Abstract
Serotonin Transporter (5HTTor SLC6A4) mRNA transcription is regulated by both genetic and epigenetic mechanisms. Unfortunately, despite intense scrutiny, the exact identity and contribution of each of these regulatory mechanisms, and their relationship to behavioral illness remain unknown. This lack of knowledge is critical because alterations in SLC6A4 function are posited to be central to a wide variety of CNS disorders. In order to address this shortcoming, we quantified 5HTTLPR genotype, SLC6A4 mRNA production and CpG methylation using biomaterial from 192 lymphoblast cell lines derived from subjects who participated in the latest wave of the Iowa Adoption Studies. We then analyzed the resulting data with respect to clinical characteristics. We confirmed prior findings that the short (s) 5HTTLPR allele is associated with lower amounts of mRNA transcription, but there was no significant effect of the “Long G” allele on mRNA transcription. We also found that CpG methylation was higher (P< 0.0008) and mRNA production (P< 0.0001) was lower in females as compared to males. Those subjects with a lifetime history of Alcohol Dependence had higher levels of SLC6A4 mRNA. There was a trend for an association of increased overall methylation with lifetime history of major depression. Finally, we confirm our prior findings that the exact levels of 5HTT mRNA expression are dependent on how it is measured. We conclude that both genetic variation and epigenetic modifications contribute to the regulation of SLC6A4 function and that more in-depth studies of the molecular mechanisms controlling gene activity and the relationship of these mechanisms to behavioral illness are indicated.
Introduction
Altered serotonergic neurotransmission is hypothesized to be central to the pathogenesis of a wide variety of CNS disorders including migraine headaches, major depression (MD), autism and alcoholism [Glover et al., 1993; Conroy et al., 2004; Feinn et al., 2005]. The serotonin transporter (5HTTor SLC6A4) is a key regulator of serotonergic neurotransmission. This gene, which is localized to 17p13, consists of 14 exons and a single promoter [Lesch et al., 1994]. Two structural elements of SLC6A4 are of particular relevance to this communication. The first is a variable nucleotide repeat (VNTR), referred to as 5HTTLPR, which is approximately 1,400 bp upstream of the transcription start site [Collier et al., 1996; Heils et al., 1996]. The second is a recently discovered CpG island that surrounds Exon 1 [Philibert et al., 2007a].
The 5HTTLPR is perhaps one of the best studied human polymorphisms. The polymorphism consists of two common alleles in European populations, a short (s) variant with 12 copies of a 22 bp repeat element and a long (l) variant, which has 14 copies of the repeat element. The properties of the s allele have been extensively examined and it has been associated with decreased mRNA transcription [Heils et al., 1996; Lesch et al., 1996; Bradley et al., 2005], decreased protein production [Stoltenberg et al., 2002] and increased vulnerability to alcohol dependence (AD) and MD as demonstrated by meta-analyses [Lotrich and Pollock, 2004; Feinn et al., 2005].
In contrast, the CpG island surrounding Exon 1 has only been recently described and its role in illness is completely unknown. However, since the amount of variance in mRNA and protein production accounted for by the 5HTTLPR is relatively modest (i.e., <10%) [Hranilovic et al., 2004; Bradley et al., 2005], it is reasonable to assume that other cis and trans acting elements in the greater SLC6A4 gene region, such as the CpG island, constitute the majority of the regulatory mechanism(s) controlling SLC6A4 activity. Because the protein product of this gene is a key target of antidepressants, the complete delineation of these and other regulatory elements in this gene could substantially advance the development of an integrated understanding of the pathogenesis of common behavioral illness.
The role of the CpG island may be particularly critical in this pathogenesis. Although it is commonly appreciated that a substantial portion of gene activity is contributed by heritable factors, over the past several years it has also become apparent that gene-environment interactions (GxE), which are defined as environmental effects whose impact is dependent on gene status, also make substantial contributions to neuropsychiatric illness [Moffitt et al., 2005]. In particular, some [Caspi et al., 2003; Kendler et al., 2005], but not all studies [Surtees et al., 2006], have suggested that GxE effects at SLC6A4 are particularly pivotal in the development of MD. Because gene elements such as CpG motifs are potentially modifiable by environmental factors, they may represent a physical substrate for these GxE effects to become manifest. If so, by characterizing the regulatory elements of SLC6A4 responsible for these interactions, it may be possible to identify vulnerable individuals and design more effective, personalized interventions for certain forms of behavioral illness and integrate the effects of these elements into a more comprehensive understanding of the biology that underlies vulnerability to neuropsychiatric illness.
One potential stumbling block to the development of this knowledge base is that technologies to assess the function of these regulatory elements in the intact human CNS do not exist. However, it is well established that lymphoblast cell lines, which are more readily obtained, are valuable tools for deciphering the effects of genetic variation on gene expression [Hranilovic et al., 2004; Bradley et al., 2005; Shukla and Dolan, 2005; Arenas et al., 2007]. In addition, we and others have recently shown that lymphoblast cell lines derived from human subjects retains transcriptional signatures reflective of the clinical status of their donors [Vawter et al., 2004; Philibert et al., 2007b,c]. Hence, by using these in vitro cell lines from well-characterized subjects as surrogates for in vivo CNS cells, it may be possible to delineate the identity of the regulatory elements and determine whether epigenetic effects on these elements affect their function in neuropsychiatric illness.
Therefore, to test the possibility, we quantified CpG residue methylation and gene expression at SLC6A4 in cell lines derived from 192 subjects who participated in the latest wave of the Iowa Adoption Studies (IAS). We then analyzed results of these biological assays with respect to the clinical data from their cognate donors to conditions with meta-analytic evidence of serotonergic dysfunction, MD and AD.
Methods
The overall methodologies and design of the IAS have been previously described [Yates et al., 1998; Philibert, 2006]. In the latest wave of the studies, the subjects were re-interviewed with an extensive environmental, cognitive and behavioral assessment battery that included the Semi-Structured Assessment for the Genetics of Alcoholism, version 2 (SSAGA-II [Bucholz et al., 1994]) and phlebotomized to provide biomaterial for cell lines and DNA. Symptom counts and categorical diagnoses for MD and AD were derived from SSAGA-II data using criteria from the DSM-IV [Association, 1994] and our previously described methods [Yates et al., 1998]. Briefly, lifetime history of AD was defined as having met three or more criteria in the current or past wave of SSAGA interviews. Lifetime history of MD was defined as having met five or more criteria of a MD episode, including low mood or anhedonia, in the current or past wave of SSAGA interviews. Current MD is defined as having low mood or anhedonia and four other symptoms at the time of the phlebotomy draw. All procedures and protocols described in this communication were approved by the University of Iowa Institutional Review Board.
DNA and RNA used in the studies were both prepared from the lymphoblast cell lines. These lymphoblast cell lines were prepared by standard EBV transformation [Klaus, 1987] and the media changed 24 hr prior to the use of the cell lines for the preparation of biomaterial. Total RNA was prepared from the lymphoblast using an Invitrogen RNA purification kit (Invitrogen, Carlsbad, CA) according to the instructions of the manufacturer [Bradley et al., 2005]. DNA was prepared for the cell lines using cold protein precipitation [Lahiri and Nurnberger, 1991]. Not all parameters were successfully measured in all cell lines. Hence, the “n” for each experiment may vary.
Genotyping for the 5HTTLPR was conducted as previously described [Bradley et al., 2005]. Genotype for the “Long G” allele described by Hu et al. [2006], was conducted as described by Stein et al. [2006].
Total RNA from each of the cell lines was quantified spectrophotometrically, then reverse transcribed into cDNA using an Applied Biosystems (ABI, Foster City, CA) cDNA archiving kit according to the manufacturer’s instructions. The resulting cDNA was then diluted, aliquotted into 384 well plates robotically, and stored at −20°C until use.
RTPCR was conducted and quantified as previously described [Bradley et al., 2005; Philibert et al., 2007b,c]. Briefly, the quantification of 5HTT was conducted using a commercially available primer probe set whose probe bridges exons 8 and 9 (ABI, Hs00169010) and a specially synthesized primer probe set whose probe recognizes transcripts containing exons 1 and 2 described previously [Philibert et al., 2007a]. The levels of these transcripts and the housekeeping control transcripts, GAPDH (ABI, Taqman® GAPDH Control Kit) and LDHA (ABI, Hs 00855332, endogenous controls), were performed using Taqman™ Universal PCR master mix and a ABI 7900 HT sequence detection system. All samples were measured in duplicate. Relative levels of each of these transcripts were determined by the comparative CT method using LDHA and GAPDH as normalizing controls after all values were transformed into Z-scores. Correlation co-efficient for sample duplicates for the 5HTT exon 1, 5HTT exon 8, GAPDH, and LDHA were 0.79, 0.98, 0.93, and 0.91, respectively.
Quantitative methylation of each of the samples was conducted by Sequenom, Inc. (San Diego, CA). Briefly, methylated cytosine residues were converted to thymidine using bisulfite modification [Ehrich et al., 2007]. The region flanking the previously identified CpG island was then PCR amplified in two contigs using the following primer sets: Set A, 5′GGGTTTTTATATG-GTTTGATTTTTAGATAG and 5′ CCTACTCCTTTATACAACCTCCCCC and Set B: 5′ GGTTATTTAGAGATTAGATTATGTGAGGGT and 5′CCTACAACAATAAACAAAAA-AACCCC. Methylation ratios for each of the residues (Methyl CpG/Total CpG) were then determined using a MassARRAY™ mass spectrometer using proprietary peak picking and spectra interpretation tools[Ehrich et al., 2005, 2007].
All data were analyzed using the JMP analytic suite, Version 5.1 (SAS Institute, Cary, SC) using Pearson’s correlation co-efficients or two tailed, regression (analysis of variance, ANOVA, ordinal logistic regression, OLR), Chi-square or t-tests tests as described in the text [Fleiss, 1981]. Symptom counts and 5HTTLPR genotype were treated as ordinal (s,s < s,l < l,l) or nominal variables when indicated. Sex and categorical diagnosis was treated as a nominal variable. Methylation ratios and normalized 5HTT mRNA expression were treated as continuous variables.
Results
The characteristics of the subjects are summarized in Table I. In total, 96 male and 96 female subjects provided biomaterial for the study. Male subjects were more likely to be older (t-test, P < 0.002) and less likely to have experienced a MD episode than the female subjects (Chi-Square, P < 0.005).
Table I.
Demographic and Clinical Characteristics of the IAS Subjects
| Male | Female | |
|---|---|---|
| N | 96 | 96 |
| Age (years) | 42.4 ± 8.5 | 38.8 ± 6.8 |
| Ethnicity | ||
| White | 88 | 91 |
| African American | 5 | 2 |
| White of Hispanic origin | 2 | 1 |
| Other | 1 | 2 |
| Lifetime history of MD | 24 | 44 |
| Lifetime history of AD | 20 | 9 |
| Current MD symptoms | ||
| 0 | 86 | 81 |
| 1 | 1 | 1 |
| 2 | 0 | 3 |
| 3 | 1 | 1 |
| 4 | 0 | 1 |
| 5 | 5 | 2 |
| 6 | 1 | 3 |
| 7 | 1 | 4 |
| 8 | 0 | 0 |
| 9 | 0 | 0 |
The distribution of genotypes at 5HTTLPR for the 192 subjects and their incidence of lifetime MD (in parentheses) are shown in Table II. The genotypes for males, females and all subjects together were in Hardy–Weinberg equilibrium.
Table II.
5HTTLPR Genotypes
| Male | Female | Total | |
|---|---|---|---|
| s,s | 18 (3) | 13 (9) | 36 |
| s,l | 47 (12) | 42 (16) | 89 |
| l,l | 31 (9) | 36 (19) | 77 |
The number in parentheses denotes the number of subjects with a lifetime history of MD.
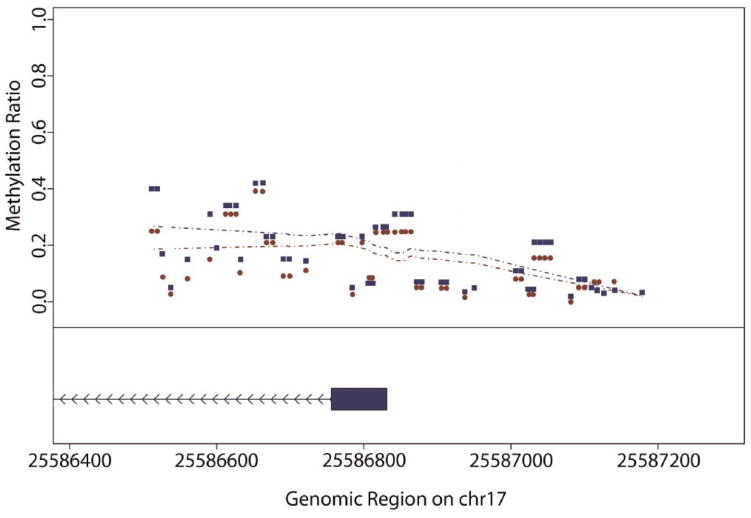
The sequence of the CpG island region and the position of each of the 71 CpG residues studied are shown in Figure 1 while the average methylation ratio for each sex at each residue is depicted in Figure 2. Due to the inability of the mass spectroscopy technique to differentiate certain peaks, the methylation scores for CpG residues 1–2, 9–11, 14–15, 16–17, 18–19, 21–23, 24–25, 28–29, 30–33, 34–39, 40–41, 42–43, 46–52, 53–54, 55–56, 57–61, 63–64, and 66–67 are reported as averaged aggregates (the order of the 71 CpG residues is specified in Figure 1). In addition, because their mass spectroscopy peaks could not be de-convoluted, CpG 16–17, 24–25 and 27; CpG 45 and 65; and CpG 66–67 and 69 have identical values. As Figure 2 illustrates, the methylation ratio increased from left to right (i.e., from CpG 1 to CpG 71) with a peak in methylation being noted near the transcription start site of exon 1. In addition, at all points surveyed, females tended to have a higher amount of methylation with the overall average methylation ratio being significantly higher in females than in males (P < 0.0008). Surprisingly, the correlation coefficients for methylation between neighboring residues are stronger in 5′ portion of the island (0.3–0.5) then in the 3′ portion of the island (0.2) with the correlation for methylation at the opposite ends of the region surveyed being very poor (Pearson’s < 0.10) (Supplemental Table I).
Figure 1.

The sequence of the 5HTT CpG island and position of the CpG residues. The base pair position according to the UCSD Genome Browser March 2006 Assembly (SLC6A4 chr17:25586414-25587412) is given on the left-hand side. All studied CpG residues are denoted in bold and blue with CpG residue 1 corresponding to bp 25586514. The sequence corresponding to SLC6A4 exon 1 is denoted by lower case letters.
Figure 2.
The average methylation at each CpG residue for each sex. The values for females are depicted by blue squares while the values for males are depicted by red circles. The position of SLC6A4 exon1 is denoted by the box with the direction of transcription being indicated by the line with arrows.
The relationship between 5HTTLPR genotype and SLC6A4 mRNA expression was explored using the commercially available primer probe set and our “in house” primer probe set that specifically recognizes transcripts containing exon 1 of SLC6A4. Consistent with our prior results [Philibert et al., 2007a], the correlation of SLC6A4 gene expression as measured by the two primer probe sets was only 0.75 (Pearson’s). The relationship between gene expression as measured by each of these primer probe sets and 5HTTLPR genotype is shown in Figure 3. When expression of the commercially available primer probe set which recognizes the sequence in exons 8 and 9 is examined, the s allele had a nominal but not a statistically significant effect upon SLC6A4 transcripts levels. In contrast, when the primer probe set specifically designed to assay the expression of transcripts containing exon 1 is used, there is a significant effect of genotype on SLC6A4 mRNA expression (ANOVA, Adjusted R square, 0.034, P < 0.02). The difference in Z scores for the exon 1 primer probe set between the s,s and l,l corresponds to a 0.84 cycles (i.e., on average, lymphoblasts with l,l genotypes produce 79% more of the mRNA that cells with the s,s genotypes produce).
Figure 3.
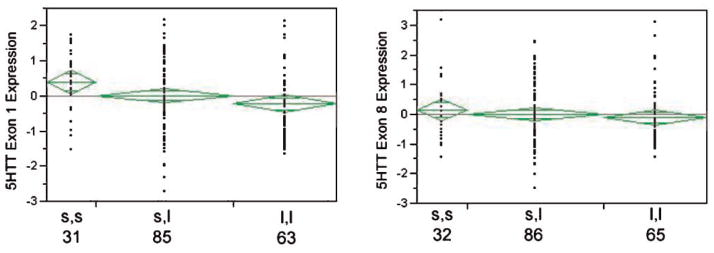
The relationship between 5HTTLPR genotype and SLC6A4 mRNA expression. Using an ordinal regression that specifies an additive effect of the s allele, there is a significant relationship between genotype and 5HTT expression as measured by the exon 1 probe (P < 0.02, ANOVA), but not the exon 8 probe (P < 0.59) [lower Z scores mean lower CT counts or more transcript]. Similarly, when contrasting just the s,s and l,l groups, the relationship is also significant for the exon 1 probe (P < 0.007, t-test), but not the exon 8 probe (P < 0.30). The n for each group is given below the genotype.
The effect of the recently described “Long G” (lG) allele was also analyzed [Hu et al., 2006]. The frequency of the “G” allele among the l alleles in the 179 subjects for whom both genotype and expression data for the exon 1 probes was available was 16% (33/211). Using an ordinal regression model that specified the following rank order s,s < s,lG < s,l < l,lG < l,l, there was no effect of the “Long G” allele on transcription using either the exon 1 probe (see data Table III, OLR, P < 0.18) or the exon 8 probe (data not shown).
Table III.
Genotype and Expression of Cell Lines Using “Triallelic” System
| Male | Female | Male exon 1 and 8 Z scores | Female exon 1 and 8 Z scores | |||
|---|---|---|---|---|---|---|
| s,s | 15 | 17 | −0.19±0.83 | 0.00±1.07 | 0.82±0.62 | 0.26 ± 0.87 |
| s,l | 35 | 33 | −0.24±0.94 | −0.10±0.85 | 0.29±1.09 | 0.21 ± 1.15 |
| s,lG | 10 | 7 | −0.03±0.98 | −0.25 ±0.83 | 0.82±0.62 | 0.04±1.43 |
| l,lG | 10 | 2 | −0.47±0.68 | −0.43±0.41 | 0.92±1.11 | 0.71 ±0.18 |
| l,l | 19 | 30 | −0.56±0.71 | −0.38±0.79 | 0.05±1.05 | 0.08±1.06 |
| lG,lG | 1 | 1 | −1.29 | −0.88 | −0.13 | −0.14 |
The total number of male and female samples does not add up to 192 because data for some points are missing.
Proband gender also had a marked effect on SLC6A4 mRNA production (Fig. 4). Lymphoblast cell lines (the z-score differences reflects an average difference of 0.87 and 0.41 cycles) from males produced 82% and 36% more of exons 1 and 8 transcript, respectively, than the lines from the female subjects.
Figure 4.
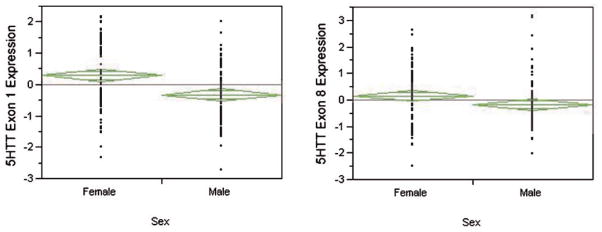
The relationship between gender and SLC6A4 mRNA expression. Males had higher levels of mRNA expression as measured by both the exon 1 probe (P < 0.0001, t-test, left panel) and the exon 8 probe (P < 0.03, t-test, right panel). The “n” for each group is 182 and 180, respectively.
The relationship between methylation and mRNA expression was then analyzed. There was no effect of average methylation on mRNA expression as determined by either primer probeset. However, since the inter-CpG residue methylation correlation coefficients were not strong across the entire island and since some residues, particularly those near the transcription start site, may be more important than others, we then examined the relationship between methylation at each residue and mRNA expression. With respect to the exon 1 probe, before correction for multiple comparison, methylation at CpG 7 and CpG 62 was significantly associated (P < 0.05, ANOVA), and with respect to the exon 8 probe, before correction for multiple comparison, methylation at CpG 30–33 and CpG 62 (P < 0.05, ANOVA) was significantly associated with mRNA production. In each case, increased amounts of methylation at these residues were associated with decreased mRNA transcription. No other significant relationships were observed.
We then turned our attention to the relationship between the lifetime history of MD or AD and both SLC6A4 methylation and gene expression. There was a trend for those without a lifetime history of MD, but not AD, to have higher amounts of total methylation than those without a history of depression (P < 0.07; ANOVA; 17.3% in those with MD vs. 15.6% in those without MD). However, when male and female samples are analyzed separately, the findings were nominally, but not significantly different (15.8% in males with a lifetime MD vs. 14.4% in those without lifetime MD, P < 0.25; ANOVA, 18.1% in females with lifetime MD vs. 17.2% in those without lifetime MD, P < 0.52). There were no significant relationships between the expression of SLC6A4 by either probe and lifetime history of MD. However, lifetime history of AD was associated with increased expression of SLC6A4 as measured by both the exon 1 (P < 0.03) and exon 8 (P < 0.04) probes (data not shown).
Finally, we examined the relationship of current depression to both SLC6A4 methylation and gene expression. No significant relationships using logistic regression or categorical cutoffs (>4 current symptoms denoting current MD) were noted, though we note that only 16 subjects met 5 or more criteria for MD at the time of the blood draw that led to the establishment of the lymphoblast lines.
Discussion
In summary, we report that SLC6A4 mRNA expression is associated with the amount of CpG island methylation and is higher in males than in females. In addition, we confirm and more precisely determine the effect size of the 5HTTLPR on mRNA expression. Finally, we provide suggestive evidence that methylation or gene expression at this locus may be associated with altered vulnerability to MD and AD.
Before discussing these results, we would like to note limitations of this study. First, the biomaterial for the studies was provided from lymphoblast cell lines, not serotonergic neurons. However, we have previously shown that these cell lines retain transcriptional signatures reflective of their cognate donors [Philibert et al., 2007b,c]. Second, the IAS subjects are an epidemiologically sound, but high-risk population that is largely white. Therefore, generalizations to other populations should be done judiciously.
The extension and confirmation of the prior findings with respect to the effect of the 5HTTLPR on mRNA expression is gratifying and more effectively restrains the effect size of this polymorphism on gene transcription and perhaps, by extension, clinical effect. The effects of the s and l variants on mRNA expression have been examined in at least four prior investigations using a total of ~150 lymphoblast cell lines [Lesch et al., 1996; Hranilovic et al., 2004; Bradley et al., 2005]. Overall, the studies found that variance at the 5HTTLPR accounted for ~4–5% of the total variance in SLC6A4 mRNA production. The current study is very much consistent with those prior findings.
The effect of gender was not expected. By itself, gender accounted for 30% of the variance in mRNA expression. This is unlikely to be an artifact for two reasons. First and foremost, all RTPCR reactions were carried out in the same plate using the same set of solutions. Second, we have re-examined our data from our earlier examination of 85 independent lymphoblast lines and found the same effect (P < 0.05, unpublished data). Hence, these differences appear to be real. Further studies will be necessary to determine whether these gender differences have significant effect on clinical status or treatment outcome with selective serotonin reuptake inhibitors.
The quantitatively different responses of the two probe sets were not entirely unexpected. In our prior study, we demonstrated a similar, but less than perfect, correlation between the SLC6A4 expression levels determined by the two probes. Whereas the differences could be artifactual, we strongly believe that they represent true differences in the amount and type of transcript measured because of the following rationale. Our primer probe set for exon 1 was specifically designed to recognize full-length transcripts that are capable of being 5′ capped and thereby stable intracellularly. In contrast, while the commercially available exon 8 probe will also measure these full-length transcripts, it will also recognize partial transcripts that result from promiscuous transcription along the gene. Since this type of transcription is generally not regulated by the promoter [Sipos and Gyurkovics, 2005; ENCODE, 2007] and these incomplete transcripts will not be properly capped or be translated into a functional protein, they, in some respects, may be biologically irrelevant. Still, there is considerable evidence that some of these promiscuous transcripts may serve other functions [ENCODE, 2007].
In a previous study using lymphoblast cell lines, Hu et al. [2005, 2006] reported that aA –G polymorphism in the l variant strongly altered the effects of this 5HTTLPR allele on mRNA expression. In addition, others have shown an effect of the Long G variant in the etiology in MD [Hu et al., 2007]. In the current study, we did not find an effect of the “Long G” allele on mRNA levels. We also examined the effect of this allele on expression in the 89 cell lines used in our prior study and did not find an effect in the cell lines either (unpublished data). Using publicly available data, Martin et al. [2007] also did not find an effect of the Long G variant on gene expression either but suggested that the role of genetic variation at this locus in regulating mRNA may be very complex. We agree and taken in total, these findings suggest that the effect size of the “long G” variant on mRNA transcription may be smaller than originally posited but suggest caution is in order.
Our main motivation for conducting this study was to examine the effect of the CpG island on mRNA production and liability to MD and AD. In our prior communication, we found evidence of genotype specific effects of methylation on mRNA production in an exploratory analysis [Philibert et al., 2007a]. In this study, which used a much more precise method of quantitating methylation and four times the number of cell lines, we did not find that effect and suggest that either this effect is small or that our previous finding in this respect were a false positive.
However, in other respects, the current findings nicely complement and expand the prior findings. First of all, in keeping with our general understanding of the effects of methylation on gene transcription, they demonstrate that greater amounts of CpG methylation are associated with lower amounts of mRNA production [Reik, 2007]. Second, the demonstration of greater amounts of methylation in females versus males and a trend for that methylation to be associated with greater vulnerability to lifetime MD directly demonstrates a mechanism through which the environment can reprogram genes to vulnerability to illness. However, it should be noted that CpG methylation is only one method through which gene transcription can be modified epigenetically. Hence, the current study should be viewed as only the beginning for future studies of this important group of phenomena.
In the current study, we did not find that SLC6A4 mRNA production was significantly associated with vulnerability to lifetime history of MD. The failure to show an effect with MD is roughly consistent with our prior study of 94 IAS subjects and our genome wide transcriptional profiling studies of MD (unpublished data and [Philibert and Madan, 2007]). However, this should not be misconstrued as asserting that altered serotonergic neurotransmission is not involved in the pathogenesis of MD. Indeed, the literature richly supports a strong role for dysfunctional serotonergic neurotransmission in the pathogenesis of MD and the current methylation data strongly suggest something interesting is happening in a longitudinal fashion. Rather, in light of recent advances in our appreciation of systems biology in the etiology of complex illnesses, these findings suggest that (1) the relationship between 5HTT mRNA expression and vulnerability to illness is likely to be exceedingly complex, (2) the 16 acutely depressed subjects analyzed herein may be too few in number to be useful in these type of studies, and (3) the direct measurement of 5HTT mRNA and protein expression in the relevant CNS neurons may be preferable.
In contrast, we did show a significant relationship between 5HTT mRNA expression and lifetime history of AD. This is consistent with some theories on the pathogenesis of AD [Enoch, 2003; Kaufman et al., 2007]. Still, we note that we have not corrected our analyses of the clinical phenotypes for multiple comparisons. Hence, these findings with respect to AD should be viewed cautiously.
In summary, we report strong effects of gender on methylation and 5HTT mRNA production and confirm prior findings of the effect of 5HTTLPR genotype on mRNA production. We suggest that confirmation and further examination of the clinical relevance of these and other findings reported herein are indicated.
Supplementary Material
Acknowledgments
This work was generously supported by a grant to Dr. Philibert (DA 015789). On behalf of Drs. Philibert and Madan, the University of Iowa has filed an intellectual property claim on the CpG island. The authors acknowledge a debt of gratitude to the late Dr. Remi Cadoret, founder of the Iowa Adoption Studies, whose vision led them to explore epigenetic effects at this locus. The authors also would like to thank Dr. Mathias Ehrich and his associates from Sequenom Inc. for their performance of the methylation assays and the provision of Figure 2.
References
- 1.Arenas M, Duley J, Sumi S, Sanderson J, Marinaki A. The ITPA c.94C > A and g.IVS2 + 21A > C sequence variants contribute to missplicing of the ITPA gene. Biochim Biophys Acta. 2007;1772(1):96–102. doi: 10.1016/j.bbadis.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Association AP. Diagnostic and statistical manual of mental disorder. 4. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 3.Bradley SL, Dodelzon K, Sandhu HK, Philibert RA. Relationship of serotonin transporter gene polymorphisms and haplotypes to mRNA transcription. Am J Med Genet Part B. 2005;136B(1):58–61. doi: 10.1002/ajmg.b.30185. [DOI] [PubMed] [Google Scholar]
- 4.Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: A report on the reliability of the SSAGA. J Stud Alcohol. 1994;55(2):149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- 5.Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: Moderation by a polymorphism in the 5-HTT gene. Science. 2003;301(5631):386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 6.Collier DA, Stober G, Li T, Heils A, Catalano M, Di Bella D, et al. A novel functional polymorphism within the promoter of the serotonin transporter gene: Possible role in susceptibility to affective disorders. Mol Psychiatry. 1996;1(6):453–460. [PubMed] [Google Scholar]
- 7.Conroy J, Meally E, Kearney G, Fitzgerald M, Gill M, Gallagher L. Serotonin transporter gene and autism: A haplotype analysis in an Irish autisticpopulation. Mol Psychiatry. 2004;9(6):587–593. doi: 10.1038/sj.mp.4001459. [DOI] [PubMed] [Google Scholar]
- 8.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci USA. 2005;102(44):15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrich M, Zoll S, Sur S, van den Boom D. A new method for accurate assessment of DNA quality after bisulfite treatment. Nucleic Acids Res. 2007;35(5):e29. doi: 10.1093/nar/gkl1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ENCODE. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enoch MA. Pharmacogenomics of alcohol response and addiction. Am J Pharmacogenomics. 2003;3(4):217–232. doi: 10.2165/00129785-200303040-00001. [DOI] [PubMed] [Google Scholar]
- 12.Feinn R, Nellissery M, Kranzler HR. Meta-analysis of the association of a functional serotonin transporter promoter polymorphism with alcohol dependence. Am J Med Genet Part B. 2005;133B(1):79–84. doi: 10.1002/ajmg.b.30132. [DOI] [PubMed] [Google Scholar]
- 13.Fleiss JL. Statistical methods for rates and proportions. 2. New York, NY: John Wiley & Sons, Inc; 1981. [Google Scholar]
- 14.Glover V, Jarman J, Sandler M. Migraine and depression: Biological aspects. J Psychiatr Res. 1993;27(2):223–231. doi: 10.1016/0022-3956(93)90010-y. [DOI] [PubMed] [Google Scholar]
- 15.Heils A, Teufel A, Petri S, Stober G, Riederer P, Bengel D, et al. Allelic variation of human serotonin transporter gene expression. J Neurochem. 1996;66(6):2621–2624. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- 16.Hranilovic D, Stefulj J, Schwab S, Borrmann-Hassenbach M, Albus M, Jernej B, et al. Serotonin transporter promoter and intron 2 polymorphisms: Relationship between allelic variants and gene expression. Biol Psychiatry. 2004;55(11):1090–1094. doi: 10.1016/j.biopsych.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 17.Hu X, Oroszi G, Chun J, Smith TL, Goldman D, Schuckit MA. An expanded evaluation of the relationship of four alleles to the level of response to alcohol and the lcoholism risk. Alcohol Clin Exp Res. 2005;29(1):8–16. doi: 10.1097/01.alc.0000150008.68473.62. [DOI] [PubMed] [Google Scholar]
- 18.Hu X-Z, Lipsky RH, Zhu G, Akhtar LA, Taubman J, Greenberg BD, et al. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet. 2006;78(5):815–826. doi: 10.1086/503850. Epub 2006 Mar 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu XZ, Rush AJ, Charney D, Wilson AF, Sorant AJ, Papanicolaou GJ, et al. Association between a functional serotonin transporter promoter polymorphism and citalopram treatment in adult outpatients with major depression. Arch Gen Psychiatry. 2007;64(7):783–792. doi: 10.1001/archpsyc.64.7.783. [DOI] [PubMed] [Google Scholar]
- 20.Kaufman J, Yang BZ, Douglas-Palumberi H, Crouse-Artus M, Lipschitz D, Krystal JH, et al. Genetic and environmental predictors of early alcohol use. Biol Psychiatry. 2007;61(11):1228–1234. doi: 10.1016/j.biopsych.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 21.Kendler KS, Kuhn JW, Vittum J, Prescott CA, Riley B. The interaction of stressful life events and a serotonin transporter polymorphism in the prediction of episodes of major depression: A replication. Arch Gen Psychiatry. 2005;62(5):529–535. doi: 10.1001/archpsyc.62.5.529. [DOI] [PubMed] [Google Scholar]
- 22.Klaus GGB. Lymphocytes: A practical approach. Oxford: IRL Press; 1987. [Google Scholar]
- 23.Lahiri DK, Nurnberger JI., Jr A rapid non -enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991;19(19):5444. doi: 10.1093/nar/19.19.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesch KP, Balling U, Gross J, Strauss K, Wolozin BL, Murphy DL, et al. Organization of the human serotonin transporter gene. J Neural Transm Gen Sect. 1994;95(2):157–162. doi: 10.1007/BF01276434. [DOI] [PubMed] [Google Scholar]
- 25.Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274(5292):1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 26.Lotrich FE, Pollock BG. Meta-analysis of serotonin transporter polymorphisms and affective disorders. Psychiatr Genet. 2004;14(3):121–129. doi: 10.1097/00041444-200409000-00001. [DOI] [PubMed] [Google Scholar]
- 27.Martin J, Cleak J, Willis-Owen SAG, Flint J, Shifman S. Mapping regulatory variants for the serotonin transporter gene based on allelic expression imbalance. Mol Psychiatry. 2007;12(5):421–422. doi: 10.1038/sj.mp.4001952. [DOI] [PubMed] [Google Scholar]
- 28.Moffitt TE, Caspi A, Rutter M. Strategy for investigating interactions between measured genes and measured environments. Arch Gen Psychiatry. 2005;62(5):473–481. doi: 10.1001/archpsyc.62.5.473. [DOI] [PubMed] [Google Scholar]
- 29.Philibert R. Merging genetic and environmental effects in the iowa adoption studies: Focus on depression. Ann Clin Psychiatry. 2006;18(4):219–222. doi: 10.1080/10401230600948399. [DOI] [PubMed] [Google Scholar]
- 30.United States (pending) 2007. The University of Iowa, assignee. Compositions and methods for detecting predisposition to a substance use disorder or to a mental illness or syndrome. [Google Scholar]
- 31.Philibert R, Madan A, Andersen A, Cadoret R, Packer H, Sandhu H. Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am J Med Genet Part B. 2007;144B(1):101–105. doi: 10.1002/ajmg.b.30414. [DOI] [PubMed] [Google Scholar]
- 32.Philibert R, Crowe RR, Ryu GY, Yoon JG, Secrest D, Sandhu H, et al. Transcriptional profiling of lymphoblast lines from subjects with panic disorder. Am J Med Genet B Neuropsychiatr Genet. 2007;144(5):674–682. doi: 10.1002/ajmg.b.30502. [DOI] [PubMed] [Google Scholar]
- 33.Philibert R, Ryu GY, Yoon JG, Sandhu H, Hollenbeck N, Barkhurst A, et al. Transcriptional profiling of subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet. 2007;144(5):683–690. doi: 10.1002/ajmg.b.30512. [DOI] [PubMed] [Google Scholar]
- 34.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 35.Shukla SJ, Dolan ME. Use of CEPH and non-CEPH lymphoblast cell lines in pharmacogenetic studies. Pharmacogenomics. 2005;6(3):303–310. doi: 10.1517/14622416.6.3.303. [DOI] [PubMed] [Google Scholar]
- 36.Sipos L, Gyurkovics H. Long-distance interactions between enhancers and promoters. The case of the Abd-B domain of the Drosophila bithorax complex. FEBS J. 2005;272(13):3253–3259. doi: 10.1111/j.1742-4658.2005.04757.x. [DOI] [PubMed] [Google Scholar]
- 37.Stein MB, Seedat S, Gelernter J. Serotonin transporter gene promoter polymorphism predicts SSRI response in generalized social anxiety disorder. Psychopharmacology. 2006;187(1):68–72. doi: 10.1007/s00213-006-0349-8. [DOI] [PubMed] [Google Scholar]
- 38.Stoltenberg SF, Twitchell GR, Hanna GL, Cook EH, Fitzgerald HE, Zucker RA, et al. Serotonin transporter promoter polymorphism, peripheral indexes of serotonin function, and personality measures in families with alcoholism. Am J Med Genet. 2002;114(2):230–234. doi: 10.1002/ajmg.10187. [DOI] [PubMed] [Google Scholar]
- 39.Surtees PG, Wainwright NW, Willis-Owen SA, Luben R, Day NE, Flint J. Social adversity, the serotonin transporter (5-HTTLPR) polymorphism and major depressive disorder. Biol Psychiatry. 2006;59(3):224–229. doi: 10.1016/j.biopsych.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Vawter MP, Ferran E, Galke B, Cooper K, Bunney WE, Byerley W. Microarray screening of lymphocyte gene expression differences in a multiplex schizophrenia pedigree. Schizophr Res. 2004;67(1):41–52. doi: 10.1016/s0920-9964(03)00151-8. [DOI] [PubMed] [Google Scholar]
- 41.Yates W, Cadoret R, Troughton E. The Iowa adoption studies methods and results. In: LaBuda M, Grigorenko E, editors. On the way to individuality: Methodological issues in behavioral genetics. Hauppauge, NY: Nova Science Publishers; 1998. pp. 95–125. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.