Abstract
Nuclear receptor (NR) coactivators are recruited to DNA by NRs, potentiating NR-dependent gene transcription. To obtain the complexity of NR-mediated gene regulation with a finite number of coactivators, the molecular properties of coactivators are dynamically modulated by posttranslational modifications (PTMs) in response to external stimuli. PTMs can regulate the molecular interactions of coactivators with transcription factors and other coactivators, as well as their cellular location, protein stability, conformation, and enzymatic activity. Therefore, dynamic regulation of the molecular properties of coactivators by PTMs allows for the complexity of NR-dependent gene expression and influences the regulation of NR-mediated physiological processes. This review focuses on recent progress in our understanding of how coactivator PTMs influence NR-mediated gene transcription and addresses their biological relevance.
Nuclear receptors (NRs) are ligand regulated transcription factors that transduce steroid, retinoid and thyroid hormone signals into hormone-regulated gene expression. A growing appreciation of the complexity of NR-mediated gene transcription led to the concept that coregulators exist (e.g., coactivators and corepressors) that modulate NR function [1–3]. Following these initial proof-of-principle experiments [4], estrogen receptor-associated protein 160 (ERAP160) was the first protein to be identified as a possible mediator of hormone-induced transcription, and the steroid receptor coactivator-1(SRC-1)/RIP160 gene was the first to be cloned as a coactivator of NR [5–7]. To date, about 300 NR coregulators have been biochemically and functionally identified [8]. Comparative structural analysis of coactivators revealed that the majority are comprised of two basic functional components. First, many coactivators have an LxxLL motif that is necessary and sufficient to permit interaction of the coactivator with the NR. In addition to LxxLL motifs, coactivators also may have other functional motifs such as RNA-interacting domains, Lin11, Isl-1 and Mec-3 (LIM)-domains and bromodomains that are important for interactions with other transcriptional factors such as, SRA [9, 10], Androgen Receptor [11], and p53 [12]. Using protein interaction motifs, coactivators generate and function as multi-coactivator interactosomes to modulate NR-mediated gene regulation. Second, coactivators possess unique intrinsic enzymatic activities required for NR-mediated gene transcription [13]. Some of the enzymatic activities inherent to NR coactivators are acetyl transferase, ubiquitin ligase, methyl transferase, small ubiquitin-like modifier (SUMO) ligase, phosphokinase, phosphatase, hydrolase, ribosylase, isomerase, helicase and pseudouridylate synthetase activities [14, 15]. In vitro and in vivo analyses have revealed that in many cases, the substrates of the enzymatic activities associated with these coactivators are histones and other coactivators present in the coactivator complex. The modulation of the enzymatic properties of coactivators affords a high level of regulatory flexibility to subtly control NR-mediated gene expression and will be discussed later.
PTMs allow for the regulation of protein properties including stability, structure, function, activity, intracellular localization and interaction with other proteins during cellular processes [16]. A range of PTMs can occur, including chemical (e.g., phosphorylation, acetylation, methylation, hydroxylation, glycosylation and nitrosylation) and structural (e.g., disulfide bridge formation and proteolytic cleavage) modifications, or the addition of small protein tags (e.g., ubiquitination and NEDDylation). Different external signals lead to distinct patterns of PTM coding on coactivators, with each pattern directing a specific cellular function in NR-mediated cellular physiology [6, 13].
Dynamic models of NR-mediated transcriptional regulation associated with PTM of coactivators
A growing number of studies have revealed that molecular mechanisms are responsible for how coactivator PTMs influence NR-mediated transcription. This review examines the transcriptional regulatory mechanisms associated with these modifications (Table 1).
Table 1.
Mechanism of Transcriptional Regulation associated with PTM of Nuclear Receptor Coactivator
| Coactivator | PTM | PTM modulator |
2nd Modification / Modulator |
Regulatory Consequence | Ref. | |
|---|---|---|---|---|---|---|
| Molecular Interaction | MED1 | Phosphorylation | ERK | Mediator complex formation | [22] | |
| MTA1 | Acetylation | P300 | Increasing interaction to RNA polymerase II complex | [20] | ||
| P300 | Methylation | CARM1 | Increasing interaction to SRC-2 | [25] | ||
| PARP-1 | Acetylation | P300/CBP | Increasing interaction to NF-kB | [49] | ||
| PARP-1 | Phosphorylation | CaMKII delta | Dissociation of TLE-1 complex | [41] | ||
| PGC-1 α | Phosphorylation | AKT2 | Increasing promoter recruitment | [39] | ||
| PGC-1 α | Phosphorylation | AMPK | Increasing promoter recruitment | [40] | ||
| PONTIN | Sumoylation | Ubc9 | Increasing interaction to beta-catenin and CBP | [45] | ||
| SRC-1 | Phosphorylation | Cdk2 | Increasing interaction to PR | [17] | ||
| SRC-1 | Sumoylation | ND | Increasing interaction to PR | [18] | ||
| SRC-3 | Methylation | CARM1 | Dissociation of SRC-3 /CARM1/CBP complex | [23, 24] | ||
| SRC-3/ACTR | Acetylation | P300/CBP | Disrupting interaction to ER | [51] | ||
| SRC-3 | Phosphorylation | Abl | Increasing interaction to Abl, ER, p300 and CARM1 | [52] | ||
| Subcellular distribution | P300 | Ubiquitination | ND | Cytoplasmic distribution | [28] | |
| PGC-1 α | Acetylation | GCN5 | Nuclear foci formation | [29] | ||
| PGC-1 α | Deacetylation | SIRT1 | Dissociation of nuclear foci | [38] | ||
| SRC-3 | Phosphorylation | EGF | Nuclear translocation | [27] | ||
| Stability | PGC-1 α | Phosphorylation | P38 & GSK3 β | Ubi1 / SCF (Cdc4) | Proteasomal degradation | [34] |
| SRC-3 | Methylation | CARM1 | Proteasomal degradation | [24] | ||
| SRC-3 | Phosphorylation | GSK3 β | Ubi / SCF(Fbw7 alpha) | Proteasomal degradation | [31] | |
| SRC-3 | Phosphorylation | PKC ζ | Inhibiting Interaction to C8 | [44] | ||
| SRC-3 | Phosphorylation | P38 MAPK | Proteasomal degradation | [53] | ||
| SRC-2 | Phosphorylation | PKA | Ubi /ND2 | Proteasomal degradation | [32] | |
| Regulation of Enzymatic activity | CARM1 | Phosphorylation | ND | Inhibition of methyltransferase | [35] | |
| EZH2 | Phosphorylation | AKT | Inhibition of methyltransferase | [36] | ||
| PARP-1 | Phosphorylation | CaMKII delta | Enhancement of poly-ribosylation | [41] | ||
| Conformation changes | SRC-3 | Phosphorylation | ND | ND / Pin-1 | Conformational Change | [37] |
Ubi1 : Ubiqutination
ND2 : Not Determined.
1. Coactivator PTMs modulate their interaction with NRs and other proteins
Comparative structural analysis has shown that most NR coactivators have an LxxLL motif that mediates the interaction between the coactivator and the NR. Therefore, regulation of this interaction by PTMs is essential for efficient NR-mediated gene transcription. For example, progesterone receptor (PR) activity is regulated by the kinase activity of cyclinA/Cyclin-dependent kinase 2 (Cdk2) during the cell cycle. Cdk2 increases PR activity by enhancing the physical interaction between SRC-1 and PR through SRC-1 phosphorylation [17]. Sumoylation of SRC-1 also has been reported to enhance the molecular interaction between SRC-1 and PR [18]. Overexpression of SUMO-1 enhances sumoylation of SRC-1 at Lys-732 and -774 residues. This sumoylation strengthens the interaction between SRC-1 and PR and enhances the activation of PR-mediated gene transcription [18].
In addition to physical interactions with NRs, PTMs of coactivators also regulate interactions with other transcriptional regulators that control NR-mediated gene transcription. For example, metastasis-associated gene 1 (MTA1) was originally identified as a protein overexpressed in metastatic carcinomas and as a transcriptional activator of Breast Cancer Amplified Sequence 3 (BCAS3), a gene that is amplified and overexpressed in breast cancers [19]. Activation of BCAS3 leads to acetylation of Lys-626 of MTA1 by p300, a modification that leads to the recruitment of RNA polymerase II complexes to the BCAS3 enhancer sequence for the activated transcription of the BCAS3 gene [20]. In this way, acetylation of MTA1 is associated with breast cancer progression through overexpression of the BCAS3 gene.
NR coactivators exist as multisubunit complexes that dynamically modulate gene transcription in response to various external stimuli. Interestingly, a recent study showed that coactivator complex dynamics are controlled by one or more of the components of the coactivator interactosome. For example, Mediator is a conserved multisubunit complex that acts as a functional interface between regulatory transcription factors and the general RNA polymerase II initiation apparatus in response to external signals [21]. Mediator 1 (MED1), one of the components of the Mediator complex, is a pivotal component involved in NR-mediated gene transcription. Phosphorylation of MED1 by extracellular signal-regulated kinase (ERK) activity enhances its binding to MED7. Therefore, MED1 phosphorylation stabilizes or enhances its association with the Mediator complex, possibly facilitating a continuing feedforward action of nuclear hormones [22]. In addition to complex formation, PTMs of coactivators also induce dissociation of coactivator complex in vivo. Dissociation of the coactivator complex plays an essential role in promoter clearance which is required for multiple-round transcription as well as for transcriptional termination. The steroid receptor coactivator 3 (SRC-3)/ACTR/AIB1/RAC3/pCIP is known to be an in vivo substrate for coactivator-associated arginine methyltransferase 1(CARM1) (Figure 1a). Methylation of SRC-3 by CARM1 serves as a molecular switch to induce dissociation of the SRC-3:CARM1 coactivator complex on estrogen receptor-dependent gene promoters [23, 24]. Besides SRC-3, CARM1 also methylates p300 at Arg-2142, located within the C-terminal region of the steroid receptor coactivator 2(SRC-2)/GRIP1/TIF2 binding domain. Similar to methylation of SRC-3, methylation of p300 inhibits coactivator complex formation by preventing the interaction between p300 and SRC-2 [25]. Therefore, the above data suggest that methylation of coactivators could be associated with promoter clearance and coactivator cycling via dissociation of the coactivator complex.
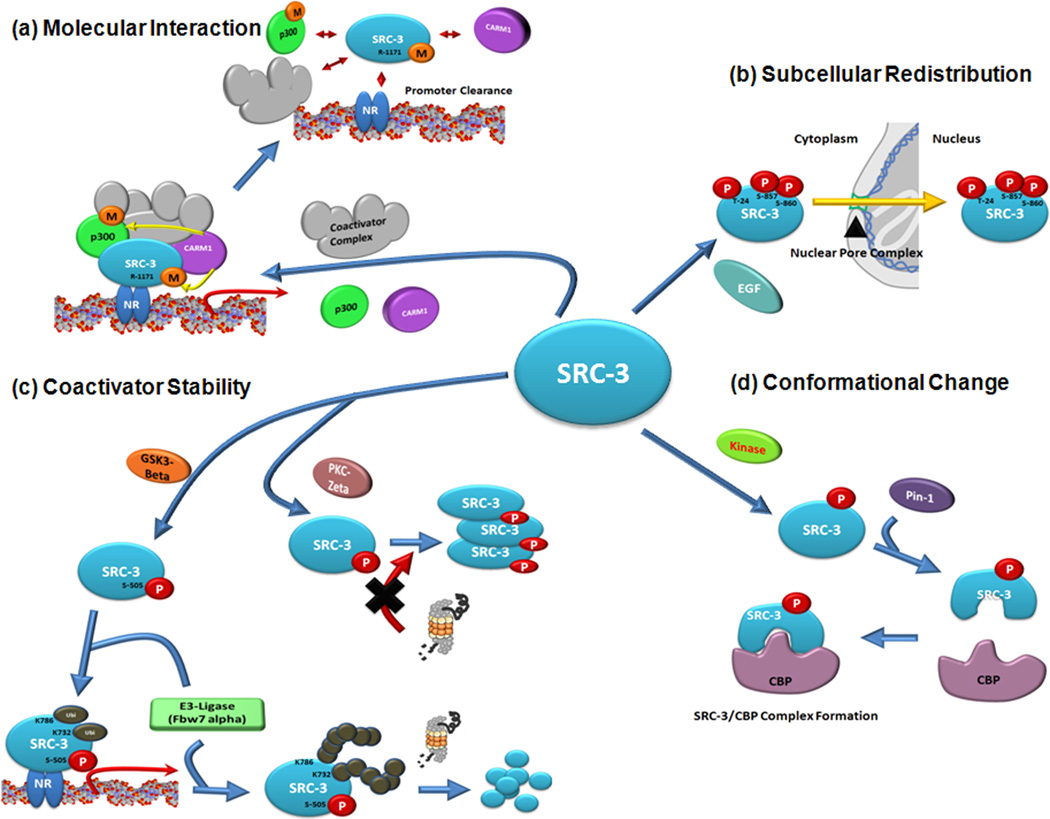
Figure 1. Dynamic regulation of molecular properties of SRC-3 by posttranslational modification (PTM).
(a) Modulation of molecular interaction. To activate nuclear receptor (NR) mediated gene transcription, SRC-3 is recruited onto NR target gene promoter regions with other coactivator complexes such as p300 and coactivator-associated arginine methyltransferase 1(CARM1) to generate pre-initiation transcription complexes. After transcription, however, methyltransferase activity of CARM1 is activated to methylate SRC-3(Arginine-1171 residue) and p300. Methylated SRC-3 loses its interaction affinity to CARM1 to be dissociated from the SRC-3/CARM1 complex. (b) Regulation of subcellular redistribution of SRC-3. Phosphorylation of tyrosine-87, serine-857 and serine-860 residues of SRC-3 by epidermal growth factor (EGF) signaling triggers subcellular trafficking of SRC-3 into the nucleus to activate SRC-3 mediated gene transcription. (c) Regulation of steroid receptor coactivator -3 (SRC-3) stability. In the presence of estradiol, serine -505 of SRC-3 is phosphorylated by glycogen synthase kinase 3β (GSK3β) in an estrogen receptor (ER) dependent manner. Lysine-732 and -786 residues of phosphorylated SRC-3 are sequentially mono-ubiquitinated by F-box and WD repeat domain-containing 7α (Fbw7α) to superactivate ER-dependent gene transcription. Eventually, functioning SRC-3 will be degraded by the proteasome when the polyubiquitin chain reaches ~4–5+ additions of ubiquitin. In contrast to GSK3β, phosphorylation of SRC-3 mediated by protein kinase C ζ prevents degradation of SRC-3 by interrupting interaction with the 20S proteasome. (d) Regulation of conformational changes. After phosphorylation of SRC-3 at Ser/Thr-Pro resides, Pin-1 recognizes phosphorylated SRC-3 to induce a conformation change that increases its interaction affinity for CBP for efficient NR mediated gene transcription.
2. Modulation of subcellular localization of coactivators by PTM
Although NRs are mainly localized in the nucleus, they are constantly shuttling between the nucleus and the cytoplasm in response to external signals [26]. Regulation of NR activity is dependent upon intracellular trafficking. Little is known, however, about the mechanism of subcellular localization and the dynamic changes in the cellular distribution of NR coregulators. Recent studies have indicated that coactivator PTMs coordinate their intracellular trafficking in response to external signals. For example, SRC-3 is a protein that extensively shuttles between the nucleus and cytoplasm, and its trafficking is regulated by phosphorylation [27]. Epidermal growth factor (EGF) stimulates SRC-3 phosphorylation and induces nuclear localization of SRC-3, enhancing SRC-3-mediated gene transcription (Figure 1b). Nuclear translocation of SRC-3 is linked with phosphorylation of SRC-3 at Thr-24, Ser-857 and Ser-860 by EGF signaling.
In addition to positive effects on gene transcription, PTM-mediated nuclear-cytoplasmic trafficking of coactivators can also result in transcriptional repression. The level of p300 in the nucleus is tightly regulated in order to maintain normal cell proliferation and development, and control of its nuclear tracking is an integral part of its function in vivo. Treatment of cells with histone deacetylase (HDAC) inhibitors renders histones in a hyperacetylated state, making the cells more transcriptionally active and less p300-dependent. In response to HDAC inhibitors, loss of p300-dependence is associated with shuttling of p300 from the nucleus to cytoplasmic inclusion bodies in a ubiquitination-dependent manner [28].
PPARγ coactivator-1 (PGC-1)α is a metabolic coactivator and has an essential role in metabolic signaling. PGC-1α exists as a multisubunit complex containing general control of amino-acid synthesis 5 (GCN5) and Tip60 acetyltransferase subcomplexes in hepatic cells [29]. In response to high glucose levels in the blood, PGC-1α is directly and specifically acetylated by GCN5, but not by Tip60. Like ubiquitination of p300, acetylated PGC-1α moves from its cognate promoter to nuclear foci containing RIP140, an NR repressor. Therefore, subcellular redistribution of PGC-1α mediated by acetylation promotes its conversion to an inactive form.
3. Cross communication of PTMs and coactivator stability
Cellular levels of NR coactivators are tightly regulated, and misregulation of these levels is associated with progression of human disease due to perturbation of normal NR-mediated gene transcriptional networks [30]. Gene amplification and hyper-activation of coactivator gene transcription are associated with aberrant levels of coactivators [8, 30]. In addition to RNA levels, PTM modulation of coactivator protein stability is important in cancer cells. For example, methylation of SRC-3 by CARM1 causes an increase in SRC-3 turnover as a result of enhanced degradation [24]. This data suggests that methylation of SRC-3 can serve as a blueprint for proteasomal degradation. In addition to methylation, phosphorylation impacts the ubiquitination-mediated degradation system, influencing coactivator stability. Phosphorylation of SRC-3 at Ser-505 by glycogen synthase kinase 3β (GSK3β) is followed by ubiquitination of SRC-3 by Skp/Culin/F-box protein complex (SCF)- F-box and WD repeat domain-containing 7α (Fbw7α) (Figure 1c) [31]. These two sequential PTMs of SRC-3 promote its eventual transcription-coupled degradation. The stability of SRC-2 protein is also regulated by the phosphorylation-dependent, ubiquitination-mediated degradation pathway. In this process, phosphorylation of SRC-2 is mediated by activated protein kinase A (PKA) [32].
Aberrant regulation of PGC-1α has been linked to diabetes, heart disease and neurodegenerative disorders. While several studies have shown that alternations in the levels of PGC-1α can result from abnormal PGC-1α mRNA expression [33], the mechanisms that determine protein levels of PGC-1α are incompletely defined. A recent study showed that PGC-1α protein levels were regulated by ubiquitin-mediated degradation via the E3 ubiquitin ligase SCFCdc4 [34]. Phosphorylation of PGC-1α by p38 mitogen activated protein kinase (MAPK) and GSK3β induces SCFCdc4 mediated PGC-1α degradation.
Collectively, regulation of coactivator stability by the ubiquitin-mediated degradation system requires communication with other PTMs, e.g. phosphorylation and methylation. Therefore, sequential PTM of coactivators is a currently accepted concept in regulating levels and turnover of these important regulators.
4. Regulation of enzymatic activities associated with coactivators by PTM
Because NR coactivators have intrinsic enzymatic activity, those activities are important for NR-mediated gene transcription. Although CARM1 methylates histones as well as other transcription factors to change their molecular properties during NR-dependent gene regulation, little is known about the upstream signaling that modulates CARM1 enzymatic activity. Recent studies have shown that Ser-228 of CARM1 is phosphorylated during mitosis. Site-specific mutation analysis revealed that the methyltransferase activity of CARM1 was negatively regulated by phosphorylation of Ser-228 [35]. However, the specific kinase that phosphorylates CARM1 is unknown. Enhancer of Zeste homolog 2 (EZH2) also is a methyltransferase that trimethylates Lys-27 in histone H3. Phosphorylation of EZH2 residue Ser-21 by AKT suppresses its methyltransferase activity by impacting its histone H3 binding affinity [36]. Therefore, regulation of coactivator enzymatic activities by PTMs provides a functional link between two major protein epigenetic modification pathways, increasing the diversity of coactivator functions.
5. Induced conformational changes of coactivators by PTM
Conformational changes in proteins are made possible by their intrinsic structural flexibility. At the molecular level, conformational changes in single polypeptides are the result of changes in peptide backbone torsional angles and side chain orientations. Such changes can result in a localized reorientation of a few residues and small torsional changes in the regional main chain, altering their molecular properties. Peptidyl-prolyl isomerase-1 (Pin1) catalyzes the isomerization of phosphorylated Ser/Thr-Pro peptide bonds to induce conformational changes in its target proteins (Figure 1d). A recent study has shown that Pin1 interacts selectively with phosphorylated SRC-3 and synergistically activates SRC-3 mediated gene transcription. In this process, Pin1 enhances the interaction of SRC-3 with CBP/p300 [37]. Even though there is no direct evidence, the above data suggest that Pin1 binds specifically to phosphorylated SRC-3 and promotes a conformational change that allows interaction between SRC-3 and CBP/p300 to occur for the enhancement of SRC-3 mediated gene expression.
Biological relevance of PTM of coactivators
The complexity of the gene expression network mediated by PTM of NR coactivators is closely correlated with regulation of cellular processes (e.g., cellular differentiation, metabolism and reproductive physiology) as well as cancer progression. Based on current studies, we will highlight selected molecular properties of coactivators that are mediated by PTM and can control cellular physiological processes and the progression of human pathologies.
1. Regulation of glucose and fatty acid metabolism by PTM of PGC-1α in liver and muscle
To adapt to different environmental conditions, tissue-specific metabolic pathways are regulated by tissue-specific metabolic coactivators in order to maintain energy and nutrient homeostasis.
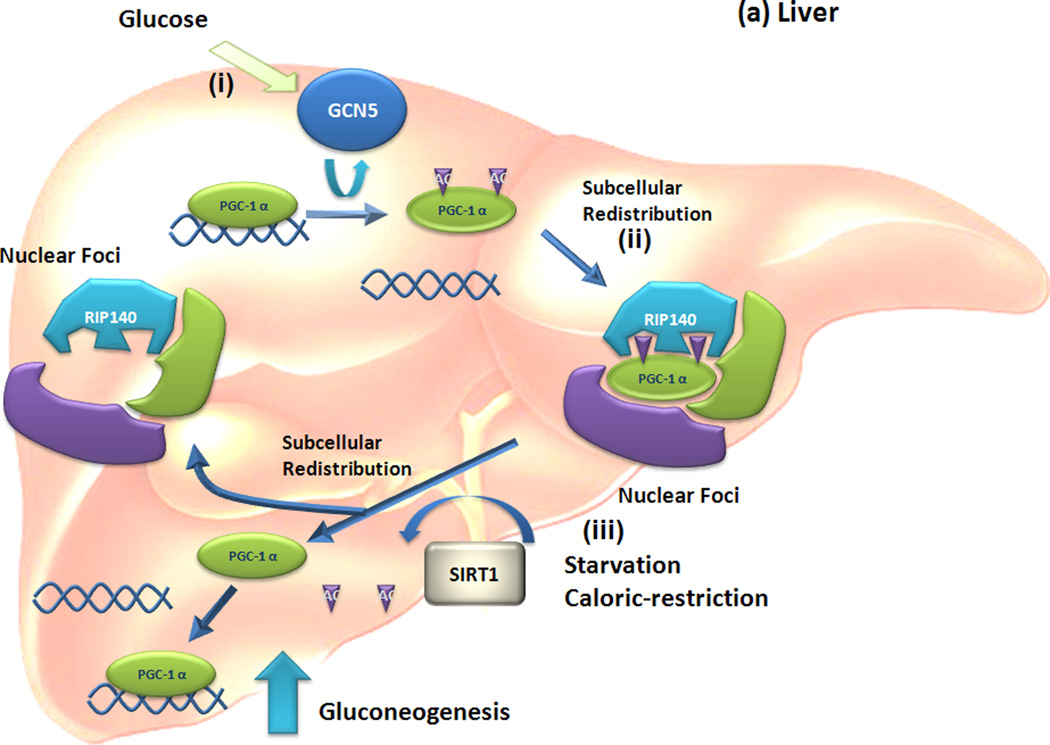
PGC-1α is a major coactivator involved in hormonal and nutrient regulation of glucose and fatty acid metabolism in liver and muscle [29]. With a normal diet, hepatic gluconeogenesis is downregulated through acetylation of PGC-1α by GCN5, which is a component of a PGC-1α containing complex [29] (Figure 2a). Acetylation of PGC-1α by GCN5 translocates PGC-1α from cognate promoter regions to nuclear foci. Although the identity of these nuclear foci is not known, they are colocalized with RIP140, a transcriptional repressor of NRs. Therefore, redistribution of acetylated PGC-1α to nuclear foci inhibits PGC-1α activity required for gluconeogenesis. During starvation or caloric restriction, however, levels of silent information regulator 2/sirtuin 1(SIRT1) deacetylase are increased in hepatocytes to deacetylate PGC-1α. In contrast to acetylation, however, deacetylation of PGC-1α by SIRT1 induces its translocation from nuclear foci to cognate gene promoter regions, inducing gene transcription for glucose-dependent fatty acid oxidation [38] (Figure 2a). Therefore, nutrient signals modulate the ratio of GCN5 to SIRT1, influencing the acetylation status of PGC-1α for the regulation of glucose and fatty acid metabolism in liver.
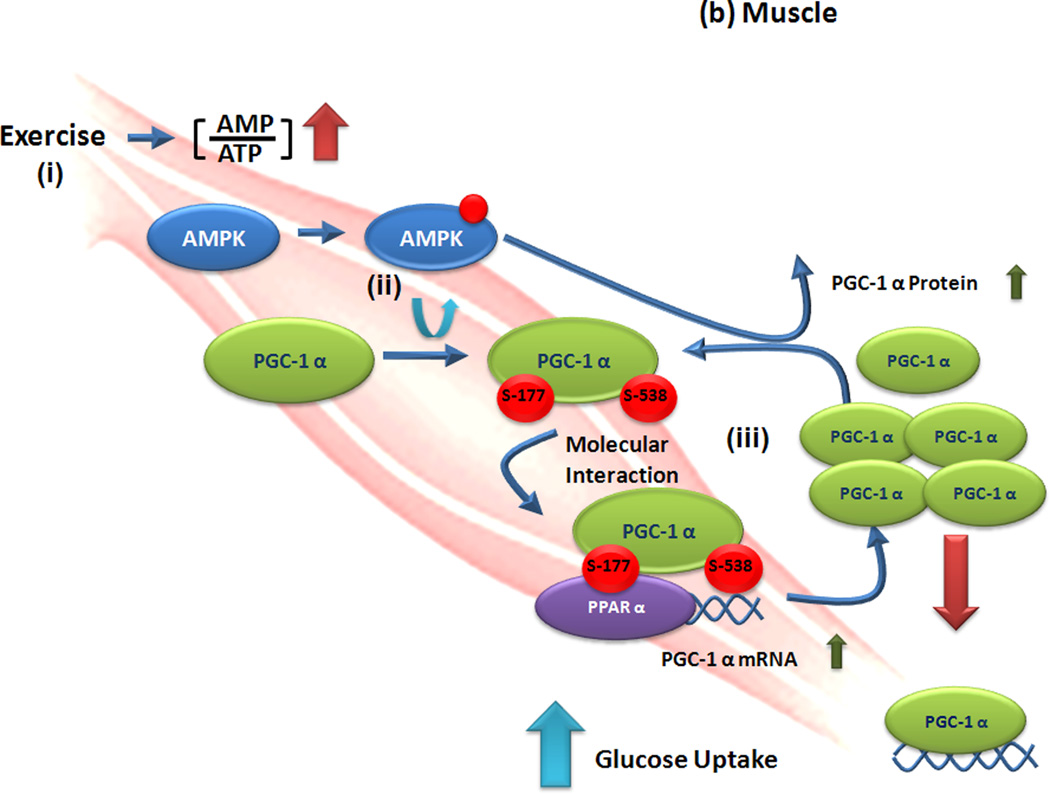
Figure 2. Regulation of glucose metabolism by tissue-specific posttranslational modification programming of PGC-1 alpha.
(a) Liver. (i) High blood glucose levels activate general control of amino-acid synthesis (GCN) 5 activity to acetylate PPARγ coactivator -1 (PGC-1) α to be translocated from active cognate promoters to (ii) nuclear foci containing receptor interacting repressor protein 140 (RIP140). Thus, gluconeogenesis mediated by PGC-1α is repressed by acetylation of PGC-1α. (iii) However, a starvation or caloric restriction condition increases activated silent information regulator 2/sirtuin 1 (SIRT1) levels in the liver. Activated SIRT1 deacetylates PGC-1α and allows its escape from inactive nuclear foci and return to its cognate promoters. In this way, altered location of PGC-1α by deacetylation induces gluconeogenesis in response to starvation. (b) Muscle. (i) Physical exercise increases the ratio of AMP to ATP and activates 5'AMP-activated protein kinase (AMPK) activity. (ii) Activated AMPK rapidly phosphorylates PGC-1α at serine-177 and – 538 to allow its recruitment onto PGC-1α promoter regions. (iii) Altered interaction on properties of PGC-1α by phosphorylation increase PGC-1α mRNA expression and elevated levels of PGC-1α increase glucose uptake.
In addition to acetylation, phosphorylation of PGC-1α also has an effect on gluconeogenesis and fatty acid metabolism in liver. In contrast to glucagon treatment, insulin treatment decreases the gluconeogenesis pathway in the liver to reduce blood glucose levels through activation of protein kinase Akt2/protein kinase B (PKB)-beta [39]. Akt2-mediated phosphorylation of PGC-1α prevents its recruitment to cognate promoters and impairs its ability to promote gluconeogenesis and fatty acid oxidation.
AMP-activated kinase (AMPK) is an important sensor of decreased energy in cells and subsequently acts to increase catabolic reactions and decrease anabolic reactions. During exercise, AMPK is activated in muscle due to the high ratio of AMP to ATP (Figure 2b). Activated AMPK leads to enhanced levels of PGC-1α through AMPK-mediated phosphorylation of PGC-1α at Thr-177 and Ser-538, increasing glucose uptake and fatty acid oxidation [40]. In this way, AMPK-mediated phosphorylation of PGC-1α alters the molecular interaction properties of PGC-1α to modulate feedforward control of PGC-1α expression in muscle during exercise (Figure 2b).
2. Phosphorylation of poly (ADP-ribose) polymerase 1 induces neuronal differentiation
The temporal and spatial regulation of gene expression in response to external and internal stimuli is a major driving factor in cellular differentiation. In most cases, regulation of gene expression is tightly associated with PTM of coactivators on cognate promoters during the differentiation process. For example, cortical progenitor cells (i.e., neural stem cells) are maintained in a pluripotent state before receiving differentiation signals. Hairy/Enhancer of spilt (HES)-1 dependent mammalian achaete-scute homologue (MASH) repression by the temporal lobe epilepsy (TLE)-1 complex is required for the maintenance of neural stem cells. In the presence of a differentiation signal (e.g., platelet-derived growth factor), Ca(++)/calmodulin-dependent protein kinase (CaMK)II delta is activated and phosphorylates the poly(ADP-ribose) polymerase-1 (PARP-1) sensor component of the groucho/TLE-corepressor complex to activate the enzymatic activity of PARP-1 [41]. Activated PARP-1 poly (ADP-ribosyl)ates components of the TLE1 complex (e.g., TLE1, Rad50, nucleolin and nucleophomin) to dissociate the corepressor complex from HES1-regulated promoters and induces recruitment of the activator complex for MASH expression. Therefore, differentiation changes the molecular interaction properties of key coactivators by PTM to form a pre-initiation complex for differentiation.
3 PTM code of coactivators for carcinogenesis
Typically, dysregulation of NR coactivator levels is associated with a higher incidence of endocrine cancer due to disruption of NR-mediated signaling in cancer cells [30]. Therefore, aberrant regulation of coactivator levels has critical clinical implications. Several potential molecular mechanisms have been proposed to address this issue. One of them is the PTM-mediated dysregulation of coactivator protein stability. SRC-3 is amplified and overexpressed in a subset of estrogen receptor-positive and -negative human breast cancers [42]. In addition to coactivators, protein kinases are overexpressed during cancer progression. Atypical PKCζ, one of the kinases frequently overexpressed in cancer cells, is an essential component of the EGF-stimulated chemotactic signaling pathway in human breast cancer cells [43]. Interestingly, estrogen-induced phosphorylation of SRC-3 by atypical PKCζ protects SRC-3 from proteasomal degradation, promoting large increases in estrogenic gene activity and proliferation of breast cancer cells through increased SRC-3 stability [44].
In addition to stability, altered molecular interactions of coactivators by PTM are also associated with the promotion of carcinogenesis. Pontin, a coactivational component of chromatin-remodeling complexes, has an essential role in regulating androgen-receptor target genes in prostate cancer. A recent study showed that 5α-dihydroxytestosterone treatments significantly increased SUMOylation of pontin at Lys-225, facilitating increased androgenic gene activity and proliferation of prostate cancer cells [45]. The increased transcriptional activity of SUMOylated pontin is due to enhancement of its binding affinity for β-catenin and CBP. Therefore, changes in the molecular interaction properties of pontin by SUMOylation accelerate prostate cancer progression through activation of androgen receptor-dependent gene transcription.
In order to prevent NR-positive cancer cell progression, selective NR modulators have been developed and applied clinically. Tamoxifen, which is a selective estrogen receptor modulator (SERM), therapy during the early phases of breast cancer improves overall survival and reduces breast cancer-induced mortality. In cases where chronic treatment is necessary, however, de novo and acquired resistance to tamoxifen remains a problem [14]. Previous studies have indicated tamoxifen treatment actively induces the interaction of SRC-3 and ER in the case of human epidermal growth factor receptor 2 (HER2) positive breast cancer cells to paradoxically activate ER-dependent gene regulation and enhance tumor progression [46].
4. Inflammatory responses modulated by PTM of coactivators
Glucocorticoids and glucocorticoid receptor (GR) regulate inflammatory processes, a crucial component in the pathophysiology of inflammatory bowel disease. In addition to GR, orphan and adopted orphan NRs also regulate inflammatory profiles in ligand-dependent and -independent manners [47]. Coactivators are also functionally involved in inflammatory responses. For example, SRC-3, CBP/p300 and CARM1 modulate NF-κB-dependent transcription. In this cross-communication, PTMs alter the molecular properties of coactivators allowing them to recognize and regulate inflammatory response pathways. For example, both PARP-1 and NF-κB have a pathophysiological role in a number of inflammatory disorders [48]. Specifically, the coactivator p300/CBP acetylates PARP-1 to stabilize its interaction with NF-κB [49]. Consequently, p300 can alter the molecular interaction properties of PARP-1 and modulate NF-κB-dependent transcriptional activity that is critical to inflammatory signal transduction.
Conclusion
Although only a limited number of NR coactivators have been characterized in great detail, each possesses the ability to modulate NR-mediated transcription and essential cellular processes. To govern the complexity of NR-mediated gene regulation in vivo, the molecular properties of NR coactivators are dynamically regulated by PTMs in response to external stimuli (Figure 1). However, questions remain to be answered for a complete understanding of NR coactivator PTM code (Box1). For example, little information is available regarding the PTM coding status of most coactivators due to current limitations in technology for detecting all changes in PTMs on a given protein in response to various external stimuli. In order to fully dissect the PTMs that exist on an entire protein, development of improved and novel methods for mass spectrometry such as Top-Down mass spectrometry [50] is necessary. In addition to improvements in mass-spectrometry technology, establishment of molecular networks between upstream signaling modules, coactivator PTM coding and the downstream molecular events mediated by modified coactivators is a critical requirement for understanding the biological relevance of each NR coactivator PTM code. Currently, however, very little information is available regarding this molecular network due to a lack of high-throughput in vivo assay systems to rapidly and precisely decipher the effects of each coactivator PTM on specific cellular process. Consequently, development of such assay systems will need to be implemented for in depth analysis of complex coactivator PTM coding and a full appreciation of the role that the coactivator PTM code plays in controlling global NR-mediated gene regulation.
Box 1. Questions regarding PTM of NR coactivators.
Do we yet know all the different kinds of posttranslational modifications involved in modulation of NR coactivators?
In cases of multiple posttranslational modifications, how often do sequential intra-communications among PTMs modify NR coactivator function?
What is the repertoire of specific PTM codes that are written onto coactivators for responses to specific external signals?
How do these specific PTM codes impact a variety of downstream cellular processes?
Acknowledgements
This work was supported by NIH grants to B.W.O. (HD07857, NIDDK-NURSA and DK59820).
Reference
- 1.McDonnell DP, et al. Identification of a negative regulatory function for steroid receptors. Proc Natl Acad Sci U S A. 1992;89:10563–10567. doi: 10.1073/pnas.89.22.10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDonnell DP, et al. In situ distinction between steroid receptor binding and transactivation at a target gene. Mol. Cell. Biol. 1991;11:4350–4355. doi: 10.1128/mcb.11.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baniahmad A, et al. The tau 4 activation domain of the thyroid hormone receptor is required for release of a putative corepressor(s) necessary for transcriptional silencing. Mol Cell Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schule R, et al. Many transcription factors interact synergistically with steroid receptors. Science. 1988;242:1418–14120. doi: 10.1126/science.3201230. [DOI] [PubMed] [Google Scholar]
- 5.Halachmi S, et al. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 6.Lonard DM, O'Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Onate SA, et al. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 8.Lanz RB, et al. Nuclear Receptor Coregulatorsin Human Diseases. In: Kumar R, O'Malley BW, editors. NR coregulator and Human Diseases. World Scientific; 2008. pp. 1–134. [Google Scholar]
- 9.Lanz RB, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 10.Hatchell EC, et al. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol Cell. 2006;22:657–668. doi: 10.1016/j.molcel.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 11.Muller JM, et al. FHL2, a novel tissue-specific coactivator of the androgen receptor. Embo J. 2000;19:359–369. doi: 10.1093/emboj/19.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mujtaba S, et al. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 13.Rosenfeld MG, et al. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 14.Jordan VC, O'Malley BW. Selective Estrogen-Receptor Modulators and Antihormonal Resistance in Breast Cancer. J Clin Oncol. 2007;25:5815–5824. doi: 10.1200/JCO.2007.11.3886. [DOI] [PubMed] [Google Scholar]
- 15.Han SJ, et al. A scoring system for the follow up study of nuclear receptor coactivator complexes. Nucl Recept Signal. 2006;4:e014. doi: 10.1621/nrs.04014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, Shang Y. Regulation of SRC family coactivators by post-translational modifications. Cell Signal. 2007;19:1101–1112. doi: 10.1016/j.cellsig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Narayanan R, et al. Cyclin-Dependent Kinase Activity Is Required for Progesterone Receptor Function: Novel Role for Cyclin A/Cdk2 as a Progesterone Receptor Coactivator. Mol. Cell. Biol. 2005;25:264–277. doi: 10.1128/MCB.25.1.264-277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chauchereau A, et al. Sumoylation of the progesterone receptor and of the steroid receptor coactivator SRC-1. J Biol Chem. 2003;278:12335–12343. doi: 10.1074/jbc.M207148200. [DOI] [PubMed] [Google Scholar]
- 19.Gururaj AE, et al. Breast cancer-amplified sequence 3, a target of metastasis-associated protein 1, contributes to tamoxifen resistance in premenopausal patients with breast cancer. Cell Cycle. 2006;5:1407–1410. doi: 10.4161/cc.5.13.2924. [DOI] [PubMed] [Google Scholar]
- 20.Gururaj AE, et al. MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proc Natl Acad Sci U S A. 2006;103:6670–6675. doi: 10.1073/pnas.0601989103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casamassimi A, Napoli C. Mediator complexes and eukaryotic transcription regulation: an overview. Biochimie. 2007;89:1439–1446. doi: 10.1016/j.biochi.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Belakavadi M, et al. MED1 phosphorylation promotes its association with mediator: implications for nuclear receptor signaling. Mol Cell Biol. 2008;28:3932–3942. doi: 10.1128/MCB.02191-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Q, et al. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naeem H, et al. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol Cell Biol. 2007;27:120–134. doi: 10.1128/MCB.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YH, et al. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci U S A. 2005;102:3611–6316. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 27.Amazit L, et al. Regulation of SRC-3 intercompartmental dynamics by estrogen receptor and phosphorylation. Mol Cell Biol. 2007;27:6913–6932. doi: 10.1128/MCB.01695-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, et al. Ubiquitin-dependent distribution of the transcriptional coactivator p300 in cytoplasmic inclusion bodies. Epigenetics. 2007;2:92–99. doi: 10.4161/epi.2.2.4326. [DOI] [PubMed] [Google Scholar]
- 29.Lerin C, et al. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 30.Lonard DM, et al. Nuclear Receptor Coregulators and Human Disease. Endocr Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- 31.Wu RC, et al. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129:1125–1140. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 32.Hoang T, et al. cAMP-dependent protein kinase regulates ubiquitin-proteasome-mediated degradation and subcellular localization of the nuclear receptor coactivator GRIP1. J Biol Chem. 2004;279:49120–49130. doi: 10.1074/jbc.M409746200. [DOI] [PubMed] [Google Scholar]
- 33.Handschin C, Spiegelman BM. Peroxisome Proliferator-Activated Receptor {gamma} Coactivator 1 Coactivators, Energy Homeostasis, and Metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 34.Olson BL, et al. SCFCdc4 acts antagonistically to the PGC-1{alpha} transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22:252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higashimoto K, et al. Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proc Natl Acad Sci U S A. 2007;104:12318–12323. doi: 10.1073/pnas.0610792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cha TL, et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 37.Yi P, et al. Peptidyl-Prolyl Isomerase 1 (Pin1) Serves as a Coactivator of Steroid Receptor by Regulating the Activity of Phosphorylated Steroid Receptor Coactivator 3 (SRC-3/AIB1) Mol. Cell. Biol. 2005;25:9687–9699. doi: 10.1128/MCB.25.21.9687-9699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerhart-Hines Z, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. Embo J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, et al. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 40.Jager S, et al. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ju BG, et al. Activating the PARP-1 sensor component of the groucho/ TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 42.Anzick SL, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 43.Sun R, et al. Protein kinase C zeta is required for epidermal growth factor-induced chemotaxis of human breast cancer cells. Cancer Res. 2005;65:1433–1441. doi: 10.1158/0008-5472.CAN-04-1163. [DOI] [PubMed] [Google Scholar]
- 44.Yi P, et al. Atypical protein kinase C regulates dual pathways for degradation of the oncogenic coactivator SRC-3/AIB1. Mol Cell. 2008;29:465–476. doi: 10.1016/j.molcel.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JH, et al. SUMOylation of pontin chromatin-remodeling complex reveals a signal integration code in prostate cancer cells. Proceedings of the National Academy of Sciences. 2007;104:20793–20798. doi: 10.1073/pnas.0710343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mc Ilroy M, et al. Tamoxifen-induced ER-{alpha}-SRC-3 interaction in HER2 positive human breast cancer; a possible mechanism for ER isoform specific recurrence. Endocr Relat Cancer. 2006;13:1135–1145. doi: 10.1677/erc.1.01222. [DOI] [PubMed] [Google Scholar]
- 47.Wang K, Wan YJ. Nuclear receptors and inflammatory diseases. Exp Biol Med (Maywood) 2008;233:496–506. doi: 10.3181/0708-MR-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korkmaz A, et al. Pathophysiological aspects of cyclophosphamide and ifosfamide induced hemorrhagic cystitis; implication of reactive oxygen and nitrogen species as well as PARP activation. Cell Biol Toxicol. 2007;23:303–312. doi: 10.1007/s10565-006-0078-0. [DOI] [PubMed] [Google Scholar]
- 49.Hassa PO, et al. Acetylation of Poly(ADP-ribose) Polymerase-1 by p300/CREB-binding Protein Regulates Coactivation of NF-{kappa}B-dependent Transcription. J. Biol. Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 50.McLafferty FW, et al. Top-down MS, a powerful complement to the high capabilities of proteolysis proteomics. Febs J. 2007;274:6256–6268. doi: 10.1111/j.1742-4658.2007.06147.x. [DOI] [PubMed] [Google Scholar]
- 51.Chen H, et al. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 52.Oh AS, et al. Tyrosine phosphorylation of the nuclear receptor coactivator AIB1/SRC-3 is enhanced by Abl kinase and is required for its activity in cancer cells. Mol. Cell. Biol. 2008 doi: 10.1128/MCB.00118-08. MCB.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gianni M, et al. P38MAPK-dependent phosphorylation and degradation of SRC-3/AIB1 and RARalpha-mediated transcription. Embo J. 2006;25:739–751. doi: 10.1038/sj.emboj.7600981. [DOI] [PMC free article] [PubMed] [Google Scholar]