Abstract
Background
Heart failure (HF) is associated with excessive extracellular matrix (ECM) deposition and abnormal ECM degradation leading to cardiac fibrosis. Connective Tissue Growth Factor (CTGF) modulates ECM production during inflammatory tissue injury, but available data on CTGF gene expression in failing human heart and its response to mechanical unloading are limited.
Methods and Results
LV tissue from patients undergoing cardiac transplantation for ischemic (ICM; n=20) and dilated (DCM; n=20) cardiomyopathies, and from nonfailing (NF; n=20) donor hearts were examined. Paired samples (n=15) from patients undergoing LV assist device (LVAD) implantation as “bridge to transplant” (34-1145 days) were also analyzed. There was more interstitial fibrosis in both ICM and DCM compared to NF hearts. Hydroxyproline concentration was also significantly increased in DCM relative to NF samples. The expression of CTGF,TGFB1, COL1-A1, COL3-A1, MMP2 and MMP9 mRNAs in ICM and DCM were also significantly elevated as compared to NF controls. Although TGFB1, CTGF, COL1-A1, and COL3-A1 mRNA levels were reduced by unloading, there was only a modest reduction in tissue fibrosis and no difference in protein-bound hydroxyproline concentration between pre- and post-LVAD tissue samples. The persistent fibrosis may be related to a concomitant reduction in MMP9 mRNA and protein levels following unloading.
Conclusions
CTGF may be a key regulator of fibrosis during maladaptive remodeling and progression to HF. Although mechanical unloading normalizes most genotypic and functional abnormalities, its effect on ECM remodeling during HF is incomplete.
Keywords: Remodeling, Heart-assist device, Gene expression, collagens
INTRODUCTION
Heart failure (HF) is a complex disorder and remains a significant cause of morbidity and mortality in both developed and developing countries. The process of left ventricular (LV) remodeling is an important indicator of morbidity and mortality in patients with HF. The most prominent changes include compensatory hypertrophy of cardiomyocytes and myocardial fibrosis.1 Myocardial fibrosis involves both increased deposition of extracellular matrix (ECM) proteins, as well as altered composition of the ECM.2 Although ECM remodeling may initially be regarded as an adaptive response to increased wall stress and neurohormonal activation, it ultimately may contribute adversely to tissue structure and function, causing increased cardiac stiffness, and may serve as a predictor of mortality in patients with HF. Therefore, it is important to identify molecular and cellular events that contribute to ECM remodeling.
The ECM of the heart includes the fibrillar collagen network, basement membranes, and proteoglycans.3-5 In both human and animal studies, changes in LV geometry and function have been associated with changes in the fibrillar collagen network.4, 5 Alterations in myocardial collagen alignment, structure and support contribute to the progressive LV dilatation and remodeling which accompanies HF. Changes in total myocardial collagen content, collagen subtypes, and collagen cross-linking are important features of ECM remodeling, and each may contribute to changes in passive myocardial stiffness.6-8
Left ventricular assist devices (LVADs) provide mechanical support for the end-stage HF patient, and are often used as a bridge to cardiac transplantation as well as for “destination therapy.” LVADs have been demonstrated to cause reverse-remodeling, which is defined as the reversal of chamber enlargement, the reduction in LV mass and the improvement in global pump function.9, 10 Mechanical unloading has been associated with normalization of diastolic chamber properties.11, 12 In addition, mechanical unloading has been associated with a trend toward normalization in cardiomyocyte function,13 calcium cycling properties,14 and gene expression.15 In addition to changes in intrinsic myocardial properties, mechanical unloading is associated with changes in the characteristics and metabolism of the ECM. However, unlike other aspects of reverse remodeling of the myocardium, the ECM changes do not uniformly reflect a return towards normal conditions.13, 16, 17
Connective Tissue Growth Factor (CTGF) is a secreted multifunctional protein that belongs to the CCN (Cyr61, CTGF, and Nov) family of growth factors.18 CTGF has been implicated in mediating fibrosis, ECM production, hypertrophy, adhesion, proliferation, differentiation, migration, angiogenesis, and apoptosis,19, 20 all of which are common features of myocardial remodeling. CTGF is highly expressed in both cardiac fibroblasts and cardiomyocytes,21, 22 and may be involved in initiating cellular processes underlying fibrosis.23, 24 For instance, CTGF was up-regulated during the pathogenesis of several fibrotic disorders, such as atherosclerosis, scleroderma, and liver and kidney fibrosis.25-28 CTGF has also been implicated in the induction of cardiac fibrosis in animal models of LV remodeling.29, 30 However, available data on CTGF gene expression in failing human heart, and its response to mechanical unloading are limited.
MATERIALS AND METHODS
Left ventricular tissue from nonfailing and failing human hearts
Samples of left ventricular (LV) tissue were obtained from Loyola University Health System’s (LUHS’s) Cardiovascular Institute Tissue Repository, and from the Gift of Hope Organ and Tissue Donor Network. The investigation conformed to the principles outlined in the Declaration of Helsinki. A detailed protocol and informed consent document were reviewed by LUHS’s Institutional Review Board prior to tissue procurement. Following informed consent, explanted LV tissue was obtained from patients undergoing heart transplantation for ischemic (ICM) and nonischemic, dilated cardiomyopathy (DCM). Tissue samples were quick-frozen in liquid N2 in the operating room and stored at −80°C. Following informed consent from organ donor family members, nonfailing (NF) donor hearts judged unsuitable for cardiac transplantation were stored in cardioplegic solution on ice and were delivered within 4h of cardiac extirpation by the Gift of Hope Organ and Tissue Donor Network. Tissue samples were then quickly frozen in liquid N2, and stored at −80°C.
Matched LV core and explanted tissue were obtained from an additional 15 patients who underwent left ventricular assist device (LVAD) implantation (HeartMate II, Thoratec Corp., Pleasanton, CA) as a bridge to transplant (DCM=11; ICM=4). All patients were in NYHA Class IV HF at the time of LVAD implantation. Unloading time ranged from 42 to 1145 days. Care was taken to obtain LV tissue from the explanted hearts directly adjacent to the placement site of the LVAD inflow cannula.
Protein-bound hydroxyproline assay
LV tissue samples (~100mg wet wt) from NF (n=20), DCM (n=20), ICM (n=20) and paired pre- and post-LVAD (n=13) patients were homogenized in lysis buffer,31 and after removal an aliquot for analysis of total protein, the homogenates were transferred to vacuum hydrolysis tubes (ThermoFisher Scientific, Rockford, IL) and hydrolyzed in 6N HCl (18h, 110°C). Hydrolysates were evaporated to dryness (Haake Buchler Evapotec Vortex Evaporator, Ft. Lee, NJ) and resuspended in 1.0ml of water. Hydroxyproline concentration was measured by the method of Woessner32 using freshly prepared, vacuum-dried L-hydroxyproline (Sigma, St. Louis, MO) in 1 mM HCl as standard. Hyproxyproline content (μg) was normalized to the total protein content (mg) determined by bicinchoninic acid protein assay (Pierce Chemical Co, Rockford IL) using bovine serum albumin as standard.
mRNA analysis
LV total RNA was extracted using TRIzol® Reagent (Life Technologies, Carlsbad, CA), and further purified with the RNeasy kit (Qiagen, Valencia, CA). On-column DNase digestion was performed with the RNase-Free DNase Set (Qiagen, Valencia, CA). RNA was quantified by absorbance at 260 nm and its integrity was determined by examining the 28S and 18S rRNA bands in ethidium bromide-stained agarose gels. CTGF, TGFB1, ANF, COL1-A1, COL3-A1, MMP2 and MMP9 mRNAs, and eukaryotic 18S rRNA were then analyzed by real-time RT-PCR, as previously described.33, 34 The mixture consisted of 10 l of sample cDNA, 1.5 l DEPC water, 12.5 l TaqMan® Universal PCR master mix, and 1 l of a primer/dual labeled probe combination specific for each gene of interest. TaqMan® and all primer/probe combinations were obtained from Applied Biosystems (Foster City, CA). PCR amplification was performed by cycling between 95°C (15s) and 60°C (6 0s) for 45 cycles, using the 6-FAM fluorophore for quantification. All samples were run in triplicate, and the results were averaged. The ΔΔCt method was then used to quantify specific mRNA levels relative to 18S rRNA.
Tissue sectioning and image analysis
Paraffin-embedded sections (6μm thick) were routinely prepared by the LUHS and Emory University Hospital Pathology Departments, and stained with Mallory-Trichrome stain. Randomly selected digital color photomicrographs were then acquired using an Olympus BH2 microscope with a 10X objective and a Nikon D200 digital camera (20 images per section). Images were then transferred to a desktop computer using Nikon Camera Pro-2 Software (Ver. 2.7.2), and analyzed for collagen content on the basis of color thresholds using ImageJ software (NIH; Bethesda, MD). A minimum threshold (50 arbitrary units) was established for background (white) and each color channel (red, orange, and blue). Interstitial fibrosis was defined as the average collagen area fraction (%) of the blue channel determined in regions of interest that excluded areas of gross scar and perivascular fibrosis.
Immunofluorescent microscopy was performed on frozen sections of failing LV myocardium. Tissue samples were embedded and frozen to −80°C in OCT. Sections (~10μm thick) were fixed in 4% (w/v) paraformaldehyde and permeabilized with 0.1% (v/v) Triton X-100. Following treatment with 10% goat serum, the sections were then sequentially stained with mouse anti-human CTGF (Santa Cruz Biotechnology; Santa Cruz, CA) and rabbit anti-chicken α-actinin (Sigma-Aldrich; St. Louis, MO) primary antibodies, followed by rhodamine-labeled goat anti-mouse or FITC-labeled goat anti-rabbit antibodies, respectively. Sections stained without primary antibodies served as controls for nonspecific staining. Sections were viewed under a Zeiss Axioskop epifluorescent microscope using Axiovision AC (Ver 4.5) software.
SDS-PAGE and Western blotting
LV tissue samples (~200mg) were homogenized in 2ml of extraction buffer, containing 10mM cacodylic acid, 150mM NaCl, 20mM ZnCl2, 1.5mM NaN3, and 0.1% Triton X-100 (pH 5.0) according to the method of Spinale, et al.35 Following centrifugation, the pellet was resuspended in extraction buffer, and the procedure was repeated in triplicate. Pooled supernatant fractions were concentrated in Pierce concentrators (20kDa molecular weight cutoff) and total protein concentration was assayed using the bicinchoninic acid protein assay. Equal amounts of extracted proteins (200μg) were separated by SDS-PAGE on 10% polyacrylamide gels. Separated proteins were transferred to nitrocellulose, and the membranes were probed with rabbit anti-human MMP9 (Cell Signaling Technology; Beverly, MA). Equal loading was confirmed by quantifying GAPDH in each sample. Primary antibody binding was detected with horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse secondary antibodies, and visualized by ECL. Band intensity was quantified using an HP Scanjet 4890 flatbed scanner and UN-SCAN-IT Gel, Ver. 6.1 software.
Statistical analysis
Results were expressed as means±SEM. Normality was assessed using the Kolmogorov-Smirnov test, and homogeneity of variance was assessed using Levene’s test. Data for multiple groups were compared by 1-way ANOVA or 1-way ANOVA on Ranks followed by the Student-Newman-Keuls test. Data from paired samples were compared by paired t-test, or Signed Rank Test, where appropriate. Differences among means were considered significant at P<0.05. Linear regression analysis was performed on 2 variables to determine correlation coefficient (R values) and the analysis of variance of the regression. Data were analyzed using the SigmaStat Statistical Software Package, Ver. 3.1 (Systat Software, San Jose, CA).
RESULTS
Fibrosis in end-stage heart failure
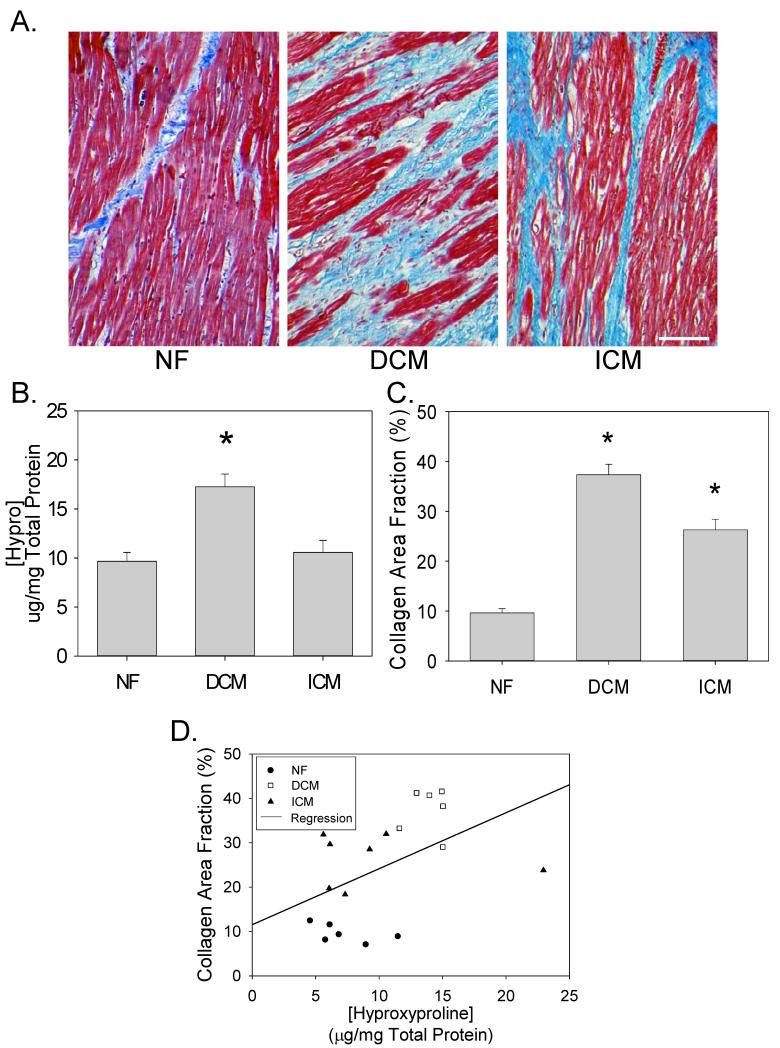
We obtained LV tissue from patients undergoing cardiac transplantation, and from nonfailing (NF) donor hearts unsuitable for transplantation. We first quantitatively analyzed LV collagen concentration in the ECM by measuring the concentration of protein-bound hydroxyproline in portions of LV tissue from NF (n=20), DCM (n=20), and ICM (n=20) patients (Figure 1B). Hydroxyproline concentration was significantly increased in DCM relative to NF samples. We then analyzed the degree of interstitial fibrosis in a subset of the same patients. Figure 1A depicts representative images of NF (n=6), DCM (n=6) and ICM (n=7) tissue sections stained with Mallory-Trichrome stain – interstitial collagens stain blue. Average collagen area fractions were quantitatively analyzed in Figure 1C. As seen in the representative images and quantitative data, LV tissue from patients with DCM had the highest collagen area fraction. Furthermore, there was a significant, albeit weak correlation (R=0.49; P=0.034) between the biochemical analysis of protein-bound hyproxproline, and the degree of interstitial fibrosis observed by quantitative image analysis (Figure 1D).
Figure 1. Fibrosis in human heart failure.
(A) Representative myocardial sections from NF, DCM and ICM patients stained with Mallory Trichrome stain – collagen stains blue. (B) LV collagen concentration in tissue homogenates was assessed by measuring the concentration of protein-bound hydroxyproline (μg/mg total protein) from NF, DCM, ICM patients (means±SEM; n=20 in each group). (C) Areas of fibrosis in Mallory-Trichrome stained histological sections were digitally analyzed for collagen area fraction (%) on the basis of color thresholds using Image J software. Data are means±SEM from 6-7 patients in each group. (D) Comparison of collagen area fraction (%) vs. hydroxyproline concentration (μg/mg total protein). *P<0.05.
CTGF, COL1-A1, and COL3-A1 expression levels in end-stage heart failure
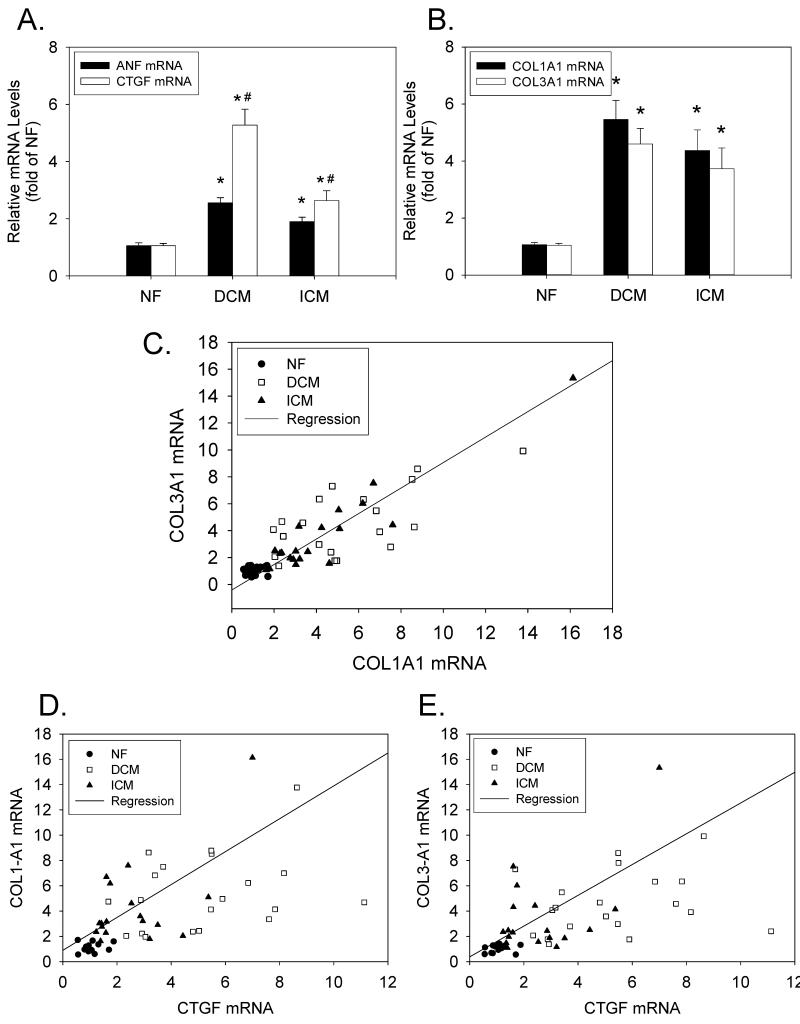
Next, we analyzed mRNA expression levels for atrial natriuretic factor (ANF) (a tissue marker of mechanical overload), and CTGF, COL1-A1 and COL3-A1 in the same NF, DCM, ICM patients (n=20 in each group). (COL1-A1 and COL3-A1 are the mRNAs encoding the α1 chains of Type I and Type III collagens, respectively.) ANF expression was significantly elevated in both DCM and ICM tissue samples (2.6±0.2- and 1.9±0.2-fold for DCM and ICM patients, respectively; P<0.05 for both) (Figure 2A). The expression of CTGF mRNA in DCM and ICM LV tissue samples was also significantly elevated as compared to NF controls (5.3±0.6- and 2.6±0.3-fold for DCM and ICM patients, respectively; P<0.05). However, the degree of CTGF up-regulation was significantly greater in DCM as compared to ICM patients (P<0.05). In addition to CTGF, the expression levels of COL1-A1 and COL3-A1 mRNAs were significantly elevated in DCM and ICM patients as compared to NF controls (Figure 2B). Furthermore, there were close correlations between mRNAs for COL1-A1 and COL3-A1 (R=0.87; P<0.001) (Figure 2C), CTGF and COL1-A1 (R=0.57; P<0.001) (Figure 2D), and CTGF and COL3-A1 (R=0.54; P<0.001) (Figure 2E).
Figure 2. CTGF, COL1-A1 and COL3-A1 expression levels in end-stage heart failure.
(A) LV tissue extracts from NF, DCM, and ICM patients (n=20 in each group) were analyzed for CTGF and ANF mRNAs (relative to 18S rRNA) by real-time RT-PCR. (B) The same LV tissue extracts were analyzed for COL1-A1 and COL3-A1 mRNAs (relative to 18S rRNA). *P<0.05 vs NF; #P<0.05 for DCM vs. ICM. (C) Comparison of COL1-A1 mRNA vs. COL3-A1 mRNA. (D) Comparison of CTGF mRNA vs. COL1-A1 mRNA. (E) Comparison of CTGF mRNA vs. COL3-A1 mRNA.
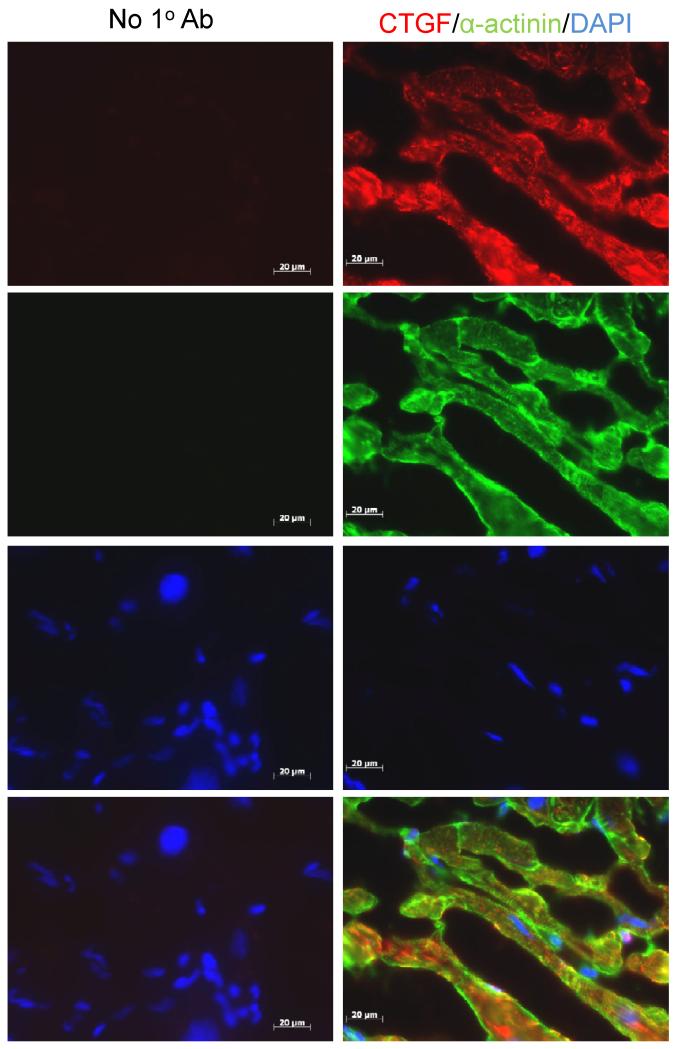
Co-immunolocalization of CTGF and α-actinin, a sarcomeric protein that is highly expressed in cardiomyocytes, was evaluated in frozen tissue sections of failing LV myocardium. As seen in Figure 3, cardiomyocytes were the predominant source of CTGF. Virtually all of the α-actinin positive cardiomyocytes stained with a CTGF-specific primary antibody. However, occasional CTGF-positive, α-actinin negative cells were identified within the interstitial space, and CTGF staining was also noted in the basement membranes surrounding individual cardiomyocytes.
Figure 3. Immunolocalization of CTGF in failing LV myocardium.
Frozen sections of LV myocardium from a patient with DCM were fixed, permeabilized and sequentially stained with mouse anti-human CTGF followed by rhodamine-labeled goat anti-mouse antibodies (red), and then rabbit anti-α-actinin followed by FITC-labeled goat anti-rabbit antibodies (green). Mounting medium containing DAPI was used to detect cell nuclei (blue). Adjacent sections were similarly stained, except for omitting the primary antibodies. Areas of co-localization appear yellow, in the bottom, merged images. Sections were viewed under a Zeiss Axioskop epifluorescent microscope using Axiovision AC (Ver 4.5) software. Scale bar=20μm.
Fibrosis in mechanically unloaded hearts
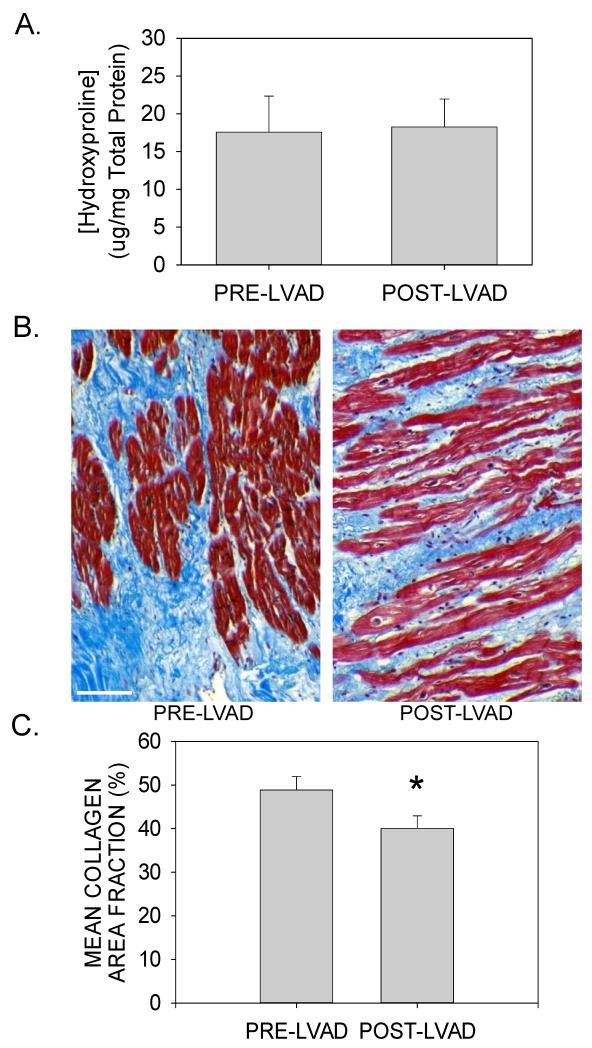
We next examined the molecular changes associated with LVAD support and how these may contribute to reverse ECM remodeling. Myocardial tissue was obtained from the apical core at the time of LVAD implantation, and from the adjacent apical area at the time of cardiac transplantation. As was the case for the explanted DCM and ICM tissue samples, there was substantial variability in the concentration of protein-bound hydroxyproline in the pre-LVAD tissue biopsies. However, mechanical unloading had no significant effect on the average hydroxyproline concentration (Figure 4A). Furthermore, individual paired samples from both diagnostic groups showed no consistent change in hydroxyproline concentration (Supplemental Figure 1A), and the duration of unloading did not appear to influence this parameter (Supplemental Figure 1B).
Figure 4. Fibrosis in mechanically unloaded hearts.
(A) LV collagen concentration in tissue homogenates was assessed by measuring the concentration of protein-bound hydroxyproline (μg/mg total protein) from pre- and post-LVAD tissue homogenates (data are means±SEM for n=13 paired samples). (B) Representative LV tissue sections from paired pre- and post-LVAD tissue sections were stained with Mallory-Trichrome stain. Scale bar = 100μm. (C) Areas of fibrosis in Mallory-Trichrome stained histological sections were digitally analyzed for collagen area fraction (%) on the basis of color thresholds using ImageJ software. Data are means±SEM from 7 patients before and after mechanical unloading; P<0.05.
Paired tissue sections from a subset of these patients (n=7) were also analyzed for the presence of interstitial fibrosis by Mallory-Trichrome staining and digital image analysis. As seen in Figures 4B and 4C, there was a small, but statistically significant reduction in mean collagen area fraction following mechanical unloading (48.9±2.9% vs. 40.1±1.1% for pre- vs. post-LVAD samples; P<0.001). Individually, all patients demonstrated a reduction in collagen area fraction (Supplemental Figure 1C), which appeared to be unrelated to the diagnosis, or the duration of unloading (Supplemental Figure 1D).
CTGF expression in mechanically unloaded human hearts
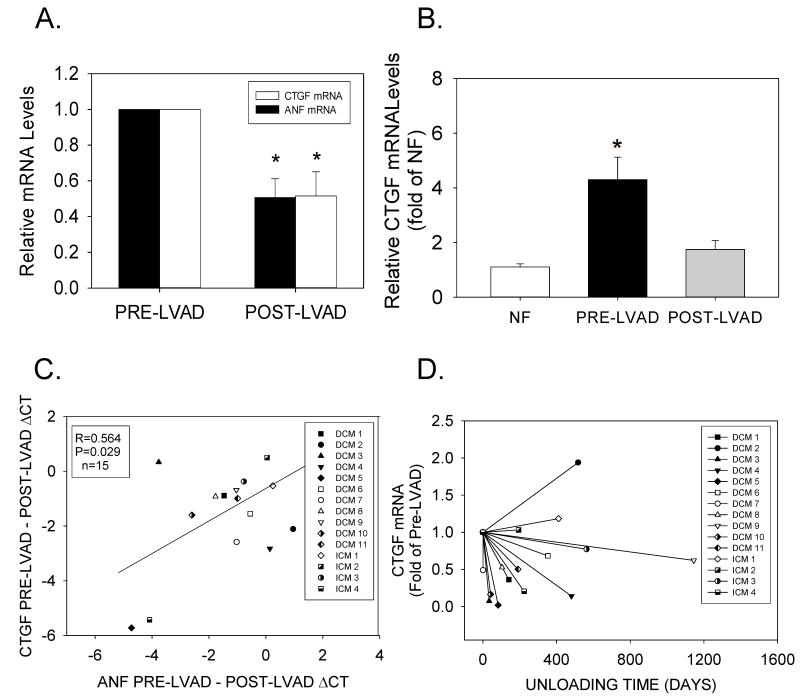
Although mechanical unloading normalizes most genotypic and phenotypic abnormalities that occur during HF, its effects on regulators of ECM production and turnover are not fully defined. One explanation for the failure of unloading to cause regression of fibrosis in HF was the continued, increased expression of profibrotic genes in the unloaded heart. Therefore, we investigated the role of LVAD support on CTGF and other pro-fibrotic genes, which may regulate ECM remodeling in HF. Although there was substantial variability in the level of CTGF expression at the time of LVAD implantation (Supplemental Figure 2), real-time RT-PCR revealed that unloading indeed significantly reduced CTGF mRNA levels in most patients (12/15) (Figures 5A), and returned average CTGF mRNA levels to those observed in NF patients (Figure 5B). The reduction in CTGF mRNA levels also correlated with the reduction in ANF mRNA (Figure 5C). CTGF mRNA down-regulation occurred in both DCM and ICM patients, but appeared unrelated to CTGF mRNA levels at the time of LVAD implantation, or the duration of unloading (Figure 5D).
Figure 5. CTGF expression in mechanically unloaded hearts.
(A) Relative CTGF and ANF mRNA levels from matched patients before and after LVAD implantation. Data are means±SEM for n=15 paired tissue samples; *P<0.05, pre- vs. post-LVAD data. (B) Relative CTGF mRNA levels in pre- and post-LVAD tissue samples (n=15 for each) were compared to the average values obtained in NF patients (n=20). *P<0.05 vs. NF. (C) Comparison between CTGF ΔCt values vs. ANF ΔCt values before and after LVAD implantation. (D) CTGF mRNA levels (normalized to pre-LVAD mRNA) vs. duration of unloading.
Transforming growth factor ß1 expression in mechanically unloaded human hearts
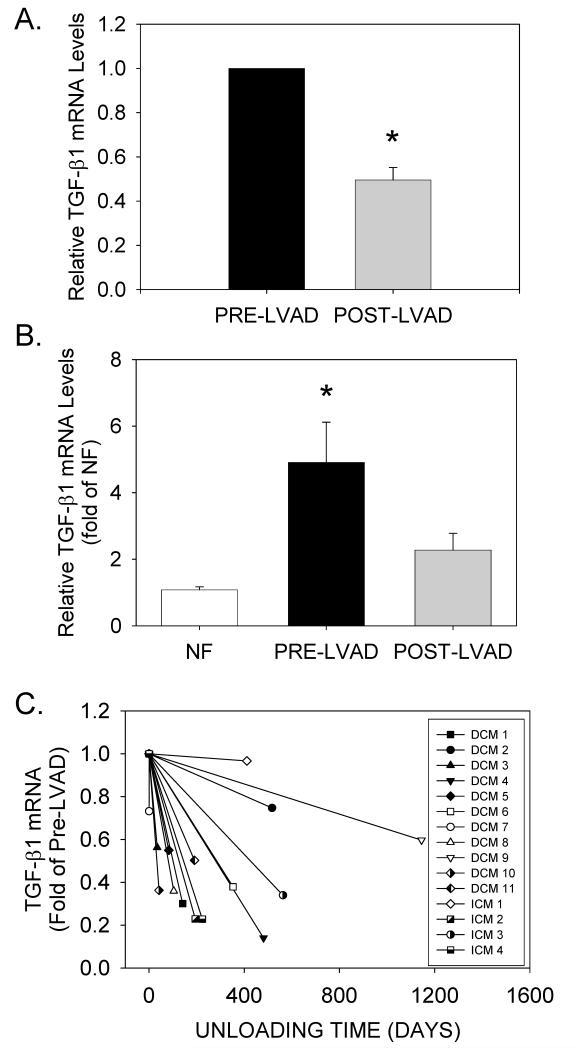
Transforming growth factor-ß1 (TGFB1) is a known regulator of both CTGF and fibrillar collagen gene expression in cardiomyocytes and cardiac fibroblasts (for review, see 36, 37). Therefore, the same samples were analyzed to determine whether unloading reduced TGFB1 mRNA levels to the same extent as CTGF. As seen in Figure 6A, unloading also significantly reduced TGFB1 mRNA levels in all patients, and returned average TGFB1 mRNA levels to those observed in NF patients (Figure 6B). TGFB1 mRNA down-regulation occurred in both DCM and ICM patients, and also appeared unrelated to levels at the time of LVAD implantation, or the duration of unloading (Figure 6C).
Figure 6. TGFB1 expression in mechanically unloaded hearts.
(A) Relative TGFB1 mRNA levels from matched patients before and after LVAD implantation. Data are means±SEM for n=15 paired tissue samples; *P<0.05, pre- vs. post-LVAD data. (B) Relative TGFB1 mRNA levels in pre- and post-LVAD tissue samples (n=15 for each) were compared to the average values obtained in NF patients (n=20). *P<0.05 vs. NF. (D) TGFB1 mRNA levels (normalized to pre-LVAD mRNA) vs. duration of unloading.
Collagen gene expression in mechanically unloaded human hearts
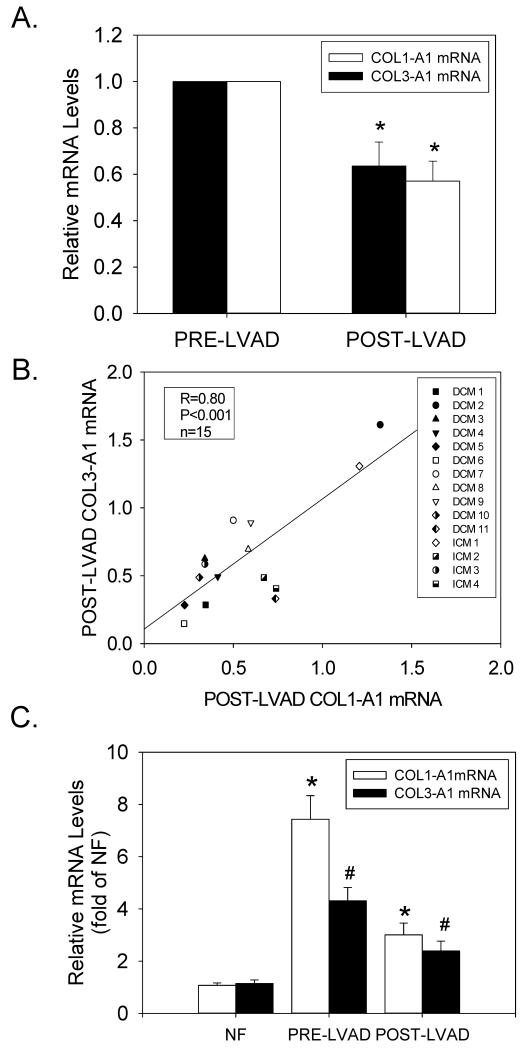
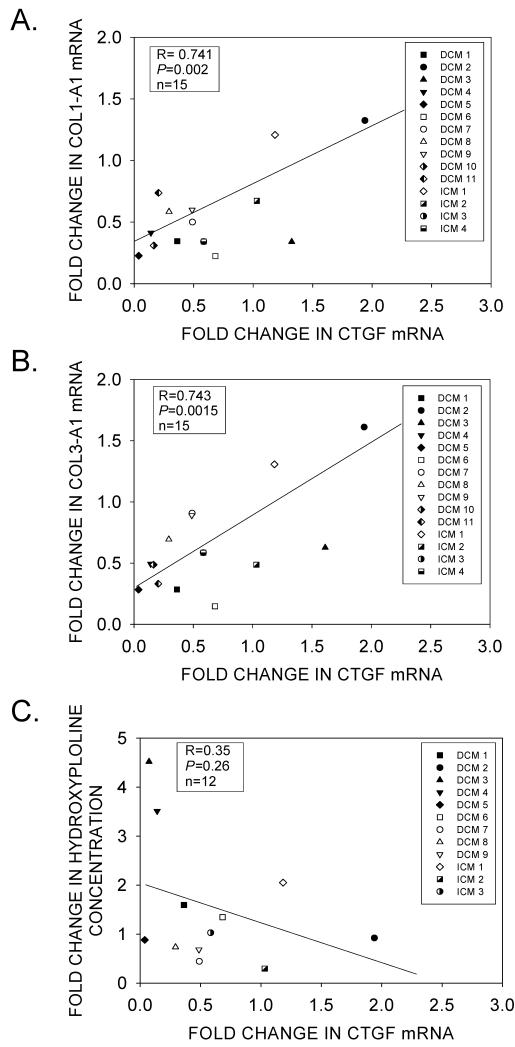
Although mechanical unloading for up to 1145 days did not significantly reduce protein-bound hydroxyproline levels (Figure 4A), we investigated whether LVAD support caused a pre-translational reduction of COL1-A1 and COL3-A1 gene expression. As shown by real-time RT-PCR, there was considerably less variability in fibrillar collagen gene expression in LV tissue samples before and after mechanical unloading (Supplemental Figure 3). Furthermore, COL1-A1 and COL3-A1 expression levels were indeed significantly reduced in post-LVAD tissue samples, indicating that mechanical unloading also regulated fibrillar collagen gene expression (Figures 7A). The degree of down-regulation was similar for both COL1-A1 and COL3-A1 (Figure 7B), and was coincident with the down-regulation of CTGF and TGFB1 mRNAs. However, unlike CTGF and TGFB1, COL1-A1 and COL3-A1 gene expression remained significantly elevated as compared to NF controls (Figure 7C). Nevertheless, there was a close correlation between the reduction in CTGF mRNA levels following unloading, and the reduction of COL1-A1 and COL3-A1 mRNAs (Figures 8A and 8B, respectively); but there was no significant correlation between the reduction in protein-bound hydroxyproline and CTGF mRNA (Figure 8C).
Figure 7. COL1-A1 and COL3-A1 expression in mechanically unloaded hearts.
(A) Relative COL1-A1 and COL3-A1 mRNA levels from matched patients before and after LVAD implantation. (B) Correlation between COL1-A1 vs. COL3-A1 down-regulation in individual patients after LVAD implantation. (C) Relative COL1-A1 and COL3-A1 mRNA levels in pre- and post-LVAD tissue samples (n=15 for each) were compared to the average values obtained in NF patients (n=20). *P<0.05 vs. NF; #P<0.05, pre- vs. post-LVAD data.
Figure 8. Correlations between CTGF mRNA levels and ECM reverse-remodeling.
(A) Comparison between the fold-reduction in CTGF and COL1-A1 mRNAs in pre- and post-LVAD patients. (B) Comparison between the fold-reduction in CTGF and COL3-A1 mRNAs in pre- and post-LVAD patients. (C) Comparison between the fold-change in CTGF mRNA levels and tissue-bound hydroxyproline in pre- and post-LVAD patients.
Matrix metalloproteinases in mechanically unloaded human hearts
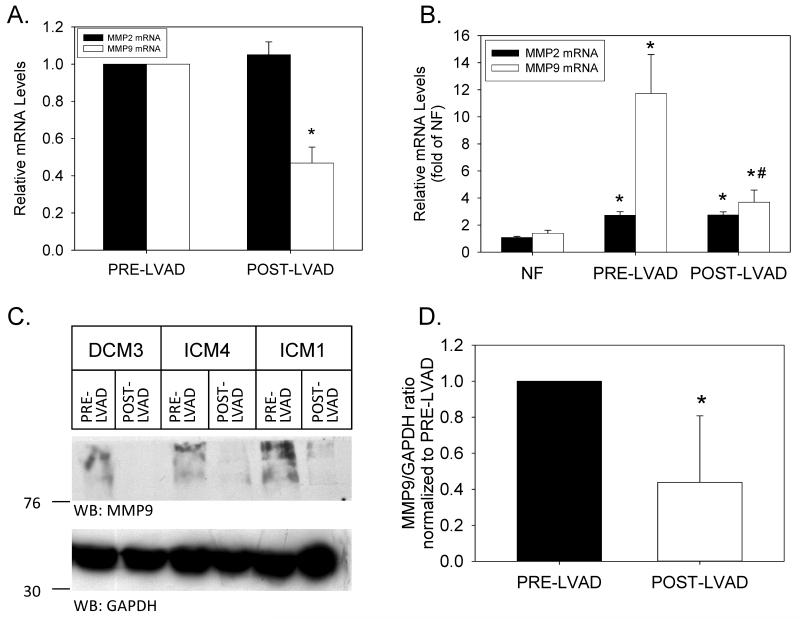
Another potential explanation for the failure of unloading to cause regression of fibrosis in HF was the simultaneous, unloading-induced down-regulation of matrix metalloproteinases responsible for the degradation of the fibrotic, cardiac ECM.38 Therefore, we first analyzed MMP2 and MMP9 mRNA levels in NF and HF samples prior to mechanical unloading. Both MMP2 and MMP9 mRNA levels were significantly elevated as compared to NF controls (2.7±0.3- and 11.7±2.9-fold for MMP2 and MMP9, respectively; P<0.05 for both). LVAD support significantly reduced MMP9 (but not MMP2) mRNA levels (Figure 9A). However, mechanical unloading did not completely return MMP9 mRNA levels to those observed in NF control hearts (Figure 9B). The pre-translational reduction in MMP9 mRNA levels caused a significant reduction in MMP9 protein expression, as analyzed by Western blotting (Figures 9C). A quantitative analysis of MMP9 protein levels (relative to GAPDH) in 13 paired pre- and post-LVAD samples is depicted in Figure 9D.
Figure 9. MMP2 and MMP9 expression in mechanically unloaded hearts.
(A) Relative MMP2 and MMP9 mRNA levels from matched tissue samples before and after LVAD implantation. Data are means±SEM for n=15 paired tissue samples; *P<0.05, pre- vs. post-LVAD data. (B) Relative MMP2 and MMP9 mRNA levels in pre- and post-LVAD tissue samples (n=15 for each) were compared to the average values obtained in NF patients (n=20). *P<0.05 vs. NF; #P<0.05 vs. PRE-LVAD. (C) Equal amounts of extracted protein (200μg per lane) from 3 representative patients before (PRE-LVAD) and after (POST-LVAD) mechanical unloading were separated by SDS-PAGE. Western blots were probed with primary antibodies specific for MMP9 (upper panel) and GAPDH (lower panel). The position of molecular weight markers is indicated to the left of each blot. (D) Quantitative analysis of immunoreactive MMP9 in paired Western blotting samples from 13 individual patients. *P<0.05.
DISCUSSION
Assessment of interstitial fibrosis in human HF
Ventricular remodeling in HF involves a complex series of events that ultimately results in substantial alterations in cardiomyocyte and fibroblast gene expression, tissue fibrosis, and worsening contractile dysfunction. Both mechanical overload and neurohormonal activation that accompany the HF state contribute to the functional deterioration, although it remains unclear what changes are reversible following mechanical unloading. In this report, we demonstrate that CTGF may be a key regulator of the interstitial fibrosis that accompanies maladaptive remodeling and progression to HF. Although mechanical unloading normalizes many of the pro-fibrotic alterations in gene expression, its effect on ECM remodeling during HF was incomplete.
As demonstrated in this and other reports, end-stage systolic HF was uniformly accompanied by substantial interstitial fibrosis. Surprisingly, we found that patients with DCM had an even greater degree of interstitial ECM deposition (as determined by collagen area fraction of tissue sections) than patients with ICM, when areas of gross scar and perivascular fibrosis were excluded from analysis. These results were then compared to the biochemical analysis of protein-bound hydroxyproline, which has been a useful measure of collagen deposition in animal models of cardiac remodeling.39, 40 However, our analysis of failing human myocardium revealed somewhat of a discordance between morphometric measurements of interstitial tissue fibrosis and protein-bound hydroxyproline, especially in ICM patients, and after ventricular unloading. Although there was a reasonable correlation between the two measurements, the analysis of tissue fibrosis using morphometry is highly dependent on tissue sampling, and is influenced by changes in cardiomyocyte size.41 Specifically, the disproportionate regression of cardiomyocyte hypertrophy can influence the relative measurement of ECM area in tissue sections, thereby underestimating the reduction in fibrosis as a result of mechanical unloading. Mallory-Trichrome staining is routinely performed by clinical laboratories and requires no special equipment, but may also underestimate the degree of fibrosis as compared to picrosirius red staining and illumination with circularly polarized light.42 Protein-bound hydroxyproline measurements are derived from a larger sample of myocardial tissue, but also have limitations. For example, the analysis does not exclude regions of fibrosis surrounding small blood vessels, or regions containing microscopic scar. Furthermore, the percentage of prolyl residues that are hydroxylated may vary within interstitial collagens depending on the activity of prolyl 4-hydroxylase, an enzyme that requires ferrous ion, alpha-ketoglutarate, and ascorbate for its activity.43
Up-regulation of CTGF gene expression in human HF
We found that LV tissue CTGF mRNA levels were up-regulated in both DCM and ICM patients, which coincided with the up-regulation of ventricular ANF, a tissue biomarker of mechanical overload. Our results confirm previous studies demonstrating that CTGF is up-regulated in both human and animal models of HF (for review, see36). In this regard, CTGF gene expression is regulated by a number of pathways that are activated during cardiac hypertrophy and HF. These include downstream effectors of Gαq-coupled receptors for angiotensin II, norepinephrine and endothelin-1,44 as well as integrin-dependent, mechanosensitive signaling pathways that involve focal adhesion kinase.4544 Indeed, preliminary studies from our laboratory46 indicate that mechanical unloading of cultured neonatal rat ventricular myocytes (by KCl-, butanedione monoxime- or nifedipine-induced contractile arrest) caused a substantial reduction in CTGF expression, suggesting that mechanosensors in cardiomyocytes also regulate its expression. Conceivably, increased cardiomyocyte CTGF expression and secretion could thereby increase fibrillar collagen gene expression by cardiac fibroblasts via a paracrine mechanism. Cardiomyocyte protein kinase C (PKC) may function as an intermediary signaling kinase in both neurohormonal and mechanical signaling. For instance, King and co-workers demonstrated that angiotensin II regulated CTGF expression in cardiomyocytes through a PKC-dependent pathway that contributed to the development of cardiac fibrosis in an animal model of diabetic cardiomyopathy.47 PKCs were also activated in response to mechanical stretch48 and stimulated contraction49 in cardiomyocytes.
Of note, we also demonstrated the coincident up-regulation of TGFB1 in our DCM and ICM patients. TGFB1 is also a known regulator of CTGF gene expression in both cardiomyocytes and cardiac fibroblasts.50 A SMAD-binding element in the CTGF promoter was necessary for the basal expression of CTGF by TGFB1,51 whereas recent studies have shown that an AP-1 site located at -624 bp of the CTGF promoter mediated CTGF’s responsiveness to endothelin-1 stimulation in cultured cardiomyocytes.52 Indeed, Xia et al.53 showed that CTGF gene transcription was cooperatively regulated by both AP-1 and SMAD binding sites, so it is reasonable to conclude that TGFB1 combined with other neurohormonal agonists that activate AP-1 caused the up-regulation of CTGF gene expression in our HF patients.
However, the extent to which CTGF merely serves as a marker of fibrosis or indeed induces interstitial fibrosis of the heart has not yet been resolved. We demonstrate that CTGF expression occurs coincidently with marked increases in the expression of Type I and Type III collagens. The modular domain structure of CTGF is known to influence the binding and subsequent signaling of TGF-ß and other growth factors that are known regulators of fibrillar collagen gene transcription in the heart and other organs.54
ECM-reverse remodeling following mechanical unloading
Unlike other aspects of reverse remodeling of the myocardium during mechanical unloading, changes in the ECM do not uniformly reflect a return towards normal conditions.13, 16, 17 Alterations in ECM and collagen metabolism after LVAD implantation have been studied by several groups, often with conflicting results. The explanation for these contradictory observations, though still unclear, may be related to differences in the causes of HF,55 duration of unloading56 as well as differences in the methodology employed.9, 57 Our results indicate that mechanical unloading not only reduced COL1-A1 and COL3-A1 mRNA levels, but also substantially reduced CTGF and TGFB1 mRNAs. Despite our small sample size, unloading time did not appear to be a factor, although changes in both gene expression and tissue fibrosis during the first 30-60 days of unloading may be qualitatively or quantitatively different from changes observed over much longer time periods. Nevertheless, we found only a modest reduction in tissue fibrosis and no significant change in protein-bound hydroxyproline. These findings are likely due in part to the very slow rate of fibrillar collagen turnover, even during pathological states that cause substantial ECM remodeling.39, 40 Furthermore, we found that unloading affected the expression of at least one matrix metalloproteinase (MMP9), which could have reduced the degree of ECM reverse-remodeling. The simultaneous, unloading-induced down-regulation of a major matrix metalloproteinase responsible for the degradation of the fibrotic, cardiac ECM provides an obvious mechanism for the lack of regression of cardiac fibrosis during LVAD support.38 Other factors may include incomplete LV unloading, right ventricular overload, persistent neurohormonal activation, and differences in collagen fibril maturation and crosslinking. These factors may have also contributed to the persistent increase in fibrillar collagen gene expression and reduced collagen degradation in post-LVAD tissue as compared to NF hearts. The persistently elevated COL1-A1 and COL3-A1 mRNAs occurred independently of reduced TGFB1 and CTGF down-regulation, and may have also contributed to the lack of regression of cardiac fibrosis during LVAD support. Nevertheless, additional studies are needed to determine if CTGF is indeed causing interstitial fibrosis during mechanical overload, and whether activation of matrix metalloproteinases can improve ECM reverse-remodeling during mechanical unloading.
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge the assistance of Carol Kartje, R.N., Jeffrey Schwartz, M.D. and the entire Heart Failure/Heart Transplant Team at Loyola University Health System for assistance in recruiting the patients and procuring the tissues used for this study.
These studies were supported in part by NIH P01 HL62426, NIH 1F32 HL096143, and a grant from the Dr. Ralph and Marian Falk Medical Research Trust.
Funding Sources: These studies were supported in part by NIH P01 HL62426, NIH 1F32 HL096143, and a grant from the Dr. Ralph and Marian Falk Medical Research Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES - none
REFERENCES
- 1.Pfeffer MA, Braunwald E. Ventricular remodeling after myocardial infarction. Experimental observations and clinical implications. Circulation. 1990;81:1161–72. doi: 10.1161/01.cir.81.4.1161. [DOI] [PubMed] [Google Scholar]
- 2.Sun Y, Zhang JQ, Zhang J, Lamparter S. Cardiac remodeling by fibrous tissue after infarction in rats. J Lab Clin Med. 2000;135:316–23. doi: 10.1067/mlc.2000.105971. [DOI] [PubMed] [Google Scholar]
- 3.Caulfield JB, Borg TK. The collagen network of the heart. Lab Invest. 1979;40:364–72. [PubMed] [Google Scholar]
- 4.Spinale FG, Crawford FA, Jr., Hewett KW, Carabello BA. Ventricular failure and cellular remodeling with chronic supraventricular tachycardia. J Thorac Cardiovasc Surg. 1991;102:874–82. [PubMed] [Google Scholar]
- 5.Rossi MA, Abreu MA, Santoro LB. Images in cardiovascular medicine. Connective tissue skeleton of the human heart: a demonstration by cell-maceration scanning electron microscope method. Circulation. 1998;97:934–5. doi: 10.1161/01.cir.97.9.934. [DOI] [PubMed] [Google Scholar]
- 6.Mann DL, Spinale FG. Activation of matrix metalloproteinases in the failing human heart: breaking the tie that binds. Circulation. 1998;98:1699–702. doi: 10.1161/01.cir.98.17.1699. [DOI] [PubMed] [Google Scholar]
- 7.Li YY, Feng YQ, Kadokami T, McTiernan CF, Draviam R, Watkins SC, et al. Myocardial extracellular matrix remodeling in transgenic mice overexpressing tumor necrosis factor α can be modulated by anti-tumor necrosis factor α therapy. Proc Natl Acad Sci USA. 2000;97:12746–51. doi: 10.1073/pnas.97.23.12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badenhorst D, Maseko M, Tsotetsi OJ, Naidoo A, Brooksbank R, Norton GR, et al. Cross-linking influences the impact of quantitative changes in myocardial collagen on cardiac stiffness and remodelling in hypertension in rats. Cardiovasc Res. 2003;57:632–41. doi: 10.1016/s0008-6363(02)00733-2. [DOI] [PubMed] [Google Scholar]
- 9.Klotz S, Jan Danser AH, Burkhoff D. Impact of left ventricular assist device (LVAD) support on the cardiac reverse remodeling process. Prog Biophys Mol Biol. 2008;97:479–96. doi: 10.1016/j.pbiomolbio.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Soppa GK, Barton PJ, Terracciano CM, Yacoub MH. Left ventricular assist device-induced molecular changes in the failing myocardium. Curr Opin Cardiol. 2008;23:206–18. doi: 10.1097/HCO.0b013e3282fc7010. [DOI] [PubMed] [Google Scholar]
- 11.Levin HR, Oz MC, Chen JM, Packer M, Rose EA, Burkhoff D. Reversal of chronic ventricular dilation in patients with end-stage cardiomyopathy by prolonged mechanical unloading. Circulation. 1995;91:2717–20. doi: 10.1161/01.cir.91.11.2717. [DOI] [PubMed] [Google Scholar]
- 12.Burkhoff D, Holmes JW, Madigan J, Barbone A, Oz MC. Left ventricular assist device-induced reverse ventricular remodeling. Prog Cardiovasc Dis. 2000;43:19–26. doi: 10.1053/pcad.2000.7190. [DOI] [PubMed] [Google Scholar]
- 13.Barbone A, Oz MC, Burkhoff D, Holmes JW. Normalized diastolic properties after left ventricular assist result from reverse remodeling of chamber geometry. Circulation. 2001;104:I229–32. doi: 10.1161/hc37t1.094914. [DOI] [PubMed] [Google Scholar]
- 14.Terracciano CM, Hardy J, Birks EJ, Khaghani A, Banner NR, Yacoub MH. Clinical recovery from end-stage heart failure using left-ventricular assist device and pharmacological therapy correlates with increased sarcoplasmic reticulum calcium content but not with regression of cellular hypertrophy. Circulation. 2004;109:2263–5. doi: 10.1161/01.CIR.0000129233.51320.92. [DOI] [PubMed] [Google Scholar]
- 15.Blaxall BC, Tschannen-Moran BM, Milano CA, Koch WJ. Differential gene expression and genomic patient stratification following left ventricular assist device support. J Am Coll Cardiol. 2003;41:1096–106. doi: 10.1016/s0735-1097(03)00043-3. [DOI] [PubMed] [Google Scholar]
- 16.Liang H, Muller J, Weng YG, Wallukat G, Fu P, Lin HS, et al. Changes in myocardial collagen content before and after left ventricular assist device application in dilated cardiomyopathy. Chin Med J. 2004;117:401–7. [PubMed] [Google Scholar]
- 17.Felkin LE, Lara-Pezzi E, George R, Yacoub MH, Birks EJ, Barton PJ. Expression of extracellular matrix genes during myocardial recovery from heart failure after left ventricular assist device support. J Heart Lung Transplant. 2009;28:117–22. doi: 10.1016/j.healun.2008.11.910. [DOI] [PubMed] [Google Scholar]
- 18.Bork P. The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett. 1993;327:125–30. doi: 10.1016/0014-5793(93)80155-n. [DOI] [PubMed] [Google Scholar]
- 19.Hishikawa K, Oemar BS, Tanner FC, Nakaki T, Luscher TF, Fujii T. Connective tissue growth factor induces apoptosis in human breast cancer cell line MCF-7. J Biol Chem. 1999;274:37461–6. doi: 10.1074/jbc.274.52.37461. [DOI] [PubMed] [Google Scholar]
- 20.Shimo T, Nakanishi T, Nishida T, Asano M, Kanyama M, Kuboki T, et al. Connective tissue growth factor induces the proliferation, migration, and tube formation of vascular endothelial cells in vitro, and angiogenesis in vivo. J Biochem. 1999;126:137–45. doi: 10.1093/oxfordjournals.jbchem.a022414. [DOI] [PubMed] [Google Scholar]
- 21.Koitabashi N, Arai M, Kogure S, Niwano K, Watanabe A, Aoki Y, et al. Increased connective tissue growth factor relative to brain natriuretic peptide as a determinant of myocardial fibrosis. Hypertension. 2007;49:1120–7. doi: 10.1161/HYPERTENSIONAHA.106.077537. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, McLennan SV, Allen TJ, Twigg SM. Regulation of pro-inflammatory and pro-fibrotic factors by CCN2/CTGF in H9c2 cardiomyocytes. J Cell Commun Signal. 2010;4:15–23. doi: 10.1007/s12079-009-0083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frazier K, Williams S, Kothapalli D, Klapper H, Grotendorst GR. Stimulation of fibroblast cell growth, matrix production, and granulation tissue formation by connective tissue growth factor. J Invest Dermatol. 1996;107:404–11. doi: 10.1111/1523-1747.ep12363389. [DOI] [PubMed] [Google Scholar]
- 24.Shi-Wen X, Leask A, Abraham D. Regulation and function of connective tissue growth factor/CCN2 in tissue repair, scarring and fibrosis. Cytokine Growth Factor Rev. 2008;19:133–44. doi: 10.1016/j.cytogfr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Igarashi A, Nashiro K, Kikuchi K, Sato S, Ihn H, Fujimoto M, et al. Connective tissue growth factor gene expression in tissue sections from localized scleroderma, keloid, and other fibrotic skin disorders. J Invest Dermatol. 1996;106:729–33. doi: 10.1111/1523-1747.ep12345771. [DOI] [PubMed] [Google Scholar]
- 26.Oemar BS, Werner A, Garnier JM, Do DD, Godoy N, Nauck M, et al. Human connective tissue growth factor is expressed in advanced atherosclerotic lesions. Circulation. 1997;95:831–9. doi: 10.1161/01.cir.95.4.831. [DOI] [PubMed] [Google Scholar]
- 27.Clarkson MR, Gupta S, Murphy M, Martin F, Godson C, Brady HR. Connective tissue growth factor: a potential stimulus for glomerulosclerosis and tubulointerstitial fibrosis in progressive renal disease. Curr Opin Nephrol Hypertens. 1999;8:543–8. doi: 10.1097/00041552-199909000-00002. [DOI] [PubMed] [Google Scholar]
- 28.Paradis V, Dargere D, Vidaud M, De Gouville AC, Huet S, Martinez V, et al. Expression of connective tissue growth factor in experimental rat and human liver fibrosis. Hepatology. 1999;30:968–76. doi: 10.1002/hep.510300425. [DOI] [PubMed] [Google Scholar]
- 29.Dean RG, Balding LC, Candido R, Burns WC, Cao Z, Twigg SM, et al. Connective tissue growth factor and cardiac fibrosis after myocardial infarction. J Histochem Cytochem. 2005;53:1245–56. doi: 10.1369/jhc.4A6560.2005. [DOI] [PubMed] [Google Scholar]
- 30.Au CG, Butler TL, Sherwood MC, Egan JR, North KN, Winlaw DS. Increased connective tissue growth factor associated with cardiac fibrosis in the mdx mouse model of dystrophic cardiomyopathy. Int J Exp Pathol. 2011;92:57–65. doi: 10.1111/j.1365-2613.2010.00750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koshman YE, Chu M, Engman SJ, Kim T, Iyengar R, Robia SL, et al. Focal adhesion kinase-related nonkinase inhibits vascular smooth muscle cell invasion by focal adhesion targeting, tyrosine 168 phosphorylation, and competition for p130Cas binding. Arterioscler Thromb Vasc Biol. 2011;31:2432–40. doi: 10.1161/ATVBAHA.111.235549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woessner JF. The determination of hydroxyproline in tissue and protein samples containing small proportions of this imino acid. Arch Biochem Biophys. 1961;93:440–7. doi: 10.1016/0003-9861(61)90291-0. [DOI] [PubMed] [Google Scholar]
- 33.Porter MJ, Heidkamp MC, Scully BT, Patel N, Martin JL, Samarel AM. Isoenzyme-selective regulation of SERCA2 gene expression by protein kinase C in neonatal rat ventricular myocytes. Am J Physiol Cell Physiol. 2003;285:C39–C47. doi: 10.1152/ajpcell.00461.2002. [DOI] [PubMed] [Google Scholar]
- 34.Heidkamp MC, Scully BT, Vijayan K, Engman SJ, Szotek EL, Samarel AM. PYK2 regulates SERCA2 gene expression in neonatal rat ventricular myocytes. Am J Physiol Cell Physiol. 2005;289:C471–82. doi: 10.1152/ajpcell.00130.2005. [DOI] [PubMed] [Google Scholar]
- 35.Spinale FG, Coker ML, Thomas CV, Walker JD, Mukherjee R, Hebbar L. Time-dependent changes in matrix metalloproteinase activity and expression during the progression of congestive heart failure: relation to ventricular and myocyte function. Circ Res. 1998;82:482–95. doi: 10.1161/01.res.82.4.482. [DOI] [PubMed] [Google Scholar]
- 36.Daniels A, van Bilsen M, Goldschmeding R, van der Vusse GJ, van Nieuwenhoven FA. Connective tissue growth factor and cardiac fibrosis. Acta Physiol. 2009;195:321–38. doi: 10.1111/j.1748-1716.2008.01936.x. [DOI] [PubMed] [Google Scholar]
- 37.Leask A. Potential therapeutic targets for cardiac fibrosis: TGFß, angiotensin, endothelin, CCN2, and PDGF, partners in fibroblast activation. Circ Res. 2010;106:1675–80. doi: 10.1161/CIRCRESAHA.110.217737. [DOI] [PubMed] [Google Scholar]
- 38.Iyer RP, Patterson NL, Fields GB, Lindsey ML. The history of matrix metalloproteinases: milestones, myths, and misperceptions. Am J Physiol Heart Circ Physiol. 303:H919–30. doi: 10.1152/ajpheart.00577.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karim MA, Ferguson AG, Wakim BT, Samarel AM. In vivo collagen turnover during development of thyroxine-induced left ventricular hypertrophy. Am J Physiol. 1991;260:C316–26. doi: 10.1152/ajpcell.1991.260.2.C316. [DOI] [PubMed] [Google Scholar]
- 40.Eleftheriades EG, Durand JB, Ferguson AG, Engelmann GL, Jones SB, Samarel AM. Regulation of procollagen metabolism in the pressure-overloaded rat heart. J Clin Invest. 1993;91:1113–22. doi: 10.1172/JCI116270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wohlschlaeger J, Schmitz KJ, Schmid C, Schmid KW, Keul P, Takeda A, et al. Reverse remodeling following insertion of left ventricular assist devices (LVAD): a review of the morphological and molecular changes. Cardiovasc Res. 2005;68:376–86. doi: 10.1016/j.cardiores.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 42.Whittaker P, Kloner RA, Boughner DR, Pickering JG. Quantitative assessment of myocardial collagen with picrosirius red staining and circularly polarized light. Basic Res Cardiol. 1994;89:397–410. doi: 10.1007/BF00788278. [DOI] [PubMed] [Google Scholar]
- 43.Eleftheriades EG, Ferguson AG, Spragia ML, Samarel AM. Prolyl hydroxylation regulates intracellular procollagen degradation in cultured rat cardiac fibroblasts. J Mol Cell Cardiol. 1995;27:1459–73. doi: 10.1016/s0022-2828(95)90095-0. [DOI] [PubMed] [Google Scholar]
- 44.Kemp TJ, Aggeli IK, Sugden PH, Clerk A. Phenylephrine and endothelin-1 upregulate connective tissue growth factor in neonatal rat cardiac myocytes. J Mol Cell Cardiol. 2004;37:603–6. doi: 10.1016/j.yjmcc.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 45.Chiquet M, Gelman L, Lutz R, Maier S. From mechanotransduction to extracellular matrix gene expression in fibroblasts. Biochim Biophys Acta. 2009;1793:911–20. doi: 10.1016/j.bbamcr.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 46.Koshman YE, Patel N, Chu M, Iyengar R, Kim T, Lewis W, et al. Mechanical load up-regulates CTGF gene expression in isolated cardiomyocytes and in animal models of heart failure. J. Mol. Cell Cardiol. 2012;53:S18. abstract. [Google Scholar]
- 47.He Z, Way KJ, Arikawa E, Chou E, Opland DM, Clermont A, et al. Differential regulation of angiotensin II-induced expression of connective tissue growth factor by protein kinase C isoforms in the myocardium. J Biol Chem. 2005;280:15719–26. doi: 10.1074/jbc.M413493200. [DOI] [PubMed] [Google Scholar]
- 48.Seko Y, Takahashi N, Tobe K, Kadowaki T, Yazaki Y. Pulsatile stretch activates mitogen-activated protein kinase (MAPK) family members and focal adhesion kinase (p125FAK) in cultured rat cardiac myocytes. Biochem Biophys Res Commun. 1999;259:8–14. doi: 10.1006/bbrc.1999.0720. [DOI] [PubMed] [Google Scholar]
- 49.Strait JB, Samarel AM. Isoenzyme-specific protein kinase C and c-Jun N-terminal kinase activation by electrically stimulated contraction of neonatal rat ventricular myocytes. J Mol Cell Cardiol. 2000;32:1553–66. doi: 10.1006/jmcc.2000.1191. [DOI] [PubMed] [Google Scholar]
- 50.Chen MM, Lam A, Abraham JA, Schreiner GF, Joly AH. CTGF expression is induced by TGF-ß in cardiac fibroblasts and cardiac myocytes: a potential role in heart fibrosis. J Mol Cell Cardiol. 2000;32:1805–19. doi: 10.1006/jmcc.2000.1215. [DOI] [PubMed] [Google Scholar]
- 51.Grotendorst GR, Okochi H, Hayashi N. A novel transforming growth factor ß response element controls the expression of the connective tissue growth factor gene. Cell Growth Differ. 1996;7:469–80. [PubMed] [Google Scholar]
- 52.Recchia AG, Filice E, Pellegrino D, Dobrina A, Cerra MC, Maggiolini M. Endothelin-1 induces connective tissue growth factor expression in cardiomyocytes. J Mol Cell Cardiol. 2009;46:352–9. doi: 10.1016/j.yjmcc.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 53.Xia W, Kong W, Wang Z, Phan TT, Lim IJ, Longaker MT, et al. Increased CCN2 transcription in keloid fibroblasts requires cooperativity between AP-1 and SMAD binding sites. Ann Surg. 2007;246:886–95. doi: 10.1097/SLA.0b013e318070d54f. [DOI] [PubMed] [Google Scholar]
- 54.Abreu JG, Ketpura NI, Reversade B, De Robertis EM. Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-ß. Nat Cell Biol. 2002;4:599–604. doi: 10.1038/ncb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li YY, Feng Y, McTiernan CF, Pei W, Moravec CS, Wang P, et al. Downregulation of matrix metalloproteinases and reduction in collagen damage in the failing human heart after support with left ventricular assist devices. Circulation. 2001;104:1147–52. doi: 10.1161/hc3501.095215. [DOI] [PubMed] [Google Scholar]
- 56.Bruggink AH, van Oosterhout MF, de Jonge N, Ivangh B, van Kuik J, Voorbij RH, et al. Reverse remodeling of the myocardial extracellular matrix after prolonged left ventricular assist device support follows a biphasic pattern. J Heart Lung Transplant. 2006;25:1091–8. doi: 10.1016/j.healun.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 57.Soppa GK, Lee J, Stagg MA, Felkin LE, Barton PJ, Siedlecka U, et al. Role and possible mechanisms of clenbuterol in enhancing reverse remodelling during mechanical unloading in murine heart failure. Cardiovasc Res. 2008;77:695–706. doi: 10.1093/cvr/cvm106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.