Abstract
Software advancements in the last several years have had a significant impact on proteomics from method development to data analysis. Herein we detail a method, which uses our in-house developed software tool termed Skyline, for empirical refinement of candidate peptides from targeted proteins. The method consists of 4 main steps from generation of a testable hypothesis, method development, peptide refinement, to peptide validation. The ultimate goal is to identify the best performing peptide in terms of ionization efficiency, reproducibility, specificity, and chromatographic characteristics to monitor as a proxy for protein abundance. It is important to emphasize that this method allows the user to perform this refinement procedure in the sample matrix and organism of interest with the instrumentation available. Finally, the method is demonstrated in a case study to determine the best peptide to monitor the abundance of surfactant protein B in lung aspirates.
Keywords: Targeted Proteomics, Selected Reaction Monitoring, SRM assays, Skyline
Introduction
Selected reaction monitoring (SRM), also referred to in the literature as multiple reaction monitoring (the latter term has been deprecated by IUPAC nomenclature)[1], is increasingly being used in the proteomics community as a method for sensitive and selective quantification of targeted species of interest across a sample set. The emergence of SRM as a powerful method in a scientist’s toolbox is largely due to advancements in instrumentation, software developments[2–4], and the limitations associated with the reproducible quantification of low abundant species with traditional data dependent analysis (DDA) proteomic workflows.[5–6] The stochastic nature of fragmentation of precursor ions in DDA leads to a bias towards the identification of more abundant species.[6] This bias is problematic for systems biology and biomarker studies (e.g., validation) where the need exists to reproducibly quantify a pre-determined set of peptides across a large sample set.
SRM has traditionally been used in the field of proteomics for absolute quantification of proteins[7–9] by digesting the proteins to peptides and then developing a quantitative assay for the measurement of the resulting peptide(s).[10],[11] More recently the technique is being used for relative quantification of 100’s of peptides in a single multiplexed assay due to its high sensitivity[12], dynamic range[13], specificity[14], and reproducibility[15]. SRM can be thought of as the mass spectrometric equivalent of a western blot[16] as it selectively targets a peptide and respective transitions believed to be representative of a specific protein from complex samples. Its principal advantages over western blots are three-fold: 1) it is significantly cheaper to develop SRM assays than to make and screen antibodies for each protein target; 2) it can be multiplexed to increase the throughput; and 3) when performed correctly, a mass spectrometry based assay can provide absolute structural specificity.
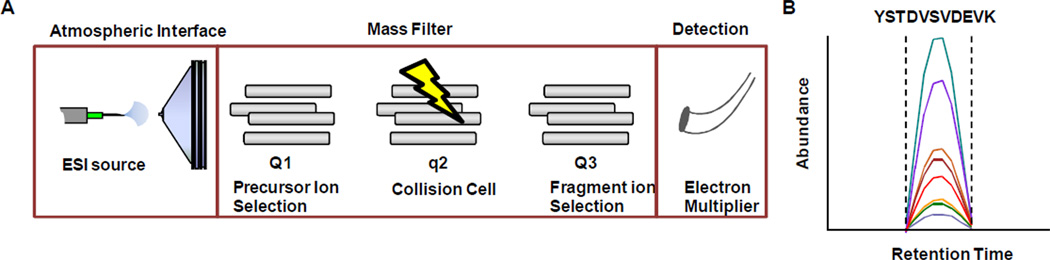
The high molecular specificity of SRM is a result of a two-stage mass filter. The first filter selects target ions for isolation in Q1 (i.e., precursor ion selection), isolated ions are then subjected to collision induced dissociation in q2, and specific fragment ions are selected for detection in Q3 (i.e., fragment ion selection). Figure 1A displays a schematic of a triple quadrupole mass spectrometer operating in selected reaction monitoring mode. A typical spectrum obtained for a given peptide is illustrated in Figure 1B in which the abundance (counts/sec) of all y fragment ions from a particular peptide were monitored for a set period (i.e., dwell time) as a function of retention time. It should be noted that other instrument platforms (i.e., ion trap, q-tof) are capable of performing SRM-like experiments; however, in these cases computer software will algorithmically extract the targeted signals from full or partial product ion scans. Although these pseudo-SRM experiments will give adequate data, the performance of these instruments (i.e., duty cycle and sensitivity) is currently less than what is achieved on a contemporary triple quadrupole.
Figure 1.
A) A schematic of a triple quadrupole operating in selected reaction monitoring mode. B) A typical spectrum created by monitoring a single peptide and all of its y-transitions
With the growing popularity of SRM assays to address complex biological problems, there is a need to actively discuss the most robust methods for development of these assays. Herein, we describe the method currently used in our laboratory for development of targeted peptide assays via SRM-MS. The ultimate goal at the conclusion of this procedure is the development of a highly specific and sensitive mass spectrometric assay to monitor the abundance of 1 or more proteins across a sample cohort.
Targeted Peptide Assay Development
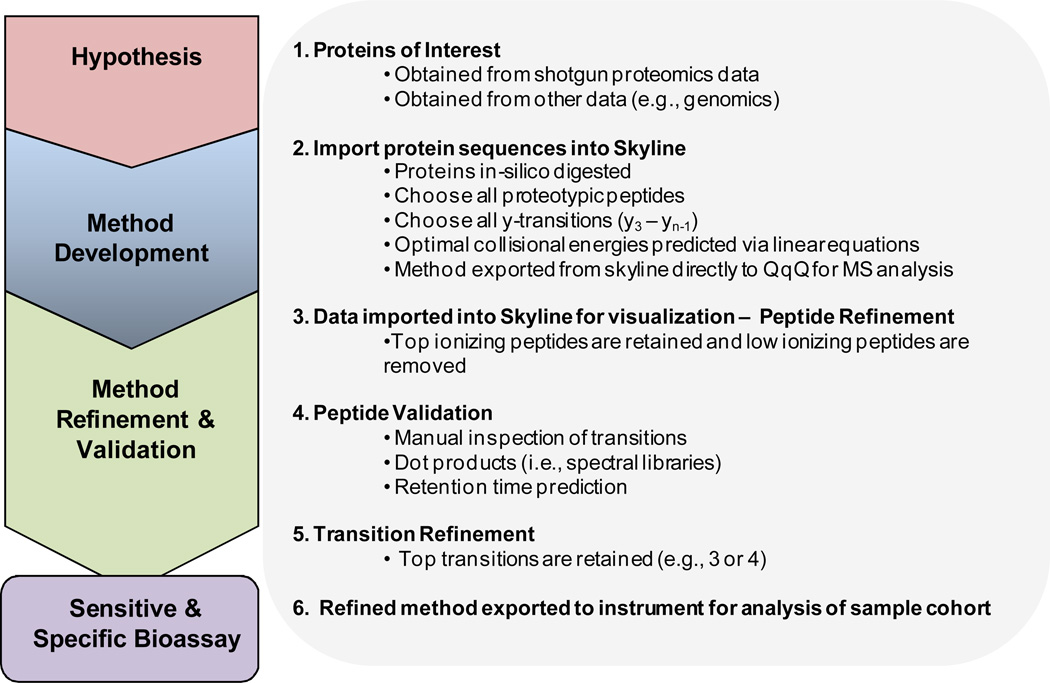
Figure 2 describes the targeted proteomics method development procedure currently used in our laboratory which consists of 4 main steps: 1) generation of a hypothesis that can be tested by quantitative protein measurements; 2) method development; 3) method refinement; and 4) method validation. This procedure relies heavily on the Skyline[2] software tool developed in the MacCoss laboratory explicitly for aiding in this targeted assay development. Skyline provides a graphical user interface in which instrument specific methods can be easily designed and exported for the instruments of the four major triple quadrupole vendors: AB SCIEX, Agilent, Thermo-Scientific and Waters. Raw data acquired with these instruments can then be imported directly into Skyline for analysis. The software package is open source, as part of the ProteoWizard project[17] and freely available from our website (http://proteome.gs.washington.edu/software/skyline).[18] Several instructional videos and tutorials are also available on the above site, including Video 2: Results Analysis and the Targeted Method Refinement tutorial, which contain complete instruction, with real data, on how to use Skyline to perform the process described in detail below.
Figure 2.
The workflow used for the development of peptide SRM assays. Four mains steps are outlined including development of a hypothesis, method development, validation and refinement.
1. Generation of Hypothesis
This step is relatively undefined and only requires that the hypothesis be testable via quantitative protein measurements. For some labs, hypotheses may be generated from discovery proteomics data (e.g., semi-quantitative data-dependent shotgun experiments). In these cases, the SRM assay is built around the observed peptides and their fragment ions. The aim of the assay is to confirm a difference in abundance between groups of samples, determine relative fold changes, or measure absolute quantities of a peptide.
Hypotheses can also be generated from other data. For example, western blots, ELISAs, genomic and transcriptomic data could be used to develop a hypothesis driven SRM experiment.
2. Method Development
After a protein or set of proteins of interest are identified, the sequences are imported into Skyline and an in-silico digestion is performed. Skyline supports various enzymes (Lys-C, chymotrypsin); however, we begin development with the commonly used trypsin and will look to other enzymes with varying specificities only if necessary. Although proteins can yield 100’s of enzymatic peptides[19] only a fraction of these are often detected[20]. In the development stage, each tryptic peptide will be monitored to determine the best species in terms of ionization efficiency and specificity. First, enzymatic peptides (in-silico digested) are pre-screened to only include protein-specific peptides – ones that are unique to a specific protein from the organism of interest. Peptides are again filtered to include those between 8–25 amino acids in length.[21–22] Skyline will exclude potential ragged end peptides (i.e., two enzymatic sites sequential to each other) as the digestion efficiency of these peptides is often irreproducible. In theory, it also advisable to avoid peptides that have amino acids that are frequently modified either in vivo (e.g., proteolysis, phosphorylation, or glycosylation) or in vitro (e.g., deamidation, methionine oxidation, alkylation).[14] In addition, if stable isotope labeled peptides are needed for the assay, then there may be additional sequence limitations imposed by the peptide synthesis methodology. However, in reality it is often impossible to abide by all of these rules. Y-type ions[9, 14, 22] are monitored due to the tendency of b-type ions to undergo secondary fragmentation[23–24] which decreases the overall signal of peptide specific transitions (transition m/z > precursor m/z). All y-ion transitions (y3 - yn-1) are monitored to improve specificity which aides in the validation step. While optimization of instrument parameters (i.e., collision energies) can lead to improved b-type ion abundance in SRM experiments, on average, the effect is not great enough for them to exceed y-type ion response.[25]
The alternative to this empirical approach to assay development is to use the large amount of public data to make informed decisions on which peptides are likely to be detected.[3, 26–33] These data repositories have proven to be a great source of information as they provide a fast way to choose or narrow a list of potential peptide targets based on previous studies. However, the empirical refinement approach described herein does offer certain advantages. First, a prerequisite for using these public repositories for assay development is that a suitable number of peptides from the targeted protein of interest have been identified previously. Next, when using shotgun proteomic data (i.e., spectral libraries) for choosing peptides, it is possible for an abundant peptide to give poor MS/MS spectra limiting its chances for identification by database search algorithms. For example, a peptide containing a proline residue may not provide a high score in a DDA experiment due to its fragmentation characteristics[34–35]; however, it could still be a desirable target for an SRM assay. The final reason is the experimental and biological variability between the conditions from which the available data was collected and which the current study is being conducted. For example peptide ionization under ESI conditions is highly dependent on several parameters[36] including solvent flow rates, sample complexity, and tip diameter, among others. These experimental conditions are difficult to replicate or may not even be known about the available data set. In addition, variations in digestion procedures, chromatographic methods and instrument platforms could alter the overall sensitivity of a particular peptide. As a result, a peptide that responds well in one laboratory potentially may not perform as well in another laboratory given simply the experimental setup and sample matrix. Also there is certain to be biological variability between the data used for choosing peptides and the current study. This problem would be much more relevant in advanced organisms (e.g., human) where genetic variation (e.g., alternative splicing and single nucleotide polymorphisms SNPs) and post translational modifications can significantly alter the protein expressed and its peptide constituents. Ultimately this could affect the presence and/or abundance of the peptide.
It should be noted that Skyline, can make use of available investigation-specific DDA data on targeted peptides of interest, as it supports the construction of spectral libraries from most major search engines. We find these spectral libraries are generally useful for choosing transitions and for validation of SRM signal – but much less helpful for selecting the best responding peptide.
Given adequate sample volume the downside to this empirical peptide evaluation approach is the time needed for both instrument analysis and data interpretation. However, Skyline streamlines method development – allowing prediction of optimal collision energies using linear equations[37] and then the direct export of instrument specific methods. In addition, the Skyline graphical interface streamlines human interpretation and validation of the data, a bottleneck frequently underestimated in its impact on successful investigation. As a result, the main rate limiting factor becomes the instrument time needed for evaluating hundreds of candidate peptides. The length of time for these experiments is overall quite small in the context of an entire study. For instance, with a reasonable dwell time of 10 ms, 250 transitions can be monitored (2.5 s duty cycle). Given an average tryptic peptide length of ~14 amino acids, 10 transitions monitored per peptide, one can monitor 25 peptides per LC-MS run. Using a short LC-MS runtime for peptide evaluation (45 min), 800 peptides can be evaluated in a 24-hour period. Depending on the size and number of proteins of interest, this evaluation stage could take a few days to a week. However, for lengthy biomarker studies where sample collection, method development, data acquisition and data analysis can take several years, it is our opinion that the cost of this time is miniscule compared to the benefit of knowing that each protein monitored has the most sensitive and specific peptide and transitions possible.
3. Method Refinement
At the start of the empirical method refinement described here, many candidate peptides will be excluded as producing a response too low to measure effectively given the experimental sample preparation. After data collection, files are imported into Skyline for refinement. In this step two main criteria are evaluated for each peptide: 1) signal intensity and 2) specificity of each transition (interference). Skyline provides a visual interface to view the intensity of each peptide from a particular protein and any interference from non-specific transitions. Peptides with low intensity and large interferences are eliminated. This process can be aided by comparing dot products between the observed intensities versus a spectral library. We employ a cut-off of ≥0.8 to identify potential targets and try to keep at least three peptides from each protein to undergo validation. Skyline provides a refinement interface that can perform operations like this en masse.
It is worth noting that in targeted proteomics positive peak identification can often wait until the peak has proven a valuable indicator. The cost of a false-positive at this phase can be expressed in two cases: 1) a protein with fewer than 3 peptides suitable for targeted measurement could be refined to continue forward with an extra misidentified peak or 2) a protein with 3 or more measurable peptides might actually be refined to replace a peptide having a correctly identified peak with a misidentified peak. This second case bears some opportunity cost, but can be made extremely unlikely to displace all correctly identified peptides for a protein. In subsequent multi-replicate analysis, consistent relative ion intensities and retention times will ensure that the same peptide is measured in all cases. Any peak, correctly or incorrectly identified, that proves a valuable indicator will be worth much greater expense in verifying its identity.
4. Method Validation
Peptide validation, however, remains a critical part of any SRM assay development. As the proteomics community continues to embrace targeted technologies for addressing biological problems there comes a need to quantitatively determine the specificity of the assay and ask the question “Is the assay measuring what it is supposed to be?”[38] Traditionally peptide validation has been performed, concurrent with absolute quantification of a single protein[7–10], by spiking an stable isotope labeled (SIL) internal standard petpide (e.g., 13C, 15N) into samples of interest. Then the similarity between retention time (i.e., co-elution) and fragmentation characteristics between the two peptides (i.e., target vs. standard) are evaluated. This method is undoubtedly the most reliable to ensure peptide identity. However, recent advances have raised the need to reproducibly measure 100’s of peptides making the cost of validating each peptide via a heavy stable isotope labeled (SIL) internal standard unreasonable. Others have used crude unlabeled peptide standards and SRM-triggered MS/MS[39] to validate peptide targets using retention time and fragmentation patterns. This method can be performed in a relatively high throughput manner and is inexpensive compared to SIL standards, although peptide synthesis for each potential target is still required.
In the case of large experiments, where 10s to 100s of peptides are monitored, we typically use a combination of spectral libraries (dot products), retention time prediction and manual inspection of elution profiles for peptide validation. In regards to manual inspection of transitions, the elution profile of each transition (y3 – yn-1) is evaluated for coelution to confirm identification. Skyline expedites this process and provides dot products from spectral libraries and correlation between predicted and observed retention time for each peptide, making it possible to review 100 peptides in minutes. The Skyline refinement interface also allows initial bulk removal of many poor matches. Following peptide validation, the final step is to keep the 3 or 4 most abundant transitions and remove the lower responding ones. Here again the Skyline refinement interface makes this a single operation. In the next section, this protocol for SRM assay development using Skyline is described in a case study.
Case Study – SP-B Detection in Neonates Diagnosed with RDS
Neonatal respiratory distress syndrome (RDS) is the most frequent respiratory cause of death in the United States during the first year of life. In 2001, RDS contributed to approximately 20% of the 28,000 neonatal deaths (i.e., 5600 deaths due to RDS).[40–42] Its etiology is directly related to the developmental deficiency of pulmonary surfactant – a phospholipids-protein film manufactured by type 2 pneumocytes that line the alveoli and maintain alveolar patency at end expiration and is essential to proper fetal-neonatal pulmonary transition.[43] Mature SP-B is involved in the distribution of phospholipids in the alveolus, affects the biological processing of other surfactant proteins (e.g., SP-C)[43–45] and; thus, is an essential component in maintaining surfactant homeostasis.[46–47] In this study, we wanted to develop an SRM assay to monitor the abundance of SP-B secreted into the lungs in infants diagnosed with RDS. First it was necessary to find protein-specific peptides from which to develop the SRM assay.
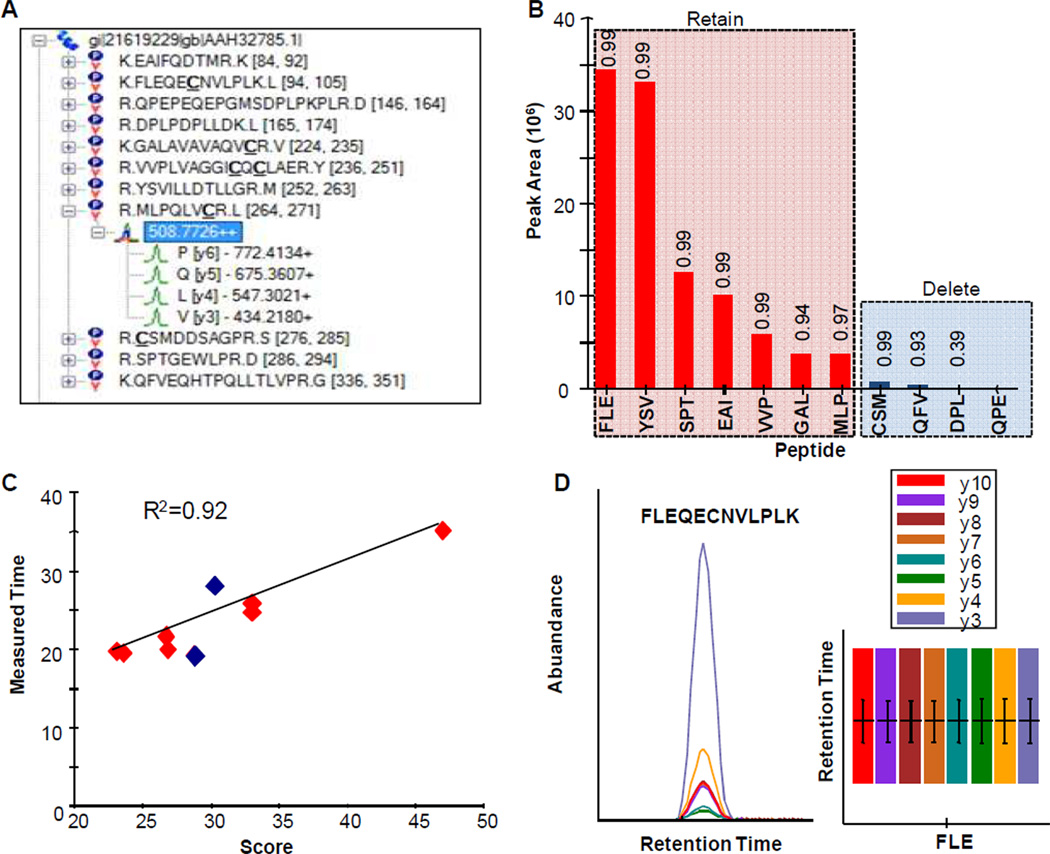
Figure 3 describes the steps taken from method development to peptide validation in order to develop a sensitive and specific bioassay. First, the FASTA sequence of SP-B was pasted into Skyline where an in-silico trypsin digestion was performed. Skyline automatically filtered the peptides to include species between 8–25 amino acids in length (user defined) and all of their y-transitions (y3 – yn-1). A background proteome database was built in Skyline from the NCBI human FASTA file to check for peptide uniqueness (i.e., peptide that is specific to only 1 protein). In this example, each tryptic peptide was found to be specific to SP-B. The method was then exported directly into a vendor-specific method file for instrument data acquisition. Figure 3B displays the abundances of the various peptides monitored. In this example there were several abundant peptides. Peptides that did not respond well were deleted and at least three peptides were retained to undergo validation using spectral libraries, retention time, and elution profiles. Figure 3C plots retention time as a function of SSRCalc[48] hydrophobicity score for the various peptides. As displayed the higher abundant species give a good linear correlation (r2>0.92) between measured retention time and hydrophobicity score. The red squares lie off the linear relationship and indicate a less confident match for the lower responding peptides (Figure 3B). In version 1.2, Skyline supports a more accurate retention time predictor where scores are based on previous measurement of individual peptides[49], making this correlation a stronger validation tool. The elution profile of all the transitions are evaluated for coelution as shown for a peptide (FLEQECNVLPLK) in the inset of Figure 3D. All transitions for this particular peptide coelute which confirms peptide identification. Finally, using the Skyline refinement interface, all but the 4 most abundant transitions were discarded. At the conclusion of these simple steps an SRM assay has been developed that is both sensitive and specific to the protein of interest. For more complex experiments where 100’s of peptides are monitored, these steps are just repeated for each one.
Figure 3.
A case study involving the development of an SRM assay to measure the abundance of SP-B in aspirate derived from neonates with respiratory distress syndrome. A) Fasta sequence of SP-B is pasted into Skyline and an in-silico digestion is performed using trypsin (user defined). All y-transitions are monitored and the transitions are exported directly into an instrument method (user defined). B) Data is imported into skyline and the abundances of the various peptides are evaluated. High responding peptides are kept while low ones are deleted. Validation is performed using dot products C) RT prediction and D) manual inspection of elution profiles. For peptide QPE no spectrum was found in the library and therefore no dot product is listed for this peptide. The dot product for DPL is significantly low. Both of these peptides also lie off the regression line shown in C (blue squares). Combined with elution profiles (data not shown) these data indicate a low probability of a positive match. For this example, these peptides would be immediately discarded due to their low response - but this is a nice example of how the combination of RT prediction, dot products, and elution profiles can be used to validate peptides.
It is important to emphasize that SRM mass spectrometry measures the abundance of peptides. Often in the literature (our lab is guilty as well) peptide abundance is directly correlated to protein abundance; however, several problems exist with this naïve assumption as well described by Duncan et al.[50] Furthermore one study demonstrated a strong correlation (r2>0.97) between C-reactive protein measured by a commercial certified ELISA and PC-IDMS – albeit the absolute values were greater than an order of magnitude different.[9] This discrepancy is possibly a result of one measurement performed on the protein level while the other is on the peptide level. Our lab has observed different absolute abundances depending on the peptide chosen for the SRM assay indicating different isoforms of the protein – a result confirmed by western blot analysis.[51]
In summary, SRM is a powerful tool to reproducibly monitor the abundance of 100s of peptides across a large sample set. Several steps exist in development of an SRM assay, though always of primary interest is which peptides to monitor as a proxy for the targeted protein. The method described here details an empirical approach to monitor all protein-specific to determine the peptides with the best response. This empirical approach is favored because the factors that contribute to strong peptide response are complicated and not well understood. Therefore prediction algorithms,[52–54] although potentially useful in narrowing the list of starting candidates, are not nearly as robust as empirical measurement. While several years ago empirical refinement would have been extremely cumbersome, the development of Skyline has helped make it a reasonable procedure on all common triple-quadrupole instruments. Currently, validation of peptide targets in SRM-MS is not standardized in the literature. The most accurate method to ensure peptide identity is synthesis of a standard; however, this method can become unreasonable when trying to validate 100s of peptide targets. For method development, we monitor all y-ions of a particular peptide and use a combination of spectral libraries (i.e., dot products), retention time prediction, and manual inspection of the elution profiles for validation. In the foreseeable future the need to measure 100–1000s peptides will warrant implementation of an automated statistical strategy to estimate false discovery rates.[55]
Acknowledgements
The authors are grateful for generous support from NIH (R01 RR032708, R01 HL082747, P41 RR011823, U24 CA126479-04S2). MSB acknowledges support from the NIH Genome Training Grant (T32 HG000035).
References
- 1.Murry KK, Boyd RK, Eberlin MN, Langley GJ. IUPAC standard definitions of term relating to mass spectrometry. IUPAC Task group MS Terms Third and Final Draft. http://mass-spec.lsu.edu/msterms/index.php/Main_Page.
- 2.MacLean B, Tomazela DM, Shulman N, Chambers M, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cham Mead JA, Bianco L, Bessant C. Free computational resources for designing selected reaction monitoring transitions. Proteomics. 2010;10:1106–1126. doi: 10.1002/pmic.200900396. [DOI] [PubMed] [Google Scholar]
- 4.Brusniak MY, Kwok ST, Christiansen M, Campbell D, et al. ATAQS: A computational software tool for high throughput transition optimization and validation for selected reaction monitoring mass spectrometry. Bmc Bioinformatics. 2011;12:78. doi: 10.1186/1471-2105-12-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoopmann MR, Merrihew GE, von Haller PD, MacCoss MJ. Post analysis data acquisition for the iterative MS/MS sampling of proteomics mixtures. J Proteome Res. 2009;8:1870–1875. doi: 10.1021/pr800828p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michalski A, Cox J, Mann M. More than 100,000 Detectable Peptide Species Elute in Single Shotgun Proteomics Runs but the Majority is Inaccessible to Data-Dependent LC–MS/MS. J. Proteome Res. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 7.Barnidge DR, Hall GD, Stocker JL, Muddiman DC. Evaluation of a cleavable stable isotope labeled synthetic peptide for absolute protein quantification using LC-MS/MS. J Proteome Res. 2004;3:658–661. doi: 10.1021/pr034124x. [DOI] [PubMed] [Google Scholar]
- 8.Barnidge DR, Goodmanson MK, Klee GG, Muddiman DC. Absolute quantification of the model biomarker prostate-specific antigen in serum by LC-Ms/MS using protein cleavage and isotope dilution mass spectrometry. J Proteome Res. 2004;3:644–652. doi: 10.1021/pr049963d. [DOI] [PubMed] [Google Scholar]
- 9.Williams DK, Muddiman DC. Absolute quantification of C-reactive protein in human plasma derived from patients with epithelial ovarian cancer utilizing protein cleavage isotope dilution mass spectrometry. J Proteome Res. 2009;8:1085–1090. doi: 10.1021/pr800922p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barr JR, Maggio VL, Patterson DG, Jr., Cooper GR, et al. Isotope dilution--mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin Chem. 1996;42:1676–1682. [PubMed] [Google Scholar]
- 11.Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35:265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Onisko B, Dynin I, Requena JR, Silva CJ, et al. Mass spectrometric detection of attomole amounts of the prion protein by nanoLC/MS/MS. J Am Soc Mass Spectrom. 2007;18:1070–1079. doi: 10.1016/j.jasms.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, et al. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Addona TA, Abbatiello SE, Schilling B, Skates SJ, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnott D, Kishiyama A, Luis EA, Ludlum SG, et al. Selective detection of membrane proteins without antibodies: a mass spectrometric version of the Western blot. Mol Cell Proteomics. 2002;1:148–156. doi: 10.1074/mcp.m100027-mcp200. [DOI] [PubMed] [Google Scholar]
- 17.Kessner D, Chambers M, Burke R, Agusand D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. https://skyline.gs.washington.edu/labkey/project/home/software/Skyline/begin.view.
- 19.Picotti P, Aebersold R, Domon B. The implications of proteolytic background for shotgun proteomics. Mol Cell Proteomics. 2007;6:1589–1598. doi: 10.1074/mcp.M700029-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Kuster B, Schirle M, Mallick P, Aebersold R. Scoring proteomes with proteotypic peptide probes. Nat Rev Mol Cell Biol. 2005;6:577–583. doi: 10.1038/nrm1683. [DOI] [PubMed] [Google Scholar]
- 21.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Prakash A, Tomazela DM, Frewen B, Maclean B, et al. Expediting the development of targeted SRM assays: using data from shotgun proteomics to automate method development. J Proteome Res. 2009;8:2733–2739. doi: 10.1021/pr801028b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paizs B, Suhai S. Fragmentation pathways of protonated peptides. Mass Spectrom Rev. 2005;24:508–548. doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]
- 24.Vachet RW, Ray KL, Glish GL. Origin of product ions in the MS/MS spectra of peptides in a quadrupole ion trap. J Am Soc Mass Spectrom. 1998;9:341–344. doi: 10.1016/S1044-0305(98)00008-7. [DOI] [PubMed] [Google Scholar]
- 25.Holstein Sherwood CA, Gafken PR, Martin DB. Collision energy optimization of b- and y-ions for multiple reaction monitoring mass spectrometry. J Proteome Res. 2011;10:231–240. doi: 10.1021/pr1004289. [DOI] [PubMed] [Google Scholar]
- 26.Frewen B, MacCoss MJ. Using BiblioSpec for creating and searching tandem MS peptide libraries. Curr Protoc Bioinformatics. 2007 doi: 10.1002/0471250953.bi1307s20. Chapter 13, Unit 13 17. [DOI] [PubMed] [Google Scholar]
- 27.Picotti P, Lam H, Campbell D, Deutsch EW, et al. A database of mass spectrometric assays for the yeast proteome. Nat Methods. 2008;5:913–914. doi: 10.1038/nmeth1108-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desiere F, Deutsch EW, King NL, Nesvizhskii AI, et al. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–D658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kandasamy K, Keerthikumar S, Goel R, Mathivanan S, et al. Human Proteinpedia: a unified discovery resource for proteomics research. Nucleic Acids Res. 2009;37:D773–D781. doi: 10.1093/nar/gkn701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathivanan S, Pandey A. Human Proteinpedia as a resource for clinical proteomics. Mol Cell Proteomics. 2008;7:2038–2047. doi: 10.1074/mcp.R800008-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 32.Jones P, Cote RG, Cho SY, Klie S, et al. PRIDE: new developments and new datasets. Nucleic Acids Res. 2008;36:D878–D883. doi: 10.1093/nar/gkm1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siepen JA, Belhajjame K, Selley JN, Embury SM, et al. ISPIDER Central: an integrated database web-server for proteomics. Nucleic Acids Res. 2008;36:W485–W490. doi: 10.1093/nar/gkn196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz BL, Bursey MM. Some proline substituent effects in the tandem mass spectrum of protonated pentaalanine. Biol Mass Spectrom. 1992;21:92–96. doi: 10.1002/bms.1200210206. [DOI] [PubMed] [Google Scholar]
- 35.Grewal RN, El Aribi H, Harrison AG, Siu KWM, Hopkinson AC. Fragmentation of Protonated Tripeptides: The Proline Effect Revisited. The Journal of Physical Chemistry B. 2004;108:4899–4908. [Google Scholar]
- 36.Fenn JB. Ion Formation from Charged Droplets - Roles of Geometry, Energy, and Time. J. Am Soc Mass Spectr. 1993;4:524–535. doi: 10.1016/1044-0305(93)85014-O. [DOI] [PubMed] [Google Scholar]
- 37.Maclean B, Tomazela DM, Abbatiello SE, Zhang S, et al. Effect of collision energy optimization on the measurement of peptides by selected reaction monitoring (SRM) mass spectrometry. Anal Chem. 2010;82:10116–10124. doi: 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman J, McKay MJ, Ashman K, Molloy MP. How specific is my SRM?: The issue of precursor and product ion redundancy. Proteomics. 2009;9:1120–1123. doi: 10.1002/pmic.200800577. [DOI] [PubMed] [Google Scholar]
- 39.Picotti P, Rinner O, Stallmach R, Dautel F, et al. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat Meth. 2010;7:43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 40.Guyer B, Freedman MA, Strobino DM, Sondik EJ. Annual summary of vital statistics: trends in the health of Americans during the 20th century. Pediatrics. 2000;106:1307–1317. doi: 10.1542/peds.106.6.1307. [DOI] [PubMed] [Google Scholar]
- 41.Arias E, MacDorman MF, Strobino DM, Guyer B. Annual summary of vital statistics--2002. Pediatrics. 2003;112:1215–1230. doi: 10.1542/peds.112.6.1215. [DOI] [PubMed] [Google Scholar]
- 42.Hoyert DL, Freedman MA, Strobino DM, Guyer B. Annual summary of vital statistics: 2000. Pediatrics. 2001;108:1241–1255. doi: 10.1542/peds.108.6.1241. [DOI] [PubMed] [Google Scholar]
- 43.Hamvas A, Cole FS, Nogee LM. Genetic disorders of surfactant proteins. Neonatology. 2007;91:311–317. doi: 10.1159/000101347. [DOI] [PubMed] [Google Scholar]
- 44.Clark JC, Wert SE, Bachurski CJ, Stahlman MT, et al. Targeted disruption of the surfactant protein B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc Natl Acad Sci U S A. 1995;92:7794–7798. doi: 10.1073/pnas.92.17.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitsett JA, Weaver TE. Hydrophobic surfactant proteins in lung function and disease. N Engl J Med. 2002;347:2141–2148. doi: 10.1056/NEJMra022387. [DOI] [PubMed] [Google Scholar]
- 46.Eijking EP, Strayer DS, van Daal GJ, Tenbrinck R, et al. In vivo and in vitro inactivation of bovine surfactant by an anti-surfactant monoclonal antibody. Eur Respir J. 1991;4:1245–1250. [PubMed] [Google Scholar]
- 47.Robertson B, Kobayashi T, Ganzuka M, Grossmann G, et al. Experimental neonatal respiratory failure induced by a monoclonal antibody to the hydrophobic surfactant-associated protein SP-B. Pediatr Res. 1991;30:239–243. doi: 10.1203/00006450-199109000-00007. [DOI] [PubMed] [Google Scholar]
- 48.Krokhin OV, Craig R, Spicer V, Ens W, et al. An Improved Model for Prediction of Retention Times of Tryptic Peptides in Ion Pair Reversed-phase HPLC. Mol Cell Proteomics. 2004;3:908–919. doi: 10.1074/mcp.M400031-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Escher C, Reiter Lea. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012 doi: 10.1002/pmic.201100463. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duncan MW, Yergey AL, Patterson SD. Quantifying proteins by mass spectrometry: the selectivity of SRM is only part of the problem. Proteomics. 2009;9:1124–1127. doi: 10.1002/pmic.200800739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomazela DM, Patterson BW, Hanson E, Spence KL, et al. Measurement of Human Surfactant Protein-B Turnover in Vivo from Tracheal Aspirates Using Targeted Proteomics. Anal Chem. 2010;82:2561–2567. doi: 10.1021/ac1001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang H, Arnold RJ, Alves P, Xun Z, et al. A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics. 2006;22:e481–e488. doi: 10.1093/bioinformatics/btl237. [DOI] [PubMed] [Google Scholar]
- 53.Mallick P, Schirle M, Chen SS, Flory MR, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 54.Webb-Robertson BJ, Cannon WR, Oehmen CS, Shah AR, et al. A support vector machine model for the prediction of proteotypic peptides for accurate mass and time proteomics. Bioinformatics. 2008;24:1503–1509. doi: 10.1093/bioinformatics/btn218. [DOI] [PubMed] [Google Scholar]
- 55.Reiter L, Rinner O, Picotti P, Huttenhain R, et al. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat Methods. 2011;8:430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]