Decades of research using purified molecules in vitro have produced a basic understanding of the enzymes and mechanisms that contribute to gene expression in eukaryotes (organisms such as animals, plants and fungi). But these results must be confirmed in living cells, and a productive approach has been to use chromatin immunoprecipitation (ChIP). This technique reveals the genomic locations of DNA-binding proteins such as those forming nucleosomes — DNA segments wrapped around a histone protein core — and transcription factors. Although these studies have produced a wealth of data, they have not always provided mechanistic insight. However, the paper by Rhee and Pugh1 on page 295 of this issue sheds light on several questions concerning the initiation of DNA’s transcription into RNA.
For ChIP, cells are first treated with a chemical that crosslinks proteins and DNA, then disrupts the cells so that their DNA is fragmented. By using specific antibodies, a protein of interest is isolated together with any bound DNA pieces, and these can then be analysed by high-throughput DNA sequencing or other techniques. This allows a quick identification of all the binding sites for a protein — such as those that form protein complexes for transcription initiation — across an entire genome.
Rhee and Pugh have previously reported2 a modification of ChIP, called ChIP-exo, in which an enzyme removes all DNA except that closest to the protein–DNA crosslink, mark- edly improving the technique’s resolution to a few DNA nucleotides. The results can be quite striking, as illustrated by the excellent correlation that the authors observe in their present study1 between the ChIP-exo crosslink sites for the transcription factors TBP (TATA-binding protein) and TFIIB, and the protein–DNA contacts seen in their crystal structures.
In their current paper, the researchers analyse promoters — sequences that specify where to begin the transcription of DNA into RNA — in cells of the yeast Saccharomyces cerevisiae, a model eukaryotic organism. Most notable are their findings regarding how the enzyme that synthesizes messenger RNA, RNA polymerase (Pol) II, is targeted to promoters. TBP is known to recognize the ‘TATA box’ — a specific DNA sequence found in many promoters — and that TBP in turn positions Pol II and its associated factors at the transcription start site (TSS). However, only some promoters, typically those that alternate between repressed and highly active states, contain an obvious TATA box sequence, which represents the optimal TBP-binding site3.
Surprisingly, Rhee and Pugh’s analysis1 of TBP-binding sites at ‘TATA-less’ promoters — more prevalent among ‘housekeeping’ genes that are expressed ubiquitously — reveals this to be a misnomer. These promoters do contain TATA boxes, but their sequences stray from the standard sequence at two or more DNA bases and so their binding to TBP is weaker. This finding echoes classic studies4 on the yeast HIS3 promoter, which contains two TATA boxes: a weak one for constitutive basal transcription and another, clearly recognizable, for maximal, regulated expression.
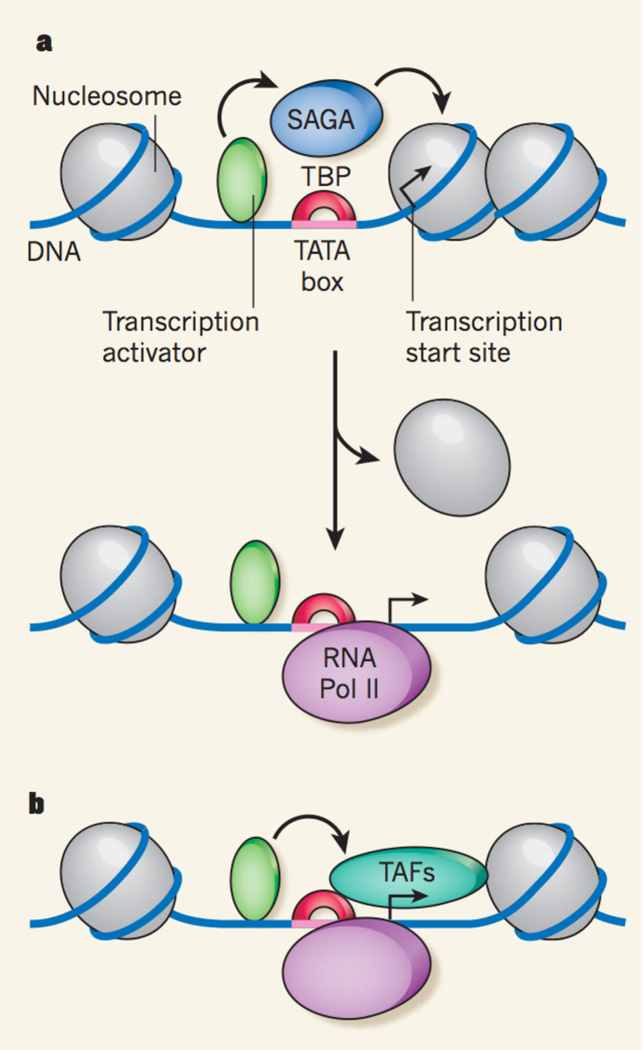
Although all Pol II promoters seem to share a common mode of binding to TBP and basal factors, the data1 help to explain a functional distinction that has been previously observed5 between the two classes of promoters. Rhee and Pugh1 report that, at promoters with obvious TATA sequences, the TSS — and even the initiation complex itself — often overlaps with the first nucleosome in the transcribed region. The expression of these genes tends to depend on the presence of the SAGA protein complex5, which, by adding acetyl groups to the nucleosome’s histones, facilitates nucleosome movement and, thus, DNA unwrapping. Therefore, the first nucleosome probably represses the gene’s transcription by blocking its TSS, and gene activation occurs when the histones are removed by targeted acetylation. Once the TSS-containing DNA is unwrapped, efficient binding of TBP, and Pol II and its associated factors, allows the promoter to be expressed at very high levels.
However, most genes have less-obvious TATA boxes, and their expression depends on other proteins known as TBP-associated factors (TAFs), which together with TBP constitute TFIID. In vitro studies6 have shown that TAFs interact with DNA sequences down- stream of the TATA box, including sequences around the TSS. These additional contacts may help to compensate for the weaker TBP binding to the DNA, but they probably have other functions. Rhee and Pugh1 find that, at those promoters to which TFIID preferentially binds, TSSs are located near the upstream boundary of the first nucleosome. Therefore, TAFs may be positioned in such a way that they contact the first nucleosome, preventing it from encroaching on the promoter and thereby allowing basal gene expression. Indeed, some TAFs form a structure resembling the nucleosome histone core7, suggesting that they might slot into position within an array of nucleosomes.
In addition to invalidating the concept of TATA-less promoters, Rhee and Pugh raise questions about two other recent hypotheses. The first proposes that Pol II promoters are intrinsically bidirectional, that is, a single TATA box can drive transcription in opposite directions. This idea seems plausible because TATA boxes are roughly palindromic, and transcript sequencing studies8,9 have shown that the TSSs of many mRNAs are close to a non-coding RNA that is transcribed in the opposite direction. How- ever, the authors’ ChIP-exo data1 show that the nucleosome-depleted regions between these divergent TSSs harbour two initiation complexes. In other words, bidirectional transcription is the result of two overlapping but divergent promoters driving transcription in opposite directions, rather than a single promoter that can fire in both directions.
The second hypothesis10 is that ‘gene looping’ — the formation of a physical linkage between the beginning and end of active genes — is mediated, in part, by TFIIB. This model is based on observed interactions between mutations in genes that encode TFIIB and 3′-end processing factors (which modify the end of mRNA precursors), as well as ChIP localization of TFIIB (but not the rest of the initiation complex) at transcription-termination regions of selected genes in yeast10. However, the pre- sent study1 and another genome-wide ChIP analysis11 failed to localize TFIIB to 3′-ends, except in the context of initiation complexes at an adjacent promoter. Therefore, the general role of TFIIB in gene looping needs further scrutiny.
It is worth noting that two transcription factors occupying the same genomic location in ChIP experiments may not actually be there at the same time in the same cell, as this technique captures a snapshot of events in a cell population. In vitro experiments are therefore needed to probe the kinetics and inter- mediates of gene expression. Rhee and Pugh1 use many biochemical and structural studies to inform the interpretation of their ChIP-exo data; the ChIP-exo data, in turn, provides an essential in vivo test for in-vitro-derived molecular models. This synergism underscores the necessity of applying both approaches to crucial questions in gene expression.
Figure 1. Two ways of starting the synthesis of messengerRNA.
Promoters are sequences within genes that specify where DNA’s transcription into RNA starts. a, Some promoters have a clearly recognizable sequence, or ‘TATA box’, to which transcription factor TBP can bind to recruit the enzyme RNA polymerase (Pol) II, which synthesizes messenger RNA. These promoters can alternate between repressed and active states. In the repressed state (top), a nucleosome (protein– DNA complex) blocks the transcription start site. Transcription activator proteins can then recruit additional proteins, such as the SAGA complex, to trigger nucleosome removal, allowing access to Pol II and therefore activating transcription (bottom). b, Most promoters lack a clearly recognizable TATA box, and their expression depends on the presence of transcription factor TFIID, a complex formed by TAF proteins and TBP. Transcription activators could recruit TAFs, which, in turn, might interact with both TBP and a nucleosome to keep the transcription start site accessible.
Footnotes
A genome-wide, high-resolution study of DNA-binding sites for proteins that transcribe DNA into RNA reveals details about how this process occurs in vivo.
References
- 1.Rhee HS, Pugh BF. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhee HS, Pugh BF. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basehoar AD, Zanton SJ, Pugh BF. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 4.Struhl K. Mol. Cell. Biol. 1986;6:3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huisinga KL, Pugh BF. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 6.Verrijzer CP, Chen JL, Yokomori K, Tjian R. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 7.Selleck W, et al. Nature Struct. Mol. Biol. 2001;8:695–700. doi: 10.1038/90408. [DOI] [PubMed] [Google Scholar]
- 8.Neil H, et al. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 9.Xu Z, et al. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh BN, Hampsey M. Mol. Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Mayer A, et al. Nature Struct. Mol. Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]