Abstract
Quantitation of human dystrophin protein in muscle biopsies is a clinically relevant endpoint for both diagnosis and response to dystrophin-replacement therapies for dystrophinopathies. A robust and accurate assay would enable the use of dystrophin as a surrogate biomarker, particularly in exploratory Phase 2 trials. Currently available methods to quantitate dystrophin rely on immunoblot or immunohistochemistry methods that are not considered robust. Here we present a mass spectrometry based approach to accurately quantitate dystrophin protein in a total protein extract from human muscle biopsies. Our approach uses a combination of stable isotope labeled dystrophin as a spike-in standard, gel electrophoresis and high precision mass spectrometry to detect and quantitate multiple peptides of dystrophin within a complex protein mixture. The method was found highly reproducible and linear over a wide dynamic range, detecting as low as 5% of dystrophin relative to the normal amount in healthy individuals.
Keywords: Duchenne, Dystrophin, Quantitation, Mass spectrometry, Stable isotope
Introduction
Dystrophin is a relatively large (427 kDa) multi-domain protein that plays an essential role in muscle fiber integrity and function. It is predominantly expressed in skeletal muscle but other isoforms have been found expressed in heart and brain [1]. Dystrophin is a fairly low abundance protein, accounting for approximately 0.002% of the total striated muscle protein [2]. Gene mutations resulting in the complete loss of dystrophin protein expression are the cause of Duchenne muscular dystrophy (DMD), the most common and severe form of muscular dystrophy [3]. Mutations resulting in the production of a truncated but partially functional dystrophin protein give rise to a milder dystrophinopathy termed Becker muscular dystrophy (BMD). In general, BMD patients have longer lives and less severe clinical presentations than DMD patients. This is mainly due to the presence of the partially functional dystrophin protein in BMD patients [4]. But, BMD patients do have a wide variety of phenotypes ranging from mild to severe most probably due the variable amounts of expressed dystrophin often seen in BMD population [5].
The need to accurately detect and quantitate dystrophin in human skeletal muscle biopsies is becoming crucial as new promising therapies aiming to restore dystrophin expression, especially in DMD patients, are entering phase II/III clinical trials [6-10]. Perhaps one of the most promising new generation therapies for DMD is ‘exon skipping’ which has been effective in restoring dystrophin expression and muscle force in both mouse and dog models of DMD [11,12]. The goal of this approach is to ‘skip’ an exon neighboring a deletion mutation during pre-mRNA splicing, thereby restoring the mRNA reading frame, resulting in the production of a shorter length, partially functional Becker-like dystrophin protein. Two pharmaceutical companies are currently in clinical trials with different exon skipping chemistries for the treatment of Duchenne muscular dystrophy [6,10].
The most widely used clinical endpoint in Duchenne muscular dystrophy clinical trials is the 6 minute walk test [13]. However, significant changes in walking speed may take a year or more to develop, and there is significant test to test variation. Thus, relying on functional outcome measures such as the 6 minute walk increase the number of patients needed for adequate statistical powering, as well as the length of trial, and also increase the cost of conducting trials.
This is a particularly significant barrier to Phase 2 trials, where a more rapid read-out showing a proof-of-principle of the therapeutic approach is desired. Quantitation of dystrophin protein is an ideal endpoint measure [14] and clinically meaningful since it is directly linked to the disease. However, current methods for dystrophin quantitation such as western blots and immunohistochemistry are notoriously difficult and inconsistent. There have been successful attempts to improve immunohistochemistry quantitation, but it remains an indirect and fairly subjective method of quantitating dystrophin [15,16]. Furthermore, all antibody based approaches are limited due to dilution issues and inability to distinguish between splice isoforms. Alternatively, mRNA quantitation was also used as an assay to evaluate the levels of exon skipped products [17]. But, mRNA expression does not always correlate with protein levels [18] and amplification saturation can be problematic. Finally, both antibody based assays and mRNA approaches as surrogate biomarkers suffer from limited reliability and sensitivity. In an effort to overcome these hurdles, we have developed a quantitative dystrophin protein assay using high precision mass spectrometry in combination with stable isotope spike-in strategy. This technique detects and quantitates dystrophin protein in small amounts of total muscle extracts and is compatible with full length or truncated Becker-like dystrophin proteins. This assay shows features of a robust surrogate biomarker sought for Phase 2 trials focused on dystrophin replacement in DMD.
Methods
Muscle biopsies
All human muscle biopsies used in this study were previously banked in our laboratory in accordance with an Institutional Review Board approved protocol at Children's National Medical Center (CNMC) and after obtaining patients written informed consent. Only excess specimens after diagnosis were used. Diagnosis was previously performed at the Center for Genetic Medicine at Children's National Medical Center. Banked specimens were deidentified and only age, gender, diagnosis and phenotype were reported to us. Total protein extracts of muscle biopsies from healthy subjects were obtained from Dr. David Rowlands at Massey University, Wellington, New Zealand.
Custom peptides
Custom dystrophin peptides LLDLLEGLTGQK, LLVEELPLR, IFLTEQPLEGLEK and filamin c peptide VYNVTYTVK with 13C6-15N2-Lysine and 13C6-15N4- Arginine were purchased from New England Peptide (Gardner, MA). Peptides were suspended in 50% molecular grade acetonitrile containing 0.1% TFA and spiked into samples prior to mass spectrometric analyses.
SILAM/SILAC mouse
Custom “heavy” C57BL6 mice, termed SILAM or SILAC Mouse (stable isotopic labeling by amino acid in a mouse/mammal) were generated in-house and handled according to Institutional Animal Care and Use Committee guidelines at the Children's National Medical Center (Approved protocol # 199-07-01). Mice were fed a custom diet containing 13C6-Lysine (Cambridge Isotope Laboratories, Andover, MA) at 1%, adhering to laboratory mouse nutritional standards. A pregnant dam was fed with ‘heavy’ diet until a litter (F1 generation) was obtained and continued through weaning of the pups. After weaning, the pups were continued on ‘heavy’ diet through breeding and generation of a F2 litter. Label incorporation was monitored for specific organs and specific proteins by mass spectrometry. Labeling efficiency was better than 96% by F2 generation, including skeletal muscle [19]. This labeling efficiency was similar to labeling efficiencies obtained by others [20]. SILAC labeled skeletal muscles were harvested, flash frozen in liquid nitrogen chilled isopentane and stored at -80°C until analysis as previously described [19].
Muscle protein preparation
Approximately 50 serial sections were prepared from each of the human muscle biopsies and SILAC mouse gastrocnemius muscle using a cryostat set at 10 μm thickness. Muscle sections were directly placed into a microcentrifuge tube on dry ice. Total protein extraction from the muscle samples was performed using a handheld homogenizer either in a RIPA buffer (ThermoScientific) or a modified Laemmli sample buffer containing 10% SDS (10% SDS, 75 mM Tris-HCl, pH 6.8, 10 mM EDTA, 20% glycerol, 50 mM DTT) [21]. Protein concentration was determined using a BCA assay (Pierce, Thermo Scientific).
Dystrophin quantitation using mass spectrometry with stable isotope spike-in strategy
Two approaches were evaluated to quantitate dystrophin in skeletal muscle extracts (Figure 1). In the first approach, called post-digestion spike-in approach hereafter, aliquots of 50 μg total protein from each muscle extract was diluted in LDS buffer (Invitrogen, Carlsbad, CA) and further fractionated by SDS-PAGE on a 3-8% Tris-Acetate midi gel (Invitrogen) for 1 hour at 160 V. Gels were stained with BioSafe commassie (BioRad) for 1 hour. The upper section of the gel (300 to 450 kDa) containing dystrophin protein and other skeletal muscle proteins was excised and processed for in-gel digestion as previously described [22] using proteomics grade trypsin (Promega, Madison, WI). Peptides were extracted from the gel and spiked with known amounts (14 nM, 40 nM, 80 nM and 160 nM) of custom synthesized stable isotope labeled dystrophin peptides as well as an internal control filamin c peptide. In the second approach, called pre- digestion spike-in approach hereafter, aliquots of 50 μg of total protein from each muscle extract were spiked with 25 μg of muscle extract prepared from 13C6-Lysine labeled SILAC mouse. The mixture was then fractionated by SDS-PAGE on a 3-8% Tris-Acetate gel and gel bands in the 300-450 kDa were processed as above. To determine the limit of detection (LOD) and limit of quantitation (LOQ) of dystrophin protein in a complex muscle extract we used a muscle extract from a DMD patient, completely lacking dystrophin protein, and mixed at different ratios with a muscle extract from a healthy donor that expresses dystrophin as follows: DMD to normal 100%/0%, 95%/5%, 90%/10%, 75%/25%, 50%/50%, 25%/75%, 10%/90%, 5%/95%, 0%/100%. The final protein content in each mixture was kept constant at 50 μg. Each sample was then spiked with 25 μg of muscle extract prepared from 13C6-Lys labeled SILAM mouse to quantitate human dystrophin. Samples were analyzed using two different types of mass spectrometry instruments. For analysis by LTQ-Orbitrap-XL (Thermo), concentrated peptides from each spiked sample above were dissolved in 10 μL of 0.1% TFA solution and 6 μL was injected via an autosampler onto a Symmetry C18 trap column (5 μm, 300 μm i.d., 23 mm, Waters) for 10 min at a flow rate of 10 μm/min, 100% A. The sample was subsequently separated by a C18 reverse-phase column (3 μm, 200A, 100 μm×15 cm, Magic C18, Michrom Bioresources) at a flow rate of 330 nL/min using an Eksigent nano-hplc system (Dublin, CA). The mobile phases consisted of water with 0.1% formic acid (A) and 90% acetonitrile (B). A 65 minute linear gradient from 5 to 40% B was employed. Eluted peptides were introduced into the mass spectrometer via a Michrom Bioresources CaptiveSpray. The spray voltage was set at 1.4 kV and the heated capillary at 200°C. The LTQ-Orbitrap-XL was operated in data-dependent mode with dynamic exclusion in which one cycle of experiments consisted of a full-MS in the Orbitrap (300-2000 m/z) survey scan in profile mode, resolution 30,000, and five subsequent MS/MS scans in the LTQ of the most intense peaks in centroid mode using collision-induced dissociation with the collision gas (helium) and normalized collision energy value set at 35%.
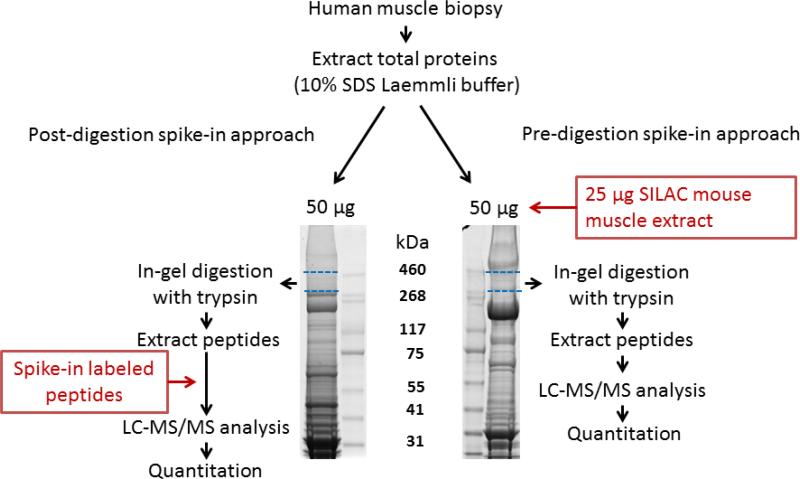
Figure 1. Overview of the experimental design used to quantitate dystrophin protein in human skeletal muscle.
Total protein extracts from muscle biopsies (50 μg aliquots) were further fractionated on SDS-PAGE using 3-8% Tris-Acetate gel, which is suitable for high molecular mass proteins. Left panel shows the post-digestion spike-in approach where the area corresponding to dystrophin migration level (dashed blue lines) was serially sliced, in-gel digested by trypsin and spiked with stable isotope labeled standard peptides (see supplemental Figure S2 for peptide sequences and their location within the dystrophin protein) for subsequent LC-MS/MS analysis and peptide quantitation. In the Pre-digestion spike-in approach (right panel), 50 μg total muscle protein extract was spiked with 25 μg of 13C6-Lysine labeled SILAC mouse muscle protein extract. The mixture was then fractionated by SDS-PAGE and the area corresponding to dystrophin migration was excised, in-gel digested and the resulting peptides were analyzed by LC-MS/MS for identification and quantitation.
For Q Exactive analysis, concentrated peptides from each spiked sample above were redissoleved in 20 μL of 90:10 water:acetonitrile with 0.1% TFA containing 2.5 fmol/μL of trainer peptide mixture. Sample were then injected via autosampler followed by separation on a HALO C18 column (75 μm ID, 10 cm, 2.7 μm) with a gradient of 0 to 35% (A: 0.1% formic acid water, B: 0.1% formic acid acetonitrile) at a flow rate of 400 nL/min on a Thermo Scientific Easy 1000 connected to a nanoESI flex source. For the peptide coverage discovery experiment, the Q Exactive was operated with a full scan (380-1500 m/z) at 70,000 resolution followed by MS/MS of the top ten most abundant ions at 17,500 resolution. For quantitative acquisition, the Q Exactive was operated with a full scan (380-1500 m/z) at 35,000 resolution followed by timed targeted MS/MS of dystrophin and internal stable isotope labeled standard peptides at 35,000 resolution.
Protein identification and quantitation
LTQ-Orbitrap XL Raw files were searched using Sequest in the Bioworks Browser (Thermo) software against a tryptic human database with 2 missed cleavages, 50 ppm mass accuracy, 1.5 Da fragment mass tolerance. Spiked peptides were identified and elution times documented. Dystrophin was quantitated by comparing the unlabeled to the spiked-in heavy isotope profile for each peptide. Q Exactive Raw files were searched with Proteome Discoverer 1.3 (ThermoFisher Scientific) using SEQUEST and the human refseq. database (release 47, ftp://ftp.ncbi.nih.gov/refseq/H_sapiens/mRNA_Prot/, with 34,340 entries). Search parameters allowed trypsin to cleave after Lysine and Arginine with 2 missed cleavages. Precursor ion mass tolerance was set to 10 ppm and fragment ion mass tolerance was 20 ppm. The dynamic modification of methionine (oxidation = 15.995 Da) and lysine (13C6 = 6.020 Da), was accepted on up to 4 residues per peptide. Within the Proteome Discoverer Software, SILAC pairs were identified using the “2 plex” workflow node, and all peptides were rescored using the Percolator algorithm node. Finally, peptides were filtered at 1% FDR. SILAC quantification from 25% and 75% normal were compared and proteins showing a consistent ratio were considered as controls. Peak areas and area ratios were calculated with Pinpoint software using recreated ion chromatograms at 25 ppm from the full scan MS/MS. The ratios of the integrated areas for endogenous peptides and [13C6 lys] labeled peptides were calculated to obtain peptide measures using multiple y-ion fragments per peptide (Supplementary Figure S1). The average of these peptide ratios determined the relative amount of each protein.
Results
High SDS containing buffer enabled efficient dystrophin extraction from skeletal muscle biopsies
Dystrophin protein is a low abundant protein, accounting for only 0.002% of total muscle protein. Since no amplification technologies exist for protein detection by mass spectrometry, efficient dystrophin extraction was essential. In addition to efficient extraction, enrichment of dystrophin protein from other more abundant muscle proteins prior to mass spectrometry was required in order to reduce the sample dynamic range and thereby ensure consistent detection of dystrophin. We evaluated a mild (RIPA buffer) and strong (10% SDS) buffer for muscle protein extraction. As expected the 10% SDS modified Laemmli buffer extracted 10 times more total protein than the RIPA buffer from the same amount of muscle tissue. Indeed, about 8 μg and 54 μg of total proteins were extracted from 1 mg of fresh muscle tissue using RIPA and 10% SDS modified Laemmli buffer, respectively. The increased extraction efficiency of the SDS buffer also increased the dynamic range of the sample by extracting more of the highly abundant skeletal muscle proteins such as myosins, titin and nebulin. To overcome the dynamic range limitations, total muscle extract from each of the RIPA and SDS preparation was further separated on a 3-8% Tris-Acetate gel. The region on the gel corresponding to the migration zone of dystrophin and/or truncated Becker-like dystrophin (300-450 kDa) was excised, and processed for mass spectrometry analysis as described above. Dystrophin protein was identified by 19 and 50 peptides when muscle sample was extracted with RIPA and 10% SDS modified Laemmli buffers, respectively (Supplementary Table S1). The location of these peptides throughout the dystrophin sequence is shown in Supplementary Figure S2.
Dystrophin quantitation by mass spectrometry
We evaluated two approaches to quantitate dystrophin protein in total muscle protein extracts (Figure 1): (i) Post-Digestion spike-in approach with stable isotope labeled custom synthesized standard dystrophin peptides; and (ii) Pre-Digestion spike-in approach with stable isotope labeled mouse muscle protein extract prior to gel separation and in-gel digestion.
Post-digestion spike-in approach
For the post-digestion spike-in approach, three stable isotope labeled dystrophin peptides were custom synthesized. The sequences of these peptides were chosen based on their consistent and reproducible detection by LC-MS/MS following in-gel digestion of SDS-PAGE fractionated muscle extracts, different retention times, amino acid content (e.g. absence of highly modifiable residues such as cysteine and methionine), as well as the location within the dystrophin sequence (e.g. distributed throughout the N-terminal, middle and C-terminal domain). Additionally, a stable isotope labeled peptide for filamin c was also custom synthesized and used as an internal standard. The sequences of the standard peptides, their retention time, observed molecular masses as well as their location within the dystrophin sequence are highlighted in Supplementary Figure S3. First the post-digestion spike-in approach was optimized for the amount of stable isotope labeled standard peptides to be used for spike-in. In-gel digested dystrophin containing bands were prepared from replicate SDS-PAGE fractionation of 50 μg of muscle protein extracts of a healthy donor and spiked with varying amounts of stable isotope labeled standard peptides (14 nM, 40 nM, 80 nM and 160 nM). Figure 2 shows MS spectra of the unlabeled peptide from the endogenous dystrophin and its corresponding heavy stable isotope spiked-in standard at varying concentrations. From this preliminary study the optimal amount of stable isotope labeled peptides to spike-in ranged between 14 and 40 nM. Spiking amounts below 14 nM resulted in poor signal to noise spectra while spiking amounts above 40 nM resulted in increased dynamic range between the standard peptide and the endogenous peptide ion intensities reducing the linear range for quantitation. To test the reproducibility of the post-digestion spike-in approach, triplicate extraction experiments performed on the same muscle biopsy obtained from a healthy donor were processed and spiked with an optimal amount of 30 nM of stable isotope labeled standard peptide mixture. Figure 3 shows the reproducibility of the triplicate assay with a CV of less than 7%. Unfortunately, the reproducibility of the experiment did not hold across the three different dystrophin targeted peptides for a titration curve of endogenous dystrophin protein including the internal control protein filamin c (Figure 4). This was attributed to the inherent variability of in-gel digestion. Therefore, the post-digestion spike-in approach suffered from high sample handling errors.
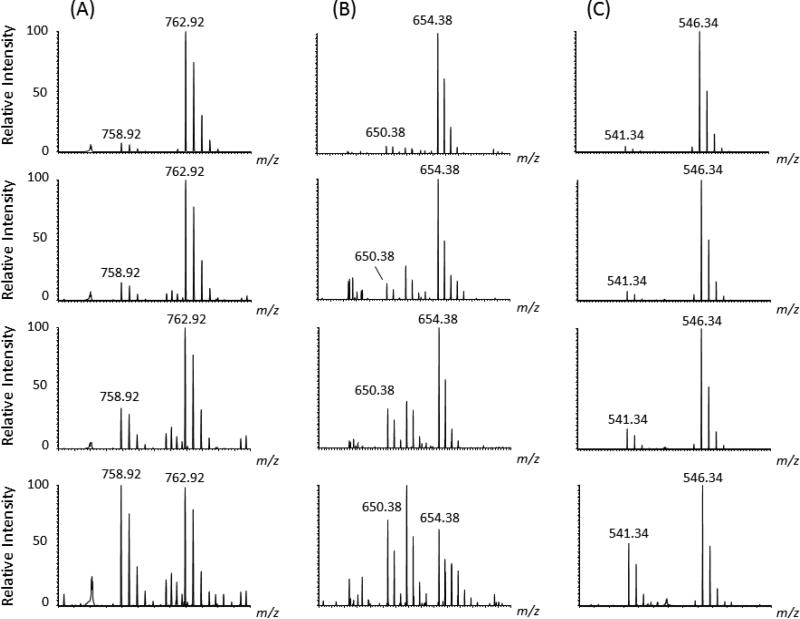
Figure 2. Representative mass spectra of spiked-in labeled standard peptides and corresponding unlabeled endogenous peptide using post-digestion spike-in approach.
(A) Mass spectra of labeled and unlabeled peptide pairs detected after spiking in-gel digested dystrophin with varying amounts of 13C6, 15N2 Lysine labeled standard peptide [IFLTEQPLEGLEK]. (B) Shows mass spectra of labeled and unlabeled peptide pairs detected after spiking in-gel digested dystrophin with varying amounts of 13C6, 15N2 Lysine labeled standard peptide [LLDLLEGLTGQK], (C) Shows mass spectra of labeled and unlabeled peptide pairs detected after spiking in-gel digested dystrophin with varying amounts of 13C6, 15N4 Lysine labeled standard [LLVEELPLR]. The concentration of each spike-in standard peptide, starting for the top to the low panels, was as follows (160 nM, 80 nM, 40 nM and 14 nM). Samples were analyzed on the LTQ-Orbitrap XL at 30,000 resolution and mass error better than 10 ppm. All ions were detected as doubly protonated species with the correct m/z values.
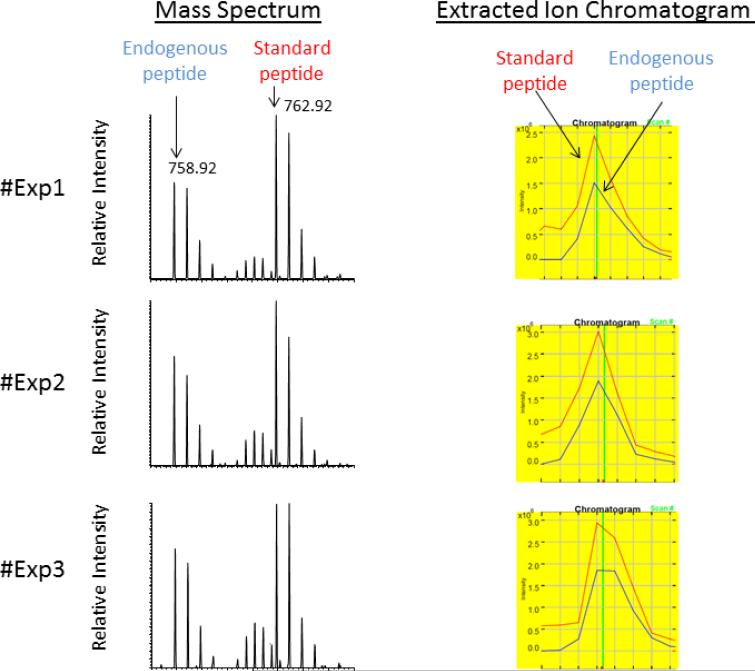
Figure 3. Technical reproducibility of the post-digestion spike-in approach showing mass spectrometry data obtained for triplicate experiments using the same extract from a muscle biopsy obtained from a healthy donor.
Three aliquots of 50 μg total muscle protein extract were separated by SDS-PAGE as described in Methods. The migration area corresponding to dystrophin was excised, in-gel digested and spiked with optimal concentration (30 nM) of stable isotope labeled standard peptides. The left panel shows MS spectra of unlabeled peptide [IFLTEQPLEGLEK] generated from endogenous dystrophin and the spike-in stable isotope labeled standard peptide [IFLTEQPLEGLEK*]. Both peptides were detected as doubly charged ions at their corresponding m/z value of 758.92 and 762.92, respectively. The right panel shows extracted ion chromatograms used for ratio measurement of unlabeled endogenous dystrophin peptide (blue trace) to the stable isotope labeled standard peptide (red trace).
Figure 4. Inter sample reproducibility of the post-digestion spike-in approach.
Three samples containing varying amount of dystrophin obtained by mixing muscle extract from a DMD and age matched healthy donor at ratio of 25/75, 50/50 and 75/25 respectively were evaluated for dystrophin quantitation using the post-digestion spike-in approach with 30 nM of stable isotope standard peptides. Histograms represent intensity ratios of endogenous dystrophin peptides and filamin c peptide to their corresponding stable isotope labeled standard peptide. P1, P2 and P3 corresponds to dystrophin peptides [LLDLLEGLTGQK], [LLVEELPLR], [IFLTEQPLEGLEK] respectively and P4 correspond to filamin c peptide [VYNVTYTVK] that was used as an internal control. It is clearly seen that some ratios did not decrease with decreasing amounts of dystrophin, especially for the peptide P1, showing high variability for this approach.
Pre-digestion spike-in approach
In order to overcome in-gel digestion variability we evaluated a pre-digestion spike-in approach using 13C6-Lysine fully labeled dystrophin protein obtained from muscle extracts prepared from a SILAC mouse. In this method the sample is spiked at the protein level prior to electrophoresis and in-gel digestion, thus minimizing variation due to sample handling and greatly improving reproducibility. Since production of a fully labeled human dystrophin protein using transfection technology and over-expression in a labeled culture system was challenging due to the low transfection rate of the largest human gene [23], use of 13C6-Lysine labeled dystrophin prepared from a SILAC mouse was employed [20,24]. Mouse dystrophin and human dystrophin have 91% sequence homology (Supplementary Figure S4). Our lab had previously generated a fully 13C6-Lysine labeled SILAC mouse [19]. Skeletal muscle from the SILAC mouse was sectioned by cryostat, total protein containing 0.002% of fully labeled dystrophin were extracted using the 10% SDS modified Laemmli buffer then used as a spike-in standard in human muscle extracts. Spiked human muscle extracts were further fractionated by SDS-PAGE and processed for mass spectrometry analysis using the Q Exactive instrument as described above. In a discovery run, 43 unique dystrophin peptides were detected with good coverage throughout the entire dystrophin sequence. Of these 43 unique peptides, five terminated in lysine and had 100% sequence homology with mouse tryptic peptides and therefore were used to quantitate dystrophin in human muscle extracts (Supplementary Figure S4). In addition to these dystrophin overlapping peptides between human and mouse, other lysine terminating peptides belonging to several other skeletal muscle proteins also had 100% sequence homology between human and mouse. This enabled the evaluation and selection of appropriate internal standards for accurate dystrophin quantitation (next section).
Limit of detection and limit of quantitation of dystrophin protein in total muscle extract
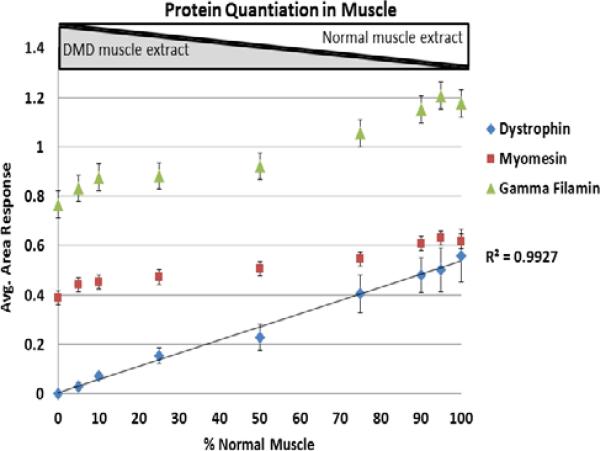
To evaluate the LOD and LOQ of the pre-digestion spike-in approach, we used targeted mass spectrometry analysis on the five dystrophin peptides detected above as well as a few internal control peptides from myomesin and filamin c. Dystrophin protein was targeted for quantitation in the following combination of normal and DMD muscle extracts: 0%/100%, 5%/95%, 10%/90%, 25/75%, 50/50%, 75/25%, 90/10%, 95/5%, 100/0%. Each mixture had a final protein content of 50 μg. The range of dystrophin protein expression, from 0% to 100% of normal, is thus artificially generated to represent possible amounts of dystrophin protein in Becker patients or in Duchenne patients receiving treatments aiming to restore dystrophin expression [6-10]. Each combination mixture above (50 μg final total protein) was then spiked with SILAC 13C6-Lysine labeled SILAC mouse muscle extract (25 μg). Samples were fractionated on a 3-8% Tris-Acetate gel, bands in the dystrophin protein region (300-450 kDa) were excised, in-gel digested and analyzed by LC-MS/MS on a Q Exactive mass spectrometer using a timed targeted MS/MS method. Unlabeled and labeled dystrophin, filmain c and myomesin peptides from both the human muscle extract and the spike-in SILAC mouse labeled extract were targeted for MS/MS analysis. Selected transition ions were used for quantitation in each sample. The average ratio of labeled to unlabeled peptide intensities for dystrophin, filamin c and myomesin were plotted against the different muscle combination mixtures resulting in a calibration curve with an R2 of 0.99 (Figure 5). While the amount of dystrophin increased with increasing % of normal muscle extract in the mixtures, two muscle specific proteins, filamin c and myomesin, remained relatively unchanged. These two proteins are therefore ideal candidates as internal controls. Note that filamin c and myomesin did show a slight increase in abundance as the % of normal increased (Figure 5), and this is expected since the amount of muscle present in dystrophic tissue is slightly decreased and is replaced by fibrotic tissue (e.g. hallmark of DMD pathogenesis) [25]. Therefore, monitoring of internal standards across the curve allows for improved accuracy and normalization.
Figure 5. Titration curve of dystrophin protein and other muscle specific proteins using pre-digestion spike-in approach.
Dystrophin protein, as well as myomesin and filamin c, were quantitated in different combination mixtures of muscle extracts from DMD and age matched healthy donors at ratios leading to 0, 5, 10, 25, 50, 75, 90, 95 and 100% of dystrophin relative to normal. Each final mixture contained 50 μg of total muscle protein and was spiked with 25 μg of 13C6-Lysine labeled SILAC mouse muscle extract. Gel bands encompassing dystrophin protein and other muscle specific proteins were excised and processed for mass spectrometry analysis as described in Methods. Transition ion intensities obtained at the MS/MS level for the targeted labeled and unlabeled peptide pairs generated from the spike-in standard and endogenous human dystrophin were used to determine the relative amount of dystrophin and other muscle proteins (myomesin and filamin c). Standard deviation at each data point represents average ratios obtained from labeled and unlabeled peptide pairs: 5 pairs for dystrophin protein (QAPIGGDFPAVQK, VLSQIDVAQK, IFLTEQPLEGLEK, TLNATGEEIIQQSSK, VHALNNVNK); 3 pairs for filamin c (SPFVVNVAPPLDLSK, EVGEHVVSVRK, HIGISFTPK) and two pairs for myomesin (VSEPVAALDPAEK, VLGGLPDWTIQEGK). These peptides have 100% sequence homology between human and mouse and were used for ratio measurements. As expected, dystrophin protein increased with increasing relative amount of normal muscle extract while myomesin and filamin c remained unchanged.
Absolute quantitation of dystrophin in becker patients
The absolute amount of dystrophin in muscle biopsies from normal donors (n = 12) and Becker patient donors (n = 5) was determined. Dystrophin in normal muscle ranged from 0.001 to 0.004% of total striated muscle, consistent with previous findings [2]. The Becker samples also showed a wide range of dystrophin expression at approximately 10% of the level found in normal muscle (Figure 6). Dystrophin protein represents approximately 0.002% of total striated muscle protein and since the amount of spike-in standard is known, we estimated the total amount of dystrophin in healthy individuals to be 75 ± 15 ng per mg of muscle while in Becker patients the amount can range from 0 to 7 ng per mg of muscle (Figure 6).
Figure 6. Calculated absolute amount of dystrophin in muscle biopsies obtained from BMD and age matched healthy controls.
Total protein extract (50 μg) from each biopsy was spiked with 25 μg of C6-Lysine labeled SILAC mouse muscle extract and processed using the pre- digestion spike-in approach (see Figure 1 and Methods). Intensity ratios of labeled and unlabeled peptide pairs were used to determine the absolute amount of dystrophin in each muscle extract.
Discussion
In skeletal muscle, dystrophin protein is located just beneath the sarcolemma connecting the cytoskeleton to the extracellular matrix via a dystrophin associated glycoprotein complex (DAC or DGC) and thus stabilizes the sarcolemma of the muscle fiber and protects it from damage during the repeated cycles of contraction and relaxation. Mutations in the dystrophin gene, resulting in decreased or lack of dystrophin expression, such as in BMD and DMD patients, results in destabilization of the DAC complex and muscle fiber degeneration.
A number of promising treatment strategies aiming to restore dystrophin expression in DMD patients are in clinical development. These include exon skipping with antisense oligonucleotide [6,10], stop codon read-through with Ataluren (PTC124) drug [7], gene therapy [26] and stem cell therapy [27]. One of the outcome measures to evaluate efficacy of these novel treatments requires accurate quantitation of restored dystrophin in muscle biopsies. Current antibody-based dystrophin assays, including both western blot and immunohistochemistry, are an indirect means by which to quantitate dystrophin, and are not considered highly reliable. Here we evaluated a promising mass spectrometry assay for accurate dystrophin quantitation in muscle. The technique uses a stable isotope labeled spike-in standard followed by high precision mass spectrometry analysis. In this study we found that spiking the muscle extracts with full length labeled muscle proteins prior to gel fractionation and in-gel digestion to be reliable and sensitive, and preferred to post-digestion peptide spike-in approach. Indeed, in-gel digestion variation rendered the latter approach less reproducible resulting in fluctuation of dystrophin measurement from run to run. Additionally, spiking samples with fully labeled muscle extracts enabled the use of other muscle specific proteins (filamin c and myomesin) as internal controls. Muscle biopsies, from normal or from patients with dystrophinopathies, are very heterogeneous containing varying proportions of myofibers, connective tissue (fibrosis) and adipose tissue (fatty replacement). Depending on the biopsy location and disease state, the proportion of myofibers (cell type expressing dystrophin) can vary significantly. Therefore, measuring myofiberspecific proteins to use as internal controls will allow normalization and precise quantitation of dystrophin protein in myofibers, and this is preferable to normalizing to total protein content of a biopsy. This was demonstrated in this study by combining DMD and normal muscle extracts at different ratios. This strategy enabled a constant protein background while the amount of dystrophin changed.
In this assay the LOD and LOQ was better than 10% (e.g. as low as 5%) of the normal amount of dystrophin, making the assay sensitive enough to measure dystrophin in clinical specimens. In this study we routinely measured dystrophin content in 50 μg total muscle protein extract from roughly 0.5 mg of frozen biopsy. This amount of muscle is readily available from diagnostic needle biopsies, which are in the range of 20 mg [28].
Despite a sequence length of 3685 amino acids, only 45 to 50 dystrophin peptides were detected by mass spectrometry analysis of 50 μg of total muscle protein extract. Increasing the amount of total muscle protein extract to 100 μg did not improve dystrophin sequence coverage. This is mainly due to the inherent low abundance of this protein in muscle (e.g. dystrophin represents only about 0.002% of total striated muscle protein) and the presence of highly abundant proteins such as the myosins, which represent up to 40% of total muscle protein. Therefore it was necessary to pre-fractionation total protein muscle extract by SDS-PAGE to isolate dystrophin from highly abundant proteins and facilitate detection and quantitation in a complex biological matrix.
Conclusions
Targeted mass spectrometry quantitation of full length or truncated Becker dystrophin protein was facilitated by stable isotope spike-in strategy using full length 13C6-Lysine labeled SILAC mouse muscle extract. The method was found to be highly reproducible, accurate and sensitive over a large dynamic range. Our aim is to apply this technique to quantitate restored dystrophin in DMD patients receiving exon skipping therapy to aid in the clinical evaluation of this promising new therapy. In the future, this technique could be refined to include quantitation of DAC complex proteins.
Supplementary Material
Acknowledgements
This work was partially supported by NIAMS 5P50AR060836-02 (Center for Research Translation of Systemic Exon-Skipping in Muscular Dystrophy), NIAMS R01AR062380 (Craig McDonald, PI), Massey University Research Fund, Wellington Medical Research Foundation Inc. and core grants NICHD/NINDS 2R24HD050846-06 (National Center for Medical Rehabilitation Research), NICHD 5P30HD040677-10 (Intellectual and Developmental Disabilities Research Center) and NIH NCATS UL1RR031988 (CTSI-CN). The authors would like to thank the following for their kind assistance: Dr. Jyoti Jaiswal, Dr. D. Ashley Hill, Dr. Mendel Tuchman and Dr. Terrence Partridge for assistance in generating the SILAM/SILAC mouse model and Dr. Akanchha Kesari for assistance in biobanking and retrieval of DMD and BMB biopsies.
Footnotes
Publisher's Disclaimer: This article was originally published in a journal published by OMICS Publishing Group, and the attached copy is provided by OMICS Publishing Group for the author's benefit and for the benefit of the author's institution, for commercial/research/educational use including without limitation use in instruction at your institution, sending it to specific colleagues that you know, and providing a copy to your institution's administrator. All other uses, reproduction and distribution, including without limitation commercial reprints, selling or licensing copies or access, or posting on open internet sites, your personal or institution's website or repository, are requested to cite properly.
Citation: Brown KJ, Marathi R, Fiorillo AA, Ciccimaro EF, Sharma S, et al. (2012) Accurate Quantitation of Dystrophin Protein in Human Skeletal Muscle Using Mass Spectrometry. J Bioanal Biomed S7: 001. doi:10.4172/1948-593X.S7-001
References
- 1.Lederfein D, Levy Z, Augier N, Mornet D, Morris G, et al. A 71-kilodalton protein is a major product of the Duchenne muscular dystrophy gene in brain and other nonmuscle tissues. Proc Natl Acad Sci. 1992;89:5346–5350. doi: 10.1073/pnas.89.12.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.van Essen AJ, Busch HFM, te Meerman GJ, ten Kate LP. Birth and population prevalence of Duchenne muscular dystrophy in The Netherlands. Hum Genet. 1992;88:258–266. doi: 10.1007/BF00197256. [DOI] [PubMed] [Google Scholar]
- 4.Bushby KMD, Gardner-Medwin D, Nicholson LVB, Johnson MA, Haggerty ID, et al. The clinical, genetic and dystrophin characteristics of Becker muscular dystrophy. J Neurol. 1993;240:105–112. doi: 10.1007/BF00858726. [DOI] [PubMed] [Google Scholar]
- 5.Anthony K, Sebahattin C, Silvia T, Giorgio T, Lucy F, et al. Dystrophin quantification and clinical correlations in Becker muscular dystrophy: implications for clinical trials. Brain. 2011;134:3547–3559. doi: 10.1093/brain/awr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sebahattin C, Virginia A, Michela G, Lucy F, Silvia T, et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose- escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finkel RS. Read-through strategies for suppression of nonsense mutations in Duchenne/ Becker muscular dystrophy: aminoglycosides and ataluren (PTC124). J Child Neurol. 2010;25:1158–1164. doi: 10.1177/0883073810371129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson SF, Crosbie RH, Miceli MC, Spencer MJ. Emerging genetic therapies to treat Duchenne muscular dystrophy. Curr Opin Neurol. 2009;22:532–538. doi: 10.1097/WCO.0b013e32832fd487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffman EP, Bronson A, Levin AA, Takeda S, Yokota T, et al. Restoring dystrophin expression in duchenne muscular dystrophy muscle progress in exon skipping and stop codon read through. Am J Pathol. 2011;179:12–22. doi: 10.1016/j.ajpath.2011.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goemans NM, Tulinius M, van den Akker JT, Burm BE, Ekhart PF, et al. Systemic administration of PRO051 in Duchenne's muscular dystrophy. N Engl J Med. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 11.Yokota T, Lu QL, Partridge T, Kobayashi M, Nakamura A, et al. Efficacy of systemic morpholinon exonskipping in Duchenne dystrophy dogs. Ann Neurol. 2009;65:667–676. doi: 10.1002/ana.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu QL, Yokota T, Takeda S, Garcia L, Muntoni F, et al. The status of exon skipping as a therapeutic approach to duchenne muscular dystrophy. Mol Ther. 2011;19:9–15. doi: 10.1038/mt.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henricson E, Abresch R, Han JJ, Nicorici A, Goude Keller E, et al. Percent predicted 6 minute walk distance in duchenne muscular dystrophy to account for maturational influences. PLoS Curr. 2012;4 doi: 10.1371/currents.RRN1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz R. Biomarkers and surrogate markers: an FDA perspective. NeuroRX. 2004;1:189–195. doi: 10.1602/neurorx.1.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arechavala-Gomeza V, Kinali M, Feng L, Brown SC, Sewry C, et al. Immunohistological intensity measurements as a tool to assess sarcolemma-associated protein expression. Neuropathol Appl Neurobiol. 2010;36:265–274. doi: 10.1111/j.1365-2990.2009.01056.x. [DOI] [PubMed] [Google Scholar]
- 16.Taylor LE, Kaminoh YJ, Rodesch CK, Flanigan KM. Quantification of Dystrophin Immunofluorescence in Dystrophinopathy Muscle Specimens. Neuropathol Appl Neurobiol. 2012;38:591–601. doi: 10.1111/j.1365-2990.2012.01250.x. [DOI] [PubMed] [Google Scholar]
- 17.Spitali P, Heemskerk H, Vossen RH, Ferlini A, den Dunnen JT, et al. Accurate quantification of dystrophin mRNA and exon skipping levels in duchenne muscular dystrophy. Lab Invest. 2010;90:1396–1402. doi: 10.1038/labinvest.2010.98. [DOI] [PubMed] [Google Scholar]
- 18.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma N, Medikayala S, Defour A, Rayavarapu S, Brown KJ. Use of quantitative membrane proteomics identifies a novel role of mitochondria in healing injured muscles. J Biol Chem. 2012;287:30455–30467. doi: 10.1074/jbc.M112.354415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman EP, Fischbeck KH, Brown RH, Johnson M, Medori R, et al. Characterization of dystrophin in musclebiopsy specimens from patients with Duchenne's or Becker's muscular dystrophy. N Engl J Med. 1988;318:1363–1368. doi: 10.1056/NEJM198805263182104. [DOI] [PubMed] [Google Scholar]
- 22.Jensen ON, Wilm M, Shevchenko A, Mann M. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol Biol. 1999;112:513–530. doi: 10.1385/1-59259-584-7:513. [DOI] [PubMed] [Google Scholar]
- 23.Tennyson CN, Klamut HJ, Worton RG. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat Genet. 1995;9:184–190. doi: 10.1038/ng0295-184. [DOI] [PubMed] [Google Scholar]
- 24.McClatchy DB, Yates JR., III Stable Isotope Labeling of Mammals (SILAM) 2008. [DOI] [PubMed]
- 25.Ardite E, Perdiguero E, Vidal B, Gutarra S, Serrano AL, et al. PAI-1-regulated miR-21 defines a novel age-associated fibrogenic pathway in muscular dystrophy. J Cell Biol. 2012;196:163–175. doi: 10.1083/jcb.201105013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendell JR, Campbell K, Rodino Klapac L, Sahenk Z, Shilling C, et al. Dystrophin immunity in Duchenne's muscular dystrophy. N Engl J Med. 2010;363:1429–1437. doi: 10.1056/NEJMoa1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel K, Morgan J. 185th ENMC International Workshop: stem/precursor cells as a therapeutic strategy for muscular dystrophies 3-5 June 2011, Naarden, The Netherlands. Neuromuscul Disord. 2012;22:447–452. doi: 10.1016/j.nmd.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Dubowitz V, Sewry CA. Muscle biopsy: a practical approach. 2007.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.