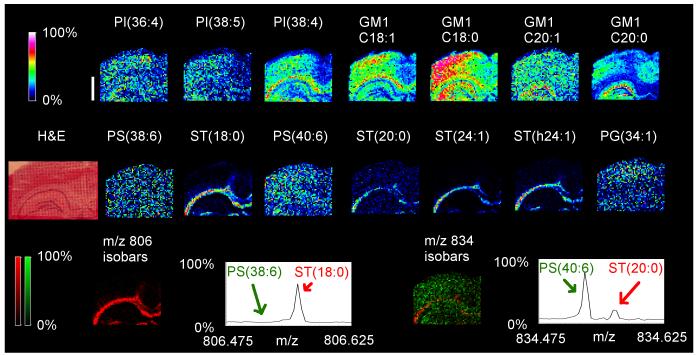
Negative-mode MALDI-MSI of naïve adult mouse brain detected a variety of anionic phospholipid species and sphingolipids as [M-H]−anions (fig. 1). At a spatial resolution of 50μm, MALDI-MSI displayed clear species-specific mapping of certain lipids to particular anatomical regions of the brain as compared with a serial H&E tissue section. MALDI-MSI indicated that many sulfatide (ST) species including ST(d18:1/24:1) were more abundant in the white matter than in the hippocampus or cortex, while the opposite was the case for several phospholipid species including PI(18:0/20:4). The ion at m/z 834.5, which is isobaric for PS(40:6) and ST(d18:1/20:0), appeared across the whole brain section while the ion at m/z 806.5, which is isobaric for PS(38:6) and ST(d18:1/18:0), was localized to the white matter. Two PI species - PI(38:4) and PI(36:4) - represented the most abundant phospholipid species detected in negative mode.
Fig 1.
MALDI-MSI images and regional spectra of adult mouse brain. “Heat map” MALDI-MSI images of PI(38:4) (m/z 885.5, panel A) and ST(24:1) with a d18:1 sphingosine (m/z 888.5, panel B) display localization to different regions of the brain. The white bar is 1000 microns and the scale displays relative intensities with respect to the abundance of each ion. These lipids correlate to specific brain structures (C) upon comparison with a serial H&E section (D). Comparison of the averaged MALDI-MSI spectrum from the entire tissue slice (E-black trace) with that of the hippocampus (E - red trace) and white matter (E - green trace) shows that many other d18:1 sulfatide species including ST(24:1) are more abundant in the white matter than the hippocampus, while the opposite is the case with several PI species including PI(38:4). Various lipids from several classes were also detected (F). Several species of ganglioside GM1 with d18:1 sphingosines exhibited different localization, depending on the composition of their long chain base. MALDI-MSI images were acquired in negative mode with an Ultraflex II.
Two serial brain sections from naïve, postnatal day 17 (pnd 17) rat were prepared and imaged in both positive and negative mode for a global analysis of phospholipid species. Several phosphatidylcholine (PC) species including PC(16:0/16:0) and PC(16:0/18:1) were detected as various adducts ([M+H]+, [M+Na]+, and [M+K]+) and these adducts were considered to be only one phospholipid species even though they occur at a different m/z values. Overall, from both modes a total of 48 species of lipids from various classes including 18 species of phospholipids were detected (table 1).
Table 1.
Number of lipid species detected by MALDI-MSI, MALDI-MS and ESI-LC-MS. These three MS methods detect different numbers of lipid species and classes (G=ganglioside). MALDI-MS and -MSI detects drastically different classes depending on instrument polarity. Lipids in most classes, especially the low abundance CL, are detected with progressively better refinement by these three MS methods. LC-MS is shown for negative mode only since the chromatographic conditions will detect species from all classes in that mode, while MALDI-MS and -MSI required both positive and negative mode for a comprehensive analysis.
| PC | PE | Cer | SM | G | ST | PG | PI | PS | PA | CL | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MALDI- MSI Positive mode |
6 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 |
| MALDI- MSI Negative mode |
0 | 0 | 0 | 0 | 7 | 10 | 1 | 5 | 4 | 0 | 10 | 37 |
| MALDI- MS extract Positive mode |
6 | 5 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 |
| MALDI-MS extract Negative mode | 0 | 0 | 0 | 0 | 12 | 10 | 1 | 7 | 1 | 0 | 31 | 65 |
| ESI-LC- MS |
29 | 27 | 7 | 8 | 5 | 18 | 16 | 16 | 16 | 13 | 70 | 225 |
2.2 Missing classes of lipids with MALDI-MSI: comparison with MALDI-MS and ESI-LC-MS of lipid extracts
Of the 48 species of lipids detected with MALDI-MSI, there were several lipids that are known to be present in the brain that were not detected using this technology. PA and PG species occur in low abundance in the brain and were not readily detected by MALDI-MSI. Another phospholipid that was under-represented/not detected was CL. There are normally over eighty detectable molecular species of brain CL, but MALDI-MSI only observed 10, and this was achieved only after the tissue was treated with phosopholipase C (table 2 and see section 2.3 below).
Table 2.
Major CL species in pnd 17 rat brain. These 70 individual molecular species were detected as [M-H]- ions by ESI-LC-MS and as [M-2H+Na]- ions at a correspondingly greater m/z value by MALDI-MS and -MSI. If a species was previously characterized by MS/MS, its acyl chain compositions are given along with the reference: [1] = (Tyurin et al., 2008); [2] = (Bayir et al., 2007). Abundance is the relative abundance among all CL species detected as determined by ESI-LC-MS.
| Species | [M-H]- m/z |
Abundance | Acyl chains | Reference |
|---|---|---|---|---|
| CL(64:4) | 1343.94 | 0.2% | (C16:1)4 | [2] |
| CL(64:3) | 1345.91 | 0.2% | ||
| CL(64:2) | 1347.92 | 0.2% | ||
| CL(66:6) | 1367.89 | 0.2% | ||
| CL(66:5) | 1369.93 | 0.3% | ||
| CL(66:4) | 1371.94 | 0.5% | (C16:1)3/(C18:1)1 | [2] |
| CL(66:3) | 1373.96 | 0.5% | (C14:0)1/(C16:1)1/(C18:1)2 | [1] |
| CL(66:2) | 1375.97 | 0.4% | ||
| CL(66:1) | 1377.97 | 0.2% | ||
| CL(68:7) | 1393.94 | 0.4% | (C16:1)3/(C20:4)1 | [2] |
| CL(68:6) | 1395.95 | 0.6% | ||
| CL(68:5) | 1397.97 | 0.9% | (C16:1)2/(C18:1)1/(C18:2)1 | [2] |
| CL(68:4) | 1399.98 | 1.5% | (C16:1)2/(C18:1)2 | [2] |
| CL(68:3) | 1401.99 | 1.2% | (C16:1)1/(C16:0)1/(C18:1)2 | [1] [2] |
| CL(68:2) | 1404.01 | 0.7% | (C16:1)1/(C16:0)1/(C18:1)1/(C18:0)1; (C16:0)2/(C18:1)2 | [1] [2] |
| CL(68:1) | 1406.02 | 0.3% | ||
| CL(70:9) | 1417.93 | 0.2% | ||
| CL(70:8) | 1419.95 | 0.7% | ||
| CL(70:7)* | 1421.97 | 1.8% | (C16:1)1/(C18:2)3 | [2] |
| CL(70:6)* | 1423.98 | 2.1% | (C16:1)1/(C18:1)1/(C18:2)2; (C16:0)1/(C16:1)1/(C18:1)1/(C20:4)1 |
[1] [2] |
| CL(70:5)* | 1426.00 | 2.6% | (C16:1)1/(C18:2)1/(C18:1)2 | [1] [2] |
| CL(70:4)* | 1428.02 | 3.1% | (C16:1)1/(C18:1)3 | [1] [2] |
| CL(70:3)* | 1430.03 | 1.7% | ||
| CL(70:2) | 1432.04 | 0.6% | (C16:0)1/(C18:0)1/(C18:1)2 | [2] |
| CL(70:1) | 1434.04 | 0.3% | ||
| CL(72:10) | 1443.95 | 0.8% | ||
| CL(72:9) | 1445.97 | 1.5% | (C16:1)1/(C18:1)1/(C18:3)1/(C20:4)1; (C16:1)1/(C16:0)1/(C18:3)1/(C22:5)1 |
[2] |
| CL(72:8)* ** | 1447.99 | 3.6% | (C18:2)4; (C16:1)1/(C18:1)1/(C18:2)1/(C20:4)1; (C16:0)1/(C18:1)1/(C18:3)1/(C20:4)1& |
[1] |
| CL(72:7)* ** | 1450.00 | 4.8% | (C18:1)1/(C18:2)3; (C16:1)1/(C18:1)1/(C18:2)1/(C20:3)1 | [1] |
| CL(72:6)* ** | 1452.02 | 4.4% | (C18:1)2/(C18:2)2; (C16:1)1/(C18:1)2/(C20:3)1 | [2] |
| CL(72:5)* ** | 1454.03 | 4.4% | (C16:0)1/(C18:1)2/(C20:3)1; (C18:1)3/(C18:2)1 | [2] |
| CL(72:4)* | 1456.05 | 3.3% | (C18:1)4; (C16:0)1/(C18:1)2/(C20:2)1 | [2] |
| CL(72:3) | 1458.05 | 1.3% | ||
| CL(72:2) | 1460.02 | 0.4% | ||
| CL(74:12) | 1467.97 | 0.4% | ||
| CL(74:11)* | 1469.97 | 1.7% | ||
| CL(74:10)* | 1471.99 | 3.1% | ||
| CL(74:9)* ** | 1474.00 | 4.5% | (C18:1)1/(C18:2)2/(C20:4)1; (C16:0)1/(C18:1)1/(C18:2)1/(C22:6)1 |
[2] |
| CL(74:8)* ** | 1476.02 | 5.1% | (C16:1)1/(C18:1)2/(C22:5)1; (C16:1)1/(C18:2)1/(C20:3)1/(C20:2)1; (C16:0)1/(C18:1)2/(C22:6)1; (C16:0)1/(C18:1)1/(C20:4)1/(C20:3)1; (C18:1)2/(C18:2)1/(C20:4)1 |
[1] [2] |
| CL(74:7)* ** | 1478.03 | 4.2% | (C18:1)3/(C20:4)1; (C16:1)1/(C18:2)1/(C20:3)1/(C20:1)1& | [2] |
| CL(74:6)* | 1480.05 | 2.3% | (C18:1)2/(C18:0)1/(C20:4)1; (C16:0)1/(C18:1)1/(C20:4)1/(C22:4)1; (C16:0)1/(C18:0)1/(C20:4)1/(C22:5)1 |
[2] |
| CL(74:5) | 1482.06 | 1.0% | ||
| CL(74:4) | 1484.03 | 0.5% | ||
| CL(76:14) | 1491.96 | 0.3% | ||
| CL(76:13) | 1493.98 | 0.7% | (C16:1)1/(C18:2)1/(C20:4)1/(C22:6)1 | [2] |
| CL(76:12)* | 1495.99 | 2.2% | (C16:1)1/(C18:2)1/(C20:3)1/(C22:6)1 | [2] |
| CL(76:11)* ** | 1498.00 | 3.3% | (C18:1)1/(C18:2)1/(C20:4)2 | [1] [2] |
| CL(76:10)* ** | 1500.02 | 3.8% | (C18:1)2/(C20:4)2; (C18:0)1/(C18:2)2/(C22:6)1 | [1] [2] |
| CL(76:9)* ** | 1502.03 | 3.1% | (C18:1)2/(C20:4)1/(C20:3)1; (C16:0)1/(C18:1)1/(C20:4)1/(C22:4)1 |
[1] [2] |
| CL(76:8)* | 1504.05 | 1.7% | (C18:1)1/(C18:2)1/(C20:3)1/(C20:2)1; (C16:0)1/(C18:2)1/(C20:2)1/(C22:4)1 |
|
| CL(76:7) | 1506.06 | 0.8% | (C16:0)1/(C18:1)1/(C20:3)1/(C22:3)1; (C16:0)1/(C18:0)1/(C20:1)1/(C22:6)1 |
|
| CL(76:6) | 1508.04 | 0.5% | ||
| CL(78:15) | 1517.97 | 0.4% | (C16:1)1/(C18:2)1/(C22:6)2 | [1] [2] |
| CL(78:14)* | 1519.99 | 0.9% | (C18:2)1/(C20:4)3 | [1] |
| CL(78:13)* | 1522.00 | 1.6% | (C18:1)1/(C18:2)1/(C20:4)1/(C22:6)1; (C18:2)1/(C20:4)3 | [2] |
| CL(78:12)* | 1524.01 | 1.9% | (C18:1)2/(C20:4)1/(C22:6)1 | [1] [2] |
| CL(78:11)* | 1526.02 | 1.5% | (C18:1)2/(C20:3)1/(C22:6)1 | [2] |
| CL(78:10) | 1528.03 | 1.0% | (C18:1)2/(C20:4)1/(C22:4)1 | [1] |
| CL(78:9) | 1530.01 | 0.7% | ||
| CL(78:8) | 1532.04 | 0.4% | (C16:1)1/(C18:1)1/(C20:3)1/(C22:3)1; (C18:1)2/(C20:0)1/(C22:6)1 |
[2] |
| CL(80:16)* | 1543.98 | 0.5% | ||
| CL(80:15)* | 1545.98 | 0.8% | (C18:1)1/(C20:4)2/(C22:6)1 | [2] |
| CL(80:14)* ** |
||||
| cl(80:13)* ** | 1550.00 | 0.9% | (c18:1)1/(c18:0)1/(c22:6)2; (c18:1)2/(c22:5)1/(c22:6)1; (c18:0)1/(c20:4)2/(c22:5)1; (c18:2)1/(c20:3)2/(c22:5)1 ; (c18:2)1/(c18:0)1/(c22:5)1/(c22:6)1 |
[1] [2] |
| cl(80:12) ** | 1551.99 | 0.7% | (c18:0)2/(c22:6)2 | [1] [2] |
| cl(80:11) | 1554.01 | 0.6% | (c18:1)2/(c22:5)1/(c22:4)1; (c18:1)1/(c18:0)1/(c22:4)1/(c22:6)1 |
[2] |
| cl(82:18) | 1567.97 | 0.3% | ||
| cl(82:17) * | 1569.96 | 0.5% | (c18:1)1/(c20:4)1/(c22:6)2 | [1] [2] |
| cl(82:16) * | 1571.96 | 0.5% | (c18:1)1/(c20:3)1/(c22:6)2 | [2] |
| cl(82:15) | 1573.96 | 0.5% |
= detected by MALDI-MS
= detected by MALDI-MSI
= minor species
To obtain a better understanding of the relatively low representation of many diverse lipid species in MALDI-MSI-maps of the brain, we compared MALDI-MSI data to those obtained by ESI-LC-MS analysis of lipid extracts obtained from serial sections from brain tissue. We reasoned that the lipid distribution of a section of tissue was approximately similar in both of its serial sections. If the lipid extract and serial-section tissue section are analyzed by MS methods (ESI-LC-MS and MALDI-MSI as appropriate), then a direct comparison can be made (Burnum et al., 2009) (Hankin and Murphy, 2010) (Koeniger et al., 2011) (Delvolve et al., 2011).
Using negative mode MALDI, similar results between MALDI-MS and MALDI-MSI were seen (table 1). The most prominent classes of lipids seen in MALDI-MS were ST, PI and gangliosides, similar to MALDI-MSI. Positive mode MALDI-MS also yielded similar results to MALDI-MSI with the only significant change being a reduction in the intensity of some salt adducts. As with MALDI-MSI, Cer(d18:1/18:0) was the only species from its class that was detected by MALDI-MS in a total lipid extract, and its intensity was divided between [M-H2O+H]+ and [M+Na]+ adducts. Although the total lipid extract significantly purifies and concentrates low abundant species such as CL, it also concentrates other phospholipid species that contribute to CL signal suppression. In general, species that were under-represented with MALDI-MSI were likewise under-represented with MALDI-MS of the total lipid extract.
A comparison of ESI-LC-MS assessments of individual molecular species of lipids from serial tissue sections to lipid species detected by MALDI-MSI is presented in Table 1. It is apparent that MALDI-MSI was able to detect (and localize) only a relatively small fraction (~25%) of the total number of lipid species identified by ESI-LC-MS. Even when the results from positive- and negative-mode MALDI-MSI are combined, several classes of lipids are missing or under-represented. While some species of PE and PS might be detected in positive mode MALDI-MS as adduct masses isobaric to certain PC species (Hsu and Turk, 2005) (Garrett et al., 2007), our LC-MS analysis did not identify any such species. Only 10 species of CL were detected by MALDI-MSI (only after treatment of the tissue with phospholipase-C (PLC), see section 2.3 below) as opposed to 70 detected by ESI-LC-MS. There are several major reasons for the more limited detection of lipids by MALDI-MSI vs. LC-MS. To attempt to improve and enrich the representation of different lipids in MALDI-MSI we used several approaches as described in sections 2.3-2.6 below.
2.3 Treatment of tissue sections with phospholipases to reduce ion suppression in MALDI-MSI
In order to enhance the number of lipid species detected, we reasoned that treatment of the tissue with PC-specific phospholipase-C (PLC) prior to matrix deposition and MALDI-MSI may be an advantageous approach. PC is a highly abundant group of lipid species that can contribute to ion suppression (Schiller et al., 2002) (Johanson et al., 2007). This enzyme cleaves the head group of PC converting it to diacylglycerol (DAG) which has not been reported to contribute to ion suppression. Indeed, treatment of naïve pnd 17 rat brain tissue with PLC prior to MALDI-MSI allowed an improvement in the detection of several CL species (figs. 2 and 3). However, this only partially resolved the issue of ion suppression. Other higher abundance lipid species that dominate the spectra in the negative mode include PI, PS and gangliosides, and these compete for negative charges with the less abundant species such as CL. In addition, some of these species such as ganglioside GM1 (d18:1/20:0) at m/z 1544.8 are in the same mass range as brain CL (m/z 1400-1600) and can significantly mask signals from isobaric CL species. Here, ESI-LC-MS has an advantage by its ability to separate individual classes of lipids, resulting in greater sensitivity for low abundance CL species.
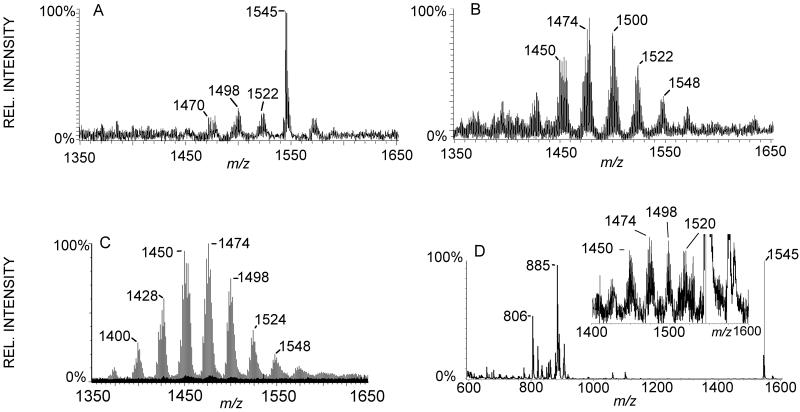
Fig 2.
Comparison MALDI-MS and ESI-LC-MS from pnd 17 rat brain. A: MALDI-MS of brain total lipid extract barely detects some CL species predominantly as [M-2H+Na]− adducts. Ganglioside GM1 at m/z 1545 obscures CL in this region. B: Split-flow LC-MS allowed collection of this CL fraction and subsequent analysis by MALDI-MS resulting in more species of CL detected, and these were mixtures of [M-H]− and [M-2H+Na]− adducts. C: ESI-LC-MS of same total lipid extract detects 70 species of CL, and these were detected as [M-H]− ions. D. On—tissue phospholipase C (PLC) treatment and subsequent washing results in MALDI-MSI detection of several species of CL predominantly as [M-H]− ions. The inset region is at 50× magnification. MALDI-MS and –MSI were acquired with an Autoflex in negative mode and ESI-LC-MS with a Q-TOF in negative mode.
Fig. 3.
MALDI-MSI of cardiolipin (CL) and other more abundant phosopholipids after on-tissue phospholipase C (PLC) treatment. The white bar is 2mm and the scale displays relative intensities with respect to the abundance of each ion. A: PI(38:4) m/z 885.5, B: ST(24:1) m/z 888.5, C: GM1(d18:1/18:0)m/z 1544.8; D: CL(72:8) m/z 1447.9, E: CL(74:7) m/z 1477.9, F: near-serial H&E section. MSI images were acquired with an Autoflex in negative mode.
2.4 Isobaric masses in MALDI-MSI: Identification of with the aid of ESI-LC-MS and high-mass resolution analyzers (FT-ICR)
Normal phase LC-MS also can be used to resolve isobaric lipid species seen in MALDI-MSI (fig. 4) by its ability to separate individual classes of phospholipids. A typical example is related to the previously mentioned (section 2.1) ions at m/z 806.5 and m/z 834.5 (negative mode) in murine brain that were isobaric for ST and PS species. Therefore each of those signals could represent PS, ST, or some combination of both. When the brain total lipid extract is separated by LC-MS, the ST and PS fractions elute at 4 min and 25 min, respectively. The spectrum of each of those fractions was examined for those two masses, and the ion at m/z 834.5 was present in both the ST and PS fractions while the ion at m/z 806.5 was present in the ST fraction with a very minimal signal seen in the PS fraction. Therefore the identity of the ion at m/z 806.5 from the MALDI image was mostly ST, while the ion at m/z 834.5 was a mixture of PS and ST at similar abundances.
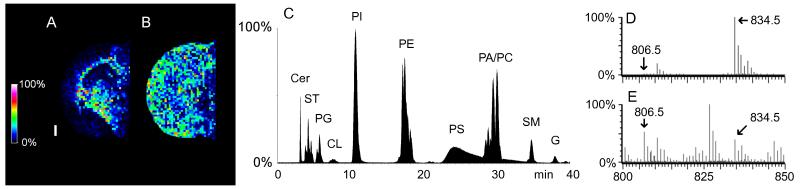
Fig. 4.
Identification of MALDI-MSI species at m/z 806.5 and m/z 834.5 with the aid of ESI-LC-MS. MALDI-MSI rat brain images of the species m/z 806.5 (A) and m/z 834.5 (B) could each be either PS or STspecies, or some combination of both. The white bar is 1 mm and the scale displays relative intensities with respect to the abundance of each ion. A total lipid extract of the serial section gives an ESI-LC-MS chromatogram (C) that separates PS and ST. LC-MS from the PS fraction (D) and ST fractions (E) show that 806.5m/z is almost entirely ST (arrows) and 834.5m/z is a mixture of PS and ST (arrows). MALDI-MS (Autoflex) and ESI-LC-MS (Q-TOF) spectra were acquired in negative mode.
Another approach to overcome the problem of isobaric masses in MALDI-MSI is through the use of high resolution FT-ICR analyzers. Although time of flight (TOF) instruments are often used for MALDI-MSI, they are not the only option. FT-ICRs, Orbitraps and other high mass resolution instruments can be fitted with MALDI sources and utilized for imaging, and such high-mass resolution spectra can be directly compared to sections analyzed with TOF instruments (Burnum et al., 2009). Performing MALDI-MSI with an FT-ICR instrument will result in considerably better mass accuracy and resolution than if a TOF instrument was used (fig. 5).
Fig. 5.
FT-ICR MSI completely resolves isobaric rat brain species. Several lipid species from various classes were detected at high mass resolution and their localizations correlated with an H&E stain of the tissue section. The white bar is 1000 microns and the scale displays relative intensities with respect to the abundance of each ion. Two sets of isobaric species (m/z 806.5 and 834.5) were completely resolved. The ions at m/z 806.497 and m/z 806.545 representing PS(38:6) and ST(d18:1/18:0) were resolved from each other, as were the ions at, m/z 834.528 and m/z 834.576 representing PS(40::6) and ST(d18:1/20:0), respectively. Each of these isobaric pairs was overlaid on the same MALDI image, but at the same absolute scale. Green pixels represented the PS species, and red represented the ST species. This confirmed that the m/z 806 species consisted almost entirely of ST(d18:1/18:0), while the m/z 834 species was mostly PS(18:0/22:6) but with a significant amount of ST(d18:1/20:0). The summation MALDI-MSI spectrum from the entire image further confirmed these results. MSI images were acquired with a Solarix in negative mode.
2.5 Limitations on spatial resolution of MALDI-MSI: oversampling for high spatial resolution
The diameter of the laser footprint usually places a limitation on the spatial resolution of MALDI-MSI of approximately 30×30 microns. However MALDI-MSI can be performed with a spatial resolution less than the diameter of the laser footprint by using oversampling-stepping (Jurchen et al., 2005). Utilizing this methodology, MSI of lipids can approach near-cellular resolution. An example illustrating the application of this to mapping lipids in the hippocampus is presented in figure 6. ST, PI, and gangliosides were still seen as the main species in negative mode, but their various co-localizations in different regions of the hippocampus were determined more precisely. ST(d18:1/24:0) was strongly localized to nearby vessels and in moderate abundance within the CA3 region, while ganglioside GM1 and PI(18:0/20:4) were almost entirely the opposite. Within the CA3 and dentate gyrus, GM1 and PI(18:0/20:4) were stronger in intensity on different layers.
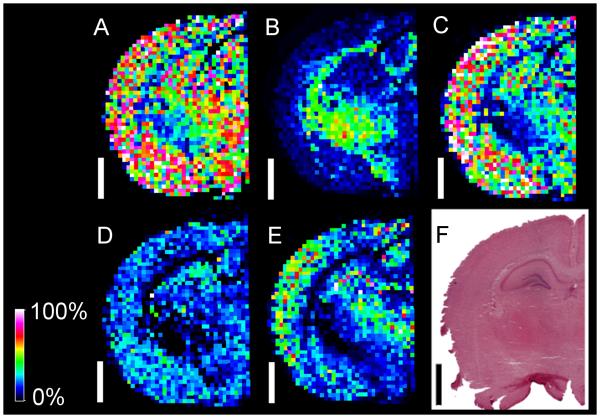
Fig. 6.
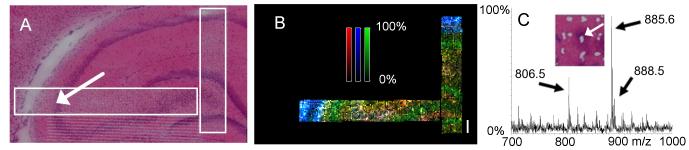
Oversampling to improve the spatial resolution of MALDI-MSI. A: Optical image of an H&E stain of rat brain after MALDI-MSI analysis of the regions of interest within the white boxes. Mass Spectra were collected with oversampling in both 20 and 10 micron rasters across the hippocampal area. B: MALDI-MSI image of three molecular species displaying distribution across fine structures within the hippocampus. The white scale bar is 200 microns. Different color pixels represent different species, and stronger pixel intensities indicate greater abundance with respect to each ion. Green: m/z 885.5 representing PI(38:4); Blue: m/z 885.5 representing ST(d18:1/24:0); Red: m/z 1544.8 representingGM1(d18:1/18:0). C: MALDI mass spectrum approaching cellular resolution (location indicated with arrows in panel A and inset region). White marks on H&E section resulted from laser ablation of tissue at the center of each position. MSI images were acquired with an Autoflex in negative mode.
Spectra approaching cellular resolution were determined by close comparison of the MALDI image to the tissue section. For these types of studies, the laser was set to dwell for repeated shots on each location resulting in the center of the beam ablating not only the matrix but also a small part of the tissue itself. Subsequent to MALDI-MSI, the actual tissue section (not a serial section in this case) was H&E stained, and the laser ablation marks were easily seen (fig. 6). Since each pixel in a MALDI image corresponds to a physical location on the tissue, the image corresponded exactly to the histological features of the tissue. Each pixel is a complete MALDI spectrum of one location. Because the tissue sections for MALDI-MSI are relatively thick (usually 8-12 microns), the analyte extraction and desorption process may involve more than one layer of cells thus setting a limit to near-celluar resolution (see section 3.2).
2.6 MALDI-MSI reveals physiologically- and pathologically-relevant changes of lipids in tissues
In spite of significant limitations inherent to current MALDI-MSI protocols, the analytical power of this emerging technology may be sufficient for revealing important site-specific changes in lipid molecular speciation in the tissues. To illustrate the potential of MALDI-MSI we present our results that have led to building lipid maps of the brain and lung tissue after several physiologically or pathologically relevant treatments. As illustrated by the data shown in figures 7, 8 and 9, we were able to reveal region-specific changes in the molecular species of lipids in brain and lung.
Fig. 7.
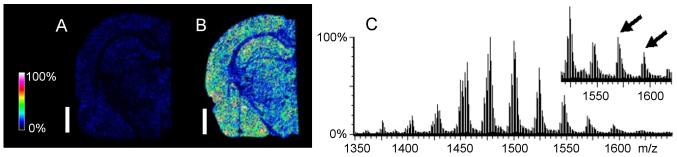
MALDI-MSI of adult rat brain tissue after DHA supplementation. Heat-map MALDI-MSI of brains from naïve (A) and DHA-dosed (B) rats showed increase in the ion at m/z 834.5 which is partly composed of PS(18:0/22:6). The white bar is 2 mm and the heat map is scaled to relative intensities with respect to the abundance of the ion at m/z 834.5. Both images are presented at the same abundance scale. As such, 100% intensity represents the highest intensity observed between both images. LC-MS profiles of CL extracts from naïve (C) and DHA-dosed (inset) serial brain sections showed an increase in DHA-containing CL marked with arrows – m/z 1570: CL(18:0)(20:4)(22:6×2); m/z 1592: CL(20:4×2) (22:6×2). Overall 29% of naïve and 38% of treated brain CL contained DHA. MALDI-MSI (Ultraflex II) and ESI-LC-MS (Q-TOF) spectra were acquired in negative mode.
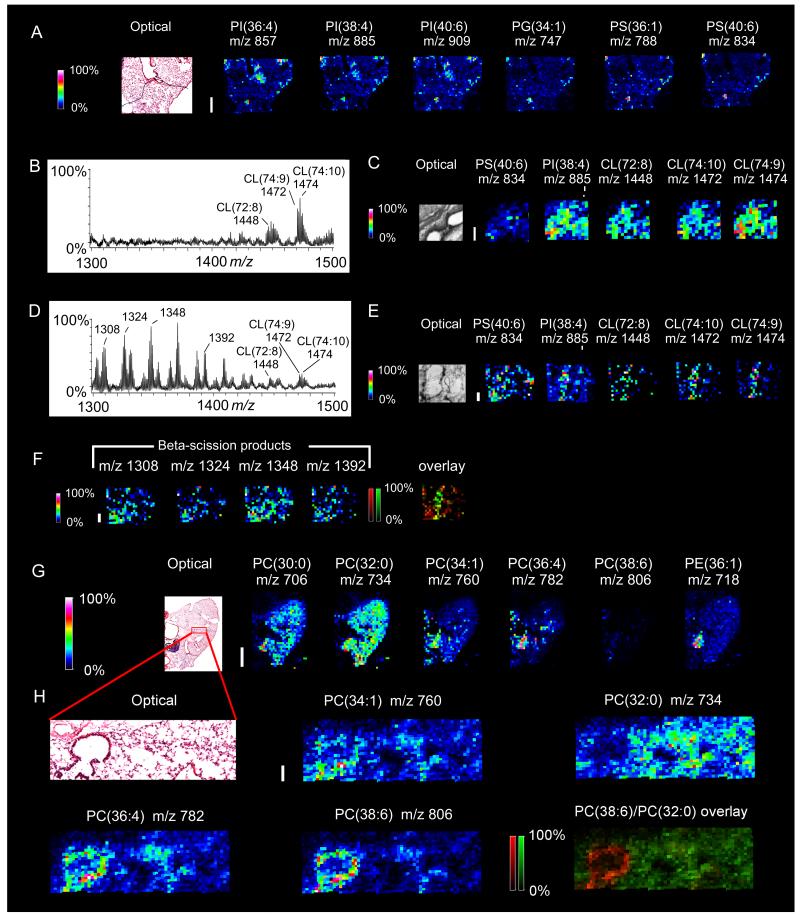
Fig. 8.
MALDI-MSI of mouse lung determines regional assignment of critical common phospholipids and oxidation products of less abundant cardiolipin in acute lung injury (ALI). MALDI-MSI of normoxic lung in negative mode (A) detected several phospholipids but not cardiolipin. MALDI-MSI was performed at 50 micron spatial resolution and the white bar is 500 microns. Averaged MALDI spectra (B) and MALDI-MSI images (C) of lung tissue section treated with PLC from normoxic mouse detected CL clusters but not oxidative species of CL. MALDI-MSI was performed at 50 micron spatial resolution and the white bar is 200 microns. ALI-model mouse lung MALDI spectra (D) and MALDI-MSI images (E,F) in negative mode demonstrate decrease in CL content with detection of ions that corresponded in mass to beta-scission fragments of oxidized CL (m/z 1308, 1324, 1348, and 1392 – see text for identification). Overlay image is of is m/z 1472 (green) and m/z 1348 (red). MALDI-MSI was performed at 50 micron spatial resolution and the white bar is 200 microns. MALDI-MSI of normoxic mouse lung in positive mode at 200 micron spatial resolution (G) detects a variety of lipids. The white bar is 2000 microns. Among the species detected were PC(16:0/16:0) and PC(16:0/22:6) as [M+H]+ ions at m/z 734.5 and m/z 806.6, respectively. MALDI-MSI with oversampling of a small area of the same lung at 20 micron resolution (H) and comparison with light microscopic H&E histology identified distinct regional distributions of these two species. Overlay of the two ions (green: m/z 734.5, red: m/z 806.6) in confirms this pattern. The white bar is 100 microns. MSI images were acquired with an Autoflex in negative mode (A), Ultraflex in negative mode (B-F), and Autoflex positive mode (G and H). All image scales depict relative intensities with respect to the abundance of each ion.
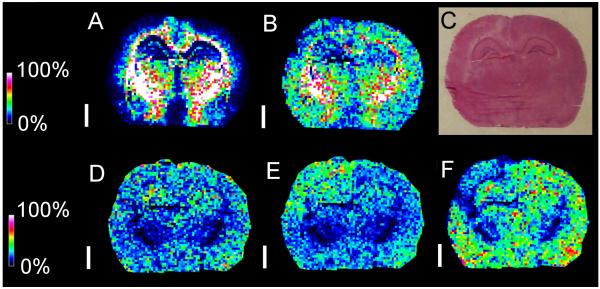
Fig. 9.
MALDI-MSI of pnd 17 rat brain after trauma. MALDI-MSI in negative mode shows different distributions of m/z 806.5 that contains nonoxidizable ST(d18:1/18:0) (A) and m/z 834.5 that containsoxidizable PS(18:0/22:6) (B) in brain after controlled cortical impact. Region of damage is shown by an arrow on serial H&E (C). The white bar is 200 microns and the scale displays relative intensities with respect to the abundance of each ion. While m/z 806.5 was detected in similar abundances between the hemispheres ipsilateral and contralateral to the point of impact, m/z 834.5 is somewhat reduced in the ipsilateral cortical and hippocampal regions with respect to the contralateral regions. MALDI-MSI in positive mode detected different abundances of PC(16:0/16:0) adducts between both hemispheres. While the [M+H]+ ion at m/z 734.5 (D) was detected in similar abundance ipsilateral and contralateral, the [M+Na]+ adduct ion at m/z 756.5 (E) was in greater abundance ipislateral while the [M+K]+ adduct ion at m/z 772.5 (F) was in greater abundance contralateral. Negative mode and positive mode MSI images were acquired with an Autoflex.
2.6.1 MALDI-MSI of brain tissue after DHA supplementation
Docosahexaenoic acid (DHA) is an essential polyunsaturated fatty acid in the central nervous system (CNS) and it has been reported to possess neuroprotective effects (Belayev et al., 2009) (Niemoller and Bazan, 2010). Both PS and CL consist of several molecular species that contain esterified DHA. The major species of DHA-containing PS in the brain is PS(18:0/22:6), and there are many diverse DHA-containing CL species that we have determined previously (Tyurin et al., 2009). We assumed that after i.p. injection of DHA as a free fatty acid the majority of supplemental DHA would become esterified into different classes of phospholipids resulting in very low levels of free DHA remaining in the brain. The identity of those phospholipids was determined and quantitatively assessed by MALDI-MSI and ESI-LC-MS (fig. 7). The effectiveness of DHA esterification into PS and CL resulted in a much greater abundance of DHA-PS and DHA-CL is illustrated by figure 7.
2.6.2 Detection of lipid peroxidation products by MALDI-MSI
Lipid peroxidation products are usually present in very low abundance and hence are particularly difficult to detect by MALDI-MSI. One can envision two major options to detect the changes associated with oxidative lipid modifications: 1) direct detection of peroxidized lipids or their oxidation products, and 2) indirect documentation of the decreased content of polyunsaturated lipids, the major substrates of lipid peroxidation. The latter protocol may benefit from cumulative monitoring of the disappearance of polyunsaturated species each of which may be converted into a plethora of oxidation products with expectedly very low levels of newly generated oxygenated molecular species.
2.6.2.1 Regional assignment of major phospholipid classes and detection of less abundant cardiolipin oxidation products by MALDI-MSI in acute lung injury (ALI)
In contrast to brain, lung presents a number of unique challenges in MALDI-MSI including: a) heterogeneous multicellular architecture places a premium on high resolution imaging to approach complexities of cellular function; and b) traditional requirement for both inflation and embedding of nonrigid lung to facilitate thin sectioning may introduce chemicals known to be incompatible with MALDI-MSI (Berry et al, 2011). Although both brain and lung CL are present in low abundances, the majority of lung CL is concentrated into less than a dozen molecular species, many of which are enriched in linoleic (C18:2) acid (Tyurina et al., 2010). Since each species of CL can produce a broad range of oxidatively modified products, starting with fewer species of CL will potentially improve the possibility of detection of their oxidized products. Accordingly, we approached aspects of uniqueness of lung phospholipid analysis by MALDI-MSI, by following lead of Berry et al (2011) and: a) preserved architecture with a modified OCT embedding agent followed by oversampling for high spatial resolution MSI; and b) acutely injuring a limited segment (upper airways) of lung of intact mice and focused on predicted changes in CL including its oxidative products (Tyurina et al, 2010; Tyurina et al, 2011).
Mouse lungs were inflated with modified OCT (Berry et al., 2011) and a section was examined by negative mode MALDI-MSI followed by light microscopy (fig. 8A). Although several species of phospholipids - PS, PI, PG - were uniformly detected, CL was not. This result was not surprising, considering the abundance of PC in lung and its ability to suppress the signals of lower-abundance phospholipids (see sections 2.2-2.3 above).
In order to analyze CL, MALDI-MSI of naive lung tissue was performed on sections treated with PLC (figs. 8B and 8C). A limited number of clusters of CL (in contrast to brain) were detected and their m/z corresponded to our recent reports (Tyurina et al., 2010) (Tyurina et al., 2011) from whole lung homogenates. After intratracheal instillation of a strong pro-oxidant system of glucose oxidase, lactoperoxidase + D-glucose (Johnson et al., 1981) that readily causes acute injury to upper airway epithelium, MALDI-MSI with PLC treatment revealed a large number of detectable oxidatively degraded (truncated) CL species (figs. 8D, 8E, and 8F). Beta-scission is the process of non-enzymatic, homolytic cleavage of peroxidated lipids (Picariello et al., 2009), and this produces a wide variety of truncated products. For example, each peroxidized linoleic (C18:2) fatty acyl chain can undergo beta scission resulting in both a truncated chain attached to the oxidized CL and the formation of 4-hydroxynonenal (4-HNE) or 4-hydroxyhexenal (4-HHE). These truncated-CLs will appear at m/z 1300-1400, a mass range that is normally devoid of lipid species. For example, TLCL (m/z 1448) with either a hydroxy-modification (+16 Da) or a peroxy-modification (+32 Da) could undergo beta scission with the formation of 4-HNE resulting in truncated species at m/z 1308 and 1324, respectively. Likewise, CL(74:10) and CL(74:9) at original m/z values 1472 and 1474 could undergo peroxy- or dihydroxy-modifications that would undergo beta scission with the formation of either 4-HNE or 4-HHE resulting in truncated species at m/z 1348 or 1392, respectively. Our previously published work indicated that lung CL species at m/z values of 1448, 1472 and 1474 contained at least one or more oxidizable fatty acyl chains (Tyurina et al., 2010). These few species of lung CL could potentially yield many oxidized species in the m/z 1300-1400 mass range. In initial MALDI-MSI experiments at a resolution of 50 μm, we detected small amounts of CL and a variety of oxidatively degraded CL in glucose oxidase treated airway epithelium (fig. 8F). To the best of our knowledge, this is the first in situ demonstration of this lower abundance PL in lung and also reveals the ability of MALDI-MSI in detecting oxidized species of CL and assigning critical anatomic location in experimental acute lung injury.
To more accurately assess regional distribution of the most common phospholipids, PC, a lung section from a naïve young-adult male mouse was examined by positive mode MALDI-MSI with both normal spatial resolution (figure 8G) and oversampling (figure 8H) followed by light microscopy. Lung phospholipids are dominated by PC and a major species, PC(16:0/16:0), contributes significantly to specialized airway lining fluid or surfactant, critical in preventing lung collapse during respiratory cycle. As noted in figure 8G, PC(16:0/16:0) with m/z 734.5 is distributed uniformly through distal lung. In contrast, the less abundant PC(16:0/22:6) with m/z 806.6 appears in a distinct compartment (fig. 8G). This localization is evident by overlaying the oversampling MALDI-MSI images of PC(16:0/16:0) (green) and PC(16:0/22:6) (red) in figure 8H. On closer examination this is associated with upper airway epithelium as confirmed by subsequent light microscopy of the tissue section. These findings are consistent with biochemical analyses of Bernhard et al (1997) who suggested that the alveolar origin of PC(16:0/16:0) was the source of this common saturated PL uniformly distributed throughout surface lining material of lung. This was in contrast to PL composition of upper airway epithelium, per se (and perhaps underlying connective tissue) that was enriched in PUFA-containing PL including PC(16:0/22:6) (Bernhard et al., 1997). The oversampling approach facilitated resolution to 20 μm allowing us to identify airway epithelium as the likely cellular source of PUFA-containing PC and conforming to the slightly lower resolution (approximately 50 μm) predictions of Berry et al (2011) that arachidonic acid- (and docosohexaenoic acid-) containing PC may be the source of precursors for important oxygenated lipid signaling pathways in lung. Imaging MS has revealed other important aspects of complex PC composition in regional components of kidney (Kaneko et al., 2011) and retina (Hayasaka et al., 2008).
2.6.2.2 Decreased content of oxidizable polyunsaturated lipids: MALDI-MSI in traumatic brain injury (TBI)
MALDI-MSI was used to analyze regional differences in PL distribution resulting from TBI. Two hours after CCI, coronal serial-sections were taken at the site of impact from pnd 17 rats for analysis in both positive and negative modes. There are two major ions in negative mode (m/z 806.5 and m/z 834.5) that are each isobaric for a PS and a ST species (fig. 9). Total lipid extracts from serial sections followed by LC-MS already have determined that the ion at m/z 806.5 is almost entirely ST(d18:1/18:0) with a small amount of oxidizable PS(16:0/22:6) while the ion at m/z 834.5 is a mixture of similar amounts of ST(d18:1/20:0) and oxidizable PS(18:0/22:6). MALDI-MSI determined that the ion at m/z 806.5 (mostly ST and localized to white matter) displayed no detectable change in the impact region. However the ion at m/z 834.5 displayed a decreased intensity in the cortical impact region but almost no change in the white matter. Other ST species did not change. Interestingly, these ST species localized predominantly to the white matter region and not the cortex, while only the ambiguous ion at m/z 834.5 displayed localization to both white matter and cortex. Therefore we concluded that the decrease in the ion at m/z 834.5 represented the loss of PS(18:0/22:6) but not its isobaric species ST(d18:1/20:0). In the brain both PS(16:0/22:6) and PS(18:0/22:6) contain a polyunsaturated DHA, and are susceptible to oxidative attack (Bayir et al., 2007). The former is in significantly less abundance than the latter. This confirms that the ion at m/z 806.5 is mostly ST, and the ion at m/z 834.5 is predominantly PS, and also indicates that PUFA-containing PS is diminished in the ipsilateral cortical and hippocampal regions in CCI. Concurrent loss of oxidizable PI(18:0/20:4) was detected in those regions as well. Although the total abundance of non-oxidizable PL species (such as PC(16:0/16:0), detected in positive mode on a serial section) did not change, as expected some displayed different adduct abundances within and outside the impact region (Hankin et al., 2011).
3. DISCUSSION
While MALDI-MS and ESI-MS represent a “gold standard” for lipid analysis at the molecular level, MALDI-MSI has recently become another critically important MS method that has added another dimension to the MS analysis of biomolecules allowing for molecular and spatial information to be correlated within a tissue section (reviewed in (Chaurand et al., 2011)). Several important MALDI-MSI papers have documented injury-associated changes in several types of lipids, including lysophosphatidylcholines, ceramides, gangliosides in brain injury models (Koizumi et al., 2010) (Hankin et al., 2011) (Whitehead et al., 2011). Murphy et al performed MALDI-MSI on lung tissue prepared with a MALDI-compatible embedding compound and demonstrated localization of PUFA-containing phospholipids to airways (Berry et al., 2011)
3.1 How many species of phospholipids do we “see” by MALDI-MSI?
Due to the high content and diversity of lipids in the CNS, brain tissue sections became one of the most common objects for MALDI-MSI (Delvolve et al., 2011). CNS tissue has the highest lipid content next to adipose tissue (Han, 2007). Groundbreaking work on lipid imaging identified 32 molecular species of phospholipids and sphingolipids in brain tissue (Jackson et al., 2007a). MALDI-MSI has been used to analyze a variety of brain lipids, including gangliosides, sulfatides, and phosphatidylcholines (Zemski-Berry et al., 2011). However there are obvious deficiencies in MALDI-MSI that warrant further methodological improvements. One of the significant limitations of MALDI-MSI is that it detects fewer species of some phospholipid classes than ESI-LC-MS (Hankin and Murphy, 2010). Our results demonstrate that ESI-LC-MS, when performed on serial sections of the brain in conjunction with MALDI-MSI, elucidates more information about lipids and their spatial localization than is possible with either technique alone. In normal brain tissue, approximately 25% of lipid molecular species detectable by ESI-LC-MS can be mapped by MALDI-MSI. Similarly, MALDI-MSI accounts for only a relatively small fraction of total brain lipids seen after DHA supplementation or TBI. In the lung, ESI-LC-MS detects ~100 molecular species of phospholipids, while only ~40 species are revealed by MALDI-MSI. Therefore, it is important to identify the major factors that contribute to the “masking” of lipids during sample preparation and performance of MALDI-MSI.
The MALDI image is generated pixel-by-pixel, by a laser rastering across the surface of a tissue. At each distinct spatial location on the tissue, the laser causes desorption and ionization of all the analytes at that location. In addition to lipids, tissue-associated salts and buffers are also desorbed. Each pixel on a MALDI image is a complete mass spectrum of everything detected at that one spatial location. A summation of many locations can provide regional spectra (for example, the brain region images in figure 1) or even a total spectrum for the entire tissue section. This total spectrum can thus be compared to the species detected from total lipid extracts of a serial section.
Instead of the location-oriented method used by MALDI-MSI, ESI-LC-MS of total lipid extracts separates lipids by their characteristic hydrophobic/hydrophilic class properties, in which the specific elution profiles are dictated by the choice of solid support and solvent system utilized. As a result, lower-abundance classes of lipids, such as CL, can be analyzed without interference from other more highly abundant lipids, thus improving the overall signal intensity of the CL species.
The issue of abundance is critical to any MS method, including MALDI-MSI. Those species that exist in higher abundance will preferentially dominate the spectrum, with the lower abundant species being barely detectable above the noise level. This is especially true with regard to lipid oxidation products. In model systems containing a purified, single lipid species, the diverse oxidation products produced will exist in readily detectable amounts. However, in biological systems, the oxidative stress insults trigger peroxidation reactions in multiple species resulting in the very minor amounts of oxidized lipid products of a particular type. These three factors (a wide variety of oxidizable molecular species, the low abundance of oxidation products, and the wide variety of possible oxidation products from each oxidizable species) make MALDI-MSI quantification of any oxidized lipid species even more challenging.
While MALDI-MSI is unique to other traditional MALDI techniques in its ability to generate both molecular and spatial information within a tissue sample, it is not surprising to find that it detects fewer species of lipids than its ESI-LC-MS counterpart. Analyte accessibility and abundance are two of the greatest challenges that MALDI-MS faces in order to perform as well as ESI-LC-MS. These issues can result in ion and signal suppressions during analysis. A variety of means have been used to overcome these challenges including improved methods of tissue preparation and MS instrumentation.
3.2 Overcoming the limitations of MALDI-MSI abundance and coverage through sample preparation
Depth of coverage for MALDI-MSI has been an ongoing question since the discovery that analytes from within the tissue interior can be extracted towards the surface during sample preparation and subsequently analyzed (Crossman et al., 2006). Phospholipid standards applied to a sample holder prior to tissue overlay have been successfully analyzed by MALDI-MSI after migration through the tissue (Landgraf et al., 2011). This demonstrates that a degree of extraction can occur either during matrix application or immediately prior to it. This extraction improves the signal intensity of phospholipids. Methods of matrix application and the addition of a recrystallization or rehydration step afterwards have been used to optimize the extraction of analytes (Bouschen et al., 2010) (Yang and Caprioli, 2011) (Thomas et al., 2012). Dry-coating of the matrix onto the tissue results in no real extraction, but this can be improved with a subsequent recrystallization step (Puolitaival et al., 2008). Some preparation methods even involve pre-coated slides in which the matrix is applied before the tissue, and is subsequently extracted to the surface (Grove et al., 2011). Partial laser ablation has been used to give some degree of depth profiling of tissues (Milasinovic et al., 2012). The ultimate extension of the analyte migration paradigm is to perform a series of lipid micro-extractions across the tissue surface, and analyze these discrete extracts by MS to generate a lipid image (Kertesz and Van Berkel, 2010) (Stegemann et al., 2011). This approach avoids the limitations of MALDI and matrix application. It also allows LC-MS to be performed at each discrete location, albeit at a significant worsening of spatial resolution as compared to MALDI-MSI (hundreds of microns as opposed to tens of microns or better with the latter).
MALDI profiling has been used to detect some lower-abundance species. It is different from MALDI-MSI in that a relatively large droplet (nanoliters) of matrix solution is applied to an area of tissue, and that whole area is analyzed by the summation of many laser shots (Jackson et al., 2005), Obviously, the spatial resolution is limited by the droplet size and is significantly worse than other techniques of matrix application such as sublimation or automated picoliter spotting (Hankin et al., 2007) (Aerni et al., 2005) (Delvolve and Woods, 2011). However MALDI profiling does strongly promote analyte extraction and incorporation into the matrix. MALDI profiling has been used for direct analysis of CL in rat tissue section (Wang et al., 2007). The tissues analyzed in that study were ones that were abundant in CL and exhibited very little molecular diversity. In contrast, CL in the brain and in oxidative injury models exhibits wide diversity (Tyurina et al., 2010) (Bayir et al., 2007).
Tissue pretreatment with a series of aqueous washes is one other method to reduce interference from tissue salts and buffers (Angel et al., 2012). Significant improvement in signal intensity and the number of lipid species observed has thus resulted from the removal of compounds that could produce in-source suppression. Phosphatidylcholine is present in large amounts in cellular membranes, and somewhat suppresses the signals of other PLs (Schiller et al., 2002). It is noteworthy that it suppresses signals in negative mode spectra even though PC is not normally detected in that mode. Another adverse effect is that in positive mode it will form a variety of homo- and hetero-dimers with other PC species and with PE, resulting in strong peaks in the same m/z 1400-1600 range where CL signals appear (Eibisch et al., 2011). As mentioned above (section 2.3), pre-treatment of samples with PLC removes the headgroup of phospholipids by cleavage at the glycerol-phosphodiester bond. Different sources of PLC will have different specificity for phospholipid substrates, and some sources (such as Clostridium bifermentans) are specific exclusively for PC (Ramrakhiani and Chand, 2011). Application of this enzyme to a tissue section will convert PC into DAG by removal of the phosphocholine headgroup, and thus help unmask signal from other, less abundant phospholipids, including CLs, in MALDI-MSI.
Imaging with tandem mass spectrometry in MS/MS or multiple-stage tandem mass spectrometry (MSn) uses fragmentation to help resolve the problem of isobaric and isomeric species (Garrett et al., 2007) (Garrett and Yost, 2010). Unfortunately MALDI-TOF can only examine one narrow mass window at a time in that mode (Wang et al., 2008). Thus it would require either re-imaging a section (Garrett et al., 2007) or preparing an additional tissue section in order to generate a comprehensive MALDI-TOF-MS/MS image. As such, it could require many sections and long instrumental run times to perform extensive MS/MS imaging. Despite this, MALDI-MS/MS and MSn is an excellent approach to identify isobaric species from MSI and quantify their compositions.
Another promising approach to more completely detect a greater variety of phospholipids is through the employment of a combination of serial sections for MALDI-MSI and ESI-LC-MS. Total lipid extracts can be performed on entire tissue section or microdissected areas thereof (Burnum et al., 2009) (Axelsen and Murphy, 2010) (Delvolve et al., 2011) (Hankin and Murphy, 2010). Tissue salts, buffers, and other non-lipid components are removed as part of the extraction. The lipid distribution of a section of tissue will be the same as both of its serial sections. With these assumptions, a direct comparison of ESI-LC-MS and MALDI-MSI data can be made to enrich the information on the diversified lipids, particularly those present in low abundance (Koeniger et al., 2011). ESI-LC-MS can be performed, where the effluent from a liquid chromatography column is fed directly into an ESI-MS instrument. Split-flow LC-MS allows fractions to be collected simultaneously with ESI-MS analysis.
3.3 Ion suppression and isobaric species
One other advantage of LC-MS is separation of isobaric and isomeric species (Minkler and Hoppel, 2010) (Kim et al., 2011). Depending on the limitations of the mass analyzer used for MALDI-MSI, isobaric species of different lipid types may not be resolved and will be detected as an ambiguous ion. Several phospholipid species are isobaric within the MALDI-TOF instrument’s mass resolution limit, hindering identification by MALDI-TOF-MSI. A similar issue is the presence of 13C isotope peaks that can be intense, due to the large amounts of carbon present in lipid species (Scherer et al., 2010). For example, in rat brain PI(16:0/18:1) at m/z 835.5 (negative mode) is significantly less abundant than PS(18:0/22:6) at m/z 834.5, and without prior separation would be masked within the A+1 isotope (m/z 835.5) of the latter.
Although MALDI-MSI cannot separate analytes prior to analysis, there are approaches that utilize separation of the analytes after they are desorbed and ionized. Differentiating ions of similar mass by ion mobility spectrometry (IMS) will greatly improve imaging data (Jackson et al., 2008). But this of course only works when there are ions to be detected - ion suppression by charge-competition at the source of ionization cannot be resolved by better separation post-ionization. Employing IMS in-line improves MALDI-TOF-MSI coverage and minimizes suppression of lower-abundance species (Jackson et al., 2007b) (Kiss and Heeren, 2011). This gives one dimension of separation after ionization, separates lipids from different classes with similar masses, and can even separate isomeric species from the same class (Woods and Jackson, 2010).
Improving the mass analyzer on the MALDI instrument is one obvious method to resolve isobaric species. ICR analyzers, Orbitraps, and other FT-MS instruments with high mass resolution have been used successfully for MALDI-MSI of lipids (Burnum et al., 2009) (Vidová et al., 2010b) (Landgraf et al., 2009). Of course, isomers cannot be resolved purely by mass resolution -- oxidizable PS(18:0/18:2) and non-oxidizable PS(18:1×2) both produce ions at m/z 786.5285. One disadvantage of FT-MS is the longer acquisition times as compared to TOF-MS. Orbitraps and ICR’s both function by trapping ions of a given m/z range, and then measuring the induced current as the ions repeatedly pass a detector plate, and this improved mass resolution requires longer measurement times (Gross, 2011). For example, the FT-MS image of a small area of a brain section (fig 5) required approximately 8 hours to acquire with as compared with 2 hours for the TOF image of an entire brain section in figure 3. However FT-MS can accumulate ions from many laser shots prior to analysis and this allows the simultaneous analysis of one imaging location while the next location is being scanned. Thus ions from only a select mass region of interest could be trapped for analysis, further improving sensitivity of FT-MS or other trapping instruments.
3.4 Matrix improvement for MALDI-MSI
MALDI-MS analysis requires the sample to be dried or otherwise turned into a condensed state, and is tolerant to some extent of tissue salts and buffers. As such, this combination makes MALDI an attractive method of tissue imaging. MALDI requires a matrix compound to be applied in large excess to the analytes of interest, although direct laser-desorption imaging of intact phospholipids from tissue samples in the absence of matrix or nanoparticles has been reported recently (Goodwin et al., 2011). The matrix is usually a small organic molecule exhibiting a strong absorption at a particular laser wavelength. A laser (usually in the UV range) is then pulsed across the sample. Just enough laser energy is applied to desorb the matrix off the surface in a plume, carrying nearby molecules with it. Rapid charge-transfer reactions take place within the plume during or immediately after desorption (Gross, 2011), resulting in the ionization and subsequent detection of phospholipid analytes. As of now, there is no one perfect matrix for lipid MALDI-MS or MSI (Wang et al., 2008) (Kim et al., 2010) (Thomas et al., 2012). As per Fuchs et al. (2010), MALDI-MSI matrices need to exhibit the following five characteristics: 1. Strong absorbance of the laser energy at its emission wavelength; 2. Ability to promote the ionization of analytes; 3. Stability under high vacuum; 4. Ability to promote the isolation of ions and minimize cluster formation; and 5. Formation of a homogenous surface layer with analyte incorporation. The matrix requirements for MSI are even more stringent than for non-imaging MALDI analyses. It is easier to deposit a homogeneous layer of matrix onto a metal MALDI target plate (for non-imaging) than it is onto a frozen tissue section. Analyte extraction from the tissue and incorporation into the matrix is essential (see section 3.2 above). Different matrix formulations have been used for different MALDI analytes (Fuchs et al., 2010) (Thomas et al., 2012). 2,5-dihydroxybenzoic acid (DHB) has often been used for lipids, but others have been reported to be useful (Stübiger and Belgacem, 2007). 9-Aminoacridine (9-AA) has been used for MALDI-MS analysis of phospholipids in the negative mode (Sun et al., 2007) (Teuber et al., 2010), including analysis of CL (Sun et al., 2008) (Lobasso et al., 2010) (Eibisch et al., 2011). In addition to organic matrix compounds, gold and silver nanoparticles have been utilized for MALDI-MSI of lipids (Jackson et al., 2007b) (Hayasaka et al., 2010). 10-N-Nonyl-acridine orange (NAO) is a commonly used fluorescent stain for cardiolipin. Photo-active compounds that selectively bind to CL such as nonyl-acridine orange may be potentially developed into useful matrices for improved MSI (Sparvero et al., 2011).
3.5 Improving the spatial resolution of MALDI-MSI
The effective utilization of MALDI-MSI for “biochemical lipid microscopy” requires sufficient spatial resolution. Analyte migration during matrix application can limit spatial resolution (see section 3.2 above). The size of the laser footprint is another critical limitation. Conventional MALDI-MSI instrumentation usually can achieve a spatial resolution of ~30×30 microns (Chaurand et al., 2011) (Urban et al., 2011) (Aizikov et al., 2011). While detailed information on lipids in different anatomical areas of organs and tissues can be obtained by these MALDI-MSI protocols, single cell or intracellular assessments are not possible. Recently several approaches have been developed to improve the spatial resolution of MALDI-MSI. One of them is based on oversampling whereby a laser beam is moved at a step of several microns with complete ablation (“bleaching”) of the matrix in the analyzed area (Jurchen et al., 2005). Although both the previous and new locations will be illuminated, there is only matrix in the new location. Thus only analytes from the new location are detected. This is repeated over a whole tissue area to give MALDI-MSI with spatial resolution approaching cellular level (less than 25μm). For this to be possible, the matrix needs to be deposited as very fine surface particles. Sublimation of organic matrices and sub-surface implantation of nanoparticle matrices have been used to achieve the highest spatial resolution (Murphy et al., 2009) (Murphy et al., 2011a) (Jackson et al., 2007b). A limitation of oversampling is that much greater acquisition times are required. As with any high spatial resolution technique, improving the spatial resolution results in more areas acquired (pixels in the image). Thus, improving the spatial resolution by a factor of two would result in four times as many discrete areas analyzed, and would increase the acquisition time by approximately a factor of four. Oversampling also requires enough laser shots to ensure the matrix in a given area is completely ablated, and this increase in number of laser shots can significantly increase the total acquisition time. For example, the entire brain section in figure 3 required approximately 2 hours to acquire, while the much smaller oversampling areas in figure 6 required 4 hours
A variant of this approach is continuous laser rastering, in which the laser is fired continuously as the sample stage moves, resulting in a continuous series of laser shots across the tissue section (Spraggins and Caprioli, 2011). This is different from other methods of MALDI-MSI where the laser is fired on discrete locations in between movements of the sample stage. One other approach is to use microinjection of a matrix solution on top of or directly into individual cells (Boggio et al., 2011). Microinjection does allow individual cells to be targeted for MALDI analysis. However its disadvantage is the difficulty in preparing a whole anatomical area for MALDI-MSI (such as the CA3 or dentate gyrus in the rat brain), while sublimation can prepare an entire tissue section in a matter of minutes.
Two other approaches allow MSI of lipids without the need for added matrix - laser desorption from tissue sections mounted to a nanostructure surface and desorption electrospray ionization (DESI). Nano-assisted laser desorption/ionization (NALDI) and Nanostructure initiator mass spectrometry (NIMS) are surface-assisted methods that use nano-structured support surfaces to absorb laser energy and thus do not require matrix to be added for desorption of analytes (Northen et al., 2007) (Greving et al., 2010). A recent NALDI-MSI work (Vidová et al., 2010a) impressed a tissue section onto the surface followed by MSI analysis of the imprint, thus identifying many prominent lipids with their spatial localization. DESI scans a sample with a spray of charged droplets, resulting in desorption and ionization of tissue analytes without a laser or matrix. This has been used to image lipids in a wide variety of tissue types (Eberlin et al., 2011a). DESI can be combined with subsequent MALDI-MSI, and this provides information on the co-localization of lipid and protein species within a single tissue section (Eberlin et al., 2011b).
Secondary-ion mass spectrometry (SIMS) is utilized for very high spatial resolution MSI (Passarelli and Winograd, 2011) (Benabdellah et al., 2010). This MS technique utilizes an ion beam as opposed to a laser-matrix combination in order to desorb and ionize analytes from a surface (Brunelle and Laprévote, 2009) (Touboul et al., 2011). Since SIMS does not require a matrix, there are no issues related to its chemical properties and the method of its deposition. An ion beam can be focused much tighter than a laser, thus the spatial resolution of SIMS is an order of magnitude better than MALDI. This has allowed single-cell imaging and the elucidation of subcellular lipid biology, as reviewed by Passarelli and Winograd (2011). For some lipid classes SIMS can produce images of comparable spectral quality to MALDI-MSI, however at much better spatial resolution (Wang et al., 2008) (Fernandez-Lima et al., 2011). SIMS can be utilized in static mode for surface analysis only or dynamic mode, and the latter allows depth profiling of tissue sections (Fletcher et al., 2011). However the high energies of the ion beam as presently used for SIMS can cause uncontrollable fragmentation of higher-mass lipid analytes (particularly CLs), and thus have been limited in practice to species of about 1000 Da or less (Debois et al., 2009).
3.6 Application of MALDI-MSI in different tissues
Comparison of progress in MALDI-MSI in lung versus brain reveals complexities and subtleties that arise from discrete structural, functional, biochemical and anatomical differences. As noted, the most progress in MALDI-MSI of lipids has been achieved with brain sections. This is primarily due to the frozen brain tissue being relatively amenable to microscopic examination after cryostat sectioning. Such preparation preserves structure without the need for embedding and fixation. In contrast, lung usually requires inflation, embedding and fixation with compounds that have proven incompatible with most forms of MALDI-MSI. In contrast to protein imaging, limited work has been done on lipid imaging of formalin-fixed tissues (Carter et al., 2011). Only recently has an embedding compound compatible with MALDI-MSI of lipids been developed for the lung (Berry et al., 2011). This methodology may be critical for a heterogeneous organ like lung devoid of large and approachable anatomic tracts of associated function as in brain. In this regard, we were gratified by our initial progress with analysis of unembedded frozen sections for preservation of tissue architecture (fig. 8). Additional specific issues in lung include the large abundance of phospholipids present as surfactant (primarily PG and PC) secreted into airways with the potential to obscure identification of more minor species. In this regard, we are encouraged by our ability to dilute such surfactants via lavage. Our focus on a low abundance phospholipid, CL, and its oxidized species truly underscores the sensitivity of MALDI-MSI. The close correlation of individual species of CL between MALDI-MSI and ESI-MS add further confidence to the approach. The capacity to discern (fig. 8) oxidation of CL within a region of the lung selectively affected by intratracheal delivery of potent free radical generating system (glucose oxidase) suggests that additional efforts in enhancing resolution (such as oversampling) may ultimately facilitate resolution to distinguish subtle biochemical events within distinct and important anatomic compartments of lung that structurally appear in close proximity with each other (e.g. endothelium and epithelium, etc.).
3.7 Conclusions
Despite its drawbacks, MALDI-MSI is a unique MS methodology that has the ability to generate both molecular and spatial information within a tissue section. While a specific biomarker, be it lipid or protein, may be evident using traditional ESI-LC-MS methods, the spatial confinements within normal or the diseased tissue will only be recognized with MALDI-MSI. We believe that we have aptly demonstrated this in several relevant paradigms addressing both the brain and the lung. The potential value of this approach in diseases such as TBI cannot be understated, given the fact that heterogeneous regional disturbances in the injured brain mandate methods that can map the lipid derangements to understand their contribution to neuronal injury and develop targeted therapies. This unique biochemical lipid microscopy may lead to new discoveries of the specific lipid biomarkers associated with area-specific alterations or damage under stress/disease conditions. As with any other newly emerging field, MALDI-MSI still has significant limitations and future optimization work aimed at improvements of its spectral and spatial resolution, sensitivity and specificity towards low abundance species of lipids is warranted.
4. MATERIALS AND METHODS
4.1 Tissue preparation
To demonstrate the capabilities of MALDI-MSI methods, we applied them to four separate experimental paradigms, namely, 1) assessment of the normal mouse brain, 2) assessment of the naïve and injured (by experimental traumatic brain injury (TBI) induced by controlled cortical impact (CCI)) immature (17 day old) rat brain, 3) assessment of the adult rat brain after i.p. injections with docosahexaenoic acid (DHA), and 4) assessment of mouse lung after acute lung injury (ALI). All procedures were pre-approved and performed according to the protocols established by the Institutional Animal Care and Use Committee of the University of Pittsburgh. Three naïve mouse brains were analyzed by MSI and LC-MS, and in each experimental condition the number of control and treated/challenged animals were as follows: CCI: 6 sham, 2 injured; DHA: 3 naïve, 2 treated. Immature (pnd 17) male Sprague Dawley rats were exposed to CCI as described previously (Bayir et al., 2007) (6-mm impactor tip, velocity of 4 ± 0.2 m/s, deformation depth of 2.5 mm, impact duration of 50 msecs) and sacrificed 2 hours after CCI. Adult Sprague Dawley rats were given DHA (16mg/kg) intraperitoneally (i.p.) for 3 days prior to sacrifice. This dose was chosen since when administered i.v. it significantly improved the neurologic outcome compared with vehicle-treated Sprague Dawley rats in a stroke model (Belayev et al., 2009). For the acute lung injury model, 6 control and 2 injured animals were studied. Acute lung injury (ALI) was created in C57B6 mice by intratracheal injection of 40μl of enzyme solution (4.4 units glucose oxidase (Sigma-Aldrich, St. Louis, MO, USA) plus 0.25 units lactoperoxidase (Sigma-Aldrich, St. Louis, MO, USA) in Hank’s balanced salt solution (HBSS, Life Technologies, Carlsbad, CA USA)) along with substrate (5.5 mM D-Glucose present in the HBSS) in anesthetized (1% isoflurane in air), intubated, spontaneously breathing mice (Johnson et al., 1981). At 3 hr after injury, mice were killed with overdose of sodium pentobarbital, chest-opened and lungs perfused through heart and inflated with phosphate-buffered saline (PBS, Life Technologies, Carlsbad, CA) by trachea. Brains or lungs were harvested from these animals and immediately frozen in liquid nitrogen with neither fixation nor embedding, and stored at −80°C until sectioning. Lungs prepared for oversampling MSI were embedded in a modified embedding compound (Berry et al., 2011) prior to freezing.
Tissue was affixed to a cryotome block with minimal Tissue-Tek OCT (Sakura FineTek USA, Torrance CA, USA). The blade and working surfaces of the cryotome were cleaned with methanol immediately prior to cutting, and at no time did the blade come into contact with OCT. Coronal brain sections of the dorsal hippocampus were cut at 10μm thickness at −19°C, while lung sections were 12μm and −22°C. Tissue sections were applied to cold histology slides pre-coated with a conductive indium tin oxide (ITO) surface (Delta Technologies LTD, Loveland, CO, USA). The slides were immediately dried under house vacuum for at least 1h and prepared for subsequent MALDI-MSI as described in section 4.2 below. Two serial sections were cut for each MALDI-MSI experiment (one each for positive and negative mode). Sections bracketing this set of two serial sections were cut and applied to plain glass slides for conventional histology and total lipid extraction. This set of sections on plain glass bracketing sections on conductive ITO slides was repeated at least three additional times, representative examples are presented for each.
4.2 LC and MS
Chloroform (HPLC grade), DHB (98+% grade) and KCl (reagent grade) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Methanol, trifluoroacetic acid (TFA), triethylamine, acetic acid were all LC-MS grade from Fisher Scientific (Pittsburgh, PA, USA). Water for LC use was LC-MS grade purchased from Fisher Scientific (Pittsburgh, PA, USA); water for other uses was purified by a milliQ system (EMD Millipore, Billerica, MA, USA). Phospholipid standards including phosphatidylcholine (PC(11:0×2)), phosphatidylserine (PS(17:0/17:0)), phosphatidylinositol (PI(16:0/16;0)), phosphatidylglycerol (PG(12:0×2)), sphinomyelin (SM(d18:1/12:0)) and cardiolipin (CL(14:0×4), CL(18:1×4), CL(18:2×4)) were purchased from Avanti Polar Lipids (Alabaster, AL, USA) and diluted to 20μM in 2:1 chloroform/methanol. Lipid standards and total lipid extracts were prepared for MALDI-MS in 2:1 chloroform: methanol by dried droplet method with 200mM DHB.
Lipid extraction from en bloc tissue and thin tissue sections on glass slides was performed with modified Folch method as described previously (Tyurina et al., 2011). Separation of individual phospholipid classes was accomplished by normal phase chromatography on a silica column (Luna 5μm, 25cm × 2 mm i.d., Phenomenex, Inc., Torrance, CA, USA) and analyzed on a Waters Premier quadrupole time-of-flight (Q-TOF) mass spectrometer (Waters, Inc., Milford, MA, USA) previously described (Samhan-Arias et al., 2012). Tuning was optimized for all lipids across the scan range and data was collected in centroid mode.
Select tissue sections were incubated prior to matrix application with 50μl of PC-specific phospholipase-C (PLC) from Clostridium bifermentans (Sigma-Aldrich, St. Louis MO, USA) at 3.3 mU/ μl for 20 min at 25°C. The PLC solution was then carefully pipetted off, followed by two brief (5 sec.) washes with ice-cold water. The tissue was vacuum dried overnight. PLC digestion of lipid standards indicated that these conditions gave a strong reduction in PC with no noticeable difference in CL (data not shown).
Matrix for MALDI-MSI was applied to sections with either solution application (Wang et al., 2008) or sublimation (Hankin et al., 2007) (Murphy et al., 2011a). Solution application of the MALDI matrix (55.0 mg/ml DHB in aqueous 70%/0.02% methanol/TFA) was accomplished using a commercial airbrush (two bursts of 1 sec. duration per minute, a cycle of 15 min. of spraying followed by 15 min. of vacuum drying repeated three times). Sublimation application of DHB was used to prepare sections for high-spatial resolution imaging. A modified sublimation chamber (Chemglass, Vineland NJ, USA) was used to deposit the matrix onto the slides as per (Murphy et al., 2011a) (350mg DHB, 4°C condenser). Prepared slides were stored at −80°C prior to analysis and allowed to equilibrate to ambient temperature for 1hr while under house vacuum immediately prior to MALDI-MSI. Serial sections were examined with hematoxylin and eosin (H&E) staining for correlation of histological features with MALDI images. In the oversampling experiment, the actual tissue section from the MALDI-MSI was subsequently used for H&E staining after remaining matrix was carefully washed off with absolute ethanol.
Time-of-flight (TOF) MALDI -MS and –MSI were performed using Bruker Ultraflex II and Autoflex TOF/TOF mass spectrometers (Bruker Daltonics, Billerica, MA, USA), and high mass resolution MSI was performed using a Solarix Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (Bruker Daltonics, Billerica, MA, USA). Negative polarity was used for anionic lipids (PS, PI, PG, CL, and phosphatidic acid (PA)) and positive polarity was used for the others. Reflector mode was used for all MALDI-TOF-MS experiments. Laser power and analyzer parameters were optimized for phospholipid detection in the m/z 400-2000 range. MALDI-MSI was carried out at a 50 μm and 200 μm rasters for normal resolution and at 10 μm and 20 μm rasters for oversampling. Normal MALDI-MSI was performed with 100 laser shots at 30% arbitrary power with partial sample random-walking every 20 shots, while oversampling used 200 laser shots at 35% power with no random-walking. In all cases the laser diameter value was set to “minimum”. MSI images and average MS spectra for regions of interest were generated with the Flex Imaging 3.0 software package (Bruker Daltonics, Billerica MA) at a window of +/- 0.2Da (+/-0.02 for FT-MS) unless stated otherwise. Extracted ion images were presented as raw pixels without interpolation. Normalization to total ion count (TIC) was not performed since it has been reported that this could, in feature-rich tissue such as brain, result in erroneous ion abundances (Deininger et al., 2011). Identification of ions as a particular lipid species was based solely on exact mass and comparison with species previously identified and characterized by MS/MS (Bayir et al., 2007) (Tyurin et al., 2008a).
Highlights.
Mapping of lipids by MALDI-MSI
Biochemical microscopy of DHA-containing lipids
MALDI-MSI of lipid oxidation products in acute lung injury
MALDI-MSI detection of lipid response to traumatic brain injury
MALDI-MSI detection of cardiolipin
Acknowledgments
This study was supported in part by grants from NIH (NS061817, HL70755, U19AIO68021, NS30318, HL65697 and ES021068); Cancer Center Support Grant (CCSG) P30 CA047904; NIOSH (OH008282) and the US Army (W81XWH-09-2-0187).
Abbreviations
- PE
Phosphatidylethanolamine
- PC
phosphatidylcholine
- CL
cardiolipin
- PS
phosphatidylserine
- PI
phosphatidylinositol
- PG
phosphatidylglycerol
- PA
phosphatidic acid
- SM
sphingomyelin
- ST
sulfatide
- Cer
ceramide
- DAG
diacylglycerol
- TOCL
1,1′,2,2′-tetraoleylcardiolipin
- TLCL
1,1′,2,2′-tetralinoleoylcardiolipin
- PL
phospholipid
- PUFA
polyunsaturated fatty acid or fatty acyl
- DHA
docosahexaenoic acid
- 4-HNE
4-hydroxynonenal
- 4-HHE
4-hydroxyhexenal
- CCI
controlled cortical impact
- CA
Cornu Ammonis
- PND
postnatal day
- ALI
acute lung injury
- TBI
traumatic brain injury
- CNS
central nervous system
- DHB
2,5-dihydroxybenzoic acid
- TFA
trifluoroacetic acid
- HBSS
Hank’s balanced salt solution
- PBS
phosphate-buffered saline
- H&E
hematoxylin and eosin
- NAO
10-N-nonyl-acridine orange
- PLC
phospholipase-C
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- MSn
multiple-stage tandem mass spectrometry
- MSI
mass spectrometry imaging
- MALDI
matrix-assisted laser desorption/ionization
- FT-ICR
Fourier transform ion cyclotron resonance
- TOF
time-of-flight
- Q-TOF
quadrupole time-of-flight
- ESI
electrospray ionization
- LC
liquid chromatography
- ITO
Indium tin oxide
- TIC
total ion count
- 9-AA
9-Aminoacridine
- SIMS
secondary ion mass spectrometry
- IMS
ion mobility spectrometry
- ISD
in-source decay
- DESI
desorption electrospray ionization
- NALDI
nano-assisted laser desorption/ionization
- NIMS
Nanostructure initiator mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author emails:
LJS, ljs39@pitt.edu; AAA, amoscatoaa@upmc.edu; CED, dixoec@upmc.edu; JBL, joseph.long@us.army.mil; PMK, kochanekpm@ccm.upmc.edu; BRP, brucep@pitt.edu; HB, bayihx@ccm.upmc.edu; VEK, kagan@pitt.edu
We have chosen to use “MALDI-MSI” although both it and “MALDI-IMS” are used by many authors in the field.
REFERENCES
- Aerni H-R, Cornett DS, Caprioli RM. Automated Acoustic Matrix Deposition for MALDI Sample Preparation. Analytical Chemistry. 2005;78:827–834. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- Aizikov K, Smith D, Chargin D, Ivanov S, Lin T, Heeren R, O’Connor P. Vacuum compatible sample positioning device for matrix assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry imaging. Rev Sci Instrum. 2011;82:054102. [Google Scholar]
- Amstalden van Hove ER, Smith DF, Heeren RMA. A concise review of mass spectrometry imaging. Journal of Chromatography A. 2010;1217:3946–3954. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- Angel PM, Spraggins JM, Baldwin HS, Caprioli RM. Enhanced Sensitivity for High Spatial Resolution Lipid Analysis by Negative Ion Mode MALDI Imaging Mass Spectrometry. Analytical Chemistry. 2012;84:1557–1564. doi: 10.1021/ac202383m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsen PH, Murphy RC. Quantitative analysis of phospholipids containing arachidonate and docosahexaenoate chains in microdissected regions of mouse brain. Journal of Lipid Research. 2010;51:660–671. doi: 10.1194/jlr.D001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayir H, Tyurin VA, Tyurina YY, Viner R, Ritov V, Amoscato AA, Zhao Q, Zhang XJ, Janesko-Feldman KL, Alexander H, Basova LV, Clark RS, Kochanek PM, Kagan VE. Selective early cardiolipin peroxidation after traumatic brain injury: an oxidative lipidomics analysis. Ann Neurol. 2007;62:154–169. doi: 10.1002/ana.21168. [DOI] [PubMed] [Google Scholar]
- Belayev L, Khoutorova L, Atkins KD, Bazan NG. Robust Docosahexaenoic Acid–Mediated Neuroprotection in a Rat Model of Transient, Focal Cerebral Ischemia. Stroke. 2009;40:3121–3126. doi: 10.1161/STROKEAHA.109.555979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabdellah F, Seyer A, Quinton L, Touboul D, Brunelle A, Laprévote O. Mass spectrometry imaging of rat brain sections: nanomolar sensitivity with MALDI versus nanometer resolution by TOF–SIMS. Analytical and Bioanalytical Chemistry. 2010;396:151–162. doi: 10.1007/s00216-009-3031-2. [DOI] [PubMed] [Google Scholar]
- Bernhard W, Haagsman HP, Tschernig T, Poets CF, Postle AD, van Eijk ME, von der Hardt H. Conductive Airway Surfactant: Surface-tension Function, Biochemical Composition, and Possible Alveolar Origin. American Journal of Respiratory Cell and Molecular Biology. 1997;17:41–50. doi: 10.1165/ajrcmb.17.1.2594. [DOI] [PubMed] [Google Scholar]
- Berry KA, Li B, Reynolds SD, Barkley RM, Gijon MA, Hankin JA, Henson PM, Murphy RC. MALDI imaging mass spectrometry of phospholipids in the mouse lung. J Lipid Res. 2011;52:1551–1560. doi: 10.1194/jlr.M015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggio KJ, Obasuyi E, Sugino K, Nelson SB, Agar NYR, Agar JN. Recent advances in single-cell MALDI mass spectrometry imaging and potential clinical impact. Expert Review of Proteomics. 2011;8:591–604. doi: 10.1586/epr.11.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouschen W, Schulz O, Eikel D, Spengler B. Matrix vapor deposition/recrystallization and dedicated spray preparation for high-resolution scanning microprobe matrix-assisted laser desorption/ionization imaging mass spectrometry (SMALDI-MS) of tissue and single cells. Rapid Communications in Mass Spectrometry. 2010;24:355–364. doi: 10.1002/rcm.4401. [DOI] [PubMed] [Google Scholar]
- Brunelle A, Laprévote O. Lipid imaging with cluster time-of-flight secondary ion mass spectrometry. Analytical and Bioanalytical Chemistry. 2009;393:31–35. doi: 10.1007/s00216-008-2367-3. [DOI] [PubMed] [Google Scholar]
- Burnum KE, Cornett DS, Puolitaival SM, Milne SB, Myers DS, Tranguch S, Brown HA, Dey SK, Caprioli RM. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. Journal of Lipid Research. 2009;50:2290–2298. doi: 10.1194/jlr.M900100-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, McLeod C, Bunch J. Imaging of Phospholipids in Formalin Fixed Rat Brain Sections by Matrix Assisted Laser Desorption/Ionization Mass Spectrometry. Journal of The American Society for Mass Spectrometry. 2011;22:1991–1998. doi: 10.1007/s13361-011-0227-4. [DOI] [PubMed] [Google Scholar]
- Chaurand P, Cornett DS, Angel PM, Caprioli RM. From whole-body sections down to cellular level, multiscale imaging of phospholipids by MALDI mass spectrometry. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.O110.004259. O110 004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossman L, McHugh NA, Hsieh Y, Korfmacher WA, Chen J. Investigation of the profiling depth in matrix-assisted laser desorption/ionization imaging mass spectrometry. RAPID COMMUNICATIONS IN MASS SPECTROMETRY. 2006;20:284–290. doi: 10.1002/rcm.2259. [DOI] [PubMed] [Google Scholar]
- Debois D, Bralet M-P, Le Naour F.o., Brunelle A, Laprevote O. In Situ Lipidomic Analysis of Nonalcoholic Fatty Liver by Cluster TOF-SIMS Imaging. Analytical Chemistry. 2009;81:2823–2831. doi: 10.1021/ac900045m. [DOI] [PubMed] [Google Scholar]
- Deininger S-O, Cornett D, Paape R, Becker M, Pineau C, Rauser S, Walch A, Wolski E. Normalization in MALDI-TOF imaging datasets of proteins: practical considerations. Analytical and Bioanalytical Chemistry. 2011;401:167–181. doi: 10.1007/s00216-011-4929-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvolve AM, Colsch B, Woods AS. Highlighting anatomical sub-structures in rat brain tissue using lipid imaging. Analytical Methods. 2011;3:1729–1736. doi: 10.1039/C1AY05107E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvolve AM, Woods AS. Optimization of automated matrix deposition for biomolecular mapping using a spotter. Journal of Mass Spectrometry. 2011;46:1046–1050. doi: 10.1002/jms.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues MRM, Reis A, Domingues P. Mass spectrometry analysis of oxidized phospholipids. Chemistry and Physics of Lipids. 2008;156:1–12. doi: 10.1016/j.chemphyslip.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Eberlin LS, Ferreira CR, Dill AL, Ifa DR, Cooks RG. Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2011a;1811:946–960. doi: 10.1016/j.bbalip.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin LS, Liu X, Ferreira CR, Santagata S, Agar NYR, Cooks RG. Desorption Electrospray Ionization then MALDI Mass Spectrometry Imaging of Lipid and Protein Distributions in Single Tissue Sections. Analytical Chemistry. 2011b;83:8366–8371. doi: 10.1021/ac202016x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibisch M, Zellmer S, Gebhardt R, Süß R, Fuchs B, Schiller J. Phosphatidylcholine dimers can be easily misinterpreted as cardiolipins in complex lipid mixtures: a matrix-assisted laser desorption/ionization time-of-flight mass spectrometric study of lipids from hepatocytes. Rapid Communications in Mass Spectrometry. 2011;25:2619–2626. doi: 10.1002/rcm.5161. [DOI] [PubMed] [Google Scholar]
- Fernandez-Lima FA, Post J, DeBord JD, Eller MJ, Verkhoturov SV, Della-Negra S, Woods AS, Schweikert EA. Analysis of native biological surfaces using a 100kV Massive Gold Cluster Source. Analytical Chemistry. 2011;83:8448–8453. doi: 10.1021/ac201481r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández JA, Ochoa B, Fresnedo O, Giralt MT, Rodríguez-Puertas R. Matrix-assisted laser desorption ionization imaging mass spectrometry in lipidomics. Anal Bioanal Chem. 2011;401:29–51. doi: 10.1007/s00216-011-4696-x. [DOI] [PubMed] [Google Scholar]
- Fletcher JS, Lockyer NP, Vickerman JC. Developments in molecular SIMS depth profiling and 3D imaging of biological systems using polyatomic primary ions. Mass Spectrometry Reviews. 2011;30:142–174. doi: 10.1002/mas.20275. [DOI] [PubMed] [Google Scholar]
- Fuchs B, Bresler K, Schiller J. Oxidative changes of lipids monitored by MALDI MS. Chemistry and Physics of Lipids. 2011;164:782–795. doi: 10.1016/j.chemphyslip.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Fuchs B, Süß R, Schiller J. An update of MALDI-TOF mass spectrometry in lipid research. Prog Lipid Res. 2010;49:450–475. doi: 10.1016/j.plipres.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Garrett T, Prieto-Conaway M, Kovtoun V, Bui H, Izgarian N, Stafford G, Yost R. Imaging of small molecules in tissue sections with a new intermediate-pressure MALDI linear ion trap mass spectrometer. Int J Mass Spectrom. 2007;260:166–176. [Google Scholar]
- Garrett TJ, Yost RA. Tandem Mass Spectrometric Methods for Phospholipid Analysis from Brain Tissue. Methods in Molecular Biology. 2010;656 doi: 10.1007/978-1-60761-746-4_12. [DOI] [PubMed] [Google Scholar]
- Girotti AW. Lipid hydroperoxide generation, turnover, and effector action in biological systems. Journal of Lipid Research. 1998;39:1529–1542. [PubMed] [Google Scholar]
- Goodwin RJA, Pitt AR, Harrison D, Weidt SK, Langridge-Smith PRR, Barrett MP, Logan Mackay C. Matrix-free mass spectrometric imaging using laser desorption ionisation Fourier transform ion cyclotron resonance mass spectrometry. Rapid Communications in Mass Spectrometry. 2011;25:969–972. doi: 10.1002/rcm.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto-Inoue N, Hayasaka T, Zaima N, Setou M. Imaging mass spectrometry for lipidomics. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2011;1811:961–969. doi: 10.1016/j.bbalip.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Greving MP, Patti GJ, Siuzdak G. Nanostructure-Initiator Mass Spectrometry Metabolite Analysis and Imaging. Analytical Chemistry. 2010;83:2–7. doi: 10.1021/ac101565f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JH. Mass Spectrometry. 2nd ed Springer-Verlag; Berlin Heidelberg: 2011. [Google Scholar]
- Grove K, Frappier S, Caprioli R. Matrix Pre-Coated MALDI MS Targets for Small Molecule Imaging in Tissues. Journal of The American Society for Mass Spectrometry. 2011;22:192–195. doi: 10.1007/s13361-010-0013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichardant M, Chen P, Liu M, Calzada C, Colas R, Véricel E, Lagarde M. Functional lipidomics of oxidized products from polyunsaturated fatty acids. Chemistry and Physics of Lipids. 2011;164:544–548. doi: 10.1016/j.chemphyslip.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Hall E, Detloff M, Johnson K, Kupina N. Peroxynitrite-mediated protein nitration and lipid peroxidation in a mouse model of traumatic brain injury. J Neurotrauma. 2004;21:9–20. doi: 10.1089/089771504772695904. [DOI] [PubMed] [Google Scholar]
- Hall ED, Vaishnav RA, Mustafa AG. Antioxidant Therapies for Traumatic Brain Injury. Neurotherapeutics: The Journal of the American Society for Experimental NeuroTherapeutics. 2010;7:51–61. doi: 10.1016/j.nurt.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. Neurolipidomics: challenges and developments. Front Biosci. 2007;12:2601–2615. doi: 10.2741/2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang K, Gross RW. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankin J, Farias S, Barkley R, Heidenreich K, Frey L, Hamazaki K, Kim H-Y, Murphy R. MALDI Mass Spectrometric Imaging of Lipids in Rat Brain Injury Models. Journal of The American Society for Mass Spectrometry. 2011;22:1014–1021. doi: 10.1007/s13361-011-0122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom. 2007;18:1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankin JA, Murphy RC. Relationship between MALDI IMS intensity and measured quantity of selected phospholipids in rat brain sections. Anal Chem. 2010;82:8476–8484. doi: 10.1021/ac101079v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka T, Goto-Inoue N, Sugiura Y, Zaima N, Nakanishi H, Ohishi K, Nakanishi S, Naito T, Taguchi R, Setou M. Matrix-assisted laser desorption/ionization quadrupole ion trap time-of-flight (MALDI-QIT-TOF)-based imaging mass spectrometry reveals a layered distribution of phospholipid molecular species in the mouse retina. Rapid Commun Mass Spectrom. 2008;22:3415–3426. doi: 10.1002/rcm.3751. [DOI] [PubMed] [Google Scholar]
- Hayasaka T, Goto-Inoue N, Zaima N, Shrivas K, Kashiwagi Y, Yamamoto M, Nakamoto M, Setou M. Imaging mass spectrometry with silver nanoparticles reveals the distribution of fatty acids in mouse retinal sections. Journal of The American Society for Mass Spectrometry. 2010;21:1446–1454. doi: 10.1016/j.jasms.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Hsu F-F, Turk J. Studies on Phosphatidylserine by Tandem Quadrupole and Multiple Stage Quadrupole Ion-Trap Mass Spectrometry with Electrospray Ionization: Structural Characterization and the Fragmentation Processes. Journal of The American Society for Mass Spectrometry. 2005;16:1510–1522. doi: 10.1016/j.jasms.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Jackson S, Ugarov M, Post J, Egan T, Langlais D, Schultz J, Woods A. A study of phospholipids by ion mobility TOFMS. Journal of The American Society for Mass Spectrometry. 2008;19:1655–1662. doi: 10.1016/j.jasms.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Wang H, Woods A. In situ structural characterization of glycerophospholipids and sulfatides in brain tissue using MALDI-MS/MS. Journal of The American Society for Mass Spectrometry. 2007a;18:17–26. doi: 10.1016/j.jasms.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Schultz JA, Woods AS. MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. Journal of Mass Spectrometry. 2007b;42:1093–1098. doi: 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SN, Wang H-YJ, Woods AS. Direct Profiling of Lipid Distribution in Brain Tissue Using MALDI-TOFMS. Analytical Chemistry. 2005;77:4523–4527. doi: 10.1021/ac050276v. [DOI] [PubMed] [Google Scholar]
- Johanson RA, Buccafusca R, Quong JN, Shaw MA, Berry GT. Phosphatidylcholine removal from brain lipid extracts expands lipid detection and enhances phosphoinositide quantification by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry Analytical Biochemistry. 2007;362:155–167. doi: 10.1016/j.ab.2006.12.026. [DOI] [PubMed] [Google Scholar]
- Johnson K, Fantone J.r., Kaplan J, Ward P. In vivo damage of rat lungs by oxygen metabolites. J Clin Invest. 1981;67:983–993. doi: 10.1172/JCI110149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurchen JC, Rubakhin SS, Sweedler JV. MALDI-MS Imaging of Features Smaller than the Size of the Laser Beam. Journal of the American Society for Mass Spectrometry. 2005;16:1654–1659. doi: 10.1016/j.jasms.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Scistunenko D, Kurnikov IV, Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapotptotic factors. Nature Chemical Biology. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Obata Y, Nishino T, Kakeya H, Miyazaki Y, Hayasaka T, Setou M, Furusu A, Kohno S. Imaging mass spectrometry analysis reveals an altered lipid distribution pattern in the tubular areas of hyper-IgA murine kidneys. Experimental and Molecular Pathology. 2011;91:614–621. doi: 10.1016/j.yexmp.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Kertesz V, Van Berkel GJ. Fully automated liquid extraction-based surface sampling and ionization using a chip-based robotic nanoelectrospray platform. Journal of Mass Spectrometry. 2010;45:252–260. doi: 10.1002/jms.1709. [DOI] [PubMed] [Google Scholar]
- Kiebish MA, Han X, Cheng H, Chuang JH, Seyfried TN. Cardiolipin and electron transport chain abnormalities in mouse brain tumor mitochondria: lipidomic evidence supporting the Warburg theory of cancer. Journal of Lipid Research. 2008;49:2545–2556. doi: 10.1194/jlr.M800319-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Minkler PE, Salomon RG, Anderson VE, Hoppel CL. Cardiolipin: characterization of distinct oxidized molecular species. J Lipid Res. 2011;52:125–135. doi: 10.1194/jlr.M010520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Shanta SR, Zhou L-H, Kim KP. Mass spectrometry based cellular phosphoinositides profiling and phospholipid analysis: A brief review. Experimental and Molecular Medicine. 2010;42:1–11. doi: 10.3858/emm.2010.42.1.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss A, Heeren R. Size, weight and position: ion mobility spectrometry and imaging MS combined. Analytical and Bioanalytical Chemistry. 2011;399:2623–2634. doi: 10.1007/s00216-010-4644-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeniger SL, Talaty N, Luo Y, Ready D, Voorbach M, Seifert T, Cepa S, Fagerland JA, Bouska J, Buck W, Johnson RW, Spanton S. A quantitation method for mass spectrometry imaging. Rapid Communications in Mass Spectrometry. 2011;25:503–510. doi: 10.1002/rcm.4891. [DOI] [PubMed] [Google Scholar]
- Koizumi S, Yamamoto S, Hayasaka T, Konishi Y, Yamaguchi-Okada M, Goto-Inoue N, Sugiura Y, Setou M, Namba H. Imaging mass spectrometry revealed the production of lyso-phosphatidylcholine in the injured ischemic rat brain. Neuroscience. 2010;168:219–225. doi: 10.1016/j.neuroscience.2010.03.056. [DOI] [PubMed] [Google Scholar]
- Korytowski W, Basova LV, Pilat A, Kernstock RM, Girotti AW. Permeabilization of the Mitochondrial Outer Membrane by Bax/Truncated Bid (tBid) Proteins as Sensitized by Cardiolipin Hydroperoxide Translocation: Mechanistic Implications for the Intrinsic Pathway of Oxidative Apoptosis. J Biol Chem. 2011;286:26334–26343. doi: 10.1074/jbc.M110.188516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarrigue M, Lavigne R, Guével B, Com E, Chaurand P, Pineau C. Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry: A Promising Technique for Reproductive Research. Biology of Reproduction. 2011 doi: 10.1095/biolreprod.111.094896. [DOI] [PubMed] [Google Scholar]
- Landgraf RR, Garrett TJ, Prieto Conaway MC, Calcutt NA, Stacpoole PW, Yost RA. Considerations for quantification of lipids in nerve tissue using matrix-assisted laser desorption/ionization mass spectrometric imaging. Rapid Communications in Mass Spectrometry. 2011;25:3178–3184. doi: 10.1002/rcm.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf RR, Prieto Conaway MC, Garrett TJ, Stacpoole PW, Yost RA. Imaging of Lipids in Spinal Cord Using Intermediate Pressure Matrix-Assisted Laser Desorption-Linear Ion Trap/Orbitrap MS. Analytical Chemistry. 2009;81:8488–8495. doi: 10.1021/ac901387u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Porter NA, Schneider C, Brash AR, Yin H. Formation of 4-hydroxynonenal from cardiolipin oxidation: Intramolecular peroxyl radical addition and decomposition. Free Radic Biol Med. 2011;50:166–178. doi: 10.1016/j.freeradbiomed.2010.10.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobasso S, Lopalco P, Angelini R, Baronio M, Fanizzi FP, Babudri F, Corcelli A. Lipidomic analysis of porcine olfactory epithelial membranes and cilia. Lipids. 2010;45:593–602. doi: 10.1007/s11745-010-3432-1. [DOI] [PubMed] [Google Scholar]
- Marcheselli VL, Hong S, Lukiw WJ, Tian XH, Gronert K, Musto A, Hardy M, Gimenez JM, Chiang N, Serhan CN, Bazan NG. Novel Docosanoids Inhibit Brain Ischemia-Reperfusion-mediated Leukocyte Infiltration and Pro-inflammatory Gene Expression. Journal of Biological Chemistry. 2003;278:43807–43817. doi: 10.1074/jbc.M305841200. [DOI] [PubMed] [Google Scholar]
- Milasinovic S, Liu Y, Bhardwaj C, Blaze MT,M, Gordon RJ, Hanley L. On the Feasibility of Depth Profiling of Animal Tissue by Ultrashort Pulse Laser Ablation. Analytical Chemistry. 2012 doi: 10.1021/ac300557a. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkler PE, Hoppel CL. Separation and characterization of cardiolipin molecular species by reverse-phase ion pair high-performance liquid chromatography-mass spectrometry. J Lipid Res. 2010;51:856–865. doi: 10.1194/jlr.D002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RC, Hankin JA, Barkley RM. Imaging of lipid species by MALDI mass spectrometry. J Lipid Res. 2009;50(Suppl):S317–322. doi: 10.1194/jlr.R800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RC, Hankin JA, Barkley RM, Zemski-Berry KA. MALDI imaging of lipids after matrix sublimation/deposition. Biochim Biophys Acta. 2011a;1811:970–975. doi: 10.1016/j.bbalip.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RC, Leiker TJ, Barkley RM. Glycerolipid and cholesterol ester analyses in biological samples by mass spectrometry. Biochim Biophys Acta. 2011b;1811:776–783. doi: 10.1016/j.bbalip.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemoller TD, Bazan NG. Docosahexaenoic acid neurolipidomics. Prostaglandins & other Lipid Mediators. 2010;91:85–89. doi: 10.1016/j.prostaglandins.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northen T, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordstrom A, Siuzdak G. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- Passarelli MK, Winograd N. Lipid imaging with time-of-flight secondary ion mass spectrometry (ToF-SIMS) Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2011;1811:976–990. doi: 10.1016/j.bbalip.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picariello G, Paduano A, Sacchi R, Addeo F. MALDI-TOF Mass Spectrometry Profiling of Polar and Nonpolar Fractions in Heated Vegetable Oils. Journal of Agricultural and Food Chemistry. 2009;57:5391–5400. doi: 10.1021/jf9008795. [DOI] [PubMed] [Google Scholar]
- Puolitaival SM, Burnum KE, Cornett DS, Caprioli RM. Solvent-Free Matrix Dry-Coating for MALDI Imaging of Phospholipids. Journal of the American Society for Mass Spectrometry. 2008;19:882–886. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrakhiani L, Chand S. Recent Progress on Phospholipases: Different Sources, Assay Methods, Industrial Potential and Pathogenicity. Applied Biochemistry and Biotechnology. 2011;164:991–1022. doi: 10.1007/s12010-011-9190-6. [DOI] [PubMed] [Google Scholar]
- Samhan-Arias AK, Ji J, Demidova OM, Sparvero LJ, Feng W, Tyurin VA, Tyurina YY, Epperly MW, Shvedova AA, Greenberger JS, Bayir H, Kagan VE, Amoscato AA. Oxidized phospholipids as biomarkers of tissue and cell damage with a focus on cardiolipin. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2012 doi: 10.1016/j.bbamem.2012.03.014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer M, Schmitz G, Liebisch G. Simultaneous Quantification of Cardiolipin, Bis(monoacylglycero)phosphate and their Precursors by Hydrophilic Interaction LC-MS/MS Including Correction of Isotopic Overlap. Analytical Chemistry. 2010;82:8794–8799. doi: 10.1021/ac1021826. [DOI] [PubMed] [Google Scholar]
- Schiller J, Süß R, Petkovic M, Zschörnig O, Arnold K. Negative-ion matrix-assisted laser desorption and ionization time-of-flight mass spectra of complex phospholipid mixtures in the presence of phosphatidylcholine: a cautionary note on peak assignment. Analytical Biochemistry. 2002;309:311–314. doi: 10.1016/s0003-2697(02)00308-1. [DOI] [PubMed] [Google Scholar]
- Schuhmann K, Herzog R, Schwudke D, Metelmann-Strupat W, Bornstein SR, Shevchenko A. Bottom-Up Shotgun Lipidomics by Higher Energy Collisional Dissociation on LTQ Orbitrap Mass Spectrometers. Anal Chem. 2011;83:5480–5487. doi: 10.1021/ac102505f. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- Singh IN, Sullivan PG, Deng Y, Mbye LH, Hall ED. Time course of post-traumatic mitochondrial oxidative damage and dysfunction in a mouse model of focal traumatic brain injury: implications for neuroprotective therapy. Journal of Cerebral Blood Flow & Metabolism. 2006;26:1407–1418. doi: 10.1038/sj.jcbfm.9600297. [DOI] [PubMed] [Google Scholar]
- Sparvero LJ, Amoscato A, Pitt BR, Bayir H, Kagan VE. Development of MALDI-TOF methodology for selective MS-analysis and imaging of cardiolipins in lipid extracts and tissues; Society of Toxicology 50th Annual Meeting; Washington, DC. 2011. [Google Scholar]
- Sparvero LJ, Amoscato AA, Kochanek PM, Pitt BR, Kagan VE, Bayir H. Mass-spectrometry based oxidative lipidomics and lipid imaging: applications in traumatic brain injury. J Neurochem. 2010;115:1322–1336. doi: 10.1111/j.1471-4159.2010.07055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spraggins J, Caprioli R. High-Speed MALDI-TOF Imaging Mass Spectrometry: Rapid Ion Image Acquisition and Considerations for Next Generation Instrumentation. Journal of The American Society for Mass Spectrometry. 2011;22:1022–1031. doi: 10.1007/s13361-011-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann C, Drozdov I, Shalhoub J, Humphries J, Ladroue C, Didangelos A, Baumert M, Allen M, Davies AH, Monaco C, Smith A, Xu Q, Mayr M. Comparative Lipidomics Profiling of Human Atherosclerotic Plaques / Clinical Perspective. Circulation: Cardiovascular Genetics. 2011;4:232–242. doi: 10.1161/CIRCGENETICS.110.959098. [DOI] [PubMed] [Google Scholar]
- Stübiger G, Belgacem O. Analysis of Lipids Using 2,4,6-Trihydroxyacetophenone as a Matrix for MALDI Mass Spectrometry. Analytical Chemistry. 2007;79:3206–3213. doi: 10.1021/ac062236c. [DOI] [PubMed] [Google Scholar]
- Sun G, Yang K, Zhao Z, Guan S, Han X, Gross RW. Shotgun Metabolomics Approach for the Analysis of Negatively Charged Water-Soluble Cellular Metabolites from Mouse Heart Tissue. Analytical Chemistry. 2007;79:6629–6640. doi: 10.1021/ac070843+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Yang K, Zhao Z, Guan S, Han X, Gross RW. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometric Analysis of Cellular Glycerophospholipids Enabled by Multiplexed Solvent Dependent Analyte-Matrix Interactions. Analytical Chemistry. 2008;80:7576–7585. doi: 10.1021/ac801200w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuber K, Schiller J, Fuchs B, Karas M, Jaskolla TW. Significant sensitivity improvements by matrix optimization: a MALDI-TOF mass spectrometric study of lipids from hen egg yolk. Chemistry and Physics of Lipids. 2010;163:552–560. doi: 10.1016/j.chemphyslip.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Thomas A, Charbonneau JL, Fournaise E, Chaurand P. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: Enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Analytical Chemistry. 2012;84 doi: 10.1021/ac2033547. [DOI] [PubMed] [Google Scholar]
- Touboul D, Brunelle A, Laprevote O. Mass spectrometry imaging: Towards a lipid microscope? Biochimie. 2011;93:113–119. doi: 10.1016/j.biochi.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Tyurin VA, Tyurina YY, Feng W, Mnuskin A, Jiang J, Tang M, Zhang X, Zhao Q, Kochanek PM, Clark RS, Bayir H, Kagan VE. Mass-spectrometric characterization of phospholipids and their primary peroxidation products in rat cortical neurons during staurosporine-induced apoptosis. J Neurochem. 2008a;107:1614–1633. doi: 10.1111/j.1471-4159.2008.05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyurin VA, Tyurina YY, Jung M-Y, Tungekar MA, Wasserloos KJ, BayIr H, Greenberger JS, Kochanek PM, Shvedova AA, Pitt B, Kagan VE. Mass-spectrometric analysis of hydroperoxy- and hydroxy-derivatives of cardiolipin and phosphatidylserine in cells and tissues induced by pro-apoptotic and pro-inflammatory stimuli. Journal of Chromatography B. 2009;877:2863–2872. doi: 10.1016/j.jchromb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyurin VA, Tyurina YY, Kochanek PM, Hamilton R, DeKosky ST, Greenberger JS, Bayir H, Kagan VE. Oxidative lipidomics of programmed cell death. Methods Enzymol. 2008b;442:375–393. doi: 10.1016/S0076-6879(08)01419-5. [DOI] [PubMed] [Google Scholar]
- Tyurin VA, Tyurina YY, Ritov VB, Lysytsya A, Amoscato AA, Kochanek PM, Hamilton R, Dekosky ST, Greenberger JS, Bayir H, Kagan VE. Oxidative lipidomics of apoptosis: quantitative assessment of phospholipid hydroperoxides in cells and tissues. Methods Mol Biol. 2010;610:353–374. doi: 10.1007/978-1-60327-029-8_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyurina YY, Kisin ER, Murray A, Tyurin VA, Kapralova VI, Sparvero LJ, Amoscato AA, Samhan-Arias AK, Swedin L, Lahesmaa R, Fadeel B, Shvedova AA, Kagan VE. Global Phospholipidomics Analysis Reveals Selective Pulmonary Peroxidation Profiles upon Inhalation of Single-Walled Carbon Nanotubes. ACS Nano. 2011;5:7342–7353. doi: 10.1021/nn202201j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyurina YY, Tyurin VA, Kaynar AM, Kapralova VI, Wasserloos K, Li J, Mosher M, Wright L, Wipf P, Watkins S, Pitt BR, Kagan VE. Oxidative lipidomics of hyperoxic acute lung injury: mass spectrometric characterization of cardiolipin and phosphatidylserine peroxidation. Am J Physiol Lung Cell Mol Physiol. 2010;299:L73–85. doi: 10.1152/ajplung.00035.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban PL, Chang C-H, Wu J-T, Chen Y-C. Microscale MALDI Imaging of Outer-Layer Lipids in Intact Egg Chambers from Drosophila melanogaster. Analytical Chemistry. 2011;83:3918–3925. doi: 10.1021/ac200481j. [DOI] [PubMed] [Google Scholar]
- Vaishnav RA, Singh IN, Miller DM, Hall ED. Lipid Peroxidation-Derived Reactive Aldehydes Directly and Differentially Impair Spinal Cord and Brain Mitochondrial Function. Journal of Neurotrauma. 2010;27:1311–1320. doi: 10.1089/neu.2009.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valko M, Morris H, Cronin M. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- Vidová V, Novak P, Strohalm M, Pol J, Havlicek V, Volny M. Laser Desorption-Ionization of Lipid Transfers: Tissue Mass Spectrometry Imaging without MALDI Matrix. Analytical Chemistry. 2010a;92:4994–4997. doi: 10.1021/ac100661h. [DOI] [PubMed] [Google Scholar]
- Vidová V, Pól J, Volný M, Novák P, Havlíček V, Wiedmer SK, Holopainen JM. Visualizing spatial lipid distribution in porcine lens by MALDI imaging high-resolution mass spectrometry. Journal of Lipid Research. 2010b;51:2295–2302. doi: 10.1194/jlr.M005488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-YJ, Jackson SN, Woods AS. Direct MALDI-MS Analysis of Cardiolipin from Rat Organs Sections. Journal of the American Society for Mass Spectrometry. 2007;18:567–577. doi: 10.1016/j.jasms.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-YJ, Post SN, Jackson J, Woods AS. A minimalist approach to MALDI imaging of glycerophospholipids and sphingolipids in rat brain sections. International Journal of Mass Spectrometry. 2008;278:143–149. doi: 10.1016/j.ijms.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH-Y, Liu CB, Wu H-W, Kuo JS. Direct profiling of phospholipids and lysophospholipids in rat brain sections after ischemic stroke. Rapid Commun. Mass Spectrom. 2010;24:2057–2064. doi: 10.1002/rcm.4620. [DOI] [PubMed] [Google Scholar]
- Weber L, Boll M, Stampfl A. Hepatotoxicity and mechanism of action of haloalkanes: carbon tetrachloride as a toxicological model. Crit Rev Toxicol. 2003;33:105–136. doi: 10.1080/713611034. [DOI] [PubMed] [Google Scholar]
- Whitehead SN, Chan KHN, Gangaraju S, Slinn J, Li J, Hou ST. Imaging Mass Spectrometry Detection of Gangliosides Species in the Mouse Brain following Transient Focal Cerebral Ischemia and Long-Term Recovery. PLoS One. 2011;6:e20808. doi: 10.1371/journal.pone.0020808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods AS, Jackson SN. The Application and Potential of Ion Mobility Mass Spectrometry in Imaging MS with a Focus on Lipids Methods Mol Biol. 2010;656:99–111. doi: 10.1007/978-1-60761-746-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Caprioli RM. Matrix Sublimation/Recrystallization for Imaging Proteins by Mass Spectrometry at High Spatial Resolution. Analytical Chemistry. 2011;83:5728–5734. doi: 10.1021/ac200998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xu L, Porter NA. Free Radical Lipid Peroxidation: Mechanisms and Analysis. Chemical Reviews. 2011;111:5944–5972. doi: 10.1021/cr200084z. [DOI] [PubMed] [Google Scholar]
- Zemski-Berry KA, Hankin JA, Barkley RM, Spraggins JM, Caprioli RM, Murphy RC. MALDI Imaging of Lipid Biochemistry in Tissues by Mass Spectrometry. Chemical Reviews. 2011;111:6491–6512. doi: 10.1021/cr200280p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-R, Scherch HM, Hall ED. Direct Measurement of Lipid Hydroperoxides in Iron-Dependent Spinal Neuronal Injury. Journal of Neurochemistry. 1996;66:355–361. doi: 10.1046/j.1471-4159.1996.66010355.x. [DOI] [PubMed] [Google Scholar]